Open Access Article
Open Access ArticleEffect of processing Verdejo grape must by UHPH using non-Saccharomyces yeasts in the absence of SO2†
Carlos
Escott
a,
Cristian
Vaquero
ab,
Juan Manuel
del Fresno
a,
Angelo
Topo
c,
Piergiorgio
Comuzzo
 c,
Carmen
Gonzalez
a and
Antonio
Morata
c,
Carmen
Gonzalez
a and
Antonio
Morata
 *a
*a
aUniversidad Politécnica de Madrid, ETSIAAB, Chemistry and Food Technology Dept., enotecUPM, Avenida Puerta de Hierro, 2, 28040, Madrid, Spain. E-mail: antonio.morata@upm.es
bUniversidad Pontificia de Comillas, Madrid Culinary Campus (MACC), Alberto Aguilera, 23, 28015, Madrid, Spain
cUniversità degli Studi di Udine, Dipartimento di Scienze Agroalimentari, Ambientali e Animali (Di4A), Via Sondrio, 2/A, 33100, Udine, Italy
First published on 16th January 2024
Abstract
Ultra-High Pressure Homogenization (UHPH) is an emerging non-thermal technology that can eliminate wild microorganisms from grape juice facilitating the use of non-competitive non-Saccharomyces yeast in fermentation to modulate the sensory profile. The use of UHPH processing in must from Verdejo variety grapes (Vitis vinifera L.) produces a more varietal profile reducing the contents of fermentative fruity varietal esters (−25–50%) and enhancing the release of volatile thiols (+25–75%). The aromatic profile of UHPH wines is clearly separated of controls by the aroma PCA. Additionally, the inactivation of oxidative enzymes by UHPH preserves a better color in wines with a paler yellow color and lower b* values. A better implantation of some non-Saccharomyces yeasts such as Lachancea thermotolerans can help to reduce the pH in wines from warm areas. Improved varietal aroma, paler wine color and, depending on the strain fermented with, lower pH help control the impact of global warming on wines.
Sustainability spotlightUHPH is a sustainable and environmentally friendly technology with very low power consumption (50% lower than HHP), even lower compared with conventional thermal treatments. The small system volume demands very moderate requirements of the process and cleaning water. Additionally, the use of steam to sanitize or sterilize the system is unnecessary because it can be done by pressure, and just hot water at 70–80 °C as technical fluid is necessary. Furthermore, the low impact of this technology in sensory and nutritional quality favors the production of healthier and less processed foods. This technology can support the United Nations' Sustainable Development Goals (SDGs), especially Goal 2 (zero hunger) and Goal 13 (climate action). |
1. Introduction
Ultra-High Pressure Homogenization (UHPH) is an emerging technology that can be considered non-thermal because of the gentle effect on sensitive molecules with impact on the sensory profile and micronutrients.1–4 The UHPH process is done by ultra-high pressure pumping at more than 200 MPa (normally 300 MPa) followed by subsequent instantaneous depressurization (<0.2 seconds) across a highly resistant valve.1,4 It has shown a protective effect on terpenes and other aroma compounds,5,6 anthocyanins7 and vitamins.8 Due to the extreme impact and shear forces in the valve the temperature can be very high (75–150 °C), but with low thermal impact because of the very short residence time, and there is a very scarce formation of thermal markers such as HMF,5 furosine,9 or carcinogenic compounds such as ethyl carbamate, the formation of which may be enhanced by temperature.10UHPH can control easily high microbial loads in liquid foods, and the inactivation of yeast populations of 6–4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1 belonging to the Saccharomyces cerevisiae (Sc) species but also to other non-Saccharomyces yeasts7,11 and 4–3
CFU mL−1 belonging to the Saccharomyces cerevisiae (Sc) species but also to other non-Saccharomyces yeasts7,11 and 4–3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log of aerobic bacteria in grape juices7,11 has been reported. Depending on the in-valve temperature also sporulate bacteria can be eliminated and it has been observed that under mild conditions (80–90 °C of in-valve temperature) the off-flavor producer, acidophilic and thermoresistant Alicyclobacillus acidoterrestris can be eliminated. Additionally, UHPH produces an effective control of enzymes,10,12 especially oxidative enzymes (PPOs), with inactivation higher than 90% and a control of browning.5,11,13 Therefore this technology can be used to produce wines with a low or null content of SO2.4,5,7
log of aerobic bacteria in grape juices7,11 has been reported. Depending on the in-valve temperature also sporulate bacteria can be eliminated and it has been observed that under mild conditions (80–90 °C of in-valve temperature) the off-flavor producer, acidophilic and thermoresistant Alicyclobacillus acidoterrestris can be eliminated. Additionally, UHPH produces an effective control of enzymes,10,12 especially oxidative enzymes (PPOs), with inactivation higher than 90% and a control of browning.5,11,13 Therefore this technology can be used to produce wines with a low or null content of SO2.4,5,7
The use of non-Saccharomyces is a current hot topic in wine biotechnology because of their ability to modify and improve the sensory profile of wines enhancing wine quality, aroma, flavor, structure, freshness and color.14–18 Several species of non-Saccharomyces have deserved special attention because of their impact on wine quality among them: Torulaspora delbrueckii (Td) by its impact on aroma, foaming properties, and recently in bioprotection,19,20Metschnikowia pulcherrima (Mp) by the expression of enzymatic activities, capacity to lower the ethanol, and bioprotective effect,21,22Lachancea termotolerans (Lt) by acidification and aroma improvement,23–25 and Hanseniaspora vineae (Hv) by aroma and impact on the body and structure.26,27 A major problem of many non-Saccharomyces yeasts is that they are weaker fermenters compared to Sc, and they have problems to compete and be prevalent in must fermentation. The use of UHPH is a way of eliminating all the competitive wild yeasts in grape juice promoting a good implantation of starters from non-Saccharomyces yeasts. Additionally, it can be done at low levels of SO2 that is harmful for many non-Saccharomyces.
Furthermore, UHPH is a sustainable and environmentally friendly technology because of the low power consumption (50% lower than HHP) and the small system volume with low requirements of the process and cleaning water (https://www.ypsicon.com/). Additionally, the use of steam to sanitize or sterilize the system is unnecessary because it can be done by pressure, and just hot water at 70–80 °C as technical fluid is necessary.
The aim of this research is to study the effect of UHPH processing on grape must of Verdejo variety and the impact on the quality of the wines produced after fermentation by several non-Saccharomyces species.
2. Materials and methods
2.1. Grape juice
The must was obtained from Verdejo blanco grapes (Vitis vinifera L.; VIVC variety number: 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949) from the DO Rueda in Valladolid, Spain. Grapes were mechanically harvested and pressed using a pneumatic press model Diemme 150 (Lugo, Ravenna, Italy) at less than 2 bar, and the yield was 60% of grape juice; after that the juice obtained was settled at low temperature (4 °C) for 2 days and processed by UHPH. No sulphites were added to the juice. The settling was performed without the use of pectolytic enzymes.
949) from the DO Rueda in Valladolid, Spain. Grapes were mechanically harvested and pressed using a pneumatic press model Diemme 150 (Lugo, Ravenna, Italy) at less than 2 bar, and the yield was 60% of grape juice; after that the juice obtained was settled at low temperature (4 °C) for 2 days and processed by UHPH. No sulphites were added to the juice. The settling was performed without the use of pectolytic enzymes.
2.2. UHPH treatment
After settling the must was processed with a 60 L h−1 UHPH device (Ypsicon Technologies, Barcelona, Spain) at 300 ± 3 MPa with an inlet temperature of 8 °C, reaching 92 °C in the UHPH valve and being refrigerated after valve depressurization and by additional continuous cooling at 15 °C. Processed juice was aseptically packed in sterilized 2 L plastic bags in the absence of O2 and stored refrigerated at 4 °C until fermentation. Control unprocessed juice was also kept at 4 °C until fermentation in 5 L plastic bottles.2.3. Fermentation and inoculation
UHPH juices and unprocessed controls were dosed at 200 mL in 250 mL ISO flasks and kept isothermally at 20 °C during fermentation. Inocula of Saccharomyces and non-Saccharomyces yeasts were grown in YPD liquid media, and the populations were synchronized by two successive passes of 2% v/v in 24 h before the final inoculation in the fermenters. Inoculation volume was 2% v/v (4 mL), producing a final population of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1. The inocula were grown at 20 °C. Fermentation was performed in triplicate. All fermentation samples of non-Saccharomyces were inoculated again on the 8th day of fermentation with S. cerevisiae (Sc7VA) until total sugar depletion to ensure the dryness of wines.
CFU mL−1. The inocula were grown at 20 °C. Fermentation was performed in triplicate. All fermentation samples of non-Saccharomyces were inoculated again on the 8th day of fermentation with S. cerevisiae (Sc7VA) until total sugar depletion to ensure the dryness of wines.
The Sc and non-Saccharomyces yeasts used were Saccharomyces cerevisiae strain 7VA (Sc7VA) selected at the enotecUPM lab (Madrid, Spain), Lachancea thermotolerans L31 (L31) selected at the enotecUPM lab, Torulaspora delbrueckii BIODIVA™ (Td) Lallemand, Hanseniaspora vineae 205 (Hv) selected by the Professor Carrau team at the Universidad de la República (Uruguay), and Metschnikowia pulcherrima M29 (M29) selected at the enotecUPM lab. All of them were kept in YPD agar and refreshed by a pass in solid YPD agar and growth at 20 °C for 48 h previously to be synchronized in the liquid YPD.
All fermentation samples from days 2, 4 and 6 were monitored for inoculated and wild yeast populations by plate seeding according to Section 2.5.
2.4. Optical microscopy of the juice
Optical microscopy was performed on a drop of sediment of the control and UHPH juices using a BA310 LED microscope (Motic, San Antonio, USA) with a 60× achromatic objective and a digital camera FULLHD Moticam 1080INT. The sediment was obtained after centrifugation at 5000 rpm for 5 minutes of 3 mL of juice in an Eppendorf tube.2.5. Yeast counts
Yeast counts were determined by plating in YPD agar for total yeasts and in lysine agar and CHROMagar™ Candida (Conda, Barcelona, Spain) for non-Saccharomyces counts. The identification was done according to the color and colony appearance in the selective and differential media. Plates were counted after 48 h at 25 °C in the dilution that had a count number in the range of 30–300 colonies.2.6. Lactic acid and enological parameters
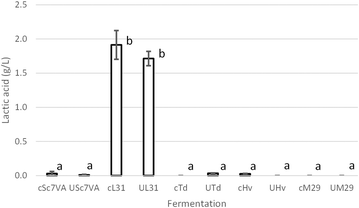
L-Lactic acid has been analyzed at the end of fermentation by enzymatic analysis using an automatic enzymatic analyzer Y25 (BioSystems, Barcelona, Spain). Previously samples were filtered with 0.45 μm membrane filters.The major compounds at the end of fermentation, such as residual sugars, organic acids, and total acidity, were measured with a FTIR OenoFoss equipment (FOSS Iberia, Barcelona, Spain). The pH was measured using a CRISON micropH 2000 pHmeter (HACH LANGE, Barcelona, Spain).
2.7. Fermentative volatiles quantified by GC-FID
Fermentative volatiles were quantified by gas chromatography with a flame ionization detector (GC-FID) using an Agilent Technologies 6850 gas chromatograph. The injector temperature was set at 250 °C, and the FID was at 300 °C. Compounds were separated on a DB-624 column (60 m × 250 μm × 1.40 μm) with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 split ratio. The quantification was performed using GC quality standards: acetaldehyde, methanol, 1-propanol, 1-butanol, 2-butanol, isobutanol, 2-methyl-1-butanol, 3-methyl-1-butanol, 2-phenylethyl acetate, 2-phenylethyl alcohol, diacetyl, ethyl acetate, isoamyl acetate, isobutyl acetate, ethyl butyrate, 3-ethoxy-1-propanol, ethyl lactate, and hexanol (Fluka, Sigma-Aldrich Corp., Buchs SG, Switzerland). The compound 4-methyl-2-pentanol was added as the internal standard, also from Fluka. The temperature program for the oven was 40 °C for five minutes, and then an increase of 10 °C per minute up to 250 °C, which was kept for five minutes, with a total running and conditioning time of 40 minutes. The carrier gas was hydrogen with a column flow of 2.2 mL min−1. One mL of sample was filtered with a 0.45 μm membrane and spiked with 100 μL of IS. The injection volume was 1 μL. The detection limit was set at 0.1 mg L−1. The volatile compounds analyzed were calibrated using a five-point calibration curve with r2 > 0.999, except 2,3-butanediol (0.991) and 2-phenylethyl alcohol (0.994).
10 split ratio. The quantification was performed using GC quality standards: acetaldehyde, methanol, 1-propanol, 1-butanol, 2-butanol, isobutanol, 2-methyl-1-butanol, 3-methyl-1-butanol, 2-phenylethyl acetate, 2-phenylethyl alcohol, diacetyl, ethyl acetate, isoamyl acetate, isobutyl acetate, ethyl butyrate, 3-ethoxy-1-propanol, ethyl lactate, and hexanol (Fluka, Sigma-Aldrich Corp., Buchs SG, Switzerland). The compound 4-methyl-2-pentanol was added as the internal standard, also from Fluka. The temperature program for the oven was 40 °C for five minutes, and then an increase of 10 °C per minute up to 250 °C, which was kept for five minutes, with a total running and conditioning time of 40 minutes. The carrier gas was hydrogen with a column flow of 2.2 mL min−1. One mL of sample was filtered with a 0.45 μm membrane and spiked with 100 μL of IS. The injection volume was 1 μL. The detection limit was set at 0.1 mg L−1. The volatile compounds analyzed were calibrated using a five-point calibration curve with r2 > 0.999, except 2,3-butanediol (0.991) and 2-phenylethyl alcohol (0.994).
2.8. Volatile compounds quantified by HS-SPME-GC-MS
Volatile compounds (VOCs) were determined by SPME-GC-MS by slightly modifying the method published by Tat et al.28 The following compounds were analysed: ethyl hexanoate, hexyl acetate, ethyl-3-hexenoate, 3-hexen-1-ol acetate, 3-hexen-1-ol, (E)-, 3-ethoxy-1-propanol, 3-hexen-1-ol, (Z)-, ethyl octanoate, acetic acid, 3-methylbutyl hexanoate, ethyl-3-octenoate, 2-ethyl-1-hexanol, benzaldehyde, linalool, 1-octanol, 2-methylpropanoic acid, γ-butyrolactone, butanoic acid, ethyl decanoate, 3-methylbutanoic acid, diethyl succinate, ethyl 9-decenoate, 3-methylthio-1-propanol, β-citronellol, ethyl phenylacetate, ethyl 4-hydroxybutanoate, β-damascenone, hexanoic acid, phenyl methanol (benzyl alcohol), octanoic acid, 4-vinylguaiacol, ethyl hexadecanoate, decanoic acid, ethyl 9-hexadecenoate, ethyl octadecanoate, and ethyl 9-octadecenoate. The instrument used was a GCMS-QP2020 NX GC-MS system (Shimadzu, Kyoto, Japan), equipped with a 2800T autosampler (HTA S.r.l., Brescia Italy). Samples were prepared as follows: 10 mL of filtered wine were introduced into 20 mL glass vials and mixed with 3 g of NaCl; 100 μL of ethyl heptanoate (0.106 g L−1 in ethanol) was added as the internal standard and vials were sealed with PTFE/silicon septa. SPME was carried out using a 2 cm long 50/30 μm DVB/Carboxen/PDMS fiber (Supelco, Bellefonte, PA, USA), at 40 °C for 15 min. Vials were pre-conditioned in an autosampler for 15 min before microextraction, to allow the thermal equilibration of the samples. Injections were performed in splitless mode, with a splitless time of 60 s. The temperatures of the injector and the transfer line were 250 °C and 240 °C respectively, while the ion source was set at 200 °C. The carrier gas was helium at a linear flow rate of 36 cm s−1. Compounds were separated on a J&W DB-Wax capillary column, with 30 m × 0.25 mm i.d. and 0.25 μm film thickness (Agilent Technologies Inc., Santa Clara, CA, USA), according to the gradient reported by Comuzzo et al. (2018).29 Electron impact mass spectra were recorded at 70 eV and the identification of volatile compounds was carried out by comparison of their mass spectra and retention times with those of standard compounds, or by comparison of the mass spectrum with those reported in the mass spectrum libraries Wiley 6 and NIST 107. Linear retention indices were also calculated on the basis of the retention times of n-alkanes and compared with those reported in the literature. Semi-quantitative analysis was performed by the internal standard method, considering a response factor equal to 1.00.2.9. Thiols quantified by LC-MS/MS
The volatile thiols 3-mercaptohexan-1-ol (3 MH), 4-mercaptopentan-2-one (4MMP), 4-mercapto-4-methylpentan-2-ol (4MMPOH), and 3-mercaptohexyl acetate (3MHA) were analysed by LC-MS/MS. Wines were extracted with modified Quechers using a mixture of ethyl acetate/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) without the clean-up step. An aliquot of this extract was analyzed by liquid chromatography coupled to mass spectrometry (LC-MS/MS). The equipment used was an Agilent 1290 Chromatograph coupled to a triple quadrupole mass spectrometer (model 6460) also from Agilent. The ionization source was electrospray type (ESI) with a gas temperature of 300 °C at a flow of 5 L min−1. The separation was carried out on a Phenomenex C18 column with the following dimensions (100 × 3 mm, 250 A, and 2.6 μm) at a temperature of 25 °C and an injection volume of 5 μL. The mobile phases were as follows: (A) 0.1% formic acid in water and (B) acetonitrile at a flow rate of 0.5 mL min−1. Detection was performed in MRM mode for each of the thiols.
1) without the clean-up step. An aliquot of this extract was analyzed by liquid chromatography coupled to mass spectrometry (LC-MS/MS). The equipment used was an Agilent 1290 Chromatograph coupled to a triple quadrupole mass spectrometer (model 6460) also from Agilent. The ionization source was electrospray type (ESI) with a gas temperature of 300 °C at a flow of 5 L min−1. The separation was carried out on a Phenomenex C18 column with the following dimensions (100 × 3 mm, 250 A, and 2.6 μm) at a temperature of 25 °C and an injection volume of 5 μL. The mobile phases were as follows: (A) 0.1% formic acid in water and (B) acetonitrile at a flow rate of 0.5 mL min−1. Detection was performed in MRM mode for each of the thiols.
2.10. Color
The DNA Phone Smart Analysis – Wine device (Parma, Italy) was used to determine the color parameters. The parameters included the CIELChuv coordinates for chroma (C) and hue (h), as well as the CIELab coordinates for lightness (L*), green-red (a*), and blue-yellow (b*). The samples were put into polymethyl methacrylate cuvettes with a 1 mm path length. The samples were filtered using 0.45 μm methyl-cellulose membranes before analysis.2.11. Statistical analysis
Statistical analysis was performed using Statgraphics Centurion v19 software (Graphics Software Systems, Rockville, MD, United States) to calculate the mean, standard deviation, analysis of variance (ANOVA), and least significant difference (LSD). Significance was set at p < 0.05.SPME-GC-MS data were elaborated by Principal Component Analysis (PCA) and One-Way ANOVA, using Statistica for Windows (version 8.0). Regarding PCA, factor loadings (FLs) were calculated by factor analysis, and the most relevant variables were selected for PCA, when marked FLs were higher than 0.7. Concerning One Way ANOVA, means and standard deviations were calculated, and significant differences were assessed by a Tukey HSD test at p < 0.05. Variances were homogeneous according to the Brown–Forsythe test.
3. Results and discussion
3.1. Results
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log on the 2nd day of fermentation in the control and UHPH processed juices except for Td which were closer to 6
log on the 2nd day of fermentation in the control and UHPH processed juices except for Td which were closer to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log. The populations of all the non-Saccharomyces were higher than 5
log. The populations of all the non-Saccharomyces were higher than 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log on day 6 of fermentation.
log on day 6 of fermentation.
| Yeasts/day of fermentation | 2 | 4 | 6 |
|---|---|---|---|
| Control Sc7VA-Inoc | 3.93 × 107 | 2.43 × 107 | 5.83 × 106 |
| UHPH Sc7VA-Inoc | 2.23 × 107 | 8.33 × 106 | 4.00 × 105 |
| Control Sc7VA-Wild | 1.83 × 106 | 1.53 × 107 | 1.33 × 105 |
| UHPH Sc7VA-Wild | nd | nd | nd |
| Control L31-Inoc | 1.40 × 107 | 3.13 × 107 | 1.01 × 107 |
| UHPH L31-Inoc | 2.17 × 107 | 7.50 × 106 | 7.33 × 105 |
| Control L31-Wild | 1.37 × 107 | 1.99 × 107 | 5.80 × 106 |
| UHPH L31-Wild | nd | nd | nd |
| Control Td-Inoc | 5.33 × 106 | 1.39 × 107 | 2.00 × 106 |
| UHPH Td-Inoc | 1.50 × 106 | 5.43 × 107 | 1.87 × 107 |
| Control Td-Wild | 3.13 × 107 | 4.73 × 107 | 8.67 × 106 |
| UHPH Td-Wild | nd | nd | nd |
| Hv-Inoc | 3.29 × 107 | 1.27 × 107 | 7.47 × 106 |
| U-Hv-Inoc | 6.77 × 106 | 9.33 × 106 | 8.33 × 105 |
| Hv-Wild | 1.63 × 107 | 1.27 × 107 | 5.73 × 106 |
| U-Hv-Wild | nd | nd | nd |
| Control M29-Inoc | 4.67 × 107 | 6.67 × 106 | 6.00 × 106 |
| UHPH M29-Inoc | 2.05 × 107 | 1.73 × 107 | 1.00 × 107 |
| Control M29-Wild | 6.13 × 106 | 2.83 × 107 | 5.67 × 106 |
| UHPH M29-Wild | nd | nd | nd |
| Ethanol (% v/v) | Total acidity (g L−1)a | pH | Volatile acidity (g L−1)b | Residual sugars (g L−1) | Malic acid (g L−1) | TPIc | Glucose (g L−1) | Fructose (g L−1) | |
|---|---|---|---|---|---|---|---|---|---|
| a Total acidity expressed in tartaric acid. b Volatile acidity in acetic acid. c TPI (total polyphenol index). | |||||||||
| cSc7VA Control | 12.9 ± 0.0bc | 4.2 ± 0.1ab | 3.7 ± 0.0c | 0.2 ± 0.0a | 1.5 ± 0.2d | 1.3 ± 0.0 cd | 12.7 ± 1.8d | 0.8 ± 0.1a | 0.9 ± 0.3bcd |
| USc7VA UHPH | 12.4 ± 0.3ab | 4.1 ± 0.1ab | 3.7 ± 0.0bc | 0.4 ± 0.0c | 1.5 ± 0.1d | 1.3 ± 0.1bc | 11.7 ± 2.0cd | 0.9 ± 0.1a | 0.8 ± 0.1bc |
| cL31 Control | 12.8 ± 0.1bc | 6.3 ± 0.2f | 3.5 ± 0.0a | 0.3 ± 0.0b | 1.1 ± 0.1ab | 1.1 ± 0.1b | 11.2 ± 1.3bcd | 0.8 ± 0.1a | 0.4 ± 0.1a |
| UL31 UHPH | 12.5 ± 0.1ab | 5.5 ± 0.2e | 3.5 ± 0.0a | 0.2 ± 0.0a | 1.0 ± 0.3a | 0.8 ± 0.1a | 5.4 ± 3.7a | 1.5 ± 0.1d | 0.4 ± 0.3a |
| cTd Control | 13.1 ± 0.0c | 4.4 ± 0.1cd | 3.7 ± 0.0bc | 0.3 ± 0.0b | 1.2 ± 0.1abc | 1.5 ± 0.0e | 9.8 ± 1.5bcd | 1.0 ± 0.1a | 0.8 ± 0.1b |
| Utd UHPH | 12.5 ± 0.0ab | 4.2 ± 0.1abc | 3.6 ± 0.0b | 0.2 ± 0.0a | 1.3 ± 0.3bcd | 1.3 ± 0.1bc | 8.5 ± 2.4abc | 1.4 ± 0.2cd | 1.0 ± 0.2bcd |
| cHv Control | 12.8 ± 0.0bc | 4.6 ± 0.2d | 3.7 ± 0.0bc | 0.4 ± 0.0c | 1.7 ± 0.1e | 1.3 ± 0.1cd | 9.8 ± 1.7bcd | 1.2 ± 0.1b | 1.1 ± 0.1d |
| UHv UHPH | 12.1 ± 0.4a | 4.0 ± 0.1a | 3.7 ± 0.0b | 0.4 ± 0.0c | 1.4 ± 0.1cd | 1.2 ± 0.1bc | 7.5 ± 2.6ab | 1.3 ± 0.2bc | 1.0 ± 0.1bcd |
| cM29 Control | 13.1 ± 0.1c | 4.5 ± 0.2cd | 3.7 ± 0.0bc | 0.3 ± 0.0b | 1.2 ± 0.1abcd | 1.4 ± 0.1de | 12.1 ± 0.2cd | 0.9 ± 0.1a | 0.8 ± 0.2bc |
| UM29 UHPH | 12.1 ± 0.6a | 4.3 ± 0.1bc | 3.7 ± 0.0c | 0.5 ± 0.0d | 1.3 ± 0.1bcd | 1.3 ± 0.1bc | 13.7 ± 3.5d | 1.3 ± 0.1bc | 1.1 ± 0.1cd |
| Sc7VA | USc7VA | L31 | UL31 | Td | UTd | Hv | UHv | M29 | UM29 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | UHPH | Control | UHPH | Control | UHPH | Control | UHPH | Control | UHPH | |
| Acetaldehyde | 101.90 ± 2.43c | 102.88 ± 7.23c | 113.14 ± 8.85cd | 29.59 ± 2.27ab | 226.25 ± 23.02f | 46.88 ± 4.59b | 97.84 ± 9.43c | 26.50 ± 5.98a | 182.49 ± 18.25e | 124.47 ± 3.52d |
| Methanol | 43.49 ± 6.21d | 30.66 ± 3.62bc | 29.29 ± 3.29abc | 23.35 ± 0.66a | 61.06 ± 6.73e | 25.22 ± 4.14ab | 56.79 ± 3.33e | 31.16 ± 1.87bc | 44.83 ± 1.82d | 35.49 ± 5.16c |
| 1-Propanol | 24.76 ± 1.75a | 33.01 ± 1.08b | 50.43 ± 1.82d | 50.66 ± 5.34d | 30.30 ± 2.71b | 62.16 ± 2.94e | 32.06 ± 1.38b | 42.08 ± 2.10c | 29.36 ± 1.40b | 45.45 ± 3.49c |
| Diacetyl | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.00 ± 0.00a | 1.41 ± 0.10b | 1.71 ± 0.16b | 1.64 ± 0.14b | 0.00 ± 0.00a | 0.00 ± 0.00a | 2.67 ± 0.51c | 0.00 ± 0.00a |
| Ethyl acetate | 18.02 ± 1.15a | 19.06 ± 2.04a | 37.19 ± 1.90bc | 32.50 ± 0.69bc | 40.86 ± 0.22cd | 30.86 ± 1.87b | 49.67 ± 9.55d | 85.39 ± 13.49e | 35.60 ± 1.99bc | 48.41 ± 5.86d |
| 2-Butanol | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.00 ± 0.00a | 2.89 ± 0.35b | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.87 ± 1.50a | 3.36 ± 0.71b | 0.00 ± 0.00a |
| Iso-butanol | 26.42 ± 1.23bc | 22.53 ± 2.25ab | 51.22 ± 1.38f | 28.14 ± 1.94c | 49.45 ± 4.24f | 34.16 ± 1.90d | 40.11 ± 1.87e | 20.07 ± 0.81a | 65.41 ± 6.93g | 47.22 ± 2.75f |
| 1-Butanol | 3.90 ± 0.05b | 0.00 ± 0.00a | 4.68 ± 0.17bcd | 5.58 ± 1.40d | 0.00 ± 0.00a | 5.13 ± 0.22cd | 0.00 ± 0.00a | 5.55 ± 0.80d | 5.53 ± 0.69d | 4.19 ± 0.18bc |
| Acetoin | 11.18 ± 1.31b | 11.26 ± 0.82b | 10.61 ± 0.42ab | 10.81 ± 0.99ab | 8.91 ± 1.19a | 11.21 ± 0.70b | 11.25 ± 0.61b | 11.74 ± 0.31b | 12.27 ± 1.43b | 12.00 ± 2.51b |
| 2-Methyl-1-butanol | 58.62 ± 2.89de | 37.20 ± 4.13a | 82.81 ± 5.49g | 55.15 ± 2.34cd | 64.24 ± 6.84ef | 62.10 ± 1.65def | 66.89 ± 4.69f | 44.01 ± 0.54ab | 68.88 ± 7.62f | 48.23 ± 0.96bc |
| 3-Methy-1-butanol | 141.50 ± 6.16c | 78.25 ± 6.27a | 187.56 ± 4.47e | 109.60 ± 1.99b | 163.37 ± 8.32d | 133.54 ± 6.30c | 144.13 ± 9.97c | 81.48 ± 1.72a | 176.88 ± 17.89de | 91.39 ± 4.96a |
| Isobutyl acetate | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.00 ± 0.00a | 3.10 ± 0.26b | 10.55 ± 0.26d | 0.00 ± 0.00a | 3.54 ± 1.14b | 3.27 ± 0.41b | 4.83 ± 0.86c | 3.63 ± 0.40b |
| Ethyl butyrate | 3.44 ± 0.53b | 4.68 ± 0.31c | 2.95 ± 0.19b | 0.00 ± 0.00a | 4.60 ± 0.82c | 4.31 ± 0.67c | 0.00 ± 0.00a | 0.00 ± 0.00a | 4.83 ± 0.40c | 0.00 ± 0.00a |
| Ethyl lactate | 15.17 ± 1.20c | 14.52 ± 0.84c | 60.60 ± 2.78e | 26.37 ± 2.92d | 10.00 ± 0.68b | 14.83 ± 0.98c | 5.53 ± 0.86a | 15.69 ± 4.91c | 12.28 ± 2.91bc | 9.32 ± 1.58ab |
| 2–3 butanediol | 484.44 ± 19.15bc | 565.98 ± 52.62cde | 362.72 ± 25.92a | 444.27 ± 34.85 ab | 370.25 ± 29.25a | 673.93 ± 29.92f | 512.73 ± 65.93bcd | 593.12 ± 25.27def | 435.27 ± 60.09 ab | 643.34 ± 86.96ef |
| Isoamyl acetate | 2.42 ± 0.67b | 0.00 ± 0.00a | 2.14 ± 0.30b | 0.00 ± 0.00a | 2.31 ± 0.34b | 0.00 ± 0.00a | 2.37 ± 0.12b | 1.90 ± 0.06b | 4.72 ± 1.06c | 2.05 ± 0.29b |
| Hexanol | 0.00 ± 0.00a | 0.00 ± 0.00a | 3.84 ± 0.12c | 3.62 ± 0.12b | 3.87 ± 0.15c | 3.88 ± 0.06c | 0.00 ± 0.00a | 3.64 ± 0.15b | 0.00 ± 0.00a | 0.00 ± 0.00a |
| 2-Phenylethanol | 47.18 ± 1.31d | 23.81 ± 1.93a | 73.39 ± 9.42e | 30.37 ± 2.88 ab | 69.35 ± 3.08e | 42.77 ± 6.10cd | 38.60 ± 4.46bcd | 23.44 ± 1.02a | 67.82 ± 8.16e | 34.52 ± 4.30bc |
| 2-Phenylethyl acetate | 5.38 ± 0.05a | 5.36 ± 0.12a | 5.51 ± 0.10a | 5.77 ± 0.03a | 7.70 ± 0.46b | 7.34 ± 0.64b | 5.43 ± 0.20a | 5.56 ± 0.34a | 7.67 ± 0.52b | 7.23 ± 0.25b |
| Higher alcohols | 302.37 ± 12.97bc | 194.79 ± 13.06a | 450.09 ± 17.99g | 279.50 ± 13.31b | 379.61 ± 18.46e | 339.86 ± 17.72d | 321.81 ± 19.68bc | 217.51 ± 4.31a | 417.25 ± 39.37f | 271.01 ± 9.22b |
| Carbonyl compounds | 11.18 ± 1.31ab | 11.26 ± 0.82ab | 10.61 ± 0.42a | 12.22 ± 1.05ab | 10.62 ± 1.04a | 12.86 ± 0.83bc | 11.25 ± 0.61ab | 11.74 ± 0.31ab | 14.94 ± 1.86c | 12.00 ± 2.51ab |
| Esters | 44.44 ± 0.33a | 43.63 ± 2.65a | 108.38 ± 0.65e | 67.74 ± 3.33cd | 76.02 ± 1.63d | 57.34 ± 1.16b | 66.54 ± 9.59bc | 111.80 ± 11.66e | 69.94 ± 5.52cd | 70.63 ± 4.82cd |
| Floral and fruity esters | 26.42 ± 0.90b | 24.57 ± 1.06b | 71.19 ± 2.53d | 35.25 ± 3.17c | 35.16 ± 1.86c | 26.48 ± 1.24b | 16.87 ± 1.42a | 26.42 ± 5.41b | 34.33 ± 3.82c | 22.22 ± 1.82b |
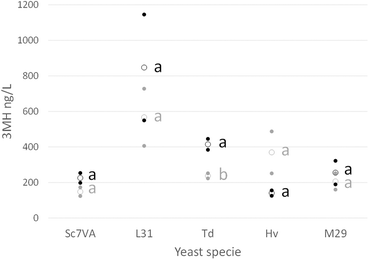
Other thiols such as 4MMP and 4MMPOH were not detected in most of the samples and concerning the 3MHA the concentrations were quite variable and did not follow a clear pattern.
| Wine | Chroma | Hue (°) |
|---|---|---|
| Sc7VA | 14.4 ± 2.7a | 91.1 ± 4.3a |
| USc7VA | 9.7 ± 1.2b | 95.5 ± 6.9a |
| L31 | 11.4 ± 0.2a | 97.8 ± 3.3a |
| UL31 | 10.2 ± 1.1a | 98.0 ± 4.0a |
| Td | 12.6 ± 1.4a | 97.4 ± 2.1a |
| UTd | 9.9 ± 0.6b | 97.6 ± 5.8a |
| Hv | 11.4 ± 1.0a | 93.4 ± 4.0a |
| UHv | 9.3 ± 1.0a | 98.0 ± 2.6a |
| M29 | 12.5 ± 0.4a | 95.5 ± 0.7a |
| UM29 | 9.3 ± 2.6b | 94.4 ± 0.3b |
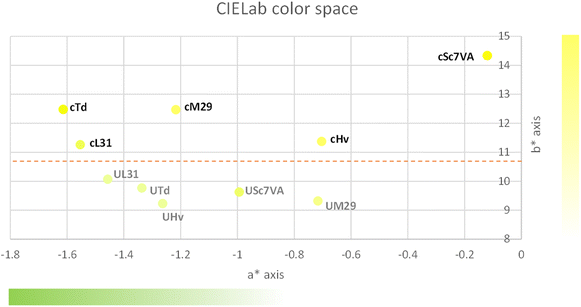
With regards to the hue (Table 4), this parameter had no significant differences between pairs of must, and the media values were slightly higher for the UHPH treated must fermented with Sc and Hv. The results observed for this parameter are consistent with the observations got for the a* axis and b* axis shown in Fig. 5, where Sc is completely isolated from the rest of the wines having the highest b* values (more yellow) and the lowest a* values (less green).
As observed in Fig. 5, the values for wines produced with must without UHPH treatment were larger on the b* axis, above the dotted line and above 11 units, which is translated into wines having more yellow appearance associated with higher levels of oxidation. On the other hand, all wines from UHPH-treated must and the wines from untreated must fermented by non-Saccharomyces yeasts had a* values larger than −0.6 in comparison to Sc. In this case, all wines except for Sc had a larger contribution of green.
3.2. Discussion
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log have been observed for Saccharomyces and non-Saccharomyces in grape juice.11 The absence of yeasts in the must after the 6th day of fermentation guarantees a better expression and prevalence of the inoculated species during the fermentation. We have observed the absence of wild yeasts in several trials during full fermentation,5,7 and it is possible to maintain unfermented musts for months or even years when sterilized by UHPH.
log have been observed for Saccharomyces and non-Saccharomyces in grape juice.11 The absence of yeasts in the must after the 6th day of fermentation guarantees a better expression and prevalence of the inoculated species during the fermentation. We have observed the absence of wild yeasts in several trials during full fermentation,5,7 and it is possible to maintain unfermented musts for months or even years when sterilized by UHPH.
We have observed a slightly lower fermentation speed in UHPH processed must inoculated with the different species than in controls [data not shown]. This can be due to the lower loads of wild yeasts or the lower availability of thiamine or some nitrogen compounds that can be nano-encapsulated and less available, but more research is necessary to support these tentative explanations.
Concerning carbonyl compounds, the excessive amount of acetoin and diacetyl can be correlated with excessive dairy notes in wines, but in this case, both compounds were at a moderate amount and without significant differences between controls and UHPH wines (3%, Table 3). Finally, the low content of acetaldehyde in UHPH wines is positive, because of the impact of such a compound on the development of flat and oxidized aromatic characters.38 Indeed, in high amounts, this compound can confer to the wine nutty aromas, which turn to green/grassy or apple-like off-flavors at even higher concentrations; in contrast, if the levels are low, it can be connected with fruity notes.38 The lower acetaldehyde content in UHPH wines may be reasonably explained due to a higher presence of wild non-Saccharomyces yeasts in controls. However, it is also reported that acetaldehyde formation during alcoholic fermentation can be negatively affected by a deficiency of thiamine, because this vitamin is a cofactor of pyruvate decarboxylase.39 When heated in water at 140 °C under pressure, thiamine may be degraded40 and, due to the in-valve temperature reached during the experiment, UHPH processing might have reduced its natural content in the grape juice. The low concentrations of acetaldehyde in UHPH wines can be complementary to the suppressive effect of thiols in the oxidative profile produced by this molecule in wines.41
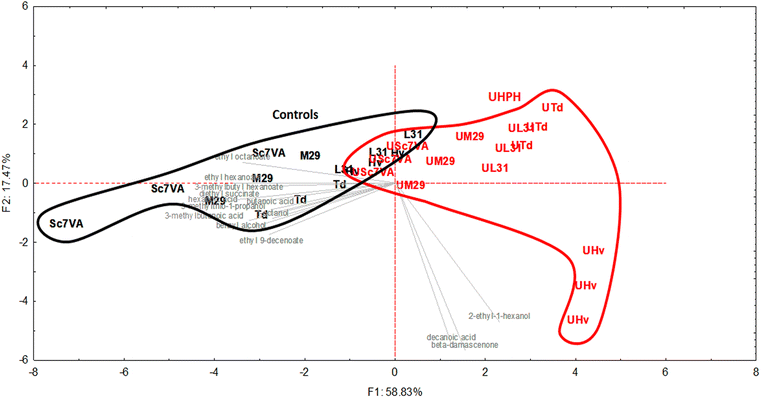
The same trend highlighted by GC-FID analysis for fermentative volatile compounds was also found for the other VOCs analyzed by SPME-GC-MS (although not always in a statistically significant way); indeed many of such compounds also have a fermentative origin. The slight decrease of different fatty acids in UHPH wines may also have a positive role on wine freshness, because of the consequent reduction of their typical pungent notes. Finally, limited to the samples fermented with H. vineae, the lower concentration of β-damascenone found in the samples processed by UHPH is an interesting feature, because this compound can modify the sensory perception of some esters. Indeed, some researchers observed that the addition of low levels of β-damascenone (0.85 μg L−1) to a model solution containing esters increased the fruity notes of the mixture, while when the concentration was higher (3.5 μg L−1), it determined the development of intense notes of raisins and dried plums.42
4. Conclusions
UHPH is a promising emerging non-thermal technology environmentally friendly with very low consumption of energy and water resources able to control microbial loads and oxidation in grape juices. The effect, when associated with the use of non-Saccharomyces yeasts, is an effective implantation of the yeast inocula and a positive metabolomic impact facilitating the preservation of varietal aroma from grapes and the improvement of the freshness if acidifying species such as L. thermotolerans are used. Some of the aroma effects of the association of UHPH and specific non-Saccharomyces species might occur only in Verdejo or maybe just in other thiolic varieties. Additionally, lower pH values together with the control of the full wild microbiome of grapes, and the inactivation of oxidative enzymes, facilitate the production of wines with very low levels of SO2. A new enology is possible with cleaner labels and a more stable sensory quality even with grapes from regions affected by global warming.Author contributions
C. Escott (investigation and methodology), C. Vaquero (investigation and methodology), J. M. del Fresno (investigation, methodology, and writing – original draft), A. Topo (investigation), P. Comuzzo (methodology, supervision, data curation, writing – original draft, and writing – review & editing), C. Gonzalez (methodology, supervision, and writing – review & editing), and A. Morata (conceptualization, funding acquisition, methodology, project administration, supervision, writing – original draft, and writing – review & editing).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by MINISTERIO DE CIENCIA E INNOVACIÓN, project ENOINNOVAPRESS—PID2021-124250OB-I00 Ministerio de Ciencia e Innovación. This work was partially supported by the State Program to Promote Scientific-Technical Research and its Transfer of the Spanish Ministry of Science and Innovation through the MALTA CONSOLIDER TEAM Research Network (RED2022-134388-T).References
- A. Zamora and B. Guamis, Food Eng. Rev., 2015, 7, 130–142, DOI:10.1007/s12393-014-9097-4.
- F. Patrignani and R. Lanciotti, Front. Microbiol., 2016, 7, 1132, DOI:10.3389/fmicb.2016.01132.
- P. Comuzzo and S. Calligaris, Beverages, 2019, 5, 56, DOI:10.3390/beverages5030056.
- A. Morata and B. Guamis, Front. Nutr., 2020, 7, 598286, DOI:10.3389/fnut.2020.598286.
- M. A. Bañuelos, I. Loira, B. Guamis, C. Escott, J. M. Del Fresno, I. Codina-Torrella, J. M. Quevedo, R. Gervilla, J. M. Rodríguez Chavarría, S. de Lamo, R. Ferrer-Gallego, R. Álvarez, C. González, J. A. Suárez-Lepe and A. Morata, Food Chem., 2020, 332, 127417, DOI:10.1016/j.foodchem.2020.127417.
- I. Codina-Torrella, J. J. Gallardo-Chacón, B. Juan, B. Guamis and A. J. Trujillo, Foods, 2023, 12, 683, DOI:10.3390/foods12040683.
- C. Vaquero, C. Escott, I. Loira, B. Guamis, J. M. del Fresno, J. M. Quevedo, R. Gervilla, S. de Lamo, R. Ferrer-Gallego, C. González, M. A. Bañuelos, J. A. Suárez-Lepe and A. Morata, Food Bioprocess Technol., 2022, 15, 620–634, DOI:10.1007/s11947-022-02766-8.
- Á. Suárez-Jacobo, C. E. Rüfer, R. Gervilla, B. Guamis, A. X. Roig-Sagués and J. Saldo, Food Chem., 2011, 127, 447–454, DOI:10.1016/j.foodchem.2010.12.152.
- V. Ferragut, M. Hernández-Herrero, M. T. Veciana-Nogués, M. Borras-Suarez, J. González-Linares, M. C. Vidal-Carou and B. Guamis, J. Sci. Food Agric., 2015, 95, 953–961, DOI:10.1002/jsfa.6769.
- G.-y. Arakawa and K.-j. Yokoi, J. Biosci. Bioeng., 2023, 136, 117–122, DOI:10.1016/j.jbiosc.2023.05.003.
- I. Loira, A. Morata, M. A. Bañuelos, A. Puig-Pujol, B. Guamis, C. González and J. A. Suárez-Lepe, Innovative Food Sci. Emerging Technol., 2018, 50, 50–56, DOI:10.1016/j.ifset.2018.10.005.
- J. Pereda, V. Ferragut, J. M. Quevedo, B. Guamis and A. J. Trujillo, Effects of Ultra-High Pressure Homogenization on microbial and physicochemical shelf life of milk, J. Dairy Sci., 2007, 90, 1081–1093, DOI:10.3168/jds.S0022-0302(07)71595-3.
- R.-M. Velázquez-Estrada, M.-M. Hernández-Herrero, B. Guamis-López and A.-X. Roig-Saguès, Int. J. Food Sci. Technol., 2019, 54, 1858–1864, DOI:10.1111/ijfs.14089.
- M. Ciani and F. Comitini, Ann. Microbiol., 2011, 61, 25–32, DOI:10.1007/s13213-010-0069-5.
- N. P. Jolly, C. Varela and I. S. Pretorius, FEMS Yeast Res., 2014, 14, 215–237, DOI:10.1111/1567-1364.12111.
- B. Padilla, J. V. Gil and P. Manzanares, Front. Microbiol., 2016, 7, 411, DOI:10.3389/fmicb.2016.00411.
- A. Morata, C. Escott, M. A. Bañuelos, I. Loira, J. M. del Fresno, C. González and J. A. Suárez-Lepe, Biomolecules, 2020, 10, 34, DOI:10.3390/biom10010034.
- L. Roudil, P. Russo, C. Berbegal, W. Albertin, G. Spano and V. Capozzi, Recent Pat. Food, Nutr. Agric., 2020, 11(1), 27–39, DOI:10.2174/2212798410666190131103713.
- M. Ramírez and R. Velázquez, Fermentation, 2018, 4, 94, DOI:10.3390/fermentation4040094.
- S. Simonin, H. Alexandre, M. Nikolantonaki, C. Coelho and R. Tourdot-Maréchal, Food Res. Int., 2018, 107, 451–461, DOI:10.1016/j.foodres.2018.02.034.
- M. Sipiczki, Appl. Environ. Microbiol., 2006, 72, 6716–6724, DOI:10.1128/AEM.01275-06.
- A. Morata, I. Loira, C. Escott, J. M. del Fresno, M. A. Bañuelos and J. A. Suárez-Lepe, Fermentation, 2019, 5, 63, DOI:10.3390/fermentation5030063.
- M. Gobbi, F. Comitini, P. Domizio, C. Romani, L. Lencioni, I. Mannazzu and M. Ciani, Food Microbiol., 2013, 33, 271–281, DOI:10.1016/j.fm.2012.10.004.
- A. Hranilovic, M. Bely, I. Masneuf-Pomarede, V. Jiranek and W. Albertin, PLoS One, 2017, 12, e0184652, DOI:10.1371/journal.pone.0184652.
- A. Morata, I. Loira, W. Tesfaye, M. A. Bañuelos, C. González and J. A. Suárez Lepe, Fermentation, 2018, 4, 53, DOI:10.3390/fermentation4030053.
- F. Viana, C. Belloch, S. Vallés and P. Manzanares, Int. J. Food Microbiol., 2011, 151, 235–240, DOI:10.1016/j.ijfoodmicro.2011.09.005.
- V. Martin, M. J. Valera, K. Medina, E. Boido and F. Carrau, Fermentation, 2018, 4, 76, DOI:10.3390/fermentation4030076.
- L. Tat, P. Comuzzo, I. Stolfo and F. Battistutta, Food Chem., 2005, 93, 361–369 CrossRef CAS.
- P. Comuzzo, M. Marconi, G. Zanella and M. Querzè, Food Chem., 2018, 264, 16–23 CrossRef CAS PubMed.
- J. N. Sauceda-Gálvez, I. Codina-Torrella, M. Martinez-Garcia, M. M. Hernández-Herrero, R. Gervilla and A. X. Roig-Sagués, LWT, 2021, 136, 110286, DOI:10.1016/j.lwt.2020.110286.
- A. Morata, I. Loira, C. González and C. Escott, Molecules, 2021, 26, 4571, DOI:10.3390/molecules26154571.
- R. Escribano-Viana, L. González-Arenzana, P. Garijo, L. Fernández, R. López, P. Santamaría and A. R. Gutiérrez, Fermentation, 2022, 8, 337, DOI:10.3390/fermentation8070337.
- A. Rapp and G. Versini, Dev. Food Sci., 1995, 37, 1659–1694, DOI:10.1016/S0167-4501(06)80257-8.
- S. Saberi, M. Cliff and H. Van Vuuren, Food Res. Int., 2012, 48, 725–735, DOI:10.1016/j.foodres.2012.06.012.
- D. Elpa, E. Durán-Guerrero, R. Castro, R. Natera and C. G. Barroso, J. Sep. Sci., 2014, 37, 1867–1872, DOI:10.1002/jssc.201400308.
- F. M. Carrau, K. Medina, L. Fariña, E. Boido, P. A. Henschke and E. Dellacassa, FEMS Yeast Res., 2008, 8, 1196–1207, DOI:10.1111/j.1567-1364.2008.00412.x.
- F. Carrau, K. Medina, L. Fariña, E. Boido and E. Dellacassa, Int. J. Food Microbiol., 2010, 143, 81–85, DOI:10.1016/j.ijfoodmicro.2010.07.024.
- I. Arias-Pérez, M. P. Sáenz-Navajas, A. de-la-Fuente-Blanco, V. Ferreira and A. Escudero, Food Chem., 2021, 361, 130081 CrossRef PubMed.
- M. Ugliano and P. A. Henschke, Yeasts and Wine Flavour, in Wine Chemistry and Biochemistry, ed. Moreno-Arribas M. V. and Polo M. C., Springer, 2009 Search PubMed.
- F. J. Francis, Wiley Encyclopedia of Food Science and Technology, John Wiley & Sons, 2nd edn, 1999 Search PubMed.
- C. Coetzee, J. Brand, D. Jacobson and W. J. Du Toit, Aust. J. Grape Wine Res., 2016, 22, 197–204, DOI:10.1111/ajgw.12206.
- A. Escudero, E. Campo, L. Fariña, J. Cacho and V. Ferreira, J. Agric. Food Chem., 2007, 55, 4501–4510 CrossRef CAS PubMed.
- K. Zott, C. Thibon, M. Bely, A. Lonvaud-Funel and D. Dubourdieu, I. Masneuf-Pomarede, Int. J. Food Microbiol., 2011, 151, 210–215, DOI:10.1016/j.ijfoodmicro.2011.08.026.
- C. Escott, C. Vaquero, I. Loira, C. López, C. González and A. Morata, Foods, 2022, 11, 3734, DOI:10.3390/foods11223734.
Footnote |
| † Electronic supplementary information (ESI) available. Discussion, limited experimental and spectral data, and crystallographic data. See DOI: https://doi.org/10.1039/d3fb00226h |
| This journal is © The Royal Society of Chemistry 2024 |