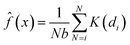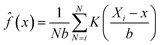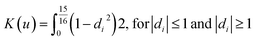Use of machine learning for monitoring the growth stages of an agricultural crop†
Received
30th June 2023
, Accepted 25th October 2023
First published on 26th October 2023
Abstract
As one of the world's major crops, oats (Avena sativa L.) require management strategies to increase their yield and quality. This study utilised an unmanned aerial vehicle (UAV) with multispectral image sensors to predict winter oats height (1.18 m at ripening stage) and yield (maximum >7.62 t per ha) using the normalised difference vegetation index (NDVI) and chlorophyll green vegetation index (CI green VI) across three different growth stages (flowering, grain filling and ripening). To corroborate the vegetation indices ground truth data on the measured crop yield, a variety of chemical soil health indicators (i.e. nitrogen, phosphorus, potassium, pH, and soil organic matter), and a crop quality indicator (β-glucan) were determined. A hierarchical multinomial logistic regression machine learning model was developed to predict the oats yield incorporating the chemical soil health indicators and crop quality indicator. The determined ‘combination model’ using the CI green VI, with 16 soil feature parameters, showed good specificity (0.87), sensitivity (0.95), and accuracy (0.93) at estimating the very high oat yield. Finally, the study provides the range of soil nutrient levels and the crop quality indicator that farmers must maintain to gain the highest oat yield at harvest. The findings of this research study will be particularly valuable as a Precision Agriculture management strategy for maximising winter oat yield and quality.
Sustainability spotlight
The ability to maintain global crop security with a focus to end hunger and promote sustainable agriculture relies on an effective approach to crop management. This paper seeks to address these issues using the specific crop of oats as its focus. Oats are one of the world's most grown cereal crops helping to feed the population. To assist farmers to produce an oat crop of the highest yield and quality requires knowledge of the importance of providing soil nutrients at the right time and in the right amount. Our research has shown that by using a UAV-MSI and a machine learning approach can provide that knowledge in terms of soil nutrients and application times to achieve the highest yield.
|
Introduction
Oats (Avena sativa L.) are Europe's fifth largest crop and the sixth most grown cereal worldwide, used extensively for both animal feed and human consumption (in breakfast cereals, beverages, bread, and infant foods), due to their high fibre and protein content.1,2 Given the global demand for oats, crop evaluation across the five phenological growth stages3 critical for informing decisions as part of a Precision Agriculture management strategy, to increase the crop yield and its quality. Crop phenotypic information can be retrieved using both manual and remote sensing techniques.4 Manual methods rely on hand-held spectrometers to collect phenotypic data in the field whereas remote sensing methods use either satellite or unmanned aerial vehicles (UAVs) to gather phenotypic crop data.5 Only a few studies have compared the performance of different UAV-based cameras with a hand-held spectrometer in crop monitoring under the same environmental conditions.4,6–10 The crops investigated include barley,7 barley, onion, potato, and rapeseed,8 maize,4 grassland,9 and the soil and vegetation of a vineyard.10 In all cases good correlation was found between the camera mounted UAV and ground-based spectral data,11 often using the normalised difference vegetation index (NDVI). Given the widespread use of UAVs by researchers in crop phenotyping, as part of a Precision Agriculture management strategy, there is a need for additional performance evaluation of UAV sensors to ground based field spectrometers in terms of different crops (e.g. oats) and evaluation of different vegetation indices (VI's).
An important aspect of soil health management is the timely and appropriate application of fertilisers to ensure favourable nutrient conditions to maximise crop yield and thereby contribute to global food security.12,13 Determination of temporal and spatial patterns of crop growth are critical for assessing fertiliser application. In many instances, the crop nitrogen status is estimated indirectly from the variables of chlorophyll content and the leaf area index as can be done using a hand-held chlorophyll meter.14–16 However, this approach is limited, as the chlorophyll meter does not capture the spatial variability present within an agricultural field. Also, these ground-based methods necessitate the collection of many samples' spectral data, which can be invasive as well as destructive to crops, in addition to being labour-intensive.17
Remote sensing techniques are a viable alternative to ground-based measurements as they provide crop reflectance and diagnostic information on crop nutrient concentration in a timely and spatially contextualised manner.17 The two main remote sensing technologies used in Precision Agriculture for soil mapping are satellites and UAVs. For instance, VI's, derived using remote sensing spectral data have been used to detect the nitrogen status in crops18 using the strong correlation between nitrogen concentration and chlorophyll content at the crop canopy. In this way chlorophyll sensitive VI's have been successfully employed to estimate chlorophyll in crops and correlated to nitrogen concentrations using normalised difference red edge (NDRE) and chlorophyll (CI) green VI in spring wheat.17 Meanwhile, a variety of methods including artificial neural network (ANN), partial least squares regression (PLSR), random forest (RF), extreme learning machine (ELM) support vector machine (SVM) and convolutional neural network (CNN) have been used to estimated crop yield and biomass.19–21 For example, four machine-learning algorithms were used to build oat biomass estimation models using a variety of VI's derived from UAV-based multispectral imagery.19 The machine learning models demonstrated provided promising results at estimating biomass as part of an oat breeding programme. An additional study used UAV-imagery and the multiple machine learning models of ANN, ELM, SVM, least absolute shrinkage and selection operator to predict leaf nitrogen content in a wheat crop.22 They concluded that the best model for predicting the nitrogen content in the wheat crop was ELM, with a correlation coefficient of R2 = 0.99. Further work from the same research group applied an RF model to predict the soil sulfur content (R2 = 0.71, RMSE = 8.86) using a hand-held visible near-infrared (VIS-NIR) reflectance spectroscopic technique.23 In addition, the group have applied a hand-held VIS-NIR spectroscopic technique combined with a PLSR model to predict soil organic carbon (R2 = 0.53, RMSE = 9.04).24
The ability to maintain global crop security with a focus to end hunger and promote sustainable agriculture relies on an effective approach to crop management. This paper seeks to address these issues using the specific crop of oats as its focus. Oats are one of the world's most grown cereal crops helping to feed the population. To assist farmers to produce an oat crop of the highest yield and quality requires knowledge of the importance of providing soil nutrients at the right time and in the right amount. The aims of this research are to investigate the phenological growth stages of a winter oats crop in North East England using spectral data from both a hand-held and unmanned aerial vehicle, alongside normal farmer interventions informed by investigative spatial and temporal soil analyses, to assess both the yield and quality of the final crop. This has been done by (a) comparing the usefulness of a hand-held multiwavelength spectrometer and an unmanned aerial vehicle with multispectral image camera, (b) determination of the soil nutrient profile of the agricultural field across three phenological growth stages, (c) determination of the final oat grain quality by determining the concentration of β-glucan, (d) manual estimation of crop yield by weighing of sub-samples, (e) use of different UAV-MSI camera derived VI's to create a crop yield estimation model using pattern analysis based on a kernel density estimation, and (f) finally to derive an machine learning method to estimate crop yield based on multiple data inputs.
Experimental
Airy Holme farm
Winter oats seed (variety, Mascani) was planted at a rate of 155 kg/hectare using a Claydon Hybrid T4 trailed drill (Rickerby, Hexham, UK) pulled by a Claas ARES 836 RZ tractor (Rickerby, Hexham, UK) to a depth of 25–30 mm on the 28 September 2020 in a 3.58-hectare field (lat. 54.880690; long. −1.915923), known locally as Copse field. The field was subject to various treatments across the five phenological growth stages from sowing of the seed to harvest, Fig. 1(a), which included pesticide treatments, Fig. 1(b) and fertilizer treatments, Fig. 1(c). Full details are provided in the ESI.† The desiccated crop was harvested on the 13 August 2021 using a Claas Lexion 570, Terra-Trac combine harvester (Rickerby, Hexham, UK). The seed is monitored by sensors (FarmTRX, Troo Corp., Ottawa, Canada), which record both the yield, and its location using GPS technology, into an on-board data logger.
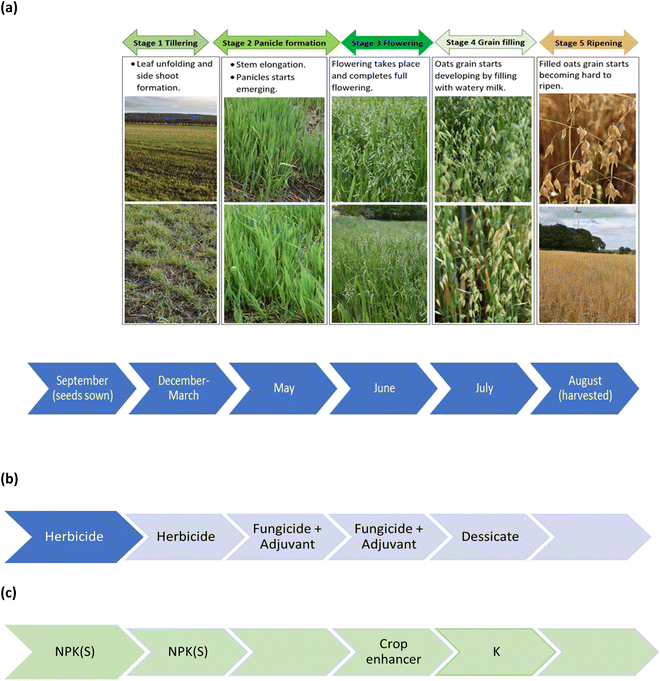 |
| | Fig. 1 Chronology of (a) phenological growth stages of oats, (b) pesticide treatments, and (c) fertilizer treatments. | |
Copse field
Initial soil analysis was done in March 2021 with data reported on the 15th March 2021 (by an independent laboratory, Lancrop Laboratories, Pocklington, UK in association with Agrovista UK Ltd, Nottingham, UK). Soil analysis indicated the following characteristics, a sandy silt loam (sand 41.3%; silt 47.3%, and clay 11.4%) with a pH of 7.5, organic matter (4.7%) and a cation exchange capacity of 19.0 meq./100 g.
Unmanned aerial vehicle
A multirotor UAV (DJI Phantom 4, supplied by Coptrz Ltd, Leeds, UK) was used with a multispectral camera, stabilized with a 3-axis gimbal, with a 5 camera-array covering the blue (450 ± 16 nm), green (560 ± 16 nm), red (650 ± 16 nm), red edge (730 ± 16 nm) and near-infrared (840 ± 26 nm) spectra with an additional camera that can also provide live images in RGB (visible) mode. The camera lenses had a field of view of 62.7°, a focal length of 5.74 mm, with the autofocus set at ∝, and an aperture of f/2.2. In all cases, the camera was angled perpendicular to the ground, with data capture occurring in hover and capture mode. Images i.e. 1554 image files per flight, were gathered over 256 waypoints and captured as 16-bit TIF files corrected for ambient radiance values. The UAV speed was 5.0 m s−1 and had an average height of 50.6 m for the 2901 m flight distance. All flights were recorded with a resolution of 2.7 cm per px, a front overlap ratio of 75%, a side overlap ratio of 60% and a course angle of 90°. Specific weather conditions relating to daytime temperature during flight, wind speed and direction (recorded using a handheld anemometer (Benetech® GM816, Amazon UK)), and UAV pilot anecdotal observations on cloud coverage are identified with specific dated data.
UAV photogrammetric processing
The multispectral UAV images were used to create an orthomosaic image (Agisoft Metashape Professional (64 bit) software v.1.7.1, Agisoft LLC, St. Petersburg, Russia). The steps for UAV photogrammetric processing were as follows. The aerial images were first merged and aligned to create a sparse point cloud by matching similar image attributes. Following that, images were precisely positioned to create a 3D point cloud based on the GPS coordinates of each image. A solid mesh model was created using the 3D point cloud. Following completion of the preceding steps, an orthomosaic image was created using the WGS 1984 Web Mercator coordinate system.
Collection of crop phenotypic data using multiwavelength spectral imaging: ground reference data
Ground truth measurements were captured using a pocket-size portable hand-held spectrometer (Spectro 1, Variable, Inc., Chattanooga, TN, USA). The Spectro 1, has an 8 mm measurement aperture which allows spectrophotometric measurements in the visible region from 400 to 700 nm at 10 nm intervals. For this experiment 9 locations were selected within the Copse field, with 5 replicate measurements per location, for spectrophotometric measurements over three different growth stages, Fig. 1(a)i.e. Stage 3: flowering (June); Stage 4: grain filling (July); and, Stage 5: full ripening (August). Prior to each measurement, the spectrometer was calibrated using a white plate according to the manufacturer's instruction. As the device is operated with the Spectro application software (Variable, Inc., Chattanooga, TN, USA) on the user's smartphone, the collected reflectance data was automatically uploaded and stored in the manufacturer's cloud storage service.
Comparison between UAV-MSI and ground reference data
For ground reference data, GRVI was calculated by using the reflectance data of green and red provided by the Spectro 1 spectrometer. The orthomosaic image generated by the Agisoft Metashape software for the UAV-MSI data are not reported in reflectance mode but as “reflectivity”. Therefore, a pseudo correction was applied that normalised the data as reflectance values between 0 and 1. This was done based on the UAV image histogram; the maximum DN value was divided with each spectral band by using the raster calculator tool in the ArcGIS Pro software, for VI calculation, Fig. S1.† The GRVI VI raster was derived by calculation of the reflectance of green and red orthomosaic image (eqn (1)). GNDVI was derived by calculation of the reflectance of green and NIR orthomosaic image (eqn (2)). NDVI was derived by calculation of the reflectance of red and NIR orthomosaic image (eqn (3)). NDRE was derived by calculation of the reflectance of red-edge and NIR orthomosaic image (eqn (4)). SAVI was derived by calculation of the reflectance of red, NIR orthomosaic image and a soil brightness correction factor (L) defined as 0.5 (eqn (5)). CI green was derived by calculation of the reflectance ratio of NIR and green orthomosaic image (eqn (6)).| | 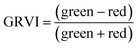 | (1) |
| | 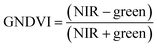 | (2) |
| | 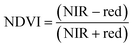 | (3) |
| | 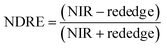 | (4) |
| | 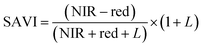 | (5) |
| | 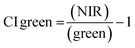 | (6) |
Soil sample collection and determination of soil health check indicators during active growing stages of oat crop
Soil samples (48) from the oats field were collected, during the first week of June, July and August 2021, from 6 rows (labelled A–F) with 8 sampling points, 30 m apart, per row in a grid format, Fig. 2(a). Surface soil samples, between 0–10 cm depth, were collected using a stainless-steel trowel. To avoid cross-contamination, the trowel was cleaned with a new antibacterial wipe, between each sample. The collected soil samples were placed in labelled collection bags (Kraft sample bags) and transported back to the laboratory for analysis of nitrogen, phosphorus and potassium, NPK, and pH using a soil Palintest kit (SKW500, Palintest UK, Kingsway, Team Valley, England) which was previously validated by the manufacturer.25 All soil samples were analysed within one week of collection, using manufacturer's instructions and prepared reagents. Nitrogen was always analysed first on damp soil, as per manufacturer's instructions, to prevent nitrogen loss due to biological activity. Results, using the SKW500, were displayed for nitrate (mg l−1), phosphorus (mg l−1), and potassium (0–450 mg l−1) using the digital photometer. Soil pH was reported, using the manufacturer's instructions, using the calibrated multiparameter pocket sensor. Additionally, soil organic matter was determined using the loss on ignition method using a muffle furnace pre-heated to 800 °C.26 Further details of the sample preparation for NPK, pH and organic matter are presented in the ESI.†
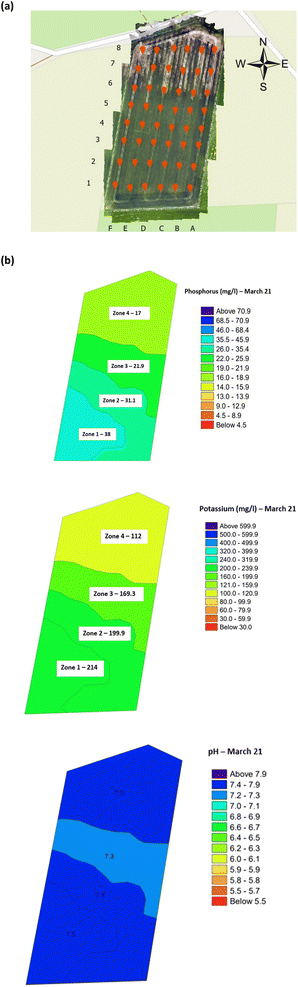 |
| | Fig. 2 Copse agricultural field planted with winter oats (September 2020–August 2021) (a) soil sample collection locations, and (b) within-field zones soil health indicator maps (March 2022). Note: the soil analysis data was abstracted from the independent laboratory results. The average concentration or pH is indicated per zone. | |
Determination of oat grain yield and its quality
The yield of oats grain was calculated by harvesting 48 × 1 m2 area of oats on 4 August 2021 when the crop was fully ripe and 9 days prior to harvest, Fig. 2(a). Harvesting was done by manually cutting the crop and carefully placing it upside down in a large brown paper sample bag prior to transportation back to the laboratory. In the laboratory the oat-husk was manually detached from the straw, followed by subsequent manual de-husking, liberating the oat grain. Sub-sampling (20 g accurately weighed) of the oat (+husk) from the 1 m2 was done to allow calculation of the yield, in t per ha, Table S1,† based on three replicates, from each of the 48 samples. The quality of the oat grain was assessed by determination of the β-glucan concentration. Full details of the extraction method, modified from27 for the determination of β-glucan and its subsequent analysis are provided in the ESI.†
Oat crop analyses using UAV-MSI camera derived data
Agisoft's 3D point clouds were extracted and used by ArcGIS Pro to generate a canopy height model (CHM), of the crop, based on time-series data relating to the phenological growth stages. The height between the ground and the top of the oat crop, CHM, was calculated in ArcGIS using the raster calculator tool, CHM = DSM - DTM.
The yield of the oat crop was estimated using the pixelated data from the digitised UAV-MSI images, using iso-cluster classification (ArcGIS pro), to generate the VI's of NDVI (eqn (3)) and CI green (eqn (6)). Previously published work,28 used a simplified pixel-based approach to estimate crop yield (t per ha) using the following equation:
| | 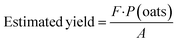 | (7) |
where
F is the extracted oat pixels/∑pixels;
P(oats) is the area of oats in the field calculated by multiplying the oat pixels by the resolution of the UAV images
i.e. (0.027 m)
2 per pixel in tonnes; and, A is the area of the field in ha. As a result, the oat yield for the 48 sampling locations was calculated using the two VI's.
The kernel density estimation (KDE) is a non-parametric spatial analysis method of estimating probability function.29 The KDE mathematical function can be defined as a kernel density estimator, ![[f with combining circumflex]](https://www.rsc.org/images/entities/i_char_0066_0302.gif) , at a location x taken from a set of data (e.g. crop yield), where x1,x2 … xn represents the number of samples (N) with an unknown probability function f(x) and defined as a kernel estimate
, at a location x taken from a set of data (e.g. crop yield), where x1,x2 … xn represents the number of samples (N) with an unknown probability function f(x) and defined as a kernel estimate ![[f with combining circumflex]](https://www.rsc.org/images/entities/i_char_0066_0302.gif) (x):
(x):
| | 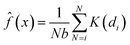 | (8) |
| | 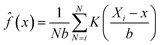 | (9) |
where
di is the distance between two points (
e.g. Xi −
x);
b the bandwidth (
b > 0, a positive number that defines the smoothness of a density plot); and
K = denotes the kernel function. The ArcGIS software employs the quartic (
bi-weight) kernel function, which is defined as:
| | 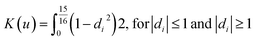 | (10) |
This process allowed thematic maps to be generated for the oat grain yield as measured in the laboratory and estimated by NDVI and Cl green VI.
Results and discussion
Soil nutrient data
Validation of the soil test kit was previously corroborated between standard laboratory methods for soil analysis with good correlation between the methods for nitrate (R2 = 0.96), phosphate (R2 = 0.95), potassium (R2 = 0.96), and pH (R2 = 0.98).25 As a result, the soil test kit is a viable method for soil analysis. In addition, previous independent soil analysis was done in March 2021 on behalf of Agrovista Co. U.K. using standard laboratory soil test methods.30 This soil data was reported in 4 zones across the agricultural field, Fig. 2(b). The reported mean (minimum and maximum) nutrient levels, across the 4 zones, are 27 mg l−1 (17–38 mg l−1) for phosphorus, 173.8 mg l−1 (112–214 mg l−1) for potassium and a pH of 7.4 (7.3–7.5) (Table S2†). No nitrate data was available. As a result, we have reported our determined soil nutrient data for the 48 samples across the 4 zones for consistency. The full results for analysis of the 48 soil sub-samples, with respect to nitrate, phosphorus, potassium, pH and soil organic matter for the months of June, July and August are reported, Table S3(a and b).† It is noted that the impact on the soil nutrients was influenced, pre-soil analyses, by application of two rounds of soil fertiliser (on 28/09/2021 and 06/03/2021). The determined soil analyses data are mapped, over June–August, for phosphorus in Fig. 3, potassium in Fig. 4, nitrate in Fig. 5 and pH in Fig. 6.
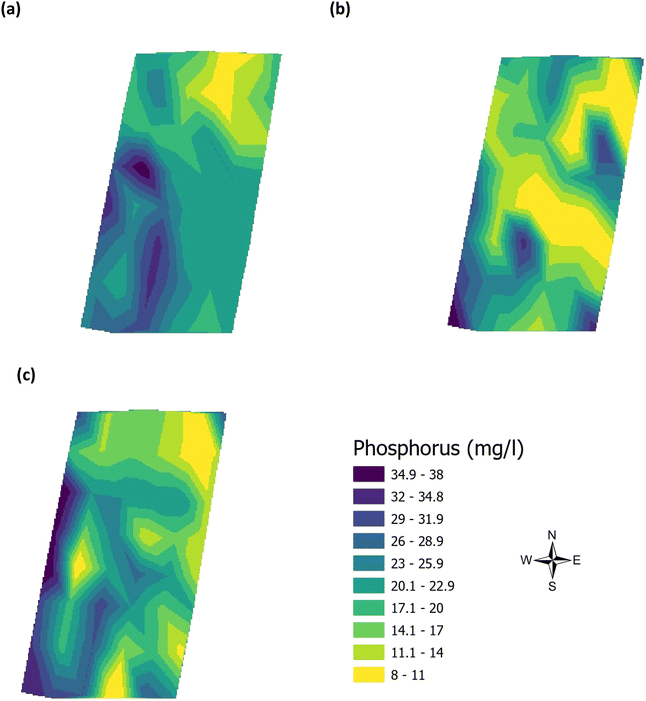 |
| | Fig. 3 Analysis of soil phosphorus (mg l−1) across the three phenological growth stages (a) June, (b) July, and (c) August 2021. | |
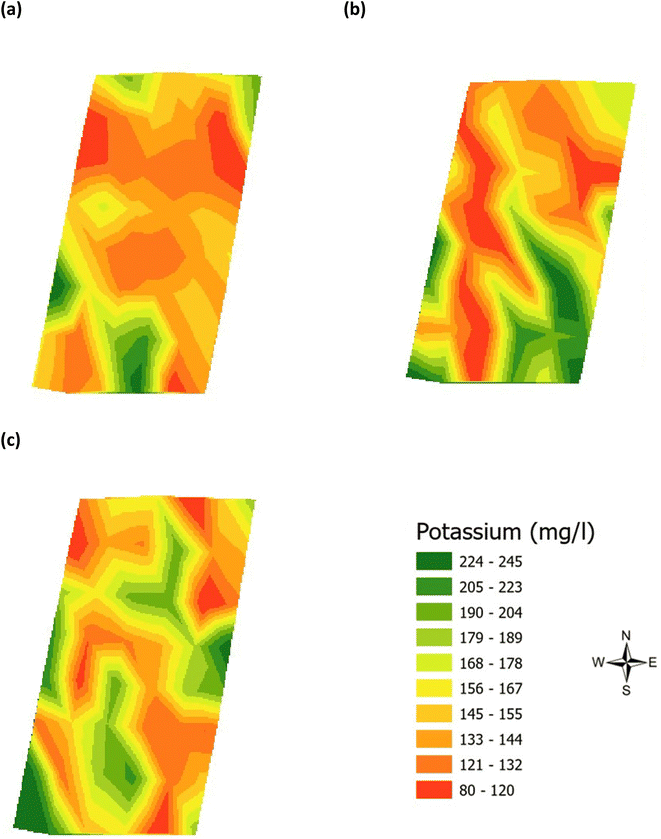 |
| | Fig. 4 Analysis of the soil potassium (mg l−1) across the three phenological growth stages (a) June, (b) July, and (c) August 2021. | |
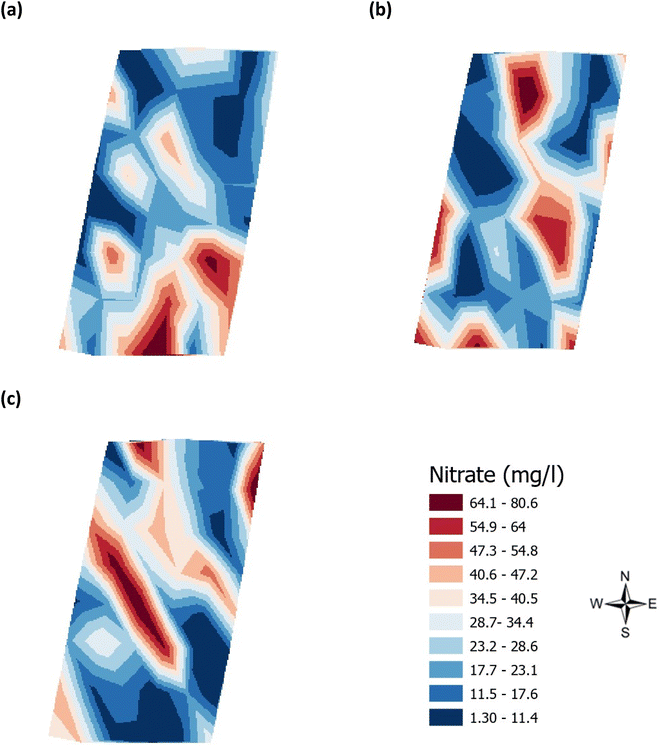 |
| | Fig. 5 Analysis of the soil nitrate (mg l−1) across the three phenological growth stages (a) June, (b) July, and (c) August 2021. | |
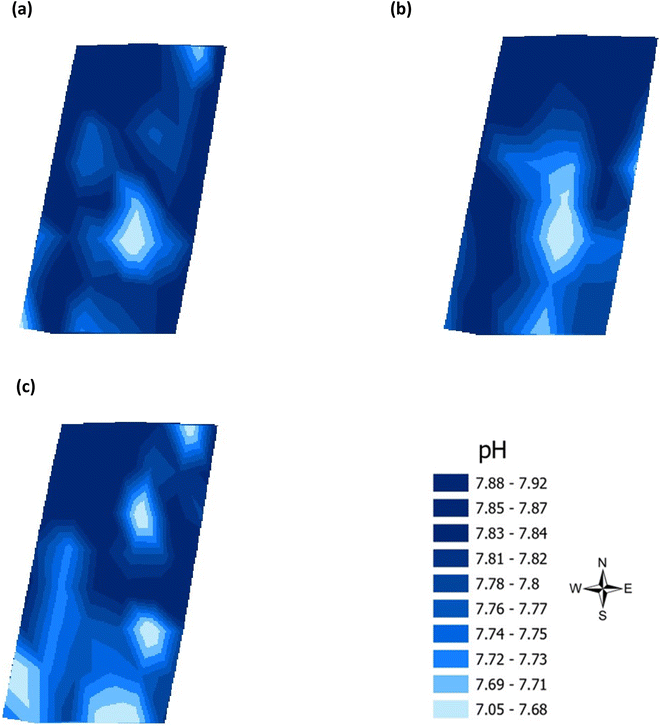 |
| | Fig. 6 Analysis of soil pH across the three phenological growth stages (a) June, (b) July, and (c) August 2021. | |
Estimation of plant height and links to soil analysis data
The UAV-MSI camera data was used to estimate the crop height using the CHM (Fig. 7). To validate the data, the height of the wall adjacent to the oats field was measured. The manually measured wall heights (1.07 m ± 0.04 m, N = 3) were statistically analysed (t-test) against the UAV-MSI camera data, using CHM, and estimated to be 1.00 ± 0.11, N = 3. The latter was averaged over three different days. There was no statistical difference between the two measurements (p-value of 0.17, at the 95% confidence interval). Therefore, the estimated oats plant height measurements, using the CHM, are appropriate for this study. The variation in canopy height across the five phenological growth stages are shown in Fig. 8. The mean height at Stage 5 (ripening) is 1.18 m which corresponds to the mean typical height of oats crop grown in the North of the UK (1.00 m).31
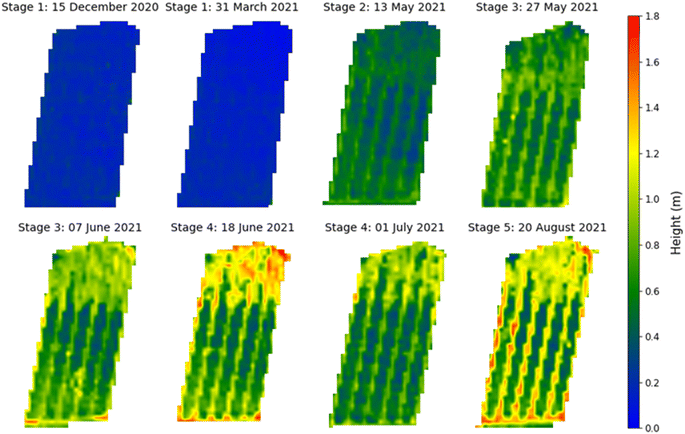 |
| | Fig. 7 Canopy height model for oat at the phenological growth stages. Note: the apparent excessive height (>1.6 m) of the crop in selected places was in fact due to the presence of invasive brome grass and wild oats, that occurred within the Copse field. | |
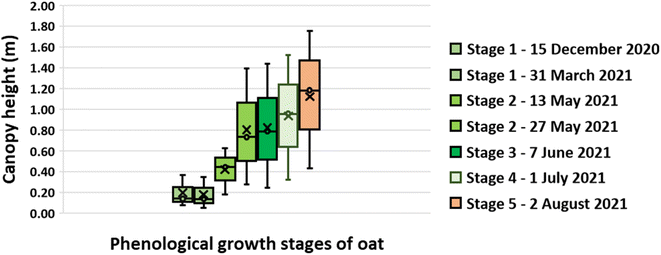 |
| | Fig. 8 Oat canopy height at phenological growth stages. Note: the limits of the box represent the upper and lower quartile of the data as assessed at the 95% confidence limit while the whiskers show the minimum and maximum heights determined. The horizontal line within the box represents the median height while the cross is the mean height. | |
The mean height of the crop has been plotted against the soil nutrient concentrations, for the four field zones, Fig. 2(b), across the five phenological growth stages from December 2020 to August 2021. For soil phosphorus levels, Fig. 9, it is noted that as the concentration in the soil decreases, a corresponding growth in the crop height occurs. Similar trends occur for soil nitrate, Fig. 10. However, the trend for soil potassium differs, Fig. 11. This is due to the addition of potassium fertiliser in June, resulting in a significant increase in soil potassium concentration, Table S3(b)† across all 4 zones.
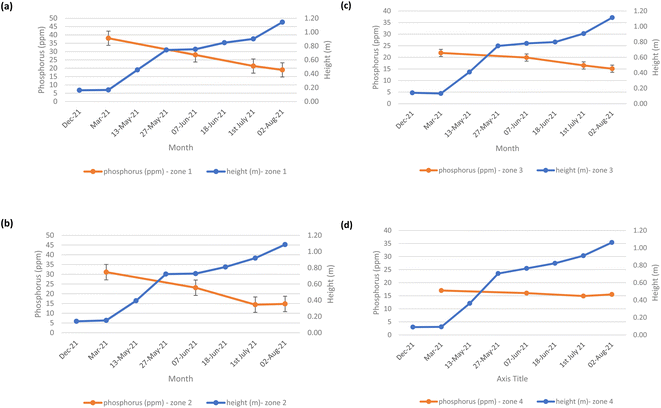 |
| | Fig. 9 Soil phosphorus (ppm) and oats height (m) in (a) zone 1, (b) zone 2, (c) zone 3, and (d) zone 4. | |
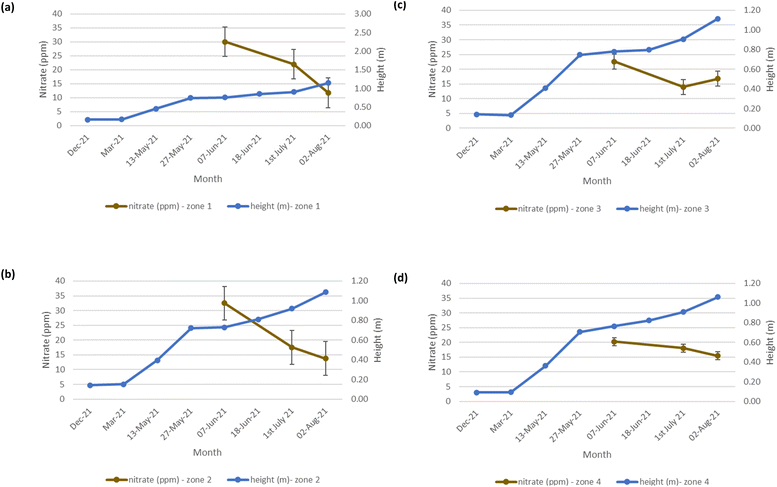 |
| | Fig. 10 Soil nitrate (ppm) and oats height (m) in (a) zone 1 (b) zone 2 (c) zone 3 (d) zone 4. Note: no soil nitrate concentration data was available for March 2021. | |
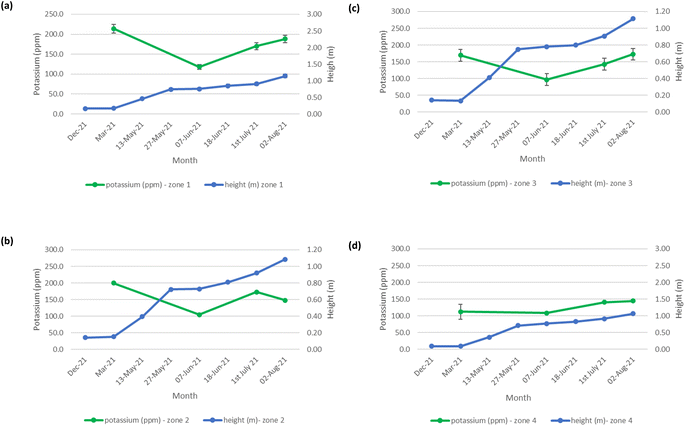 |
| | Fig. 11 Soil potassium (ppm) and oats height (m) in (a) zone 1, (b) zone 2, (c) zone 3, and (d) zone 4. | |
β-Glucan concentration of oat grain
Validation of the β-glucan determination, determined as glucose, was assessed across its four extraction stages, and reported, Table S4(a).† The mean extraction efficiency, assessed by spiking glucose (15 mM) on oat extracts, was as follows: Stage 1 (before alkali extraction): mean 84.4% (85.2%; 83.5%); Stage 2 (before acid neutralisation): mean 90% ± 5% (n = 6); Stage 3 (before freeze drying): 95% ± 6% (n = 6); and, Stage 4 (before acid hydrolysis): mean 105% ± 12% (n = 6). In addition, the conversion efficiency of β-glucan to glucan was assessed by spiking a sample with β-glucan (15 mM); this was determined to be 102% ± 8% (N = 6), Table S4(b).† The analytical performance data, using this colorimetric assay method, was determined as follows: for glucose, a lower limit of detection (LLOD) of 34 mg l−1 and a limit of quantitation (LOQ) of 102 mg l−1. Precision was determined to be 9.5% at the low concentration (0.1 mM glucose) and 1.9% at the higher concentration (40 mM glucose). The equivalent LLOD for β-glucan was 30 mg l−1 with an LOQ of 92 mg l−1, Table S4(c).† The LLOD and LOQ were determined using the standard curve method: LLOD = (3.3σ)/S and LOQ = (10σ)/S, where σ is the standard deviation and S is the slope of the curve.32 The full results for analysis of the 48 oat grain sub-samples for their β-glucan content in August are reported in Table S3.† It is noted however, that the β-glucan content of winter oat Mascani grown in the north of the UK ranges from 3.7% to 4.2%, with a mean of 3.9%.28 In our study however, the β-glucan content ranges between 0.37–2.26%. Differences in the β-glucan content of oats can, however, vary between cultivator, growing location, storage, and processing conditions.2
Evaluation of vegetation indices from the UAV-MSI and ground reference data
A comparison of the spectral reflectance derived from multispectral and multiwavelength spectral data (derived from the UAV-MSI camera and hand-held spectrometer, respectively), over three months of the growth stages, is shown in Fig. 12. It is noted that the multispectral data (derived from the UAV-MSI camera) always had higher reflectance values; a trend also reported by others.33 It was postulated that the reasons could be due to the differences in flight height; a higher altitude between the sampling location and the remote sensing platform, which could have an impact on the quality of the data. Others have postulated,10 that the variability can be justified due to the opposite data acquisition approaches adopted by the two methods; the data collected by the hand-held spectrometer is proximal and static, whereas the UAV-MSI data collection is remote and dynamic. Furthermore, reflectance values can be affected by the illumination geometry i.e. the time of day when the data was collected.34
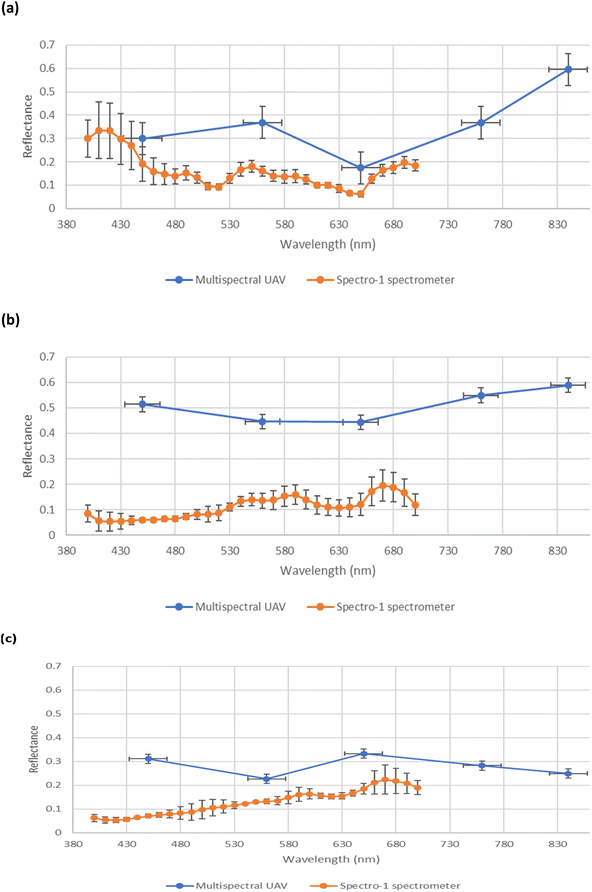 |
| | Fig. 12 Comparison of spectral profile in reflectance between multispectral UAV (blue) and multiwavelength spectral ground reference data using a Spectro-1 spectrometer (orange) across three growth stages of oats. (a) June (Stage 3 – flowering), (b) July (Stage 4 – grain filling), and (c) August (Stage 5 – ripening). | |
Spectral VI's were generated from data derived from both the UAV-MSI camera and hand-held spectrometer, noting that the latter can only operate in the visible region, and hence is limited to GRVI only (eqn (1)). A statistical comparison (Student's t-test) of UAV-MSI and ground reference spectral data with respect to generation of VI's is represented in Table S5.† If p < 0.05, the VI is statistically significant indicating there is a difference between the VI derived from the UAV-MSI camera data and the ground reference data, or vice versa. Statistical differences (p < 0.05) between the GRVI generated values from both approaches are noted in June and July, no such differences are noted in August. However, the extended range of spectral bands that can be used to generate a diverse range of VI's (eqn (1)–(6)) makes the data from the UAV-MSI camera more informative. Generally, the numerical values for the VI's follow the trend (highest to lowest): CI green VI > NDVI > SAVI > GNDVI > NDRE > GRVI. The results, Table S5,† highlight the most suitable VI's as a phenology indicator for the oats crop to be GRVI, NDVI and the CI green VI. Firstly, this is since the index values of the GNDVI, SAVI, and NDRE do not vary significantly across phenological growth stages; hence it is challenging to differentiate the oats crops in the different phenological growth stages. Conversely, GRVI, NDVI and CI green VI values have been shown to vary significantly across the phenological growth stages, Table S5.† Finally, when the oats were initially green and then fully ripe, the NDVI and CI green VI values varied widely between high and low values, indicating that these indices could be used to differentiate oats at different phenological growth stages. As a result, the NDVI and CI green VI were used to examine the relationship between soil health indicators and yield estimation over the phenological growth stages of oats.
Correlation between estimated and actual oats grain yield
The estimated oats grain yield was determined at the peak of flowering (stage 3, Fig. 1). Iso-cluster classification was used to classify the VI maps into three clusters: soil surface, grasslands, and oats crop, Fig. 13. Oats were classified using NDVI values between 0.6–0.8 and CI green VI values between 2–7, grasslands between 0.59–0.12 (NDVI) and 1.8–0.5 (CI green VI), and soil surfaces were classified as <0.12 (NDVI) and <0.5 (CI green VI). As a result, the final oats grain yield (in August) was estimated by calculating the pixel areas of the oats crop visible at the top of the panicle and classified by iso-cluster classification for the NDVI and CI green VI maps, Fig. 14. Moderate correlation of determination, determined using linear regression, was found between the estimated oats grain yield and the actual oats grain yield for NDVI (R2 = 0.74) and CI green VI (R2 = 0.70). It was noted that the NDVI iso-cluster classified image in Fig. 13(b), demonstrated better classification of the crop from surrounding grasslands and soil surfaces than the CI green VI. Therefore, this could explain the slightly better correlation between estimated and actual oats grain yield by NDVI in comparison to CI green VI. Also, other researchers have indicated that NDVI has a stronger correlation when estimating white oat grain yield in comparison to other VI's.35,36
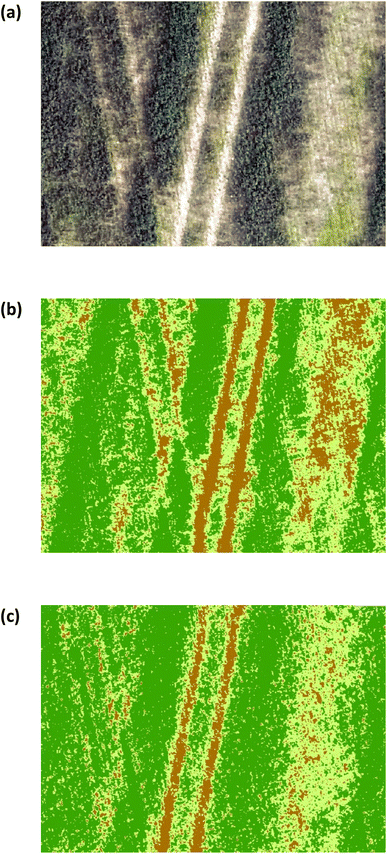 |
| | Fig. 13 (a) RGB image (b) NDVI iso-clustered image (c) CI green VI iso-clustered image. Note: dark green represents oats crop. Light green represents grasslands. Brown represents soil surface. | |
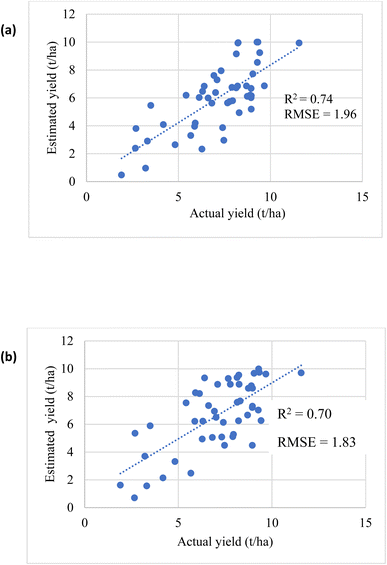 |
| | Fig. 14 Correlation between estimated and actual yield (a) NDVI (b) CI green VI. | |
Evaluation of thematic yield maps by kernel density estimation
Yield maps using estimated and actual oats grain yield were generated by the statistical pattern analysis method of KDE. The oats grain yield map variation was highlighted as follows: low (red = 0–4.97 t per ha), medium (orange = 4.97–6.18 t per ha), high (yellow = 6.18–7.11 t per ha) and very high (green > 7.62 t per ha) yield areas. It is noted (Fig. 15(a–c)) that the estimated oats grain yield maps by NDVI, in Fig. 15(b), and the CI green VI in Fig. 15(c), represent visually similar patterns to the actual oats grain yield measured in the laboratory, Fig. 15(a). Also, a significant proportion of the lower yield areas identified, in both the estimated and actual yield maps, are at the top (north end) of the field. This was visually noted on the regular visits to the site over the duration of the project. Furthermore, the yield was confirmed by the combine harvester on-board tracking system, Fig. 15(d). In comparison to the grain yield map generated by the combine harvester on-board system, Fig. 15(d), the yield maps generated by the KDE method show improved spatial resolution, Fig. 15(a–c). As a result, it was concluded that the use of VI iso-cluster classification can successfully estimate oats grain yield, using the KDE method, two months prior to harvesting of the crop.
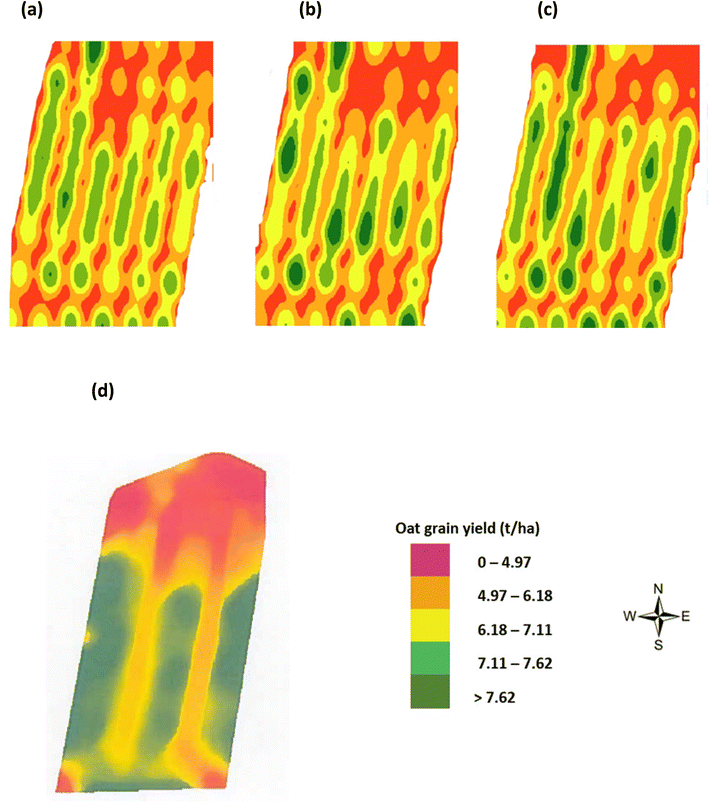 |
| | Fig. 15 Oats grain yield maps by KDE (a) actual yield as measured in laboratory (in August), (b) estimated yield by NDVI (in June), (c) estimated yield by CI green VI (in June), and (d) from on-board combine harvester yield monitor (FarmTRX system) (in August). | |
Hierarchical multinomial logistic regression model
A hierarchical multinomial logistic regression model was built (Matlab, version R2021a), for a given set of independent variables, to predict the probabilities of the possible outcomes of a categorical dependent variable i.e. oat grain yield, categorised into five classes, Fig. 15(d). The independent variables were the soil concentration of nitrate, potassium, and phosphorus, as well as the soil pH and SOM as determined in the period June–August and the β-glucan concentration, in August only. The significance of building a model using the measured yield, and the UAV-MSI estimated yields using NDVI and CI green VI alongside soil indicators is that it helps identify the range of soil properties that are required to be obtained and maintained to obtain the desired yield. This in turn provides the users (farmers and agronomists) who are operating a Precision Agriculture management strategy the information required to maintain the soil nutrient concentration by appropriate intervention, by the application of fertilisers. A flow chart, Fig. 16, outlines the key steps required to build this machine learning model. Firstly, the measured yield in Table S1,† as well as the NDVI and CI green VI estimated yields, for the 48 locations in Table 1, were recoded into the five yield classes. After recoding, only 40–50% similarity was evidenced in the measured yield, as well as the NDVI and CI green VI estimated yields, Table 2. Since 48 data points are insufficient for developing an effective model and validating its accuracy, the dataset was augmented using six different approaches to generate sufficient data: multivariate linear interpolation of datapoints for each yield class; random selection of datapoints from a normal distribution fitted to the sample dataset (consisting of original and interpolated datapoints for each yield class); selection of datapoints from a bias corrected normal distribution fitted to the sample dataset; random selection of datapoints from a lognormal distribution fitted to the sample dataset; random selection of datapoints from a bias corrected lognormal distribution fitted to the sample dataset; and random selection of datapoints from a skewed normal distribution fitted to the sample dataset.
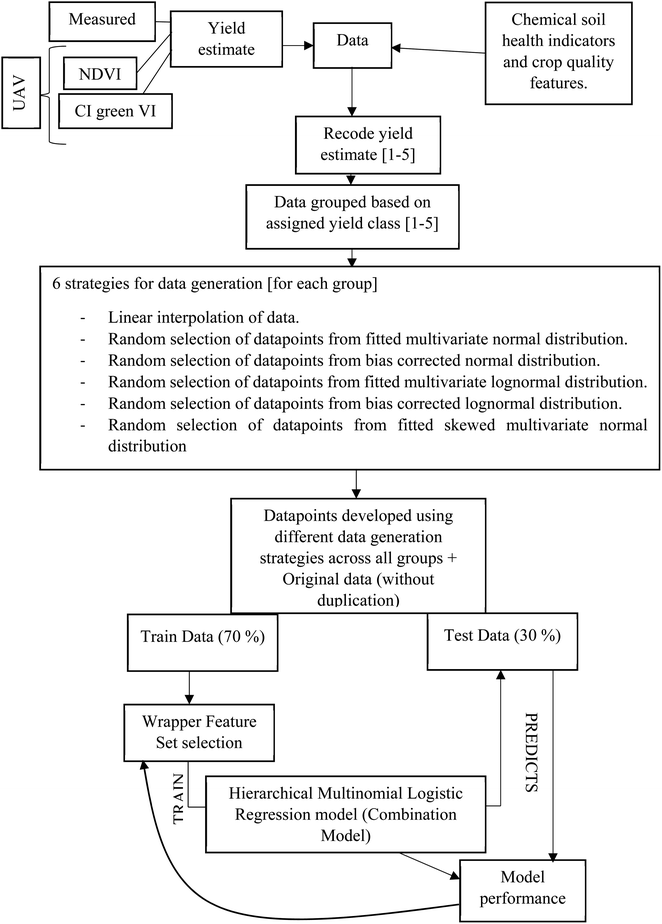 |
| | Fig. 16 Flow chart representing data generation, model training and performance evaluation. | |
Table 1 NDVI and CI green VI basic statistical parameters across phenological growth stages at 48 location points
| Phenological growth stage |
NDVI |
CI green VI |
| Mean ± SD |
Min |
Max |
N
|
Mean ± SD |
Min |
Max |
N
|
| Stage 3 – flowering (June) |
0.64 ± 0.083 |
0.46 |
0.81 |
48 |
2.63 ± 0.82 |
1.01 |
4.62 |
48 |
| Stage 4 – grain filling (July) |
0.41 ± 0.13 |
0.18 |
0.64 |
48 |
1.64 ± 0.71 |
0.27 |
2.91 |
48 |
| Stage 5 – ripening (August) |
0.34 ± 0.11 |
0.11 |
0.57 |
48 |
1.55 ± 0.73 |
0.31 |
3.15 |
48 |
| Mean: June, July and August |
0.46 ± 0.17 |
0.11 |
0.81 |
144 |
1.94 ± 0.88 |
0.27 |
4.62 |
144 |
Table 2 Similarity in the recoded class assigned to the measured yield estimate, NDVI estimated yield and CI green VI estimated yield
| Recoded yield classes |
Measured yield |
Estimated yield by NDVI |
Estimated yield by CI green VI |
| Measured yield |
1 |
0.4167 |
0.4792 |
| Estimated yield by NDVI |
0.4167 |
1 |
0.4583 |
| Estimated yield by CI green VI |
0.4792 |
0.4583 |
1 |
Firstly, for each of the yield approaches, data was grouped into the 5 yield classes. Then, multivariate datapoints were assigned to each class, and then linearly interpolated to have a substantial number of datapoints in each class to train and test a classification model and to fit a multivariate probability distribution. An example for the CI green VI is shown in Fig. 17. As the linearly interpolated data assumes a constant rate of change between variables it is limited to the sample dataset's range of feature values. As a result, the linearly interpolated data across the five yield classes does not accurately reflect the population from which the data points have been derived. Hence, in the absence of a representative sample dataset, the linearly interpolated dataset along with the original datapoints for each yield class, was used to fit the five parametric distributions. Multivariate parametric distributions were fitted separately to the sample dataset for each yield class in each yield approach. It is hypothesized, that if sufficient datapoints were collected for each yield class then the data would follow a normal distribution. Therefore, a multivariate normal distribution was fitted to datapoints for each yield class and a total of five multivariate normal distributions were developed for each yield approach i.e., measured and the NDVI and CI green VI estimated yields, and shown in Fig. 18. By neglecting datapoints consisting of any negative values, a set of 100 datapoints were randomly selected from each multivariate distribution. Due to the presence of some extreme values i.e., outliers, in the data, the covariance and mean of the fitted distribution are frequently overestimated and/or biased. The Orthogonalized Gnanadesikan Kettenring algorithm,37 was therefore applied to the dataset, since this corrects for the overestimation of parameters in the distribution fitted to the sample dataset. The corrected mean and covariance value for each multivariate normal distribution was generated for each of the 5 yield classes, Fig. 19. This allowed a set of 100 random points, with no negative values in any of the 16 features, to be selected for each yield class.
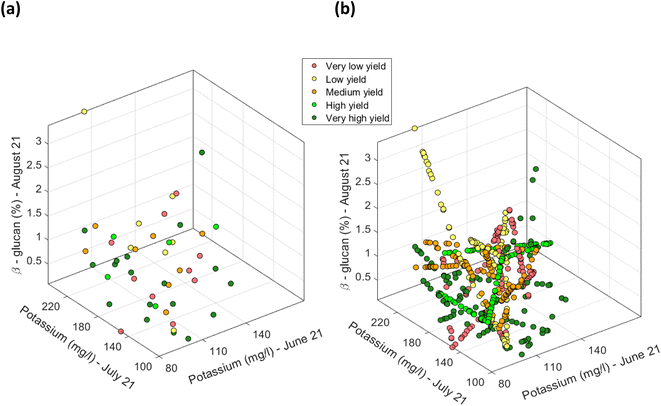 |
| | Fig. 17 Scatter plot of CI green VI estimated yield for selected features of potassium (mg l−1) (in June), potassium (mg l−1) (in July) and % β-glucan (in August). (a) Using the actual 48 determined datapoints, and (b) the linearly interpolated datapoints along with the original 48 determined datapoints [note: datapoints defined in terms of the soil nutrients were grouped based on the recoded five yield classes. Datapoints pertaining to each yield class were separately interpolated]. | |
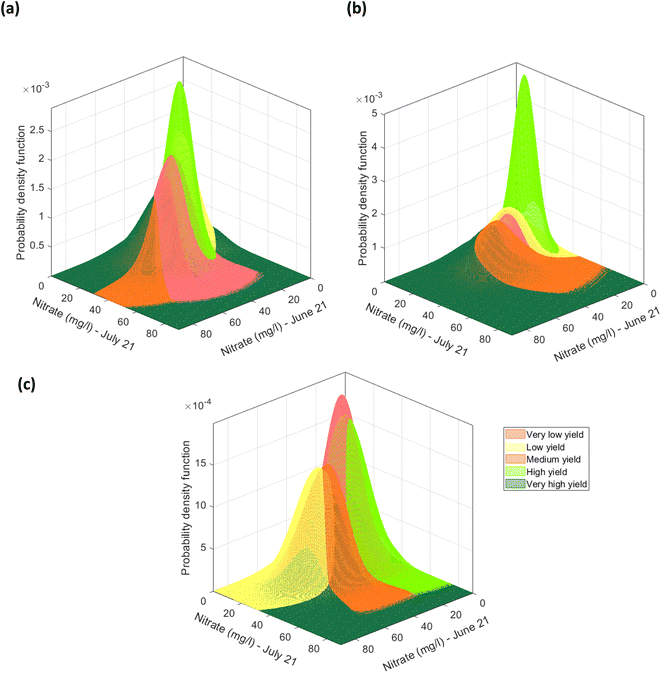 |
| | Fig. 18 Multivariate normal distribution for nitrate (mg l−1) (in June) and nitrate (mg l−1) (in July) fitted to the datapoints grouped using the (a) recoded measured yield, (b) recoded NDVI estimated yield, (c) the recoded CI green VI estimated yield. | |
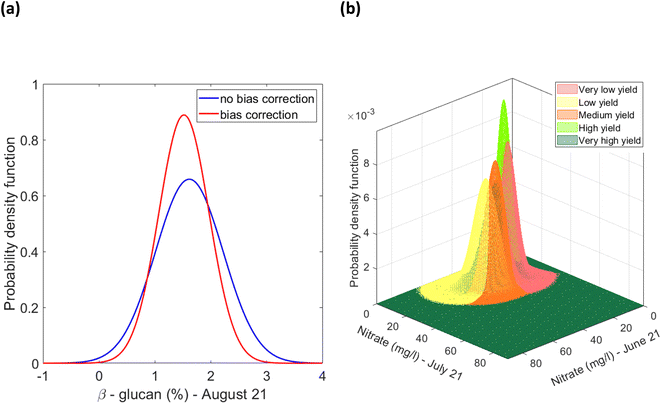 |
| | Fig. 19 (a) Example of univariate distribution of β-glucan (%) (in August) for the low yield estimate using the CI green VI method, (b) example of multivariate overestimation/bias corrected normal distributions of nitrate (mg l−1) (in June and July) fitted to datapoints pertaining to each yield class based on the CI green VI yield estimate [note: the univariate representation shows the effect of the orthogonalized Gnanadesikan Kettenring method on the distribution of the normal distribution fitted to the variable. The original distribution is represented in blue. The overestimation of variance owing to the presence of outliers is compensated by the orthogonalized Gnanadesikan Kettenring method]. | |
The 16 features i.e., nitrate, potassium, phosphate, pH, SOM in June–August, alongside the % β-glucan content in August, representing the soil characteristics and crop quality do not contain any negative values, Table S3.† Due to this characteristic of the sample dataset, a multivariate lognormal distribution was fitted to the sample data for each of the 5 yield classes, in the three datasets pertaining to the yield estimation approach, Fig. 20(a). The fitted multivariate lognormal distributions were used to randomly select 100 datapoints for each yield class. Furthermore, after correction for overestimation and/or bias the mean and covariance, were used to fit lognormal distributions, as shown in Fig. 20(b), and 100 datapoints were randomly selected from each yield estimation approach. After a closer examination of the 48 datapoints it was revealed that the variables can have absolute zero values, for example, the absence of nitrate in August, Table S3.† Hence, the dataset was adjusted by adding a value of 1.001 to fit a lognormal distribution. In view of the sample data characteristics, a skewed normal distribution was fitted to the sample dataset for each yield class, as shown in Fig. 20(c). From the skewed normal distribution, a set of 100 datapoints with no negative variable values were selected.
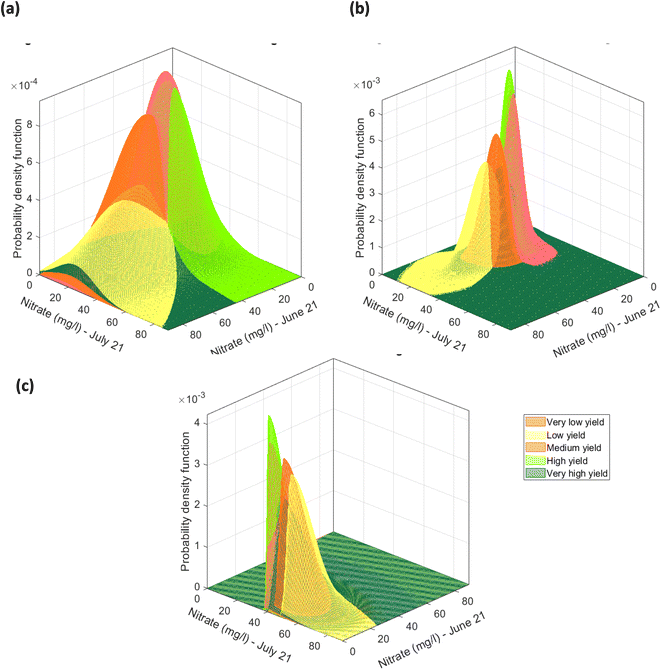 |
| | Fig. 20 Multivariate distribution for CI green VI yield estimate representing the feature nitrate (mg l−1) (in June and July) (a) multivariate log normal distributions fitted to datapoints grouped to the 5 classes of the CI green VI yield estimate, (b) multivariate lognormal distribution corrected for overestimation due to outliers by the orthogonalized Gnanadesikan Kettenring method, and (c) skewed normal distributions fitted to datapoints grouped based on the CI green VI yield estimate. | |
Then, the datasets generated using six different approaches for each of the 5 yield classes was used separately to train and test hierarchical multinomial logistic regression models. The goodness of fit of the trained model and its performance on the test data set, was investigated using the following metrics: trained model deviance over an intercept only model (assessed at the 0.05 significance level); McFadden's pseudo R2; and the accuracy, sensitivity, and specificity of the model.
The variability (or proportion of variation) in yield that can be explained by the trained model is expressed using McFadden's pseudo-R2 approach,38 and if the given model perfectly fits the data, the McFadden's R2 value will be close to 1.39 Specificity indicates whether the model correctly identifies a datapoint as not belonging to a specific class. Whereas sensitivity indicates whether the model correctly identifies whether a datapoint belongs to a specific class. Finally, accuracy identifies the correctly predicted total number of datapoints by the model. Hence, all generated datapoints and original samples were used to train and test the performance of a single hierarchical multinomial logistic regression model. The generalised format of the regression model for the different yields are shown in the ESI.†
To prevent localized clustering of datapoints that have the same yield class assigned, or were generated from the same distribution, the datapoints were randomly shuffled. Then, 70% of the unique datapoints in the dataset were used to train the model, and the remaining 30% were used to test the model. As the recoded yield class assigned to the manually measured yield and the estimated yields using NDVI and CI green VI represented about 40–50% similarity, shown in Table 2, three hierarchical multinomial logistic regression models were separately trained and tested. The hierarchical multinomial logistic regression models trained on the data developed using the six different approaches will hereafter be referred to as a ‘combination model’.
From the training data, sets of variables/features i.e., soil nutrients including nitrate, phosphorus, potassium, pH, SOM and the β-glucan concentration, that had a correlation lower than 0.4 (arbitrarily attributed), were selected and the logistic regression model was trained and tested using a dataset defined by only these sets of variables. The prediction accuracy of the models trained on the selected feature sets was compared among themselves and with a model that is trained on a dataset that takes into consideration all the 16 variables/features. This was done to estimate the effect of multi-co-linearity i.e., features bringing in similar or overlapping information, based on classifier performance.
For the datasets generated, based on the estimated yield using NDVI and the CI green VI, the combination model using all 16 features i.e. nitrate, phosphorus, potassium, pH and SOM across June–August and the β-glucan in August, was found to have the best prediction accuracy, Table 3. However, this soil feature set selection is dependent on the training set and the prediction performance of the trained model on the test set data used in the study. While for the measured yield data a subset of 9 features i.e. potassium and phosphorus in June, nitrate, potassium, phosphorus and SOM in July and nitrate, potassium and β-glucan in August, was found to perform best, Table 3. These 9 features are found to be relevant for all the three yield models, separately trained and tested, using the multinomial logistic regression model. As shown in Table 3, the hierarchical multinomial logistic regression model for the CI green VI estimated yield data provides the best performance in terms of specificity, sensitivity, and accuracy. The final trained hierarchial multinomial logistic regression model for the different yields is provided in the ESI,† along with their variable and coefficients, Table S6.† The specificity, sensitivity and accuracy on the trained measured yield data model, yield estimated using NDVI and CI green VI data models are provided in Table S7.† The performance of the models separately fitted to the linearly interpolated datapoints and the randomly selected points from different fitted parametric distributions for the measured yield, the yields estimated using NDVI and CI green VI are also provided in Table S8.†
Table 3 Performance of hierarchical multinomial logistic regression model (i.e., combination model) trained on measured yield data, and the estimated yield data using the NDVI and CI green VIa
| Combination model – measured yield |
|
The performance is defined based on the goodness of fit of the model to the training data and the model's ability to make predictions on unseen/test data
|
| Significance of model developed over intercept only model |
χ
2 (36) = 4.6701 × 103, p = 0 < 0.05 [based only on training data] |
| McFadden's R2 |
0.9476 [based only on training data] |
| Test data |
Specificity |
Sensitivity |
Accuracy |
| Class 1 – very low yield (0–4.97 t per ha) |
0.56 |
0.98 |
0.89 |
| Class 2 – low yield (4.97–6.18 t per ha) |
0.75 |
0.94 |
0.91 |
| Class 3 – medium yield (6.18–7.11 t per ha) |
0.76 |
0.97 |
0.93 |
| Class 4 – high yield (7.11–7.62 t per ha) |
0.89 |
0.88 |
0.88 |
| Class 5 – very high yield (>7.62 t per ha) |
0.72 |
0.90 |
0.86 |
| Overall |
|
|
0.74 |
| Combination model – NDVI yield |
| Significance of model developed over intercept only model |
χ
2 (64) = 4.9506 × 103, p = 0 < 0.05 [based only on training data] |
| McFadden's R2 |
0.5594 [based only on training data] |
| Test data |
Specificity |
Sensitivity |
Accuracy |
| Class 1 – very low yield (0–4.97 t per ha) |
0.13 |
1.00 |
0.83 |
| Class 2 – low yield (4.97–6.18 t per ha) |
0.36 |
0.89 |
0.78 |
| Class 3 – medium yield (6.18–7.11 t per ha) |
0.81 |
0.80 |
0.80 |
| Class 4 – high yield (7.11–7.62 t per ha) |
1.00 |
0.86 |
0.89 |
| Class 5 – very high yield (>7.62 t per ha) |
0.73 |
0.97 |
0.92 |
| Overall |
|
|
0.61 |
| Combination model – CI green VI yield |
| Significance of model developed over intercept only model |
χ
2 (64) = 5.7487 × 103, p = 0 < 0.05 [based only on training data] |
| McFadden's R2 |
1.00 [based only on training data] |
| Test data |
Specificity |
Sensitivity |
Accuracy |
| Class 1 – very low yield (0–4.97 t per ha) |
0.58 |
0.99 |
0.91 |
| Class 2 – low yield (4.97–6.18 t per ha) |
0.47 |
0.94 |
0.84 |
| Class 3 – medium yield (6.18–7.11 t per ha) |
0.95 |
0.86 |
0.88 |
| Class 4 – high yield (7.11–7.62 t per ha) |
1.00 |
0.96 |
0.97 |
| Class 5 – very high yield (>7.62 t per ha) |
0.87 |
0.95 |
0.93 |
| Overall |
|
|
0.76 |
In terms of the usability of the model as part of a Precision Agriculture management strategy, based on the nutrient content, soil organic matter and pH of the soil in June–August and crop quality, as assessed by the β-glucan concentration in August, the ‘combination model’ using the CI green VI can assist farmers, and agronomists, to estimate the oat crop yield, across the 3 phenological growing seasons (months). This is since the chosen 16 soil feature parameters show good, specificity, sensitivity, and accuracy at estimating the oat crop yield over the five different classes, Table 3. However, this probabilistic machine learning model only holds if the crops aren't affected by any natural or adverse conditions in the months before harvest. To further improve the proposed yield prediction model and the proportional soil nutrient concentrations, as shown in Table 4, more data collection and further experimentation would be required. However, this ‘combination model’, and particularly the CI green VI, Table 3, could be used for future studies in alternate crops to identify if the chemical soil health indicators and crop quality features aid in estimation of crop yield. This important development of predicting crop yield, using machine learning, can be extremely useful in supporting the World Health Organization's food security and nutrition programme for communities around the world.40
Table 4 Proportional concentration range of soil nutrients that resulted in very high yield across all 3 methodsa
| Class 5 – very high yield (>7.62 t per ha) |
|
NA = not applicable.
|
| Phenological growth stage |
Stage 3: flowering |
Stage 4: grain filling |
Stage 5: ripening |
| Month |
June |
July |
August |
| All yield estimates |
| Nitrate (mg l−1) |
16–81 |
6.6–38 |
3.5–35 |
| Phosphorous (mg l−1) |
20–33 |
13–27 |
7–20 |
| Potassium (mg l−1) |
90–165 |
105–235 |
125–190 |
| pH |
7.46–7.84 |
7.31–7.83 |
7.65–7.87 |
| SOM (%) |
9.2–10.8 |
9.1–11.7 |
7.8–10.5 |
| β-Glucan (%) |
NA |
0.37–2.26 |
Conclusion
This research has demonstrated the important of using vegetation indices as predictors of crop yield ahead of harvesting. It has additionally shown that using an unmanned aerial vehicle with a multispectral image camera, that can operate in the near infra-red and visible spectral region, is more effective at obtaining the necessary spectral data compared to hyperspectral data in the visible spectral region only. The use of an unmanned aerial vehicle also allows rapid data collection compared to a hand-held spectrometer. By integrating chemical soil health indicators and crop quality into a hierarchical multinomial logistic regression has allowed an effective model to be developed to predict the highest crop yield. This is an important component of an effective Precision Agriculture management strategy designed to maximum crop yield. Adoption of the generic framework of this research can be implemented in local, regional, and global contexts to inform farmers of the necessary actions to maximise crop yield. The adoption of the framework does require however, both soil nutrients levels to be monitored and controlled pesticide application to maximise yield. Additionally, the effective deployment of a low cost commercially available UAV with MSI camera does allow effective crop monitoring.
Author contributions
Conceptualization: C. E. N., J. R. M., J. J. P., J. R. D. Data curation: S. A., N. B., C. E. N., S. R. R., J. R. M., J. J. P., J. R. D. Formal analysis: S. A., N. B., S. R. R., J. R. D. Funding acquisition: J. J. P., J. R. D. Investigation: S. A., N. B., C. E. N., J. R. M., J. J. P., J. R. D. Methodology: S. A., N. B., C. E. N., S. R. R., J. R. M., J. J. P., J. R. D. Project administration: C. E. N., J. J. P., J. R. D. Resources: N. B., C. E. N., J. R. M., J. J. P., J. R. D. Supervision: N. B., C. E. N., J. J. P., J. R. D. Writing – original draft: S. A., N. B., J. R. D. Writing – review & editing: S. A., N. B., C. E. N., S. R. R., J. R. M., J. J. P., J. R. D.
Conflicts of interest
The authors have no conflict of interest to declare.
Acknowledgements
Access to visit and monitor the farm at Kiln Pitt Hill, Consett DH8 9SL, was granted by John Miller and family (T & AE Miller). Support from Palintest UK, Kingsway, Team Valley, England is also gratefully acknowledged.
References
-
S. M. Tosh and S. S. Miller, Oats, in Encyclopedia of Food and Health, 2018, pp. 119–125 Search PubMed.
- D. Paudel, B. Dhungana, M. Caffe and P. Krishnan, A Review of Health-Beneficial Properties of Oats, Foods, 2021, 10(11), 2591, DOI:10.3390/foods10112591.
- J. C. Zadoks, T. T. Chang and C. F. Konzak, A decimal code for the growth stages of cereals, Weed Res., 1974, 14(6), 415–421 CrossRef.
- L. Deng, Z. Mao, X. Li, Z. Hu, F. Duan and Y. Yan, UAV-based multispectral remote sensing for precision agriculture: A comparison between different cameras, ISPRS J. Photogramm. Remote Sens., 2018, 146, 124–136, DOI:10.1016/j.isprsjprs.2018.09.008.
- G. Yang, J. Liu, C. Zhao, Z. Li, Y. Huang, H. Yu and H. Yang, Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives, Front. Plant Sci., 2017, 8, 1111, DOI:10.3389/fpls.2017.01111.
- S. A. Burney and H. Tariq, K-means cluster analysis for image segmentation, Int. J. Comput. Appl. Technol., 2014, 96(4), 0975–8887, DOI:10.5120/16779-6360.
- G. Bareth, H. Aasen, J. Bendig, M. L. Gnyp, A. Bolten, A. Jung and J. Soukkamäki, Leichte und UAV-getragene hyperspektrale, bildgebende Kameras zur Beobachtung von landwirtschaftlichen Pflanzenbeständen: spektraler Vergleich mit einem tragbaren Feldspektrometer, Photogramm. Fernerkundung, Geoinformation, 2015, 2015(1), 69–79, DOI:10.1127/pfg/2015/0256.
- S. Nebiker, N. Lack, M. Abächerli and S. Läderach, Light-Weight Multispectral Uav Sensors and Their Capabilities for Predicting Grain Yield and Detecting Plant Diseases, Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci., 2016, XLI-B1, 963–970, DOI:10.5194/isprsarchives-XLI-B1-963-2016.
- H. Lu, T. Fan, P. Ghimire and L. Deng, Experimental Evaluation and Consistency Comparison of UAV Multispectral Minisensors, Remote Sens., 2020, 12(16), 2542, DOI:10.3390/rs12162542.
- S. F. Di Gennaro, C. Nati, R. Dainelli, L. Pastonchi, A. Berton, P. Toscano and A. Matese, An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards, Forests, 2020, 11(3), 308, DOI:10.3390/f11030308.
- M. Bascietto, E. Santangelo and C. Beni, Spatial Variations of Vegetation Index from Remote Sensing Linked to Soil Colloidal Status, Land, 2021, 10(1), 80, DOI:10.3390/land10010080.
- W. H. Maes and K. Steppe, Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture, Trends Plant Sci., 2019, 24(2), 152–164, DOI:10.1016/j.tplants.2018.11.007.
- J. D. A. Barbosa, R. T. D. Faria, A. P. Coelho, A. B. Dalri and L. F. Palaretti, Nitrogen fertilization management in white oat using spectral indices, Pesqui. Agropecu. Trop., 2020, 50, e64924, DOI:10.1590/1983-40632020v5064924.
- S. X. Chang and D. J. Robison, Nondestructive and rapid estimation of hardwood foliar nitrogen status using the SPAD-502 chlorophyll meter, For. Ecol. Manage., 2003, 181(3), 331–338, DOI:10.1016/s0378-1127(03)00004-5.
- J. A. Hawkins, J. E. Sawyer, D. W. Barker and J. P. Lundvall, Using Relative Chlorophyll Meter Values to Determine Nitrogen Application Rates for Corn, Agron. J., 2007, 99(4), 1034–1040, DOI:10.2134/agronj2006.0309.
- H. García-Martínez, H. Flores-Magdaleno, R. Ascencio-Hernández, A. Khalil-Gardezi, L. Tijerina-Chávez, O. R. Mancilla-Villa and M. A. Vázquez-Peña, Corn Grain Yield Estimation from Vegetation Indices, Canopy Cover, Plant Density, and a Neural Network Using Multispectral and RGB Images Acquired with Unmanned Aerial Vehicles, Agriculture, 2020, 10(7), 277, DOI:10.3390/agriculture10070277.
- O. S. Walsh, S. Shafian, J. M. Marshall, C. Jackson, J. R. McClintick-Chess, S. M. Blanscet and W. L. Walsh, Assessment of UAV Based Vegetation Indices for Nitrogen Concentration Estimation in Spring Wheat, Adv. Remote Sens., 2018, 7(02), 71–90, DOI:10.4236/ars.2018.72006.
- M. Schlemmer, A. Gitelson, J. Schepers, R. Ferguson, Y. Peng, J. Shanahan and D. Rundquist, Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels, Int. J. Appl. Earth Obs. Geoinf., 2013, 25, 47–54, DOI:10.1016/j.jag.2013.04.003.
- P. Sharma, L. Leigh, J. Chang, M. Maimaitijiang and M. Caffe, Above-Ground Biomass Estimation in Oats Using UAV Remote Sensing and Machine Learning, Sensors, 2022, 22(2), 601, DOI:10.3390/s22020601.
- K. Y. Li, R. Sampaio de Lima, N. G. Burnside, E. Vahtmäe, T. Kutser and K. Sepp, Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation, Remote Sens., 2022, 14(5), 1114, DOI:10.3390/rs14051114.
- J. Csajbók, E. Buday-Bódi, A. Nagy, Z. Z. Fehér, A. Tamás, I. C. Virág and E. Kutasy, Multispectral Analysis of Small Plots Based on Field and Remote Sensing Surveys—A Comparative Evaluation, Sustainability, 2022, 14(6), 3339, DOI:10.3390/su14063339.
- R. N. Sahoo, S. Gakhar, R. G. Rejith, R. Ranjan, M. C. Meena, A. Dey, J. Mukherjee, R. Dhakar, S. Arya, A. Daas and S. Babu, Unmanned Aerial Vehicle (UAV)–Based Imaging Spectroscopy for Predicting Wheat Leaf Nitrogen, Photogramm. Eng. Remote Sens., 2023, 89(2), 107–116, DOI:10.14358/PERS.22-00089R2.
- B. P. Mondal, R. N. Sahoo, N. Ahmed, R. K. Singh, B. Das, N. Mridha and S. Gakhar, Rapid prediction of soil available sulphur using visible near-infrared reflectance spectroscopy, Indian J. Agric. Sci., 2021, 91(9), 1328–1332, DOI:10.56093/ijas.v91i9.116080.
- B. Das, D. Chakraborty, V. K. Singh, D. Das, R. N. Sahoo, P. Aggarwal, D. Murgaokar and B. P. Mondal, Partial least square regression based machine learning models for soil organic carbon prediction using visible–near infrared spectroscopy, Geoderma Reg., 2023, 33, e00628, DOI:10.1016/j.geodrs.2023.e00628.
-
S. Eddy, S. R. Johnston, Comparison of Palintest Soil Analysis to External Laboratory Analysis, Palintest Ltd., 2009 Search PubMed.
-
D. L. Rowell, Soil Science Methods and Applications, Longman, Harlow, 1994 Search PubMed.
- A. Bzducha-Wróbel, S. Błażejak and K. Tkacz, Cell wall structure of selected yeast species as a factor of magnesium binding ability, Eur. Food Res. Technol., 2012, 235, 355–366, DOI:10.1007/s00217-012-1761-4.
- P. B. Shirsath, V. K. Sehgal and P. K. Aggarwal, Downscaling Regional Crop Yields to Local Scale Using Remote Sensing, Agriculture, 2020, 10(3), 58, DOI:10.3390/agriculture10030058.
- Y. C. Chen, A tutorial on kernel density estimation and recent advances, Biostat. Epidemiol., 2017, 1(1), 161–187, DOI:10.1080/24709360.2017.1396742.
-
AHDB, Measuring oil Nutrients, pH and Organic Matter, 2023, https://ahdb.org.uk/knowledge-library/measuring-soil-nutrients-ph-and-organic-matter, last accessed on 30/6/2023 Search PubMed.
-
Quaker, Oats Growth Guide, 2019, https://www.pepsico.co.uk/docs/librariesprovider22/default-document-library/quaker-oat-growth-guide-june-2019.pdf?sfvrsn=6c8d5164_4, last accessed on 30/6/2023 Search PubMed.
-
ICH, ICH guideline Validation of Analytical Procedures—Test and Methodology, 1995, https://www.ema.europa.eu/en/ich-q2r2-validation-analytical-procedures-scientific-guideline, last accessed on 30/6/2023 Search PubMed.
- B. Mamaghani and C. Salvaggio, Multispectral Sensor Calibration and Characterization
for sUAS Remote Sensing, Sensors, 2019, 19(20), 4453, DOI:10.3390/s19204453.
- D. Stow, C. Nichol, T. Wade, J. Assmann, G. Simpson and C. Helfter, Illumination Geometry and Flying Height Influence Surface Reflectance and NDVI Derived from Multispectral UAS Imagery, Drones, 2019, 3(3), 55, DOI:10.3390/drones3030055.
- A. P. Coelho, R. T. Faria, F. T. Leal, J. D. A. Barbosa and D. L. Rosalen, Validation of white oat yield estimation models using vegetation indices, Bragantia, 2020, 79(2), 236–241, DOI:10.1590/1678-4499.20190387.
- A. P. Coelho, R. T. Faria, F. T. Leal, J. D. A. Barbosa, A. B. Dalri and D. L. Rosalen, Estimation of irrigated oats yield using spectral indices, Agric. Water Manage., 2019, 223, 105700, DOI:10.1016/j.agwat.2019.105700.
- R. A. Maronna and R. H. Zamar, Robust estimates of location and dispersion for high-dimensional datasets, Technometrics, 2002, 44(4), 307–317, DOI:10.1198/004017002188618509.
-
D. McFadden, Conditional logit analysis of qualitative choice behavior, in Frontiers in Econometrics, ed. P. Zarembka, Academic Press, New York, 1973, pp. 104–142 Search PubMed.
-
T. J. Smith and C. M. McKenna, A comparison of logistic regression pseudo R2 indices, Multiple Linear Regression Viewpoints, 2013, vol. 39, 2, pp. 17–26 Search PubMed.
-
WHO, The State of Food Security and Nutrition in the World 2021: Transforming Food Systems for Food Security, Improved Nutrition and Affordable Healthy Diets for All, Food & Agriculture Org, Geneva, 2021, https://www.who.int/publications/m/item/the-state-of-food-security-and-nutrition-in-the-world-2021, last accessed on 30/6/2023 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional information on the mathematical coding of the hierarchial multinomial logistic regression model can be found: https://figshare.com/articles/dataset/oats_yield_estimator_code/23608593/1. See DOI: https://doi.org/10.1039/d3fb00101f |
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Nabanita
Basu
,
Nabanita
Basu
 ,
Catherine E.
Nicholson
,
Catherine E.
Nicholson
 ,
Simon R.
Rutter
,
Simon R.
Rutter
 ,
John R.
Marshall
,
John R.
Marshall
 ,
Justin J.
Perry
,
Justin J.
Perry
 and
John R.
Dean
and
John R.
Dean
 *
*

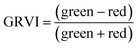
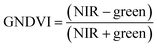
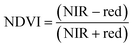
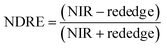
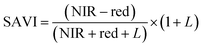
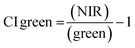
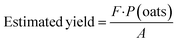
![[f with combining circumflex]](https://www.rsc.org/images/entities/i_char_0066_0302.gif) , at a location x taken from a set of data (e.g. crop yield), where x1,x2 … xn represents the number of samples (N) with an unknown probability function f(x) and defined as a kernel estimate
, at a location x taken from a set of data (e.g. crop yield), where x1,x2 … xn represents the number of samples (N) with an unknown probability function f(x) and defined as a kernel estimate ![[f with combining circumflex]](https://www.rsc.org/images/entities/i_char_0066_0302.gif) (x):
(x):