Bifidobacterium animalis subsp. lactis boosts neonatal immunity: unravelling systemic defences against Salmonella†
Chunxiu
Lin‡
ab,
Yugui
Lin‡
c,
Shunhe
Wang
abd,
Jialiang
Wang
ab,
Xuhua
Mao
e,
Yonghua
Zhou
f,
Hao
Zhang
 abdg,
Wei
Chen
abdg,
Wei
Chen
 abd and
Gang
Wang
abd and
Gang
Wang
 *abdg
*abdg
aState Key Laboratory of Food Science and Resources, Jiangnan University, Wuxi 214122, P. R. China. E-mail: wanggang@jiangnan.edu.cn; Tel: +86-510-85912155
bSchool of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China
cMicrobiology Laboratory, Zhongshan Bo'ai Hospital, Southern Medical University, Zhongshan 528400, P. R. China
dNational Engineering Research Center for Functional Food, Jiangnan University, Wuxi 214122, P. R. China
eDepartment of Clinical Laboratory, Yixing People's Hospital, Wuxi 214200, P. R. China
fKey Laboratory of National Health Commission on Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, P. R. China
g(Yangzhou) Institute of Food Biotechnology, Jiangnan University, Yangzhou 225004, P. R. China
First published on 29th November 2023
Abstract
Bifidobacterium animalis subsp. lactis may be a useful probiotic intervention for regulating neonatal intestinal immune responses and counteracting Salmonella infection. However, recent research has focused on intestinal immunity, leaving uncertainties regarding the central, peripheral, and neural immune responses in neonates. Therefore, this study investigated the role and mechanisms of B. animalis subsp. lactis in the systemic immune responses of neonatal rats following Salmonella infection. Through extremely early pretreatment with B. animalis subsp. lactis (6 hours postnatal), the neonatal rat gut microbiota was effectively reshaped, especially the Bifidobacterium community. In the rats pretreated with B. animalis subsp. lactis, Salmonella was less prevalent in the blood, liver, spleen, and intestines following infection. The intervention promoted T lymphocyte subset balance in the spleen and thymus and fostered neurodevelopment and neuroimmune balance in the brain. Furthermore, metabolic profiling showed a strong correlation between the metabolites in the serum and colon, supporting the view that B. animalis subsp. lactis pretreatment influences the systemic immune response by modifying the composition and metabolism of the gut microbiota. Overall, the results imply that B. animalis subsp. lactis pretreatment, through the coordinated regulation of colonic and serum metabolites, influences the systemic immune responses of neonatal rats against Salmonella infection.
1 Introduction
Salmonella is a genus of opportunistic pathogenic bacteria widely present in nature, especially some pathogenic serotypes such as Salmonella typhi, Salmonella paratyphi, and invasive non-typhoidal Salmonella (mainly Salmonella enterica serovar Typhimurium (S. typhimurium) and S. enteritidis), which cause intestinal infections in both humans and animals.1 Due to their immature intestinal immunity and intestinal barrier, infants and young children who are infected with Salmonella can experience severe complications such as sepsis and toxaemia, which pose a significant threat to their life and health.2,3 In resource-limited areas (e.g., sub-Saharan Africa), the mortality rate of infants and young children resulting from Salmonella infection is particularly high due to limited healthcare facilities and poor sanitation.2,3 Despite some progress, the treatment and prevention of Salmonella infections in infants and young children remain challenging. In particular, antibiotic resistance in Salmonella infections remains a serious challenge.4 Additionally, the lack of widely used vaccines specifically targeting Salmonella further complicates the management and control of infections.5Research evidence increasingly suggests that improving the gut microbiota is crucial for treating and preventing Salmonella infection in neonates.6 The introduction of probiotics has provided novel insights and solutions in this field. In response to the severe health threat posed by neonatal Salmonella infection, abundant research has revealed the beneficial impact of probiotics in alleviating infection symptoms, enhancing immune responses, and inhibiting Salmonella colonisation in the intestines.7–11 Among various probiotics, the genus Bifidobacterium has garnered significant research attention due to its immunomodulatory effects on the neonatal immune system.12–14 In particular, Bifidobacterium animalis subsp. lactis, an important and widespread representative of the genus,15 has attracted considerable interest in the study of the neonatal gut microbiota due to its excellent safety profile and range of beneficial effects on intestinal diseases such as severe necrotising enterocolitis, sepsis, and infant colic.16,17 As such, it is considered an attractive probiotic strain for infants. In particular, the commercial B. animalis subsp. lactis strains BB-12, HN019, and Bi-07 have received approval from the U.S. Food and Drug Administration for use as essential components in infant formula milk.18 Research has confirmed that B. animalis subsp. lactis can regulate the intestinal immune balance and improve the gut barrier function in neonatal rats, effectively reducing the risk of Salmonella infection.19 However, a comprehensive understanding of the underlying mechanisms by which B. animalis subsp. lactis acts against Salmonella infection during early life stages is currently lacking. Moreover, its effects on the central, peripheral, and neural immune responses in neonates are still uncertain.
This study aimed to address these knowledge gaps and investigate the key role and mechanisms of B. animalis subsp. lactis in the systemic immune response of neonatal rats infected with Salmonella. To simulate early intervention with probiotics, we pretreated neonatal model rats with B. animalis subsp. lactis in the extremely early postnatal period. By conducting metabolomics analyses and exploring the correlation between serum and colonic content metabolites, we aimed to gain deeper insights into this probiotic's potential mechanisms. This study underscores the potent probiotic effect of B. animalis subsp. lactis intervention in enhancing the systemic immune response of neonatal rats against Salmonella infection. Understanding the multifaceted immunomodulatory effects and mechanisms of B. animalis subsp. lactis will contribute to the development of novel strategies for combating neonatal infections and improving early-life immunity.
2 Materials and methods
2.1 Bacterial strain isolation and culture
With the goal of reducing any research bias brought on by regional and demographic differences, this study isolated and identified five strains of B. animalis subsp. lactis (BJHD3M6 CDS, S10 (SDJN2M②1032), SC-YA-1-M1 CDS, HuNan2016 222 T71 CDS and FNMGHHHT2M2 CDS) from a diverse cohort of healthy individuals in China, encompassing various regions, genders, and age groups (as indicated in Table S1†). During the collection of fecal samples, particular attention was paid to the well-being and comfort of the participants, and strict adherence to ethical requirements was ensured with written informed consent obtained from either the patients or their legal guardians. All of the aforementioned B. animalis subsp. lactis strains, as well as the S. typhimurium SL1344 strain with streptomycin resistance, have been preserved in the Culture Collection of Food Microorganisms at Jiangnan University.The process of isolating, screening, and identifying bifidobacteria was conducted as described previously.19,20 The assessment of the B. animalis subsp. lactis species was carried out as detailed in the earlier reference.19,21 In this study, the B. animalis subsp. lactis strains were cultured in modified de Man, Rogosa, and Sharpe broth supplemented with 0.05% (w/v) L-cysteine under anaerobic conditions (80% N2, 10% CO2, and 10% H2) at 37 °C. Additionally, the strain of S. typhimurium SL1344 was cultured in Luria–Bertani broth at 37 °C with shaking at 200 rpm for 12 hours. The gradient dilution method was used to determine the number of bacterial cells. For oral administration, freshly cultured bacterial cells were collected by centrifugation (5000g for 10 minutes at 4 °C) and suspended in a 10% skimmed milk solution. The five B. animalis subsp. lactis strains were mixed in equal proportions and stored in sterilized tubes at a final concentration of 1 × 1010 colony-forming units (CFU) per mL at −80 °C until use. Similarly, the strain of S. typhimurium SL1344 was prepared and stored in sterilized tubes at a final concentration of 1 × 106 CFU per mL at −80 °C until use.
2.2 Animal experiment
All animal experiments and procedures were approved by the Experimental Animal Ethics Committee of the Jiangsu Institute of Parasitic Diseases, China (JIPD-lACUC_2021095). The design and conduct of these animal experiments followed the ARRIVE guidelines, the Guiding Opinions on the Treatment of Laboratory Animals issued by the Ministry of Science and Technology of the People's Republic of China, and the Laboratory Animal-Guideline for Ethical Review of Animal Welfare issued by the National Standard GB/T35892-2018 of the People's Republic of China. Adult Wistar rats, aged 8 weeks, were obtained from Charles River Laboratory (Beijing, China) and housed in specific pathogen-free facilities. Experimental neonatal rats were generated through timed mating in-house, and the day of birth was designated as postnatal day 0 (P0). The newborn rats were randomly assigned, and their birth times did not differ by more than 6 hours. The B. animalis subsp. lactis group received daily gastric intubation of skim milk containing B. animalis subsp. lactis suspension (1 × 1010 CFU per mL), while the control and model groups received skim milk without B. animalis subsp. lactis suspension. This gavage regimen continued until P8. Subsequently, at P6, we induced infection with S. typhimurium SL1344 (1 × 106 CFU per mL) and sacrificed the pups 48 hours post-infection to assess their immune-related markers. The dosage of all bacterial strains was calculated based on the body weight of the newborn rats and administered at 8 μL per g body weight. The treatment protocol for each group and the experimental timeline are detailed in Fig. 1A.In the experiment, blood, brain, thymus, liver, spleen, colon, and contents from different segments of the intestine were collected. All samples were duplicated for further processing. Upon removal, the thymus, spleen, and brain were immediately weighed, and the relative organ weight was calculated as a percentage of organ weight to body weight. For histological assessment, the distal colon and brain samples were immersed in formalin for fixation, followed by embedding in paraffin, and subsequently sectioned into 5 μm-thick slices. To maintain bacterial and cellular viability, half of the blood, brain, thymus, liver, spleen, and contents from different segments of the intestine were promptly diluted in cold phosphate buffered saline (PBS). The other half of the samples were stored at −80 °C to be utilized in subsequent experiments. Furthermore, any suspected contaminants were safely treated after the conclusion of the experiment.
2.3 Cytokine/biochemical assays
An enzyme-linked immunosorbent assay (ELISA) was employed to measure the concentrations of various cytokines in both colon and brain tissue, including tumor necrosis factor-alpha (TNF-α), interleukins (IL-1β, IL-6, IL-10, IL-22, and IL-17A), as well as the concentration of diamine oxidase (DAO) in serum. The longitudinally opened colon and cerebral cortex tissue was rinsed and homogenized in ice-cold PBS at a concentration of 10% w/v. Subsequently, centrifugation was carried out at 4 °C at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes to collect the supernatant. All ELISA assays were performed in accordance with the manufacturer's instructions provided by Mlbio, Shanghai, China. To ensure accurate comparisons, the protein content of the tissue supernatant was determined using the bicinchoninic acid protein assay kit from Beyotime Biotechnology, Shanghai, China, which served as a normalization method.
000g for 15 minutes to collect the supernatant. All ELISA assays were performed in accordance with the manufacturer's instructions provided by Mlbio, Shanghai, China. To ensure accurate comparisons, the protein content of the tissue supernatant was determined using the bicinchoninic acid protein assay kit from Beyotime Biotechnology, Shanghai, China, which served as a normalization method.
2.4 Haematoxylin and eosin (HE) and Nissl staining
Colon and brain sections were stained with HE and Nissl, respectively, and digitally scanned using a digital slice scanner (Pannoramic MIDI, 3DHISTECH, Hungary). Colon tissue damage was blindly assessed using an established scoring system, which included the evaluation of epithelial loss, crypt injury, goblet cell depletion, and inflammatory cell infiltration. The degree of tissue damage was scored based on previously reported criteria.22 The average number of cells per square millimeter in the brain tissue was calculated by the experimental researchers using ImageJ 1.8.0 software (National Institutes of Health, Bethesda, MD, USA) through blind analysis of Nissl-stained brain sections.2.5 Immunohistochemistry (IHC)/immunofluorescence (IF) staining
Colon sections were incubated overnight at 4 °C with primary antibodies against specific proteins, including anti-PCNA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, ab92552, Abcam, Shanghai, China) and anti-BDNF (1
200, ab92552, Abcam, Shanghai, China) and anti-BDNF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, ab108319, Abcam, Shanghai, China). Subsequently, they were incubated with the secondary antibody HRP-labeled goat anti-rabbit IgG (H + L) (1
200, ab108319, Abcam, Shanghai, China). Subsequently, they were incubated with the secondary antibody HRP-labeled goat anti-rabbit IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Sangon Biotech, Shanghai, China). Similarly, brain sections were incubated overnight at 4 °C with a primary antibody targeting a specific protein, anti-Iba1 (1
500, Sangon Biotech, Shanghai, China). Similarly, brain sections were incubated overnight at 4 °C with a primary antibody targeting a specific protein, anti-Iba1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, ab178847, Abcam, Shanghai, China), followed by a Cy3-labeled goat anti-rabbit secondary antibody at 1/500 dilution (Sangon Biotech, Shanghai, China). The nuclei were stained with DAPI (Beyotime, Shanghai, China). The digital slice scanner was used to capture staining images. Protein expression levels were quantified by measuring the mean intensity in five randomly selected crypts from each sample using ImageJ software.
400, ab178847, Abcam, Shanghai, China), followed by a Cy3-labeled goat anti-rabbit secondary antibody at 1/500 dilution (Sangon Biotech, Shanghai, China). The nuclei were stained with DAPI (Beyotime, Shanghai, China). The digital slice scanner was used to capture staining images. Protein expression levels were quantified by measuring the mean intensity in five randomly selected crypts from each sample using ImageJ software.
2.6 Apoptosis detection
Cell apoptosis was assessed using the TdT-mediated dUTP-biotin nick end labeling (TUNEL) FITC apoptosis detection kit from Vazyme Biotech Co. Ltd (Nanjing, China) as per the manufacturer's instructions. Colon tissue sections were treated with protein kinase K (20 μg ml−1) for 15 minutes and stained with the FITC-12-dUTP labeling mixture at room temperature for 60 minutes. Negative control samples lacked the FITC-12-dUTP labeling mixture, while positive control samples were pre-treated with 20 U ml−1 DNase I before incubation with the FITC-12-dUTP labeling mixture. At least 200 cells from five randomly selected crypts per sample were counted to quantify the positive TUNEL signals and calculate the percentage of apoptotic cells.2.7 Colon content 16S rRNA sequencing and bioinformatic analysis
Microbial genomic DNA was extracted from colon content samples using the FastDNA® spin kit (MP Biomedicals, Santa Ana, CA) following the manufacturer's instructions, as previously described.20 The V3–V4 region of bacterial 16S rRNA was amplified using universal primers (341F and 806R) and the 2× Taq PCR master mix (TaKaRa, Dalian, China). After PCR product purification with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany), the resulting amplicons were pooled and subjected to paired-end sequencing on the MiSeq PE300 platform (Illumina, San Diego, CA, USA). The obtained sequencing data were analyzed using MicrobiomeAnalyst (https://www.microbiomeanalyst.ca/, accessed on July 30, 2023) for microbial composition determination and bioinformatics analysis.2.8 Bifidobacterial genome sequencing and bioinformatics analysis
To analyze the Bifidobacterium species composition, a ∼500 bp fragment of the Bifidobacterium GroEL gene (Bif-GroEL) was amplified using the primers Bif-groEL-F and Bif-groEL-R.20 The PCR reaction and procedure were performed according to the previous reference.20 All PCR products were purified using the QIAquick gel extraction kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions. DNA amplicon libraries were prepared using the TruSeq nano DNA LT kit (Illumina, San Diego, CA) and sequenced on the Illumina MiSeq platform with the MiSeq reagent kit v3 (600 cycles-PE, Illumina, San Diego, CA) according to the provided instructions.2.9 Determination of bacterial loads
Based on the literature,8 the Salmonella load in various tissue samples of neonatal rats was calculated. Briefly, intestinal contents (from the cecum, colon, ileum, jejunum, or duodenum), visceral organs (liver, spleen, thymus and brain) and blood were homogenized using cold PBS. For the intestinal contents, we utilized the MacConkey agar plates (containing 50 μg ml−1 streptomycin) with a series of dilutions and the plate counting method to determine bacterial CFU per g after 24 hours of culture. As for visceral organs and blood samples, our focus was on observing the presence of bacteria without quantification.2.10 Detection of T-lymphocyte subsets
The fresh spleen and thymus samples were placed in pre-cooled PBS, mechanically minced, and filtered through a 100 μm nylon mesh to prepare a single-cell suspension of 1 × 107 cells. To investigate the cytokine secretion of T cells, cells were placed at 37 °C in 5% CO2 and continuously stimulated for 6 hours with phorbol 12-myristate 13-acetate (50 ng mL−1), ionomycin (1 μg mL−1), and monensin (5 μg mL−1) (all from Sigma-Aldrich, USA). For surface staining, cells were co-incubated with antibodies diluted in PBS at recommended concentrations: ANTI-RAT CD4 OX35 SB436 (eBioscience), ANTI-RAT CD25 (OX39) APC (eBioscience), Alexa Fluor® 488 anti-rat CD3 (Biolegend), and ANTI-RAT CD8A OX8 SB600 (eBioscience). Subsequently, for intracellular and intranuclear antibody staining, cells were fixed and permeabilized in advance using the intracellular fixation & permeabilization buffer set for intracellular staining and the Foxp3/transcription factor staining buffer set (both from eBioscience) for intranuclear staining. Intracellular antibodies included ANTI-MO/RAT IL-17A (EBIO17B7) PE-CYANINE 7 (eBioscience), PE anti-rat IL-4 (Biolegend), and Alexa Fluor® 647 anti-rat IFN-γ (Biolegend); intranuclear antibody included ANTI-MOUSE/RAT FOXP3 (FJK-16S) PE (eBioscience). Following standard cell staining procedures, cells were acquired using an Attune NxT acoustic focusing cytometer (Model AFC2 Thermo, CA, USA), and data were analyzed using FlowJo software (Tree Star, Ashland, OR, USA). T lymphocyte subsets were defined as follows: Th (CD3+ CD8−) cells; Tc (CD3+ CD8+) cells; Treg (CD4+ CD25+ Foxp3+) cells; Th1 (CD3+ CD8− IFN-γ+) cells; Th2 (CD3+ CD8− IL-4+) cells; Th17 (CD3+ CD8− IL-17+) cells. Results were expressed as the percentage of positive cells within the lymphocyte population selected based on their forward-scatter cells and side-scatter characteristics, or within a specific selected population.2.11 Neurological reflex testing
Three neurobehavioral reflexes—negative geotaxis, cliff avoidance, and surface righting reflex—were assessed. The assessment protocol was adapted from previous studies with slight modifications.23 Each pup underwent three trials, and the average value was used for analysis.For negative geotaxis, pups were positioned head-down on a 30° inclined surface, and the time taken for a 180° upright flip from the starting position was recorded. If the pup took over 60 seconds to complete the flip, the test was stopped, and the time was recorded as 60 seconds.
In the cliff avoidance test, pups were positioned at the edge of an experimental table with their noses and front paws hanging over the edge. Their responses, whether they retreated or turned sideways to avoid falling off the “cliff”, were recorded. Performance was scored on a five-point scale, with 5 indicating excellent performance and 0 representing complete failure.
Regarding the surface righting reflex, pups were gently placed on their backs, and their ability to flip onto their abdomen was observed and recorded. Performance was scored on a five-point scale, with 5 indicating excellent performance when the pup quickly flipped and started exploring its surroundings, while 0 represented a complete failure.
2.12 Metabolomics profiling
Colonic contents and serum for metabolite analysis were extracted following Zhu et al.'s protocol.24 To ensure quality control (QC), a pooled QC sample was created by combining 10 μL from each sample. The sample analysis was performed using an ItiMateU-3000 ultraperformance liquid chromatography (UPLC) system (Thermo Fisher Scientific, MA, USA) coupled with a high-resolution Q-exactive mass spectrometer (QE/MS) (Thermo Fisher Scientific, MA, USA). The operating parameters of the UPLC-QE/MS system were previously reported.24 For metabolite identification, the data were compared against the human metabolome database, mzCloud, and ChemSpider databases. Metabolic pathway analysis was conducted using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/, accessed on July 30, 2023) to elucidate the involved metabolic pathways, utilizing Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.2.13 Statistical analysis
The data are presented as the mean ± standard error of the mean. To assess the significance, a one-way analysis of variance was performed. Post hoc multiple comparisons were conducted using either Dunnett's (2-sided) test for homogeneous variance or the Dunnett T3 test for non-homogeneous variance. Specifically, we compared the model group with the control group and the B. animalis subsp. lactis pretreatment group with the model group, respectively. Statistical analysis was conducted using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). Significance was considered at p < 0.05, denoted as (#p < 0.05, ##p < 0.01, ###p < 0.001) versus control, and *p < 0.05, **p < 0.01, ***p < 0.001 versus model, respectively.3 Results
3.1 B. animalis subsp. lactis pretreatment facilitates neonatal rats in combating S. typhimurium SL1344 infection by modulating intestinal immune homeostasis
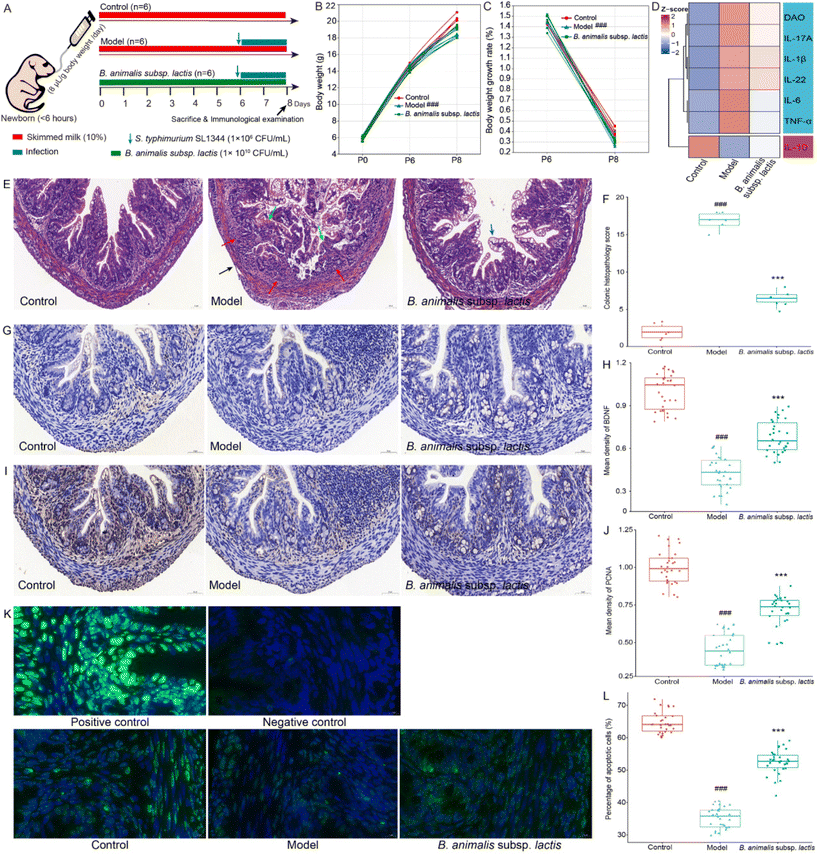
Based on previously published experimental data,19 we conducted a preliminary study and determined that the infection dose of 1 × 106 CFU per mL of S. typhimurium SL1344 was suitable for our experiments (pre-experimental data not shown). It was suggested that the chosen infection dose allowed ample scope to investigate the impact of B. animalis subsp. lactis pretreatment on the modulation of Salmonella infection in neonatal rats. Moreover, no mortality was observed among the neonatal pups during the two-day infection experiment. The specific experimental procedures and details are illustrated in Fig. 1A.Clearly, all groups of newborn rats exhibited similar body weights on P0 (birth) (Fig. 1B). Consistent with previous research,19 the pre-intervention with B. animalis subsp. lactis before infection (P0 to P6) did not affect the body weight of newborn rats, and their weight gain rates remained unchanged (Fig. 1B and C). However, on P6, after exposure to S. typhimurium SL1344, the model group of pups showed a significant reduction in body weight compared to the control group, with a weight gain rate loss of 10.91% after 48 hours (Fig. 1B and C). Although the effect was not significant, the pre-intervention with B. animalis subsp. lactis partially mitigated the weight gain rate loss by 4.09% (Fig. 1B and C). Following exposure to S. typhimurium infection, an increasing trend was noted in the levels of pro-inflammatory cytokines (IL-17A, IL-1β, IL-22, IL-6, and TNF-α), while a decreasing trend was observed in the levels of the anti-inflammatory cytokine (IL-10), as depicted in Fig. 1D. Additionally, the serum level of DAO, a clinical indicator of intestinal mucosal function,25 showed a significant elevation (Fig. 1D). However, pretreatment with B. animalis subsp. lactis significantly attenuated the alterations in colonic pro-inflammatory and anti-inflammatory cytokine levels induced by S. typhimurium infection, and concurrently mitigated the increase in serum DAO level (Fig. 1D).
The results of colonic tissue structure observed through HE staining clearly demonstrate that in newborn rats pre-treated with B. animalis subsp. lactis, the severity of colonic tissue damage caused by S. typhimurium infection was significantly alleviated. Compared to the uninfected group, the model group exhibited evident crypt destruction (indicated by green arrows), extensive infiltration of inflammatory cells (indicated by red arrows), and even lesions reaching the submucosa, muscular layer, and serosal layer (indicated by black arrows) (Fig. 1E and F). In contrast, newborn rats that underwent B. animalis subsp. lactis pretreatment and were subsequently challenged with S. typhimurium infection showed a significant activation of goblet cells (indicated by blue arrows) (Fig. 1E), which synthesized and secreted mucoprotein, forming a robust mucosal barrier to protect the underlying epithelial cells.26 Notably, without B. animalis subsp. lactis pretreatment, such a defense strategy seemed to be ineffective in response to S. typhimurium infection (Fig. 1E). In the model group, it is evident that the structure of goblet cells is significantly disrupted, with some even becoming damaged, showing a distinct pattern from that observed in the B. animalis subsp. lactis group (Fig. 1E). It suggests that the activated goblet cells in the model group may not actually be involved in mucosal protection but rather represent a response to the failure of protective measures in the face of a substantial infectious challenge. Moreover, Salmonella infection not only led to impaired intestinal barrier function in the neonatal hosts but also disrupted the intestinal nervous system, as evidenced by the decreased brain-derived neurotrophic factor (BDNF) expression levels (a crucial neurotrophic factor,27Fig. 1G and H). However, the pretreatment with B. animalis subsp. lactis significantly reversed this unfavorable situation, prominently maintaining BDNF levels, although not fully restoring them to normal levels (Fig. 1G and H).
To further understand the benefits of B. animalis subsp. lactis on the intestinal development of neonatal rats, we used proliferating cell nuclear antigen (PCNA) staining to assess the impact of infection on the proliferation of intestinal cells28 and employed TUNEL staining to investigate the regulatory effect of infection on intestinal cell apoptosis.29 The results clearly showed that Salmonella infection led to a significant decrease in PCNA staining intensity (Fig. 1I and J) and the number of TUNEL-positive cells (Fig. 1K and L) in the intestines of neonatal rats, indicating severe cellular damage and disrupted regeneration processes. However, in neonatal rats pre-treated with B. animalis subsp. lactis, the impact of Salmonella infection on intestinal development was reduced. Notably, B. animalis subsp. lactis not only significantly maintained the level of intestinal cell proliferation (Fig. 1I and J) but also preserved the level of cell apoptosis (Fig. 1K and L), though not completely restoring it to the natural state.
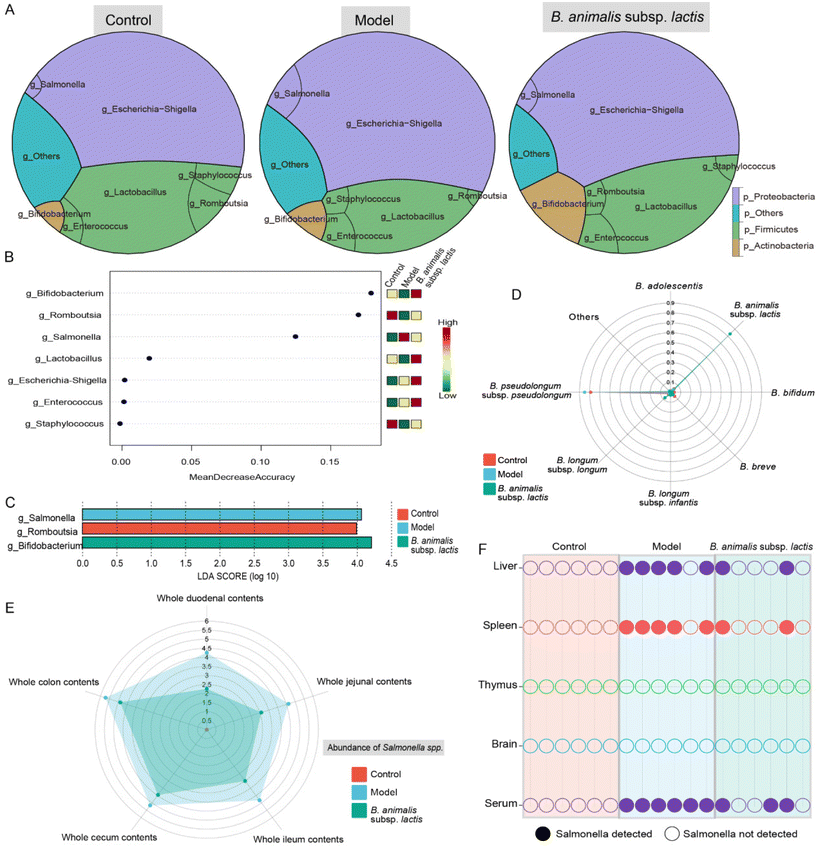
3.2 B. animalis subsp. lactis pretreatment enhances neonatal rats’ resilience against S. typhimurium SL1344 infection by improving gut microbiota and reducing Salmonella load in intestinal contents and viscera
To investigate the potential role of B. animalis subsp. lactis in modulating the neonatal rat intestinal microbiota to combat S. typhimurium infection, we analysed the colonic microbial network structure using 16S rRNA gene sequencing. As previously observed in neonatal rats,19 Proteobacteria and Firmicutes were identified as the major phyla shaping the neonatal rat intestinal microbial network (Fig. 2A), consistent with findings in human infants.30 Notably, both B. animalis subsp. lactis pretreatment and S. typhimurium infection considerably influenced the neonatal rat intestinal microbiota, reshaping the network structure of the intestinal microbial barrier. Specifically, infection with S. typhimurium resulted in a notable increase in the abundance of Proteobacteria, with Salmonella seemingly playing a crucial role in the gut (Fig. 2A). Additionally, B. animalis subsp. lactis-treated neonatal rats showed a significant increase in the abundance of Actinobacteria, which was primarily driven by the dominance of Bifidobacterium (Fig. 2A). These findings were further supported by linear discriminant analysis effect size (LEfSe) and random forest analyses, which identified Salmonella and Bifidobacterium as significantly enriched biomarkers in the model group and the B. animalis subsp. lactis group, respectively (Fig. 2B and C).Furthermore, the enrichment of the genus Bifidobacterium prompted us to investigate its species-level composition. The results revealed that B. animalis subsp. lactis pretreatment significantly altered the composition of Bifidobacterium in the neonatal rat intestine. Under natural conditions, B. pseudolongum dominated the gut microbiota of neonatal rats (Fig. 2D). However, following B. animalis subsp. lactis pretreatment, a pattern characterised by the predominance of B. animalis subsp. lactis was established (Fig. 2D). We further investigated the contents of Salmonella in different segments of the intestines of neonatal rats. As expected, the model group exhibited the highest level of Salmonella colonisation in all intestinal segments, while pretreatment with B. animalis subsp. lactis significantly reduced Salmonella colonisation (the Salmonella content in the control group was below the detection limit) (Fig. 2E). Notably, the colon and cecum serve as the main sites for Salmonella and Bifidobacteria colonisation,31,32 and B. animalis subsp. lactis exhibit a robust inhibitory effect on Salmonella in these areas. Notwithstanding, B. animalis subsp. lactis's inhibitory impact on Salmonella colonisation is not confined to the colon and cecum alone. Other segments of the intestine, including the duodenum, jejunum, and ileum, displayed a more significant inhibitory effect, which might be attributed to Salmonella's weak colonisation capability in these regions. Overall, the observation reflects the ameliorative effect of B. animalis subsp. lactis on the entire intestinal microecology. Furthermore, tissue invasion by Salmonella through the bloodstream, involving the liver and spleen, was observed. This finding was supported by the detection of Salmonella in serum samples and in five of six liver and spleen samples from the model group (Fig. 2F). However, in the B. animalis subsp. lactis pretreatment group, the detection rates of Salmonella in the corresponding samples were significantly reduced (Fig. 2F). Notably, we did not detect Salmonella in the thymus or brain in any of the experimental groups (Fig. 2F), suggesting that Salmonella had not directly invaded these organs. However, it is crucial to acknowledge that the absence of detected infection in these organs may also be attributable to limitations in the sensitivity of our detection method. Nonetheless, the results indicated that, if present, the infection load in these organs was very low. Additionally, it is important to acknowledge that the risk of infection may also arise in sites not directly invaded, as bacterial surface antigens or toxins can be released into the bloodstream or lymphatic system.33
3.3 B. animalis subsp. lactis pretreatment assists neonatal rats in combating S. typhimurium SL1344 infection by regulating peripheral immune homeostasis
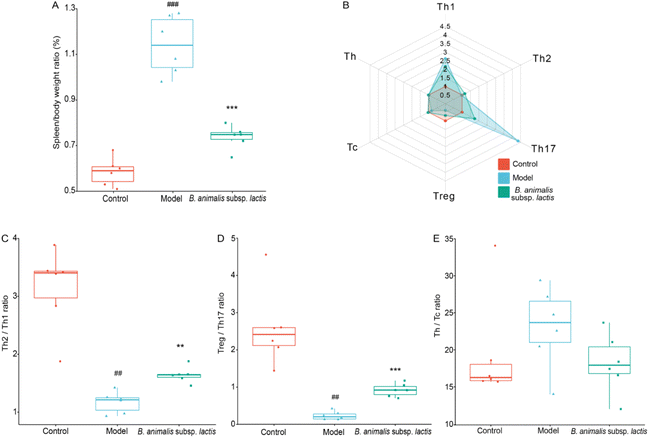
The results above suggested that the spleen, as a crucial peripheral immune organ,34 might have become a primary target immune organ in neonatal rats after S. typhimurium SL1344 invaded the bloodstream (Fig. 2F). We observed that the model group exhibited obvious splenomegaly, while B. animalis subsp. lactis pretreatment significantly suppressed this phenomenon (Fig. 3A). Consequently, we further explored the immune balance in the spleen, with a particular focus on different T lymphocyte subpopulations, including the pro-inflammatory T helper cell (Th) 1, Th17, and cytotoxic T lymphocyte (Tc) subsets, as well as the regulatory Th2, regulatory T cell (Treg) and Th cell subsets (representative flow cytometry dot plots are shown in Fig. S1†), which play key roles in immune activation, cytotoxicity, and inflammation regulation.35 T cell subsets represent distinct types of immunoregulation, and delicate and intricate interplay between subsets, such as the Treg–Th17, Th2–Th1, and Th–Tc balances, during the development and functional regulation of the neonatal immune system is crucial for ensuring the appropriateness and coordination of immune responses.36S. typhimurium SL1344 infection significantly increased the frequencies of Th1 and Th17 cells in the neonatal rat spleen, while markedly reducing the frequencies of Treg; no significant effects on the frequencies of Th2, Tc, and Th cells were observed (Fig. 3B). Increased frequencies of Th1 and Th17 cells might play a crucial role in the early immune response to infection, while decreases in Treg might weaken immune suppression, leading to inappropriate enhancement of the immune response.35 In the T cell subpopulation balance analysis, S. typhimurium SL1344 infection shifted the Th2–Th1 immune balance towards Th1 dominance in the spleens of the neonatal rats (Fig. 3C), while the Treg–Th17 balance was significantly shifted towards Th17 differentiation (Fig. 3D). S. typhimurium SL1344 infection did not appear to disrupt the Th–Tc cell balance (Fig. 3E). Notably, pretreatment with B. animalis subsp. lactis significantly reduced the immune balance shift induced by S. typhimurium SL1344 infection, reducing the gap between the control group and the model group (Fig. 3B–D). These findings suggest that B. animalis subsp. lactis pretreatment helps to restore the spleen's anti-inflammatory state during neonatal rats’ response to S. typhimurium SL1344 infection, emphasising the potential role of B. animalis subsp. lactis in maintaining the immune balance in the spleen.
3.4 B. animalis subsp. lactis pretreatment supports neonatal rats in combating S. typhimurium SL1344 infection through modulation of central immune homeostasis
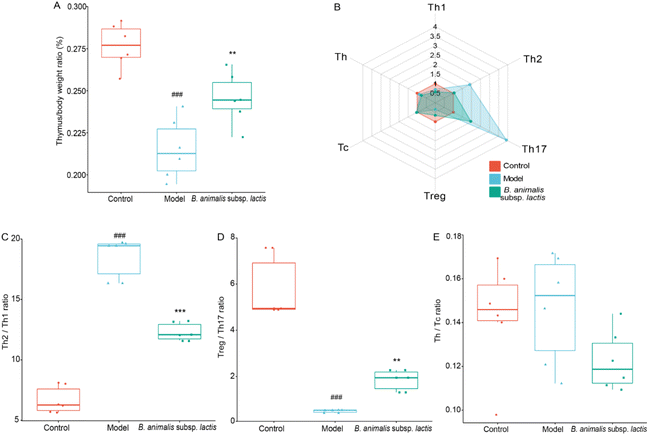
The results obtained above indicated that the thymus of the neonatal rats was not prominently invaded by S. typhimurium SL1344 (Fig. 2F). However, this bacterium might exert adverse effects on thymic immune development in neonatal hosts through mechanisms such as influencing immune cell migration and activation as well as disrupting immune tolerance.37 Notably, S. typhimurium SL1344 infection indeed led to impaired thymic development in the neonatal rats, resulting in a reduction in the thymus-to-body weight ratio during the early developmental stage (Fig. 4A), when it would be expected to increase.38 However, pretreatment with B. animalis subsp. lactis significantly attenuated this reduction in the thymus-to-body weight ratio (Fig. 4A), thereby mitigating the detrimental impact of the infection on thymic development.Upon further analysis of the thymic T cell subpopulation frequencies (representative flow cytometry dot plots are shown in Fig. S2†), we observed significant alterations in response to Salmonella infection, both in terms of quantity and diversity of T cell types (Fig. 4B). It seems that the development and maturation of T cells in the thymus were not hindered, as previously demonstrated.39 Similar to the observations in the spleen, S. typhimurium SL1344 infection led to an increase in the frequency of Th17 cells and a decrease in that of Treg cells in the thymuses of neonatal rats (Fig. 4B). This disruption resulted in an imbalance between Treg and Th17 cells, which, however, could be attenuated by pretreatment with B. animalis subsp. lactis (Fig. 4B and D). Meanwhile, the frequencies of Th and Tc cells remained unaffected, indicating that the balance between these subsets remained stable (Fig. 4B and E).
In contrast to the observations in the spleen, Salmonella infection caused a significant decrease in the frequency of Th1 cells and an increase in the frequency of Th2 cells in the neonatal rat thymus (Fig. 4B). While B. animalis subsp. lactis treatment did not completely reverse the reduction in Th1 cells induced by infection, it substantially restored the frequency of Th2 cells to a level comparable to that of the uninfected group (Fig. 4B). This pattern of response suggested that the infection-induced Th2–Th1 imbalance in the thymus could be successfully counteracted by B. animalis subsp. lactis, although not fully restored to the levels observed in the thymus of a naturally uninfected neonatal rat (Fig. 4C).
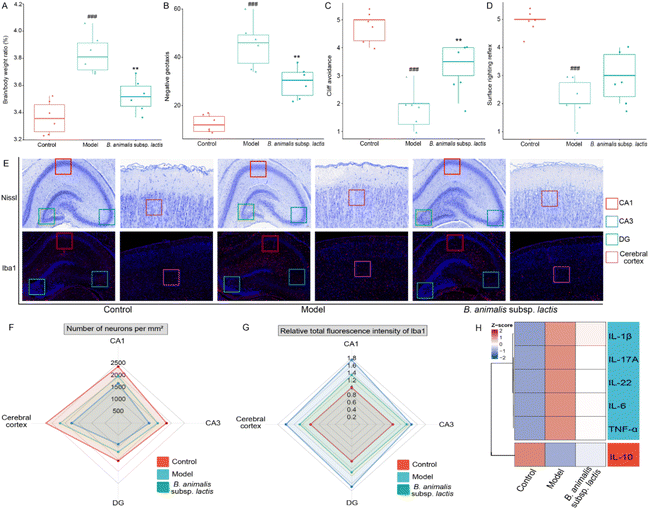
3.5 B. animalis subsp. lactis pretreatment aids neonatal rats in combating S. typhimurium SL1344 infection by modulating neuroimmune homeostasis
Given the above results, the neuroimmune responses in the brains of the neonatal rats were investigated in-depth. Infection with S. typhimurium SL1344 led to a clear increase in the brain-to-body weight ratio of the rats (Fig. 5A). Notably, supplementation with B. animalis subsp. lactis partially mitigated this increase, although it did not fully restore the ratio to normal levels (Fig. 5A). Additionally, in the investigation of neonatal reflex development, promising results were obtained. While no significant improvement was observed in the surface righting reflex (Fig. 5D), B. animalis subsp. lactis exhibited an overall potential for enhancement, specifically regarding improved negative geotaxis (Fig. 5B) and cliff avoidance (Fig. 5C).To further explore the beneficial effects of B. animalis subsp. lactis on the neuroprotection of neonatal rats challenged by S. typhimurium SL1344, we digitally analysed Nissl-stained brain sections from specific regions (representative images shown in Fig. 5E), namely the CA1 area (indicated by the red box), CA3 area (indicated by the blue box), DG area (indicated by the green box), and cortex (indicated by the purple box), to quantify the neuronal numbers and densities. The results clearly demonstrated that exposure to S. typhimurium SL1344 significantly reduced the numbers of neurons in various brain regions (Fig. 5F). However, the addition of B. animalis subsp. lactis effectively counteracted this impact, bridging the gap between the model and control groups (Fig. 5F).
Furthermore, an in-depth examination of the neuroimmune inflammatory response to S. typhimurium SL1344 in the brain was carried out through IF staining to detect the microglial marker ionised calcium-binding adapter molecule 1 (Iba1). Microglial cells, as resident immune cells in the brain, rapidly detect neuronal damage or injury and become activated to engulf and clear dead cells.40 Under the condition of S. typhimurium SL1344 infection, the Iba1 staining intensity in various specific brain regions of the neonatal rats was significantly enhanced (Fig. 5E and G), indicating the activation of neuroimmune cells. However, pretreatment with B. animalis subsp. lactis improved the condition of the infected neonatal rats, as evidenced by the attenuation of Iba1 activation (Fig. 5E and G). The finding is consistent with our investigation of cytokines in the brain cortex, suggesting that B. animalis subsp. lactis pretreatment helps to maintain a balance between pro-inflammatory and anti-inflammatory responses (Fig. 5H).
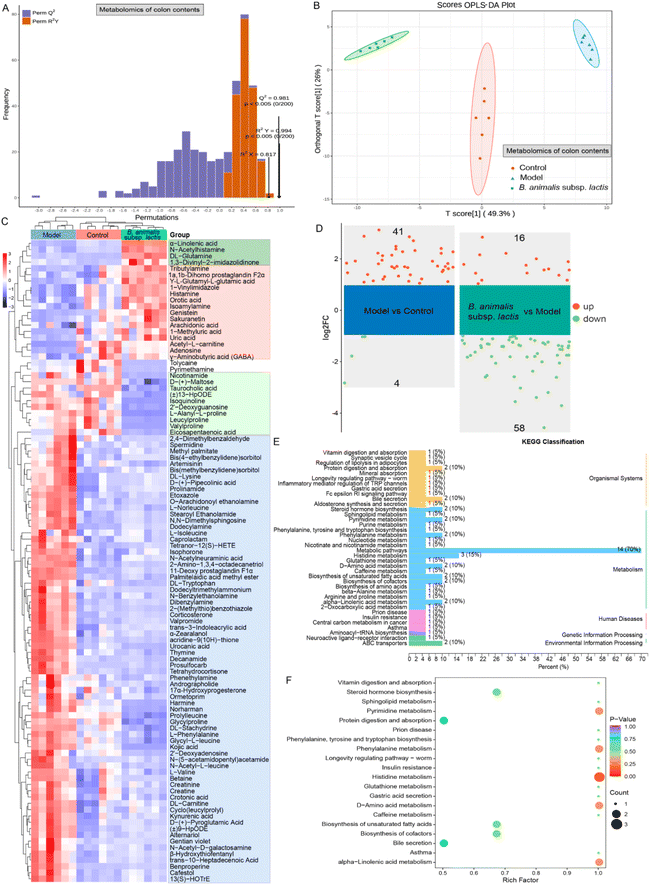
3.6 Use of colonic content metabolomics to unveil the impact of B. animalis subsp. lactis pretreatment on neonatal rats’ ability to combat S. typhimurium infection
To gain a deeper understanding of the impact of B. animalis subsp. lactis pretreatment on neonatal rats’ ability to combat S. typhimurium infection, we conducted metabolomics analysis of the colonic content. To ensure reliability, we first subjected all of the experimental colonic content samples and QC samples to principal component analysis (PCA) to assess the stability of the metabolomic profiles. The PCA results showed distinct clustering of the QC samples in both the positive and negative electrospray ionisation modes (Fig. S3A and B†). This result indicated strong instrument stability and high data reliability throughout the testing process. Furthermore, we employed orthogonal partial least-squares discriminant analysis (OPLS-DA) for data processing and validated the model through 200 permutation tests to ensure the accurate differentiation of different samples without overfitting (Fig. 6A).We further observed the clustering pattern of metabolites in each experimental group, and all groups exhibited significant separation (Fig. 6B). This separation strongly indicated significant changes in the colonic content metabolome. To gain deeper insights into these changes, we performed a differential analysis and KEGG annotation enrichment analysis on metabolites with variable importance in the projection (VIP) values greater than 1, resulting in a total of 102 metabolites being involved (Fig. 6C). Among the changes induced by Salmonella infection, 71 metabolites were widely activated (as shown in the blue box, Fig. 6C). These metabolites are primarily associated with carnitine synthesis, biotin metabolism, methionine metabolism, steroidogenesis, glycine and serine metabolism and beta oxidation of very long chain fatty acids in KEGG pathways. Promisingly, however, B. animalis subsp. lactis intervention demonstrated a certain degree of inhibition, i.e., it mitigated the activation levels of these metabolites induced by the infection (Fig. 6C). Additionally, Salmonella infection led to the downregulation of 17 metabolites, while B. animalis subsp. lactis intervention reversed the changes in 15 of these metabolites (as indicated by the red box, Fig. 6C). These metabolites are mainly associated with several relevant KEGG metabolic pathways, including purine metabolism, beta oxidation of very long chain fatty acids, alpha linolenic acid and linoleic acid metabolism, betaine metabolism, caffeine metabolism, oxidation of branched chain fatty acids. Moreover, we observed that B. animalis subsp. lactis intervention led to the production of four metabolites (indicated by the dark green box) and reduced the levels of 10 metabolites (indicated by the light green box) (Fig. 6C). Importantly, these regulatory changes were independent of S. typhimurium SL1344 infection (Fig. 6C). The affected metabolites primarily involved various KEGG metabolic pathways, including alpha linolenic acid and linoleic acid metabolism, nicotinate and nicotinamide metabolism, phenylacetate metabolism, purine metabolism, urea cycle, starch and sucrose metabolism.
Furthermore, we conducted a volcano plot analysis of the potential differentially regulated metabolites using stringent criteria (fold change (FC) > 2 and p < 0.05) to further deepen our understanding of the underlying molecular mechanism by which B. animalis subsp. lactis pretreatment enhanced resistance against S. typhimurium infection in neonatal rats. In comparison with the natural state, exposure to Salmonella led to a significant upregulation of 41 metabolites in the neonatal rat intestines, while four metabolites were markedly downregulated (Fig. 6D). Nonetheless, following pretreatment with B. animalis subsp. lactis, the intestines of neonatal rats regulated 74 active metabolites in response to Salmonella infection challenge. Among these, 58 intestinal metabolites were broadly downregulated, while 16 metabolites were activated (Fig. 6D). These metabolites primarily belong to the ‘metabolism’ category in the KEGG classification, and are specifically identified as ‘metabolic pathways’ (Fig. 6E). To gain deeper insights into the functions of these differentially regulated metabolites, we performed KEGG pathway annotation and analysis of all of the screened differentially expressed metabolites to determine the main metabolic and signalling pathways involving the B. animalis subsp. lactis-derived metabolites. In total, we annotated 20 metabolic pathways, with a focus on histidine metabolism, followed by pyrimidine metabolism, phenylalanine metabolism, D-amino acid metabolism, and alpha-linolenic acid metabolism (Fig. 6F).
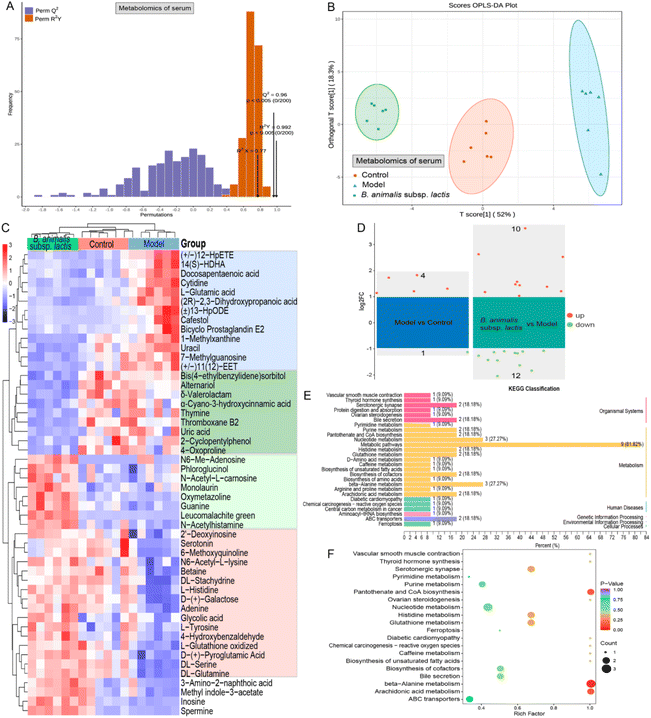
3.7 Serum metabolomics revealed systemic immunomodulatory effects of B. animalis subsp. lactis pretreatment against S. typhimurium infection in neonatal rats
As Salmonella can enter the bloodstream, and given that intestinal metabolites might leak into the bloodstream, we used serum metabolomics to gain a deeper understanding of the systemic immune regulatory effects of B. animalis subsp. lactis pretreatment on neonatal rats combating S. typhimurium infection. First, we confirmed the stability of the serum metabolomic profiles, the reliability of the OPLS-DA model, and the distinct separation of samples from different experimental groups, ensuring the credibility of our study (Fig. S4† and Fig. 7A, B).Similar to the results of the colonic content metabolomics analysis, numerous serum metabolites were found to have undergone changes in response to S. typhimurium infection in neonatal rats. As depicted in Fig. 7C, 13 potential metabolites (the blue box) were upregulated during the infection process, while 16 metabolites (the red box) were downregulated. These alterations primarily involved metabolic pathways such as ammonia recycling, beta-alanine metabolism, purine metabolism, pyrimidine metabolism, phenylalanine and tyrosine metabolism, and urea cycle, according to KEGG annotation enrichment analysis. Notably, B. animalis subsp. lactis pretreatment effectively reversed these changes (Fig. 7C). Furthermore, B. animalis subsp. lactis pretreatment induced additional changes that were independent of the infection process: it significantly suppressed the levels of nine serum metabolites (indicated by the dark green box) and increased the levels of eight metabolites (indicated by the light green box) (Fig. 7C). These changes primarily affected KEGG metabolic pathways related to purine metabolism, pyrimidine metabolism, and arachidonic acid metabolism.
When employing more stringent criteria (FC > 2 and p < 0.05) for further screening, we identified only five potential differentially regulated metabolites that differed significantly between the model group and the control group (Fig. 7D). The more minor differences observed in the serum metabolites than in the intestinal metabolites may be primarily attributable to the following factors. First, the Salmonella load in the serum is much lower than in the intestine, reducing the disruptive effects of bacterial and metabolic products on the host. Second, the circulatory system, being a relatively conservative and closed environment compared with the open intestinal system, possesses robust self-regulatory capabilities. Nevertheless, it is evident that B. animalis subsp. lactis pretreatment exerted regulatory effects on the serum metabolome of neonatal rats infected with Salmonella, involving 10 upregulated and 12 downregulated metabolites (Fig. 7D). KEGG classification analysis revealed that these 22 potential serum biomarkers derived from B. animalis subsp. lactis were concentrated in several specific metabolic pathways (Fig. 7E). The affected metabolic pathways included beta-alanine metabolism, pantothenate and CoA biosynthesis, and arachidonic acid metabolism (Fig. 7F).
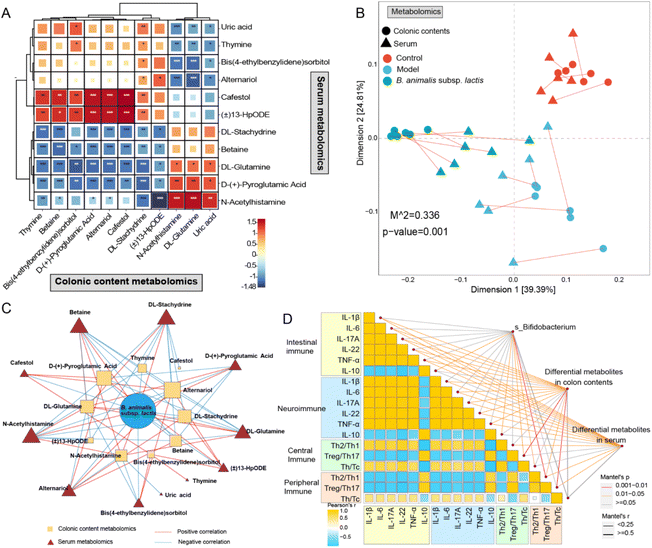
Based on the above analysis, we discovered 11 differentially regulated metabolites and seven distinct metabolic pathways that were shared between the colonic content metabolome and the serum metabolome. Moreover, the identified shared differential metabolites exhibited extensive and profound correlations between the two metabolic compartments according to Pearson's correlation analysis (Fig. 8A). This finding prompted us to further investigate their correlations. As anticipated, a strong correlation was observed between the colonic content metabolome and the serum metabolome based on Procrustes analysis (Fig. 8B). In addition, we observed an intricate network of correlations between the relative abundance of B. animalis subsp. lactis and the levels of differential metabolites shared between the colon contents and serum (Fig. 8C). This result reinforces the notion that the beneficial effects of B. animalis subsp. lactis on neonatal rats infected with S. typhimurium might be mediated through the gut and subsequently affect systemic immunity. Fig. 8D presents the Spearman correlation coefficients of immune-related indicators in specific organs, as well as the Mantel correlation coefficients between the aforementioned immune variables and the Bifidobacterium species composition, colonic content metabolome, and serum metabolome. We observed extensive and profound associations (Fig. 8C). In summary, these results not only reveal the close connection between the colonic content metabolome and the serum metabolome but also emphasise the regulatory role of B. animalis subsp. lactis in influencing the systemic immunity of neonatal rats against S. typhimurium infection.
4 Discussion
Salmonella infection poses a serious threat to neonates, and the issue of Salmonella antibiotic resistance is becoming increasingly prominent.2,3 Research has provided evidence supporting B. animalis subsp. lactis as a potential early probiotic intervention to regulate the intestinal immune balance and barrier function against Salmonella infection in neonates.19 However, the impact of B. animalis subsp. lactis on central, peripheral, and neural immunity, and the underlying mechanisms, remain uncertain. Therefore, this study aimed to comprehensively investigate the pivotal role of B. animalis subsp. lactis in the systemic immune response against Salmonella infection and explore its potential mechanisms. The results demonstrated that B. animalis subsp. lactis pretreatment influenced the systemic immune response by modulating the composition and metabolism of the gut microbiota, extending its regulatory effects beyond the intestines.In our experimental procedures, we followed previous studies19 and carefully selected a dose of 1 × 106 CFU per mL of S. typhimurium SL1344 to establish a reliable and stable infection model. However, the uniqueness of our study lies in the early intervention with B. animalis subsp. lactis, which started within 6 hours of the birth of neonatal rats. The purpose of this early intervention was to better create a stable, B. animalis subsp. lactis-dominated, beneficial gut microbiota during the initial stages of the neonatal rats’ lives, enabling us to comprehensively investigate the impact of B. animalis subsp. lactis on the infant host's immune imprint against Salmonella infection. We successfully achieved this concept. Our findings demonstrated that B. animalis subsp. lactis indeed restructured the gut microbial community of neonatal rats, with particularly significant effects on the Bifidobacterium population. The dominant Bifidobacterium species in the neonatal rat gut clearly shifted from B. pseudolongum to B. animalis subsp. lactis after oral gavage of B. animalis subsp. lactis, accompanied by a marked increase in the overall relative abundance of Bifidobacterium. The effectiveness of this intervention was further confirmed by random forest analysis, which revealed that Bifidobacterium surpassed other genera, even Salmonella, as the top-ranking differentiating taxon among the groups, with significantly higher mean decrease accuracy scores than those found in previous research.19 Moreover, B. animalis subsp. lactis pretreatment not only reduced the relative abundance of Salmonella in the colonic contents but also decreased the Salmonella load in various segments of the intestine in neonatal rats.
Considering that the gut is the primary target of Salmonella infection41 and serves as the colonisation site for B. animalis subsp. lactis,31 we followed previous studies19 and further investigated how B. animalis subsp. lactis pretreatment mitigated the effects of S. typhimurium SL1344 infection in neonatal rats and affected the cytokine balance and mucosal barrier integrity. To gain deeper insights into the intestinal immune modulation mediated by B. animalis subsp. lactis-induced intestinal early development during the defence against S. typhimurium infection in neonatal rats, we conducted a series of histological observations and assessed the expression of proteins related to neuromodulation and barrier development in the gut. Through HE staining, we observed that B. animalis subsp. lactis significantly promoted the development of goblet cells in the colon. This activation facilitated the production of mucins, thereby increasing the thickness of the mucosal layer, which serves as the primary physical barrier against Salmonella invasion in the intestines of neonatal rats.42 This finding highlights the critical role of the mucosal layer in the intestinal immune mechanism of B. animalis subsp. lactis against S. typhimurium infection in neonatal rats.19 Moreover, based on the enhanced levels of BDNF expression and cell proliferation in intestinal cells induced by B. animalis subsp. lactis pretreatment, our study not only corroborates prior discoveries concerning intestinal immune equilibrium but also highlights the significance of gut neuromodulation and barrier development within this context. Previous research has suggested that BDNF can inhibit TLR4 signaling, which, in turn, diminishes the loss of enteric glia triggered by TLR4 signaling.43 Given that S. typhimurium infection activates TLR4 signaling,19 it's conceivable that the anti-Salmonella impact of B. animalis subsp. lactis might be linked to its capacity to mitigate the decline in BDNF.
In neonatal rats, S. typhimurium infection has been demonstrated to induce compromised immune organ development and neuronal reflex function, along with inflicting severe disruption upon the integrity of the colonic barrier. Previous studies have demonstrated the highly invasive nature of S. typhimurium SL1344, which can invade organs such as the liver and spleen, leading to systemic infection.8 Therefore, we hypothesised that this pathogen might breach the intestinal barrier in neonatal rats, enter the bloodstream, and disseminate to organs like the liver, spleen, thymus, and brain, triggering extra-intestinal inflammatory responses. In line with our hypothesis, we detected Salmonella in serum, liver, and spleen samples from the model group. Evidently, pretreatment with B. animalis subsp. lactis not only reduced the content of Salmonella within the intestine but also diminished its burden in both intestinal and systemic tissues. It suggests that the direct reduction of the Salmonella load in various body sites might be one of the primary mechanisms through which B. animalis subsp. lactis exerts its anti-infective effects. Moreover, B. animalis subsp. lactis exhibited multifaceted regulation of the neonatal rat immune system against attack from Salmonella invasion or exposure to bacterial components, involving the spleen, thymus, and brain, which correspond to peripheral, central, and neural immunity, respectively. In the spleen and thymus, B. animalis subsp. lactis intervention facilitated the restoration of balance between different T lymphocyte subpopulations (Treg–Th17 and Th2–Th1) under infection, thereby maintaining immune homeostasis. The immune modulation observed in the spleen and thymus correlates with the trends in cytokines in the intestinal tract. It suggests that the pre-intervention with B. animalis subsp. lactis counteracts the excessive immune activation response triggered by Salmonella infection. The finding further underscores the imprinting effect of B. animalis subsp. lactis on the neonatal host's immune system, assisting in restraining excessive immune activation and maintaining immune balance. The specificity of this immune modulation was further validated in our study. The differences observed in the Th1 and Th2 cell responses between the spleen and thymus might be attributable to the fact that the thymus is not a direct target of S. typhimurium SL1344. Another important factor to consider is the difference in immune development and immune function of T cells between the thymus and the spleen. It is well-established that T cells are thymus-dependent lymphocytes that primarily develop and mature in the thymus. Therefore, the phenotypic differences observed in T cells in the thymus and spleen in response to Salmonella infection may be attributed to their distinct development and differentiation processes, which in turn impact their maturity and functional phenotype.44–48 Thus, the thymus plays a crucial role in neonatal immune development and tends to promote Th1 immune responses over Th2 responses, which favours early immune maturation.36 The intricate immune regulation mediated by B. animalis subsp. lactis implies its finely tuned ability to intervene in the host's immune processes, thereby avoiding undue impacts on systemic metabolism. Such immune modulatory effects also extended to neurodevelopment and neuroimmune homeostasis in neonatal rats. Recent research has shown that specific bifidobacterial strains maintain homeostasis in both the neonatal gut and peripheral immune systems, suppressing excessive immune reactions.12–14 We believe that B. animalis subsp. lactis should be considered among these beneficial bifidobacterial strains.
Through metabolomics studies, we uncovered the underlying molecular basis of B. animalis subsp. lactis pretreatment in mediating systemic immune regulation in neonatal rats infected with S. typhimurium. Given the direct association between the gut and Salmonella infection, investigating the changes in metabolites can directly reflect the infection status of neonatal rats and the beneficial effects of B. animalis subsp. lactis supplementation. Therefore, our analysis of the metabolomic profile of colon contents revealed that the substantial changes induced by Salmonella infection were effectively attenuated by B. animalis subsp. lactis pretreatment. Furthermore, B. animalis subsp. lactis was found to produce and suppress certain specific metabolites, such as α-linolenic acid and DL-glutamine. The conversion of α-linolenic acid is a crucial process for supplying arachidonic acid and docosahexaenoic acid, which are essential for the growth and development of infants.49 While DL-glutamine is classified as a ‘non-essential’ amino acid, its supplementation can be highly beneficial for the health of neonates, especially premature infants or those with underdeveloped intestines.50 A recent study demonstrated that DL-glutamine enhances the resistance of cholangiocytes to natural killer cell-mediated cytotoxicity, thereby ameliorating the symptoms of infection-induced biliary atresia and liver injury.51 To gain a deeper understanding of how B. animalis subsp. lactis modulates the systemic immune response against Salmonella infection in neonatal rats, we conducted paired serum metabolomics studies. Although the changes in serum metabolomics induced by Salmonella were relatively modest compared with those observed in the colon contents, B. animalis subsp. lactis still exerted significant regulatory effects. B. animalis subsp. lactis pretreatment extensively modulated the metabolomic profiles of both the colon contents and serum in neonatal rats through several shared metabolic pathways, such as pyrimidine metabolism, histidine metabolism, glutathione metabolism, caffeine metabolism, biosynthesis of unsaturated fatty acids, and bile secretion, which are known to be vital to early-life metabolism and cell development.52–55 Notably, we also observed a distinct correlation between the metabolomic profiles of the colon contents and serum, further reinforcing the conclusion that B. animalis subsp. lactis pretreatment impacts systemic immunity by regulating the composition and metabolism of the gut microbiota. Indeed, our pivotal findings indicate that B. animalis subsp. lactis plays a critical role in shaping the reciprocal interaction between the colon content and serum metabolomic profiles, thereby influencing the overall immune response in neonatal rats during S. typhimurium 1344 infection.
Research has extensively explored the beneficial effects of B. animalis subsp. lactis on the intestinal immune system. However, our study reveals that for neonatal hosts, the benefits of B. animalis subsp. lactis extend beyond the intestinal immune response, encompassing the central, peripheral, and neural immune systems. The mechanisms involved include metabolites produced by the gut microbiota, such as unsaturated fatty acids. These metabolites may serve as crucial mediators through which B. animalis subsp. lactis influences the neonatal host's immune system, subsequently impacting the serum metabolome. Hence, it is plausible that B. animalis subsp. lactis may exert a more extensive and systematic regulatory influence on the neonatal host's immune system than previously anticipated in earlier research. It should be noted that the current study primarily focuses on the systemic immune response and has not extensively explored local mucosal reactions. For instance, a more in-depth investigation of the mesenteric lymph nodes in their role as intestinal immune effector sites could provide valuable insights. Additionally, integrating markers of T cell maturation and different developmental stages into future research, especially when investigating the presence of anomalies or delays in the immune activity of neonatal hosts during infections, will enhance the generation of a more comprehensive dataset. Moreover, the potential impact of thymic developmental inhibition induced by Salmonella infection on thymic immune activity during the recovery phase or after infection clearance is an intriguing and concerning issue. In conclusion, these findings offer novel insights into the immunomodulatory mechanisms of B. animalis subsp. lactis and provide promising prospects for the future development of gut microbiota-based immunotherapies.
Abbreviations
| B. animalis subsp. lactis | Bifidobacterium animalis subsp. lactis |
| Bif-GroEL | Bifidobacterium GroEL gene |
| BDNF | Brain-derived neurotrophic factor |
| CFU | Colony-forming units |
| Tc | Cytotoxic T lymphocyte |
| DAO | Diamine oxidase |
| ESI | Electrospray ionization |
| ELISA | Enzyme-linked immunosorbent assay |
| FC | Fold change |
| HE | Hematoxylin–eosin |
| IF | Immunofluorescence |
| IHC | Immunohistochemistry |
| IL | Interleukin |
| Iba1 | Ionized calcium-binding adapter molecule 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LEfSe | Linear discriminant analysis effect size |
| OPLS-DA | Orthogonal partial least squares discriminant analysis |
| PBS | Phosphate buffer solution |
| P | Postnatal day |
| PCA | Principal component analysis |
| PCNA | Proliferating cell nuclear antigen |
| QE/MS | Q-exactive mass spectrometer |
| QC | Quality control |
| Treg | Regulatory T cell |
| S. typhimurium | Salmonella enterica serovar Typhimurium |
| TUNEL | TdT-mediated dUTP-biotin nick end labeling |
| Th | T helper cell |
| TNF-α | Tumor necrosis factor-alpha |
| UPLC | Ultraperformance liquid-chromatography |
| VIP | Variable important in the projection |
Data availability
All data generated or analyzed during this study are included in this published article and its ESI files.Author contributions
Chunxiu Lin: investigation, formal analysis, writing – original draft, visualization. Yugui Lin: investigation, formal analysis, writing – original draft. Shunhe Wang: methodology. Jialiang Wang: investigation. Xuhua Mao: methodology, resources. Yonghua Zhou: methodology, resources. Hao Zhang: conceptualization, methodology, software, data curation. Wei Chen: resources, data curation, writing – review & editing, supervision, funding acquisition. Gang Wang: conceptualization, validation, resources, data curation, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 32272332, 31972052, 32021005, 31820103010), the Fundamental Research Funds for the Central Universities (JUSRP622020, JUSRP22006, JUSRP51501), the Program of Collaborative Innovation Centre of Food Safety and Quality Control in Jiangsu Province, Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX22_2391).References
- G. L. Popa and M. I. Popa, Salmonella, spp. infection - a continuous threat worldwide, Germs, 2021, 11, 88–96 CrossRef PubMed.
- M. A. Gordon, N. A. Feasey, T. S. Nyirenda and S. M. Graham, Nontyphoid Salmonella disease, in Hunter's Trop, Med. Emerg. Infect. Dis, 10th edn, 2020, pp. 500–506 Search PubMed.
- S. Graham, Salmonellosis in children in developing and developed countries and populations, Curr. Opin. Infect. Dis., 2002, 15, 507–512 CrossRef.
- A. Aleksandrowicz, E. Carolak, A. Dutkiewicz, A. Blachut, W. Waszczuk and K. Grzymajlo, Better together-Salmonella biofilm-associated antibiotic resistance, Gut Microbes, 2023, 15, 2229937 CrossRef PubMed.
- F. Micoli, F. Bagnoli, R. Rappuoli and D. Serruto, The role of vaccines in combatting antimicrobial resistance, Nat. Rev. Microbiol., 2021, 19, 287–302 CrossRef CAS.
- A. W. L. Rogers, R. M. Tsolis and A. J. Baumler, Salmonella versus the microbiome, Microbiol. Mol. Biol. Rev., 2021, 85, e00027–e00019 CrossRef CAS PubMed.
- J. Liu, H. Liu, H. Liu, Y. Teng, N. Qin, X. Ren and X. Xia, Live and pasteurized Akkermansia muciniphila, decrease susceptibility to Salmonella Typhimurium infection in mice, J. Adv. Res., 2023, 52, 89–102 CrossRef CAS PubMed.
- J. Liu, Z. Gu, W. Lu, D. Hu, X. Zhao, H. Huang, H. Zhang, J. Zhao and W. Chen, Akkermansia muciniphila, decrease susceptibility to Salmonella Typhimurium infection in mice, J. Adv. Res., 2018, 9, 2787–2795 CAS.
- J. Liu, Z. Gu, H. Zhang, J. Zhao and W. Chen, Preventive effects of Lactobacillus plantarum ST-III against Salmonella infection, LWT – Food Sci. Technol., 2019, 105, 200–205 CrossRef CAS.
- J. Liu, D. Hu, Y. Chen, H. Huang, H. Zhang, J. Zhao, Z. Gu and W. Chen, Strain-specific properties of Lactobacillus plantarum for prevention of Salmonella infection, Food Funct., 2018, 9, 3673–3682 RSC.
- Y.-F. Huang, P.-Y. Liu, Y.-Y. Chen, B.-R. Nong, I.-F. Huang, K.-S. Hsieh and K.-T. Chen, Three-combination probiotics therapy in children with Salmonella and Rotavirus gastroenteritis, J. Clin. Gastroenterol., 2014, 48, 37–42 CrossRef.
- M. F. Laursen, M. Sakanaka, N. von Burg, U. Morbe, D. Andersen, J. M. Moll, C. T. Pekmez, A. Rivollier, K. F. Michaelsen, C. Molgaard, M. V. Lind, L. O. Dragsted, T. Katayama, H. L. Frandsen, A. M. Vinggaard, M. I. Bahl, S. Brix, W. Agace, T. R. Licht and H. M. Roager, Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut, Nat. Microbiol., 2021, 6, 1367–1382 CrossRef CAS.
- B. M. Henrick, L. Rodriguez, T. Lakshmikanth, C. Pou, E. Henckel, A. Arzoomand, A. Olin, J. Wang, J. Mikes, Z. Tan, Y. Chen, A. M. Ehrlich, A. K. Bernhardsson, C. H. Mugabo, Y. Ambrosiani, A. Gustafsson, S. Chew, H. K. Brown, J. Prambs, K. Bohlin, R. D. Mitchell, M. A. Underwood, J. T. Smilowitz, J. B. German, S. A. Frese and P. Brodin, Bifidobacteria-mediated immune system imprinting early in life, Cell, 2021, 184, 3884–3898 CrossRef CAS PubMed.
- J. Samara, S. Moossavi, B. Alshaikh, V. A. Ortega, V. K. Pettersen, T. Ferdous, S. L. Hoops, A. Soraisham, J. Vayalumkal, D. Dersch-Mills, J. S. Gerber, S. Mukhopadhyay, K. Puopolo, T. A. Tompkins, D. Knights, J. Walter, H. Amin and M.-C. Arrieta, Supplementation with a probiotic mixture accelerates gut microbiome maturation and reduces intestinal inflammation in extremely preterm infants, Cell Host Microbe, 2022, 30, 696–711 CrossRef CAS.
- C. B. Wong, T. Odamaki and J. Xiao, Insights into the reason of human-residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits, FEMS Microbiol. Rev., 2020, 44, 369–385 CrossRef CAS PubMed.
- R. L. Morgan, G. A. Preidis, P. C. Kashyap, A. Weizman and B. Sadeghirad, Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: A systematic review and network meta-analysis of randomized trials, Gastroenterology, 2020, 159, 467–480 CrossRef CAS PubMed.
- R. Nocerino, F. De Filippis, G. Cecere, A. Marino, M. Micillo, C. Di Scala, C. de Caro, A. Calignano, C. Bruno, L. Paparo, A. M. Iannicelli, L. Cosenza, Y. Maddalena, G. della Gatta, S. Coppola, L. Carucci, D. Ercolini and R. B. Canani, The therapeutic efficacy of Bifidobacterium animalis subsp. Lactis, BB-12® in infant colic: A randomized, double-blind, placebo-controlled trial, Aliment. Pharmacol. Ther., 2020, 51, 110–120 CrossRef CAS PubMed.
- S. Ku, S. Yang, H. H. Lee, D. Choe, T. Johnston, G. E. Ji and M. S. Park, Biosafety assessment of Bifidobacterium animalis subsp. Lactis AD011 used for human consumption as a probiotic microorganism, Food Control, 2020, 117, 106985 CrossRef CAS.
- Y. Lin, Z. Xie, Z. Li, C. Yuan, C. Zhang, Y. Li, K. Xie and K. Wang, Assessment of the role and mechanism of Bifidobacterium animalis subsp. lactis, isolated from neonates’ feces in protecting neonatal rats from Salmonella infection, Microb. Pathog., 2023, 174, 105935 CrossRef CAS PubMed.
- B. Yang, M. Ding, Y. Chen, F. Han, C. Yang, J. Zhao, P. Malard, C. Stanton, R. P. Ross, H. Zhang and W. Chen, Development of gut microbiota and bifidobacterial communities of neonates in the first 6 weeks and their inheritance from mother, Gut Microbes, 2021, 13, 1–13 Search PubMed.
- L. Hu, W. Lu, L. Wang, M. Pan, H. Zhang, J. Zhao and W. Chen, Assessment of Bifidobacterium species using groEL, gene on the basis of Illumina MiSeq high-throughput sequencing, Genes, 2017, 8, 336 CrossRef PubMed.
- C. Lin, Y. Zheng, J. Lu, H. Zhang, G. Wang and W. Chen, Differential reinforcement of intestinal barrier function by various Lactobacillus reuteri, strains in mice with DSS-induced acute colitis, Life Sci., 2023, 314, 121309 CrossRef CAS.
- C. J. Heyser, Assessment of Developmental Milestones in Rodents, Curr. Protoc. Neurosci., 2003, 25, 8.18.1–8.18.15 Search PubMed.
- G. Zhu, M. Guo, J. Zhao, H. Zhang, G. Wang and W. Chen, Integrative metabolomic characterization reveals the mediating effect of Bifidobacterium breve on amino acid metabolism in a mouse model of Alzheimer's disease, Nutrients, 2022, 14, 735 CrossRef CAS PubMed.
- Z. Tan, Y. Ou, W. Cai, Y. Zheng, H. Li, Y. Mao, S. Zhou and J. Tu, Advances in the clinical application of histamine and diamine oxidase (DAO) activity: A review, Catalysts, 2023, 13, 48 CrossRef CAS.
- M. E. V. Johansson and G. C. Hansson, Immunological aspects of intestinal mucus and mucins, Nat. Rev. Immunol., 2016, 16, 639–649 CrossRef CAS PubMed.
- A. Joly, F. Leulier and F. De Vadder, Microbial modulation of the development and physiology of the enteric nervous system, Trends Microbiol., 2021, 29, 686–699 CrossRef CAS PubMed.
- W. Strzalka and A. Ziemienowicz, Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation, Ann. Bot., 2011, 107, 1127–1140 CrossRef CAS PubMed.
- R. Mirzayans and D. Murray, Do TUNEL and other apoptosis assays detect cell death in preclinical studies?, Int. J. Mol. Sci., 2020, 21, 9090 CrossRef CAS.
- J. E. Koenig, A. Spor, N. Scalfone, A. D. Fricker, J. Stombaugh, R. Knight, L. T. Angenent and R. E. Ley, Succession of microbial consortia in the developing infant gut microbiome, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4578–4585 CrossRef.
- C. Lin, Y. Lin, H. Zhang, G. Wang, J. Zhao, H. Zhang and W. Chen, Intestinal “Infant-type” bifidobacteria mediate immune system development in the first 1000 days of life, Nutrients, 2022, 14, 1498 CrossRef CAS.
- K. Ehrhardt and G. A. Grassl, Mouse model to study Salmonella-induced colitis, Methods Mol. Biol., 2022, 2427, 201–213 CrossRef CAS PubMed.
- E. Lemichez, M. Lecuit, X. Nassif and S. Bourdoulous, Breaking the wall: targeting of the endothelium by pathogenic bacteria, Nat. Rev. Microbiol., 2010, 8, 93–104 CrossRef CAS PubMed.
- H. Yu, Y. Cai, A. Zhong, Y. Zhang, J. Zhang and S. Xu, The “dialogue” between central and peripheral immunity after ischemic stroke: Focus on spleen, Front. Immunol., 2021, 12, 792522 CrossRef CAS PubMed.
- M. A. Van Herck, J. Weyler, W. J. Kwanten, E. L. Dirinck, B. Y. De Winter, S. M. Francque and L. Vonghia, The differential roles of T cells in non-alcoholic fatty liver disease and obesity, Front. Immunol., 2019, 10, 82 CrossRef CAS.
- K. Donald and B. B. Finlay, Early-life interactions between the microbiota and immune system: Impact on immune system development and atopic disease, Nat. Rev. Immunol., 2023, 23, 735–748 CrossRef CAS.
- J. R. Kurtz, J. A. Goggins and J. B. McLachlan, Salmonella infection: Interplay between the bacteria and host immune system, Immunol. Lett., 2017, 190, 42–50 CrossRef CAS.
- A. B. Lansdown, Histological observations on thymic development in fetal and newborn mammals subject to intrauterine growth retardation, Biol. Neonate, 1977, 31, 252–259 CrossRef CAS.
- J. P. Leyva-Rangel, M. de Los Angeles Hernandez-Cueto, C.-S. Galan-Enriquez, M. Lopez-Medina and V. Ortiz-Navarrete, Bacterial clearance reverses a skewed T-cell repertoire induced by Salmonella infection, Immun., Inflammation Dis., 2015, 3, 209–223 CrossRef CAS.
- H. Zheng, C. Zhang, J. Zhang and L. Duan, “Sentinel or accomplice”: Gut microbiota and microglia crosstalk in disorders of gut-brain interaction, Protein Cell, 2023, 14, 726–742 CrossRef PubMed.
- J. E. Galan, Salmonella Typhimurium and inflammation: A pathogen-centric affair, Nat. Rev. Microbiol., 2021, 19, 716–725 CrossRef CAS PubMed.
- J. K. Gustafsson and M. E. V. Johansson, The role of goblet cells and mucus in intestinal homeostasis, Nat. Rev. Gastroenterol. Hepatol., 2022, 19, 785–803 CrossRef.
- M. L. Kovler, A. J. G. Salazar, W. B. Fulton, P. Lu, Y. Yamaguchi, Q. Zhou, M. Sampah, A. Ishiyama, T. Prindle Jr., S. Wang, H. Jia, P. Wipf, C. P. Sodhi and D. J. Hackam, Toll-like receptor 4-mediated enteric glia loss is critical for the development of necrotizing enterocolitis, Sci. Transl. Med., 2010, 8, 93–104 CrossRef.
- H.-S. Lee, H.-R. Kim, E.-H. Lee, M. H. Jang, S.-B. Kim, J.-W. Park, J.-Y. Seoh and Y.-J. Jung, Characterization of CCR9 expression and thymus-expressed chemokine responsiveness of the murine thymus, spleen and mesenteric lymph node, Immunobiology, 2012, 217, 402–411 CrossRef CAS.
- G. Cui, T. Hara, S. Simmons, K. Wagatsuma, A. Abe, H. Miyachi, S. Kitano, M. Ishii, S. Tani-ichi and K. Ikuta, Characterization of the IL-15 niche in primary and secondary lymphoid organs in vivo, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1915–1920 CrossRef CAS PubMed.
- B. Vidana, N. Majo, M. Perez, M. Montoya, J. Martorell and J. Martinez, Immune system cells in healthy ferrets: an immunohistochemical study, Vet. Pathol., 2014, 51, 775–786 CrossRef CAS PubMed.
- D. M. Richards, M. Delacher, Y. Goldfarb, D. Kaegebein, A.-C. Hofer, J. Abramson and M. Feuerer, Treg cell differentiation: from thymus to peripheral tissue, Prog. Mol. Biol. Transl. Sci., 2015, 136, 175–205 Search PubMed.
- J. Bai, L. Zhang, Z. Zhao, N. Li, B. Wang and L. Yang, Expression of melatonin receptors and CD4 in the ovine thymus, lymph node, spleen and liver during early pregnancy, Immunology, 2020, 160, 52–63 CrossRef CAS.
- C. Campoy, A. M. Chisaguano Tonato, A. de la Garza Puentes, M. Sáenz de Pipaón, E. Verduci, B. Koletzko, I. González Casanova, E. Larqué, R. Valenzuela, J. M. Moreno Villares and A. Gil, Controversy about the critical role of long-chain polyunsaturated fatty acids, arachidonic acid (ARA) and docosahexaenoic acid (DHA), during infancy, Nutr. Hosp., 2021, 38, 1101–1112 CAS.
- L. Zheng-Hong, W. Dan-Hua and D. Mei, Effect of parenteral glutamine supplementation in premature infants, Chin. Med. J., 2007, 120, 140–144 Search PubMed.
- J. J. Jee, L. Yang, P. Shivakumar, P.-P. Xu, R. Mourya, U. Thanekar, P. Yu, Y. Zhu, Y. Pan, H. Wang, X. Duan, Y. Ye, B. Wang, Z. Jin, Y. Liu, Z. Cao, M. Watanabe-Chailland, L. E. Romick-Rosendale, M. Wagner, L. Fei, Z. Luo, N. J. Ollberding, S.-T. Tang and J. A. Bezerra, Maternal regulation of biliary disease in neonates via gut microbial metabolites, Nat. Commun., 2022, 13, 18 CrossRef CAS.
- T. Chen, Y. Qin, M. Chen, Y. Zhang, X. Wang, T. Dong, G. Chen, X. Sun, T. Lu, R. A. White, P. Ye, H. M. Tun and Y. Xia, Gestational diabetes mellitus is associated with the neonatal gut microbiota and metabolome, BMC Med., 2021, 19, 120 CrossRef CAS PubMed.
- F. Piersigilli, T. T. Lam, P. Vernocchi, A. Quagliariello, L. Putignani, Z. H. Aghai and V. Bhandari, Identification of new biomarkers of bronchopulmonary dysplasia using metabolomics, Metabolomics, 2019, 15, 20 CrossRef CAS PubMed.
- S. Y. Lim, C. B. May, P. N. Johnson and J. L. Miller, Caffeine dosing in premature neonates: Impact of birth weight on a pharmacokinetic simulation study, Pediatr. Res., 2023, 93, 696–700 CrossRef CAS PubMed.
- H. Li, L. Fu, X. Chen, H. Xu, Q. Jing, C. Yang, Z. Wan and Y. Chen, Gut microbiota and metabolome description of antibiotic-treated neonates from parturients with intrauterine infection, Front. Cell. Infect. Microbiol., 2022, 12, 817832 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo03686c |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |