Open Access Article
Open Access ArticleNewly synthesized palladium(II) complexes with dialkyl esters of (S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzil))acetic acid: in vitro investigation of biological activities and HSA/DNA binding†
Kemal
Ćorović
af,
Danijela Lj.
Stojković
 *b,
Đorđe S.
Petrović
c,
Sandra S.
Jovičić Milić
b,
Maja B.
Đukić
c,
Ivana D.
Radojević
d,
Ivana
Raković
e,
Milena
Jurišević
fg,
Nevena
Gajović
g,
Marina
Jovanović
gh,
Jovana
Marinković
g,
Ivan
Jovanović
*b,
Đorđe S.
Petrović
c,
Sandra S.
Jovičić Milić
b,
Maja B.
Đukić
c,
Ivana D.
Radojević
d,
Ivana
Raković
e,
Milena
Jurišević
fg,
Nevena
Gajović
g,
Marina
Jovanović
gh,
Jovana
Marinković
g,
Ivan
Jovanović
 g and
Bojan
Stojanović
g and
Bojan
Stojanović
 gi
gi
aCommunity Health Center Tutin, Department of Emergency Medicine, Bogoljuba Čukića 12, 36320 Tutin, Republic of Serbia
bUniversity of Kragujevac, Institute for Information Technologies, Department of Science, Jovana Cvijića bb, 34000 Kragujevac, Republic of Serbia. E-mail: danijela.stojkovic@kg.ac.rs
cUniversity of Kragujevac, Faculty of Science, Department of Chemistry, Radoja Domanovića 12, 34000 Kragujevac, Republic of Serbia
dUniversity of Kragujevac, Faculty of Science, Department of Biology and Ecology, Radoja Domanovića 12, 34000 Kragujevac, Republic of Serbia
eUniversity of Kragujevac, Faculty of Medical Sciences, Department of Infectious Diseases, Svetozara Markovića 69, 34000 Kragujevac, Republic of Serbia
fUniversity of Kragujevac, Faculty of Medical Sciences, Department of Pharmacy, Svetozara Markovića 69, 34000 Kragujevac, Republic of Serbia
gUniversity of Kragujevac, Faculty of Medical Sciences, Center for Molecular Medicine and Stem Cell Research, Svetozara Markovića 69, 34000 Kragujevac, Republic of Serbia
hUniversity of Kragujevac, Faculty of Medical Sciences, Department of Otorinolaringology, Svetozara Markovića 69, 34000 Kragujevac, Republic of Serbia
iUniversity of Kragujevac, Faculty of Medical Sciences, Department of Surgery, Svetozara Markovića 69, 34000 Kragujevac, Republic of Serbia
First published on 22nd April 2024
Abstract
The four new ligands, dialkyl esters of (S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzil))acetic acid (R2-S,S-pddtyr·2HCl) (R = ethyl (L1), propyl (L2), butyl (L3), and pentyl (L4)) and corresponding palladium(II) complexes have been synthesized and characterized by microanalysis, infrared, 1H NMR and 13C NMR spectroscopy. In vitro cytotoxicity was evaluated using the MTT assay on four tumor cell lines, including mouse mammary (4T1) and colon (CT26), and human mammary (MDA-MD-468) and colon (HCT116), as well as non-tumor mouse mesenchymal stem cells. Using fluorescence spectroscopy were investigated the interactions of new palladium(II) complexes [PdCl2(R2-S,S-pddtyr)]; (R = ethyl (C1), propyl (C2), butyl (C3), and pentyl (C4)) with calf thymus human serum albumin (HSA) and DNA (CT-DNA). The high values of the binding constants, Kb, and the Stern–Volmer quenching constant, KSV, show the good binding of all complexes for HSA and CT-DNA. The mentioned ligands and complexes were also tested on in vitro antimicrobial activity against 11 microorganisms. Testing was performed by the microdilution method, where the minimum inhibitory concentration (MMC) and the minimum microbicidal concentration (MMC) were determined.
Introduction
With discovery of cisplatin,1 chemotherapy with metal-based drugs represents one of the most widely used strategies in clinical practice for cancer therapy.Therefore, the discovery of new potential anticancer compounds has become one of the most important goals in medicinal chemistry, and for these reasons, medicinal chemistry has developed rapidly, especially in the field of drug design and development.2 The main reason is to obtain new potential drugs or to modify the parent compounds with different structural modifications to achieve the desired profile of biological activity and less limitations due to toxicity and resistance. Non-platinum complexes may exhibit anticancer activity and toxic side effects that differ significantly from those of platinum-based drugs, which are expected to have different chemical behavior, hydrolysis rates, and mechanisms of action. Based on the structural analogy between platinum(II) and palladium(II) complexes, there is great interest in the study of palladium(II) complexes as potential anticancer drugs.3 The application of Pd(II) complexes in medicine is very limited, and the only application is 103Pd as a radioactive isotope in the treatment of rapidly growing high-grade prostate cancer.4,5 Initially, palladium complexes showed lower antitumor activity in vitro than platinum(II) complexes, which could be related to the more labile nature of palladium(II) compared to platinum(II) complexes.6,7 The choice of ligands greatly affects the anticancer activity of the complex, because they can modify the reactivity and lipophilicity.8 Some palladium(II) complexes have been found to have high cytotoxicity comparable to cisplatin.9,10 Due to the proven cytotoxicity of these palladium(II) compounds, a large number of new palladium(II) complexes are continuously being synthesized. As a result of these findings, N,N′ bidentate esters, R2pdda-type ligands, are now mainly used in our studies. Recently, we have reported on the synthesis and characterization of the palladium(II) complexes with R2pdda type esters of (S,S)-propylenediamine-N,N′-di-2-(2-benzyl)acetic acid.11
Now, this study is focused on in vitro the biological activity from newly palladium(II) complexes with dialkyl esters of O,O′-diethyl- (L1); O,O′-dipropyl- (L2); O,O′-dibutyl- (L3); O,O′-dipentyl- (L4) (S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzil))acetic acid dihydrochloride and their corresponding palladium(II) complexes: dichloro-(O,O′)-diethyl-(S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzyl))-acetato-palladium(II)-complex [PdCl2(det-S,S-pddtyr)], (C1), dichloro-(O,O′)-dipropyl-(S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzyl))-acetato-palladium(II)-complex [PdCl2(dpr-S,S-pddtyr)], (C2), dichloro-(O,O′)-dibutyl-(S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzyl))-acetato-palladium(II) complex [PdCl2(dbu-S,S-pddtyr)], (C3), dichloro-(O,O′)-dipentyl-(S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzyl))-acetato-palladium(II)-complex [PdCl2(dpe-S,S-pddtyr)] (C4).
The cytotoxicity study revealed cytotoxic effects of these compounds, particularly the L4 ligand, against carcinoma cells with lower toxicity towards non-cancerous cells. L4 ligand as well as C4 complex, induced apoptosis in HCT116 cells, demonstrated by increased early and late-stage apoptosis and caspase-3 expression, and decreased Bcl-2 expression. A notable decline in Ki67 expression and cell cycle arrest in the G0/G1 phase was also observed, indicating their potential in inhibiting cell proliferation.
Also, in this work, the interactions of palladium(II) complexes with HSA and DNA were investigated. The fluorescence spectra were recorded to examine the structural changes of HSA induced by the addition of complexes and to determine their binding constants (Kb) and the number of binding sites (n) for this biomolecule. To examine the ability of the compound to displace ethidium bromide (EB) from the EB–DNA complex, the interactions of the complex with DNA in the presence of EB were examined by using fluorescence spectroscopy.
The antimicrobial test confirmed that L4 shows the most significant activity against most tested Gram-positive bacteria and yeasts, especially Rhodotorula mucilaginosa, where the activity is in the range of the positive control. The same ligand L4 and the complex C2 show good antimicrobial activity on E. coli ATCC 25922.
Results and discussion
Chemistry
Esters L1–L4 were synthetized by using similar well known procedure (Scheme 1).11,12 The complexes were obtained by direct reaction of K2[PdCl4] with corresponding esters (Scheme 2).13,14 These esters are soluble in water and dimethylsulfoxide while complexes are soluble in dimethylsulfoxide and dimethylformamide. The structure of new ligands and corresponding palladium(II) complexes were assumed based on the elemental analysis. | ||
| Scheme 2 General procedure for the synthesis of palladium(II) complexes (C1–C4) with numeration used for NMR data. | ||
Spectroscopic analysis
IR and NMR (1H and 13C) spectra of isolated ligands and complexes were used to determine the coordination mode of dialkyl esters to the palladium(II)-ion. The obtained spectral data were in good agreement with the proposed structures of complexes (Scheme 2).A characteristic strong absorption band for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of esters was found in the infrared spectra of the ligands (1737, 1737, 1740 and 1738) and complexes (1727, 1729, 1726 and 1723), as well as the specific absorption bands of ν(C–O) of the ligands (L1–L4: 1234, 1230, 1243 and 1235) and of corresponding complexes (C1–C4: 1237, 1237, 1239 and 1236), clearly indicated that there was no coordination via oxygen atom. The position of absorption bands for secondary amino groups of complexes C1–C4: ν(R2N-H) was found at 3212, 3213, 3212 and 3210 cm−1 indicating the coordination of the ligands through the nitrogen atoms. Other specific bands were found at similar positions for corresponding ligands and complexes, thus indicating no other atom was coordinated to the palladium center.
O stretching vibration of esters was found in the infrared spectra of the ligands (1737, 1737, 1740 and 1738) and complexes (1727, 1729, 1726 and 1723), as well as the specific absorption bands of ν(C–O) of the ligands (L1–L4: 1234, 1230, 1243 and 1235) and of corresponding complexes (C1–C4: 1237, 1237, 1239 and 1236), clearly indicated that there was no coordination via oxygen atom. The position of absorption bands for secondary amino groups of complexes C1–C4: ν(R2N-H) was found at 3212, 3213, 3212 and 3210 cm−1 indicating the coordination of the ligands through the nitrogen atoms. Other specific bands were found at similar positions for corresponding ligands and complexes, thus indicating no other atom was coordinated to the palladium center.
The signals in the 1H NMR spectra originating from the ester protons of the complexes and the ligands showed similar chemical shifts, suggesting that there is no coordination between the palladium(II) ion and the oxygen from the mentioned group. In the 1H NMR spectra, the signals of hydrogen atoms from secondary amino groups appeared between 5.15–5.79 ppm for C1–C4, while in the ligands mentioned signals occurred at around 10 ppm from NH2+ amino salts. The signals of protons from the propylenediamine bridge (C9H2) displayed coordination-induced shifts in the spectra of complexes (up to 0.6 ppm). In 13C NMR spectra, ester carbon atom resonances were found as expected position, at 168 ppm in ligands and at 170 ppm in complexes, which verifying that oxygen is not ligating atom. All the signals of the carbon atoms from alcoholic residues in both ligands and complexes were found on similar chemical shifts. Six aromatic carbon atoms showed resonances at around 125 ppm (C4), 115 ppm (C6a, C6b), 130 ppm (C5a, C5b) and 156 ppm (C7). The chemical shifts of C2 atom in complexes were shifted by 5 ppm compared with signals of same carbon atom in the ligands. Also, due to coordination, there are a shifts in the signal of the C8 and C9 carbon atoms from the propylenediamine bridge about 7 ppm and 8 ppm. Mentioned above indicates the coordination of palladium(II) ions via two nitrogen atoms.
HSA binding studies
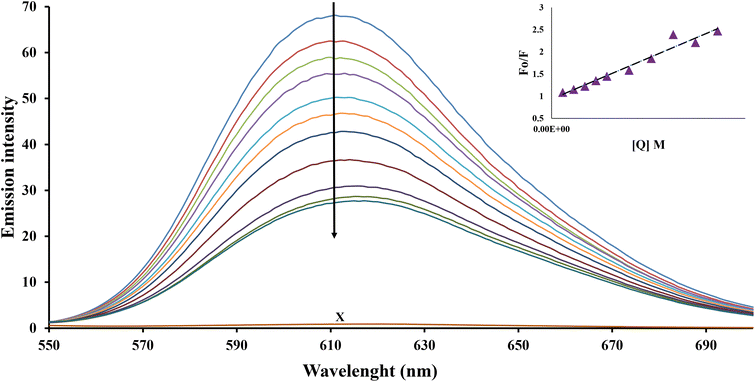
It has been proved that the most important role of the serum albumins is the transportation of metal ions and metal complexes as well as other biologically active compounds in the blood.15,16 The investigation of binding interactions between the potentially active compounds and HSA can be important in exploring their potential biological activity and application.17,18 To investigate the structural changes in HSA caused by the addition Pd complexes and determine the quenching constants (kq), the binding constant (K) and the number of binding sites (n) for the complex formed between complex and HSA, fluorescence spectra were measured.Interaction of Pd(II) complexes with HSA by fluorescence spectroscopy
The recorded fluorescence spectra of HSA with different concentrations of complexes C1–C4 are shown in Fig. 1 and Fig. S1–S3 (ESI†). As shown in Fig. 1 and Fig. S1–S3 (ESI†), adding a complex of Pd(II) to the HSA solution, HSA fluorescence intensity, with an increase of complex concentration, decreased gradually. That means the result suggests that complex can interact with HSA and quench its internal fluorescence.The KSV and quenching constants (kq) of the interactions of the compounds with the albumins were calculated (Table 1) from the Stern–Volmer quenching equation (eqn (1)) (given in experimental), where the fluorescence lifetime of tryptophan in HSA was taken as τ0 = 10−8 s. As can be seen in Table 1, the quenching constants (>1012 M−1 s−1) are higher than the different quenching types for the biopolymer fluorescence (1010 M−1 s−1), suggesting that a new conjugate was formed between each complex and HSA and the interaction of complexes with albumins take place through a static quenching mechanism.
| Complex | K SV (M−1) | k q (M−1 s−1) |
R
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
K b (M−1) | n |
R
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
|---|---|---|---|---|---|---|
| a R is the correlation coefficient. | ||||||
| C1 | 6.37 ± 0.26 × 104 | 6.37 ± 0.26 × 1012 | 0.99 | 1.82 ± 0.15 × 105 | 1.10 | 0.99 |
| C2 | 3.02 ± 0.33 × 104 | 3.02 ± 0.33 × 1012 | 0.92 | 1.42 ± 0.18 × 104 | 0.91 | 0.98 |
| C3 | 3.59 ± 0.29 × 104 | 3.59 ± 0.29 × 1012 | 0.96 | 5.90 ± 0.17 × 103 | 0.80 | 0.98 |
| C4 | 1.45 ± 0.09 × 105 | 1.45 ± 0.09 × 1013 | 0.97 | 4.37 ± 0.02 × 105 | 1.10 | 0.98 |
The values of Kb (association binding constant) and n (number of binding sites per albumin) for the C1–C4 were obtained from the intercept and slope of the plots of log(F0 − F)/F versus log[Q] (Fig. 2), using the equation (eqn (1)) (see experimental) and they are given in Table 1.
The HSA-binding constants showed that there is a strong binding force between complex and HSA, which implies that HSA can transfer investigated complexes toward potential biotargets. The calculated value of n is around one for all complexes, indicating the existence of just a single binding site in HSA.
DNA binding studies
The determination of the interactions between small molecules and DNA is important in pharmacology when evaluating the potential of new antitumor complexes,19 and therefore interactions between DNA and the synthesized complexes should be investigated. Studies indicate that transition metal complexes bind covalently with DNA by intercalation, groove binding, and external electrostatic binding.20 The mode and strength for the binding of the synthesized complexes of palladium(II) to CT-DNA were studied with the fluorescence spectroscopic method.Interaction of Pd(II) complexes with DNA by fluorescence spectroscopy
Ethidium bromide (3,8-diamino-5-ethyl-6-phenylphenanthridiniumbromide) belongs to the group of intercalation indicators because it builds fluorescent complexes with nucleic acids. Changes after the addition of the test complexes observed in the fluorescent spectrum of the EB–DNA system are used to study their interaction with DNA.21,22 To examine the ability of the compound to displace ethidium bromide (EB) from the EB–DNA complex, the interactions of new complexes of paladium(II) with DNA in the presence of EB were examined by using fluorescence spectroscopy.The emission spectra of DNA in the absence and presence of complexes, at room temperature in solution, have been recorded for increasing amounts of complexes C1–C4 and were shown in Fig. 3 (C1) and in ESI (Fig. S4–S6† for C2–C4). The fluorescence changes are results that indicate that the complex interacted with DNA. The addition of complexes at diverse r values results in a significant decrease in the intensity of the emission band of the DNA–EB system at 612 nm indicating the competition of the complexes with EB in binding to DNA. The observed quenching of DNA–EB fluorescence for all tested compounds suggests that they displace EB from the DNA–EB complex, and they can interact with DNA, but the certain mode of binding still cannot be defined.
The quenching parameter can be analyzed according to the Stern–Volmer eqn (1) given in Experimental. The calculated constants shown in Table 2, indicate that all the complexes can insert between DNA base pairs.
| Complex | K SV (M−1) | k q (M−1 s−1) |
R
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
K b (M−1) | n |
R
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
|---|---|---|---|---|---|---|
| a R is the correlation coefficient. | ||||||
| C1 | 2.64 ± 0.20 × 104 | 2.64 ± 0.20 × 1012 | 0.96 | 6.80 ± 0.16 × 104 | 1.10 | 0.99 |
| C2 | 9.61 ± 0.15 × 103 | 9.61 ± 0.15 × 1011 | 0.92 | 1.50 ± 0.08 × 107 | 1.79 | 0.95 |
| C3 | 1.20 ± 0.07 × 104 | 1.20 ± 0.07 × 1012 | 0.97 | 5.42 ± 0.02 × 104 | 1.15 | 0.97 |
| C4 | 1.02 ± 0.06 × 105 | 1.02 ± 0.06 × 1013 | 0.98 | 1.83 ± 0.02 × 105 | 1.08 | 0.95 |
Viscosity
Using the viscometric method, possible intercalation, or non-intercalation of compounds into DNA nucleobases can be determined.23 It is known that intercalation results in DNA elongation, due to the separation of base pairs at the intercalation site, which at the same time leads to an increase in the relative specific viscosity of such solutions.24 The results of viscosity measurements of complexes C1–C4 were indicated that complexes are not performing intercalation between the DNA bases, and which probably bind to minor/major grooves. The obtained measurement values were shown in Fig. 4. | ||
| Fig. 4 Viscosity assays in solution in a DMSO (5%)/PBS pH 7.4 mixture buffered solution with [complex]/[DNA] ratios. | ||
Assessment of cytotoxic cffects of synthesized ligands and palladium(II) complexes on carcinoma cells using MTT assay
The cytotoxicity of four newly synthesized ligands – dialkyl esters of (S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzil))acetic acid dihydrochloride (R2-S,S-pddtyr·2HCl) (designated as R = ethyl (L1), propyl (L2), butyl (L3), and pentyl (L4)) – and their corresponding palladium(II) complexes [PdCl2(R2-S,S-pddtyr)]; (R = ethyl (C1), propyl (C2), butyl (C3), and pentyl (C4)) was evaluated using MTT assay procedures. This assessment was conducted on a variety of cell lines: mouse (4T1) and human (MDA-MB-468) breast carcinoma cells, mouse (CT26) and human (HCT116) colon carcinoma cells, and non-cancerous mouse mesenchymal stem cells (mMSC). These cell lines were subjected to exposure of the synthesized complexes for 24 hours, at concentrations ranging from 7.8 to 500 μM. The results indicated that both ligands (designated as L1–L4) and complexes (designated as C1–C4) displayed modest cytotoxic effects, reducing viability in both human and mouse carcinoma cells (as shown in Fig. 5). Additionally, the complexes and ligands exhibited dose-dependent cytotoxic activity against breast cancer cell lines (4T1 in mice and MDA-MB-468 in humans) and colorectal cancer cells (CT26 in mice and HCT116 in humans), as depicted in Fig. 5E. Correspondingly, previous research has shown that palladium(II) complexes with certain esters of (S,S)-ethylenediamine-N,N′-di-(2,2′-di(4-hydroxy-benzyl))-acetic acid and their ligand precursors also exhibited dose-dependent cytotoxic effects towards various human cancer cell lines, including MDA-MB-231 (breast cancer), A549 (lung cancer), and CLL lymphocytes (chronic lymphocytic leukemia cells).25In the process of developing new drugs with low toxicity, it is crucial to evaluate their antiproliferative activity against non-cancerous cells in vitro. For this purpose, the toxicity of the tested compounds to normal mouse mesenchymal stem cells (mMSC) was assessed. The study found that all the tested ligands and complexes diminished the viability of non-cancerous cells (mMSC) in a dose-dependent manner, as illustrated in Fig. 5E. Notably, the C4 complex exhibited a lower cytotoxic effect on mMSC at concentrations ranging from 7.8 to 63.5 μM when compared to its effect on cancer cells, including the 4T1 and CT26 mouse carcinoma cells, as well as the MDA-MB-468 and HCT116 human carcinoma cells. The reduced cytotoxicity of the C4 complex towards mMSC, coupled with its ability to decrease the viability of 4T1, MDA-MB-468, CT26 and HCT116 tumor cells, suggests a potential selective cytotoxicity of this new complex against tumor cell lines. These observations imply that the C4 complex might be better tolerated in live organisms. On the other hand, the L4 ligand demonstrates minimal cytotoxic effects on mMSC at lower concentrations, ranging from 7.8 to 31.25 μM. However, at higher concentrations, there is a significant reduction in the viability percentage of mMSC.
In this study, the cytotoxic efficacy of C1–C4 complexes and L1–L4 ligands was thoroughly investigated by quantifying their half-maximal inhibitory concentration (IC50) values. The evaluation focused on several cell lines, including mouse (4T1) and human (MDA-MB-468) breast carcinoma cells, mouse (CT26) and human (HCT116) colon carcinoma cells, and non-cancerous mouse mesenchymal stem cells (mMSC). The IC50 values, which represent the concentration required to inhibit 50% of cell viability, are meticulously detailed in Table 3. Analysis of these values revealed that the ligand L4 displayed significant cytotoxic effects, outperforming other ligands (L1–L3) and the palladium complexes (C1–C4). In contrast, cisplatin (CDDP), a standard chemotherapeutic agent, exhibited more potent cytotoxic effects on these tumor cells compared to the C1–C4 complexes. However, it is important to note that cisplatin also showed increased cytotoxicity towards mouse mesenchymal stem cells, as detailed in Table 3. Based on these findings from the MTT assay, the C4 complex and L4 ligand were selected for further molecular analyses of their effects on cancer cells.
| Agents | IC50 ± SD (μM) (24 h) | ||||
|---|---|---|---|---|---|
| 4T1 | MDA-MD-468 | CT26 | HCT116 | mMSC | |
| C1 | 185 ± 10 | >500 | 405 ± 5 | 321 ± 6 | 469 ± 4 |
| C2 | 274 ± 5 | >500 | 360 ± 6 | 234 ± 6 | 254 ± 9 |
| C3 | 281 ± 8 | 459 ± 8 | 471 ± 4 | 176 ± 3 | >500 |
| C4 | 291 ± 9 | >500 | 345 ± 7 | 223 ± 8 | 448 ± 4 |
| L1 | >500 | 298 ± 7 | >500 | 190 ± 3 | 431 ± 9 |
| L2 | 273 ± 9 | 100 ± 1 | 260 ± 9 | 62 ± 2 | 247 ± 3 |
| L3 | 54 ± 4 | 71 ± 0.53 | 90 ± 4 | 19 ± 3 | 47 ± 0.23 |
| L4 | 47 ± 0.09 | <7.8 | 56 ± 1 | <7.8 | 24 ± 2 |
| CDDP | 17 ± 1 | 13 ± 1 | 20 ± 1 | 18 ± 0.36 | 14 ± 2 |
Assessment of apoptotic induction by C4 complex and L4 ligand in carcinoma cells
Apoptosis, or programmed cell death, is a critical and highly controlled process which selectively eliminates specific cells without harming adjacent healthy cells. This targeted elimination is especially beneficial in treating various carcinomas by focusing on the diseased cells while preserving healthy ones.26 The capacity of the C4 complex and L4 ligand to prompt apoptosis in carcinoma cells was scrutinized using flow cytometric analysis. This method employed Annexin V FITC and propidium iodide staining to discern apoptotic cells. Annexin V FITC binds to phosphatidylserine, which is externalized in early apoptosis, while propidium iodide permeates and stains cells only in the later stages of apoptosis or necrosis.27 The study's results, illustrated in Fig. 6, revealed that a substantial fraction of HCT116 cells entered late apoptosis after 24 hours of treatment with the C4 complex and L4 ligand. Similarly, a significant number of HTC116 cells were observed in the initial stages of apoptosis following comparable treatment periods, in contrast to untreated cells. Furthermore, a greater proportion of HCT116 cells displayed late-stage apoptotic characteristics after a 24-hour period with these compounds (p < 0.05) compared to the untreated group, as detailed in Fig. 6A. These results align with findings from a study conducted by Zhou M. et al., which also demonstrated the impact of palladium complexes in inducing apoptosis.28The extrinsic apoptotic pathway primarily involves death receptors leading to the activation of caspase-3.29 The intrinsic or mitochondrial apoptotic pathway, on the other hand, is triggered by an imbalance between proapoptotic and antiapoptotic proteins, one of which is Bcl-2. Bcl-2, a critical antiapoptotic protein, plays a significant role in regulating cell death by maintaining mitochondrial membrane integrity and preventing the release of cytochrome c, a key step in apoptosis induction. An imbalance in Bcl-2 levels affects mitochondrial membrane permeability, leading to the release of cytochrome c, which activates caspase-3 and consequently triggers apoptosis.30 Our study's results demonstrated that treating HCT116 cells with IC50 concentrations of the tested compounds, C4 and L4, resulted in a notable decrease in the percentage of Bcl-2 positive HCT116 cells (as shown in Fig. 6B) compared to untreated cells. Conversely, there was a marked increase in the percentage of cells expressing active caspase-3 in HCT116 cells treated with the C4 complex or L4 ligand, as illustrated in Fig. 6C. The treatment with the C4 complex and L4 ligand resulted in an increased percentage of cancer cells in both early and late stages of apoptosis and an elevated percentage of caspase-3 positive cancer cells, while simultaneously decreasing the expression of critical anti-apoptotic components like Bcl-2. These findings suggest that the C4 and L4 complex can potentially trigger cancer cell death via the apoptotic pathway. Additionally, related research has shown that new palladium(II) complexes with certain derivatives of 2-aminothiazoles also lead to the inhibition of the antiapoptotic Bcl-2 molecule.31 This finding highlights a common mechanism where palladium(II) complexes, through their structural variations, effectively reduce the expression of antiapoptotic proteins like Bcl-2, thereby promoting apoptosis in cancer cells. Furthermore, similar results were reported by Espino J. et al., who found that the percentage of active caspase-3 was elevated in human promyelocytic leukemia HL-60 cells following treatment with a newly synthesized thiazoline-based palladium(II) complex, further supporting the potential efficacy of such compounds in inducing apoptosis in cancer cells.32
Comprehensive evaluation of proliferation and cell cycle dynamics
Ki67 is a well-acknowledged protein associated with cell proliferation, acting as a DNA-binding marker primarily expressed in actively dividing cells, and absent in quiescent ones. It serves as a reliable indicator of cell proliferation, being present throughout the active stages of the cell cycle (G1, S, G2, and M phases) but not during the resting phase (G0). The expression level of Ki67 is directly linked to the rate of cell proliferation, making it an invaluable tool for gauging the proliferative activity of cancer cells and the impact of anticancer treatments.33In this study, the antiproliferative effects of the C4 complex and L4 ligand on HCT116 cells were assessed by measuring Ki67 expression levels. The findings revealed a significant decrease in Ki67 expression in HCT116 cells treated with these compounds, compared to untreated control cells, as depicted in Fig. 7A. This reduction in Ki67 expression signifies a notable decline in cell proliferation, highlighting the potential potency of these treatments in curtailing cancer cell growth. Additionally, a study by Frenich A. et al. reported similar findings in the context of dinuclear palladium(II) complexes with benzodiazines as bridging ligands. In their research, it was observed that the percentage of Ki67-positive CT26 cells was significantly reduced when treated with these palladium complexes.34 This parallel finding further corroborates the potential of palladium(II) complexes in reducing cell proliferation and underscores the utility of Ki67 as a reliable biomarker for assessing the efficacy of anticancer treatments.
This study investigated the cell cycle arrest in HCT116 colorectal carcinoma cells following treatment with the potent compounds C4 and L4 to better understand the cellular mechanisms behind their anticancer effects. HCT116 cells were exposed to these compounds for a 24-hour period. The results indicated that the newly synthesized C4 complex and L4 ligand effectively induced a G0/G1 phase arrest in the treated cells, as shown in Fig. 7B. This arrest is likely due to molecular changes within the cancer cells after treatment. Additionally, a significant reduction in the synthesis (S) phase, crucial for DNA replication, was noted following treatment with tested compounds, as depicted in Fig. 7B. This decrease in the S phase highlights the effectiveness of these compounds in impeding cell cycle progression. These observations are consistent with findings from a similarly designed study examining new palladium(II) complexes with propylenediamine derivatives of phenylalanine. In that research, it was found that the ligand L4 and the corresponding C4 complex facilitated cell cycle arrest in the G0/G1 phase, further supporting the potential of these complexes in exerting antiproliferative effects on cancer cells.11
Cyclins are crucial in regulating the cell cycle, functioning as a family of proteins that govern cell progression through the activation of cyclin-dependent kinases (CDKs).35
Cyclin D3, in particular, is an oncogene and serves as one of the initial internal regulators of the cell cycle. It plays a key role in transitioning cells from the G1 to the S phase, thus its regulation is vital in cancer biology.36 In this study, the impact of the C4 complex and L4 ligands on cyclin D expression in HCT 116 cells was investigated. Cells were treated with an IC50 concentration of the tested compounds for 24 hours. The results, as depicted in Fig. 7C, showed that there was a decrease in cyclin D expression following treatment with the complex/ligand. However, this decrease was not statistically significant. This reduced expression of cyclin D indicates a potential mechanism by which these compounds inhibit cell proliferation. Furthermore, it has been demonstrated in related studies that new palladium(II) complexes with derivatives of 2-aminothiazoles also lead to an inhibition of cyclin D expression, further implicating a decrease in cyclin-D as a common pathway in the antiproliferative effects of these complexes.31
Antimicrobial activity
The results of in vitro testing antimicrobial activities of the synthesized ligands and corresponding palladium(II) complexes, tetracycline, and fluconazole, are shown in Tables 4 and 5. The solvent (10% DMSO) did not inhibit the growth of the tested microorganisms. The intensity of antimicrobial action varied depending on the species of microorganism and on the type of substances. In general, the tested compounds show selective and moderate activity. MIC values for ligands were obtained from 15.63 to >1000 μg mL−1 and for complexes from 62.50 to >1000 μg mL−1.| Tested compounds | L1 | C1 | L2 | C2 | L3 | C3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | MICa | MMCb | MIC | MMC | MIC | MMC | MIC | MMC | MIC | MMC | MIC | MIC |
| a MIC values (μg mL−1) – means inhibitory activity. b MMC values (μg mL−1) – means microbicidal activity. | ||||||||||||
| Bacillus subtilis | 500 | 1000 | 500 | >1000 | 250 | 1000 | 500 | >1000 | 62.5 | 500 | 500 | >1000 |
| B. subtilis ATCC 6633 | 500 | 1000 | 250 | 500 | 500 | 500 | 250 | 500 | 62.5 | 125 | 250 | 500 |
| Staphylococcus aureus | >1000 | >1000 | 1000 | 1000 | >1000 | >1000 | 1000 | >1000 | >1000 | >1000 | 1000 | 1000 |
| S. aureus ATCC 25923 | 500 | 1000 | 1000 | >1000 | 500 | 1000 | 1000 | >1000 | 125 | 1000 | 1000 | >1000 |
| Escherichia coli | 1000 | >1000 | 1000 | 1000 | 1000 | >1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 |
| E. coli ATCC 25922 | 500 | 1000 | 250 | 1000 | 500 | 1000 | 62.5 | 500 | 250 | 1000 | 250 | 500 |
| Proteus mirabilis ATCC 12453 | 1000 | >1000 | 1000 | 1000 | >1000 | >1000 | 1000 | 1000 | >1000 | >1000 | 1000 | 1000 |
| Salmonella enterica | >1000 | >1000 | 1000 | 1000 | 1000 | >1000 | 1000 | 1000 | 1000 | >1000 | 1000 | 1000 |
| Candida albicans ATCC 10231 | 1000 | >1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 |
| Rhodotorula mucilaginosa | 500 | 500 | 250 | 500 | 500 | 500 | 250 | 500 | 500 | 500 | 250 | 1000 |
| Saccharomyces boulardii | >1000 | >1000 | 1000 | >1000 | 500 | 1000 | 1000 | 1000 | 250 | 250 | 1000 | 1000 |
| Tested compounds | L4 | C4 | Tetracycline | Fluconazole | ||||
|---|---|---|---|---|---|---|---|---|
| Species | MICa | MMCb | MIC | MMC | MIC | MMC | MIC | MMC |
| a MIC values (μg mL−1) – means inhibitory activity. b MMC values (μg mL−1) – means microbicidal activity. | ||||||||
| Bacillus subtilis | 15.63 | 500 | 500 | 1000 | 0.11 | 1.95 | — | — |
| B. subtilis ATCC 6633 | 31.25 | 62.50 | 125 | 250 | 1.95 | 15.63 | — | — |
| Staphylococcus aureus | >1000 | >1000 | 1000 | 1000 | 0.98 | 15.63 | — | — |
| S. aureus ATCC 25923 | 250 | 500 | 1000 | 1000 | 0.22 | 3.75 | — | — |
| Escherichia coli | 1000 | >1000 | 1000 | 1000 | 15.63 | 31.25 | — | — |
| E. coli ATCC 25922 | 62.50 | 1000 | 500 | 1000 | 15.63 | 31.25 | — | — |
| Proteus mirabilis ATCC 12453 | >1000 | >1000 | 1000 | 1000 | 125 | 125 | — | — |
| Salmonella enterica | 1000 | >1000 | 1000 | 1000 | 15.63 | 31.25 | — | — |
| Candida albicans ATCC 10231 | 250 | 250 | 1000 | 1000 | — | — | 31.25 | 1000 |
| Rhodotorula mucilaginosa | 62.5 | 250 | 500 | 500 | — | — | 62.5 | 1000 |
| Saccharomyces boulardii | 125 | 125 | 1000 | >1000 | — | — | 7.81 | 31.25 |
The activity of the ligands and the corresponding complexes are generally similar. In several cases, the ligands work better than the complex itself, but regularity was not observed.11 Complexes C1 and C2 have better antimicrobial activity than their ligands L1 and L2, while in C3 and C4 the situation is reversed. However, the differences in action are not significant.37L4 shows the best effect on Gram-positive bacteria (Staphylococcus aureus ATCC 25923 and Bacillus subtilis standard and clinical strain). The same ligand also shows good activity on yeasts (62.50–250 μg mL−1). Rhodotorula mucilaginosa is most sensitive to influence. This sensitivity is in the range of the fluconazole positive control. There is a clear difference in the effect of the tested substances on Gram-positive and Gram-negative bacteria.11,37,38 The influence on G-bacteria was low (Table 4) with exception C2 and L4 (MICs 62.50 μg mL−1) on E. coli ATCC 25922. And other studies show that some ligands and palladium complexes show strong antibacterial activity against E. coli.39,40
Earlier research showed that some palladium(II) complexes with alkyl esters of (S,S)-ethylenediamine-N,N′-di-2-propanoic acid demonstrated the significant antifungal activity against pathogenic fungi Aspergillus flavus and Aspergillus fumigatus. Like our study, these complexes demonstrated moderate antibacterial activity.41 Some palladium(II) complexes with alkyl esters of (S,S)-ethylenediamine-N,N′-di-2-(3-methyl)-butanoic acid show selective and moderate activity.37 There are studies that indicate a difference in antimicrobial activity between ligands and complexes and it is always higher with palladium(II) complexes which is not in agreement with our research.39,41
Like our research some dialkyl esters of ethylenediamine-N,N′-di-S,S-(2,2′-dibenzyl)acetic acid and their platinum(IV) complexes showed different degree of antimicrobial activity in relation to the tested species. This ligands demonstrated better activity against majority of tested fungi than corresponding platinum(IV) complexes like our study.42 Also, the platinum(IV) complexes manifested significant or moderate activity against Gram-positive bacteria and low activity against the Gram-negative bacteria.42
Similar to our study, it was observed that the palladium complexes with different esters of ethylenediamine or (S,S)-propylenediamine as ligands showed mostly lower antimicrobial activity compared to commercial antibiotics.39,41,42
Experimental and discussion
Chemistry
Elemental microanalyses for C, H and N were done by standard methods at the Institute for Information Technologies, University of Kragujevac, Republic of Serbia.
Infrared spectra were recorded by PerkinElmer FT-IR spectrophotometer spectrum two, using KBr pellet technique in the range 400–4000 cm−1. 1H and 13C NMR spectra were recorded by a Varian Gemini-2000 (200 MHz) spectrometer in DMSO-d6 for both ligands precursors and palladium(II) complexes.
Human serum albumin (HSA), highly polymerized calf thymus DNA (CT-DNA), ethidium bromide (EB) and DMSO were purchased from Sigma-Aldrich and used as received. The solutions of HSA, DNA and EB were prepared by dissolving in a phosphate buffer (PBS, 5 × 10−2 mol dm−3, pH 7.4) and 0.15 mol dm−3 NaCl after which it was stirred at 277 K for 3 days and kept no longer than a week. The concentration of the stock solution of CT-DNA was determined by UV absorption at 260 nm using a molar absorption coefficient of 6600 dm3 mol−1 cm.43 The purity of the DNA was checked which provided a ratio of the UV absorbance at 260/280 nm was of 1.8–1.9, which indicates that DNA was adequately free from protein.44 For the preparation of all solutions was used double distilled water.
Emission measurements were carried out by using an RF-1501 PC spectrofluorometer (Shimadzu, Japan). The excitation wavelength was fixed, and the emission range was adjusted before measurements, with the excitation and emission slit widths set at 10 nm. Viscosity was measured using a Anton Paar Density Meter, DMA 4100 M viscometer.
General procedure for the synthesis of O,O′-dialkyl esters of (S,S)-propylenediamine-N,N′-di-(2,2′-di-(4-hydroxy-benzil))acetic acid, (R2-S,S-pddtyr·2HCl)
5 g (9.82 mmol) of H2-S,S-pddtyr·2HCl·2H2O was added into 40 mL of dry alcohol (ethanol, n-propanol, n-butanol or n-pentanol) previously saturated with gas HCl during 3 hours and the mixture was refluxed for 12 hours.12 Then the ligands were determined according to the similar already described procedure.11 After cooling at room temperature, the reaction mixture evaporated to dryness on a rotary vacuum evaporator and the ocher precipitate was left overnight. Then ethyl acetate was added to the ester and after the treatment in an ultrasonic bath, a suspension was obtained and separated by centrifugation. The resulting precipitate was washed with ether. In Scheme 1 is presented general procedure for the synthesis of new ligands (L1–L4).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2980–2850 (ν(CH)), 1737 (ν(C
CH))ar, 2980–2850 (ν(CH)), 1737 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1614, 1516, 1467, 1446 (ν(C
O)), 1614, 1516, 1467, 1446 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1234 (νas(C–O)), 1175(νs(C–O)), 851, 830, 781 (γar(
C)ar), 1234 (νas(C–O)), 1175(νs(C–O)), 851, 830, 781 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)).
CH)).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2939–2879 (ν(CH)), 1737 (ν(C
CH))ar, 2939–2879 (ν(CH)), 1737 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1615, 1595, 1517, 1463, 1448 (ν(C
O)), 1615, 1595, 1517, 1463, 1448 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1230 (νas(C–O)), 1175 (νs(C–O)), 928, 827, 781 (γar(
C)ar), 1230 (νas(C–O)), 1175 (νs(C–O)), 928, 827, 781 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)).
CH)).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2936–2873 (ν(CH)), 1740 (ν(C
CH))ar, 2936–2873 (ν(CH)), 1740 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1615, 1596, 1464, 1448 (ν(C
O)), 1615, 1596, 1464, 1448 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1243 (νas(C–O)), 1176 (νs(C–O)), 934, 830, 784 (γar(
C)ar), 1243 (νas(C–O)), 1176 (νs(C–O)), 934, 830, 784 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)).
CH)).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2934–2871 (ν(CH)), 1738 (ν(C
CH))ar, 2934–2871 (ν(CH)), 1738 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1614, 1595, 1466 (ν(C
O)), 1614, 1595, 1466 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1235 (νas(C–O)), 1175 (νs(C–O)), 958, 829, 780 (γar(
C)ar), 1235 (νas(C–O)), 1175 (νs(C–O)), 958, 829, 780 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)).
CH)).
General procedure for the synthesis of complexes (C1–C4)
In water solution of K2[PdCl4] (0.1 g, 0.306 mmol) was added equimolar amount of the L1 (0.1626 g, 0.306 mmol), L2 (0.1712 g, 0.306 mmol), L3 (0.1797 g, 0.306 mmol) or L4 (0.1884 g, 0.306 mmol) in small portions. Previously esters were dissolved in water and neutralized with the LiOH (in molar ration 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). After complete addition of the corresponding dissolved ligand, the mixture was stirred for 3 hours. After this period, ocher/yellow precipitate of the complex (C1–C4) was obtained, then filtered off and dried on air. General procedure for the synthesis of complexes (C1–C4) is presented in Scheme 2.
2). After complete addition of the corresponding dissolved ligand, the mixture was stirred for 3 hours. After this period, ocher/yellow precipitate of the complex (C1–C4) was obtained, then filtered off and dried on air. General procedure for the synthesis of complexes (C1–C4) is presented in Scheme 2.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 3020–2937 (ν(CH)), 1727 (ν(C
CH))ar, 3020–2937 (ν(CH)), 1727 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1612, 1444 (ν(C
O)), 1612, 1444 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1237 (νas(C–O)), 1176 (νs(C–O)), 962, 827, 735 (γar(
C)ar), 1237 (νas(C–O)), 1176 (νs(C–O)), 962, 827, 735 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)), 540 (Pd–N).
CH)), 540 (Pd–N).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2970–2881 (ν(CH)), 1729 (ν(C
CH))ar, 2970–2881 (ν(CH)), 1729 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1613, 1445 (ν(C
O)), 1613, 1445 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1237 (νas(C–O)), 1176 (νs(C–O)), 931, 827, 735 (γar(
C)ar), 1237 (νas(C–O)), 1176 (νs(C–O)), 931, 827, 735 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)), 543 (Pd–N).
CH)), 543 (Pd–N).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2960–2872 (ν(CH)), 1726 (ν(C
CH))ar, 2960–2872 (ν(CH)), 1726 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1612, 1446 (ν(C
O)), 1612, 1446 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1239 (νas(C–O)), 1175 (νs(C–O)), 936, 826, 736 (γar(
C)ar), 1239 (νas(C–O)), 1175 (νs(C–O)), 936, 826, 736 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)), 540 (Pd–N).
CH)), 540 (Pd–N).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH))ar, 2956–2867 (ν(CH)), 1723 (ν(C
CH))ar, 2956–2867 (ν(CH)), 1723 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1612, 1444 (ν(C
O)), 1612, 1444 (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)ar), 1236 (νas(C–O)), 1176 (νs(C–O)), 957, 825, 732 (γar(
C)ar), 1236 (νas(C–O)), 1176 (νs(C–O)), 957, 825, 732 (γar(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH)), 541 (Pd–N).
CH)), 541 (Pd–N).
HSA and DNA binding study
 | (1) |
The binding constant (Kb) and the binding stoichiometry (n) of HSA-compounds system can be estimated by the following eqn (2)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 using the fluorescence intensity data:
45 using the fluorescence intensity data:
 | (2) |
The values of K and n were obtained from the intercept and slope of the plots of log(F0 − F)/F versus log[Q].
DNA-binding experiments
All the experiments involving CT-DNA were studied by emission spectroscopy at room temperature in PBS buffer solution (pH 7.4). The complexes were dissolved in a mixed solvent of 5% DMSO and 95% PBS buffer for all the experiments. The ethidium bromide displacement experiments were carried out with fluorescence measurements. In all the experiments, the DNA ([DNA] = 2.27 × 10−3 mol dm3) was pre-treated with ethidium bromide ([EB] = 2.0 × 10−5 mol dm3) for 30 min. Then to these solutions were added a solution of the test compounds at different concentrations (0–6 × 10−5 mol dm3), and the change in the fluorescence intensity was measured. The excitation and the emission wavelength were 520 nm and 610–615 nm, respectively.The Stern–Volmer constant KSV is described by the Stern–Volmer relation (eqn (1)), similarly as described above for HSA binding studies.
Viscosity measurements
Changes in DNA viscosity was measured at room temperature in the presence of increasing concentrations of tested compounds (C1–C4). Density values were obtained by direct reading and were shown graphically.Cytotoxic study
MTT assay
This study focused on assessing the cytotoxic effects of newly synthesized ligands and complexes, in addition to cisplatin, employing the MTT assay technique based on established protocols.47 Cells in the exponential growth phase were harvested from culture flasks, quantified, and then seeded in 96-well culture plates at a density of 5 × 103 cells per well. The treatment phase involved exposing these cells to varied concentrations of the synthesized complexes, ligands, and cisplatin, ranging from 7.8 to 500 μM, over a period of 24 hours, using fresh complete medium as a control. The cytotoxicity results were quantified relative to the control group, thereby providing a comparative measure of the compounds’ cytotoxic effects. Moreover, IC50 values, indicative of the concentration necessary to inhibit 50% of cell viability, were calculated utilizing Microsoft Office Excel 2010.Analysis of apoptotic response in cells
This study conducted an evaluation of apoptosis in treated cells using the Annexin V and propidium iodide double staining assay, adhering to the methodologies established in previous research.48 This technique was instrumental in determining the percentage of apoptotic cells after a 24-hour treatment with the newly synthesized ligand L4 and its corresponding palladium(II) complex, C4. In subsequent experiments, cells were fixed and permeabilized using a buffer provided by BD Bioscience, Heidelberg, Germany. The cells were then incubated with antibodies targeting Bcl-2 and caspase-3, sourced from Thermo Fisher Scientific, Cambridge, MA, USA, in line with protocols from prior studies.49 The analysis of these treated cells was performed using a FACS Calibur flow cytometer, with data processing carried out using FlowJo software.Investigating the impact on cell cycle dynamics
This research delved into the potential effects of the newly synthesized L4 ligand and C4 complex on the cell cycle dynamics of HCT 116 tumor cells, with a focus on identifying changes in Ki67 expression and cell cycle progression. The study involved a comparative analysis between cells treated with IC50 concentrations of these compounds and a control group of untreated cells. Following a 24-hour incubation period post-treatment, the cells were subjected to trypsinization, triple washing in PBS, and counting. The cells in each sample tube were then stained with Ki67-specific antibodies (eBioscience, San Diego, USA) and 1 μL of Vybrant® DyeCycleTM Ruby stain (Thermo Fisher Scientific, Inc.). After permeabilization, the cells were incubated with antibodies targeting Cyclin D (Thermo Fisher Scientific, Inc.) for cell cycle analysis. The samples were subsequently examined using a FACS Calibur flow cytometer, with a minimum of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample being analyzed. The data analysis was performed using FlowJo vX.0.7 software, facilitating a thorough assessment of the cell cycle phases.
000 events per sample being analyzed. The data analysis was performed using FlowJo vX.0.7 software, facilitating a thorough assessment of the cell cycle phases.
The binding constant (Kb) and the binding stoichiometry (n) of HSA-compounds system can be estimated by the following eqn (2)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 using the fluorescence intensity data:
48 using the fluorescence intensity data:
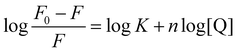 | (2) |
The values of K and n were obtained from the intercept and slope of the plots of log(F0 − F)/F versus log[Q].
Materials
In vitro antimicrobial assay
The antimicrobial activity of the ligands and complexes was tested against 11 microorganisms. The experiment involved 8 strains of bacteria (four standard strains and four isolates: Bacillus subtilis, B. subtilis ATCC 6633, Staphylococcus aureus, S. aureus ATCC 25923, Proteus mirabilis ATCC 12453, Escherichia coli, E. coli ATCC 25922, Salmonella enterica). Also, three yeast species were tested (Candida albicans ATCC 10231, Rhodothorula mucilaginosa, Saccharomyces boulardii). All isolates were a generous gift from the Institute of Public Health, Kragujevac. The other microorganisms were provided from the collection held by the Microbiology Laboratory Faculty of Science, University of Kragujevac.
Suspension preparation
The bacterial suspensions were prepared by the direct colony method. The turbidity of the initial suspension was adjusted using densitometer (DEN-1, BioSan, Latvia). When adjusted to the turbidity of the 0.5 McFarland's standard50 the bacteria suspension contains about 108 colony forming units (CFU) per mL and the suspension of yeast contains 106 CFU per mL. Ten-fold dilutions of the initial suspension were additionally prepared into sterile 0.85% saline. Bacterial inoculi were obtained from bacterial cultures incubated for 24 h at 310 K on Müller-Hinton agar substrate and brought up by dilution according to the 0.5 McFarland standard to approximately 106 CFU per mL. Yeast inoculi incubated for 48 h at 299 K on Tryptone soya agar substrate and brought up by dilution according to the 0.5 McFarland standard to approximately 104 CFU per mL.Microdilution method
Antimicrobial activity was tested by determining the minimum inhibitory concentrations (MIC) and minimum microbicidal concentration (MMC) using the microdilution plate method with resazurin.51 The 96-well plates were prepared by dispensing 100 μL of nutrient broth, Mueller-Hinton broth for bacteria and Tryptone soya broth for yeast, into each well. A 100 μL aliquot from the stock solution of the tested compound (with a concentration of 2000 μg mL−1) was added into the first row of the plate. Then, twofold serial dilutions were performed by using a multichannel pipette. The obtained concentration range was from 1000 to 7.8 μg mL−1. The method is described in detail in the reported paper.52Tetracycline and fluconazole were used as a positive control. 10% DMSO (as solvent control test) was recorded not to inhibit the growth of microorganisms. Each test included growth control and sterility control. All the tests were performed in duplicate, and the MICs were constant. Minimum microbicidal concentrations were determined by plating 10 μL of samples from wells where no indicator color change, or no mycelia growth was recorded, on nutrient agar medium. At the end of the incubation period the lowest concentration with no growth (no colony) was defined as the minimum microbicidal concentration.
Conclusion
Four new ligands L1–L4 and their corresponding palladium(II) complexes C1–C4 were synthesized and characterized by IR, 1H NMR, 13C NMR spectroscopy and elemental analysis. All spectroscopic data indicate that ligands were coordinated for palladium(II) ion via nitrogen donor atoms. In this comprehensive study, the cytotoxic and antiproliferative effects of novel ligands and their corresponding palladium(II) complexes were evaluated on various carcinoma and non-cancerous cell lines. The results highlighted the L4 ligand as particularly potent, exhibiting significant cytotoxicity against carcinoma cells while showing lower toxicity towards non-cancerous cells. L4 ligand, as well as C3 complex, successfully triggered apoptosis in HCT116 cells, evidenced by a rise in both early and late-stage apoptosis, enhanced expression of active caspase-3, and a reduction in the expression of the anti-apoptotic protein Bcl-2. Additionally, a marked reduction in Ki67 expression and cell cycle arrest in the G0/G1 phase underscored their potential in inhibiting cell proliferation. Despite these promising results, it is crucial to acknowledge the need for additional in vitro and in vivo research to fully understand the mechanisms of action, optimize the therapeutic efficacy, and evaluate the safety profile of these compounds. This further investigation is essential to pave the way for potential clinical applications and the development of effective cancer treatments.The DNA interaction and protein binding properties of the new complexes were evaluated by fluorescence spectroscopies. All the complexes show a good binding affinity to HSA protein giving relatively high binding constants. The present study of interaction with CT-DNA shows that palladium(II) complexes interact with DNA, and by additional viscosity measurements were indicated that complexes are not performing intercalation between the DNA bases, and which probably bind to minor/major grooves.
The intensity of antimicrobial action varied depending on the species of microorganism and on the type of substances. In general, the tested compounds show selective and moderate activity of all tested substances, ligand L4 shows the best antimicrobial activity against most tested Gram-positive bacteria and yeasts. The same ligand L4 and the complex C2 show good antimicrobial activity on E. coli ATCC 25922.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Faculty of Medical Sciences, University of Kragujevac (MP02/19, JP15/19, JP11/18, JP02/20, JP07/23, JP09/23) and the Serbian Ministry of Science, Technological Development and Innovation (agreements no. 451-03-66/2024-03/200378, no 451-03-65/2024-03/200122, 451-03-66/2024-03/200122 and no 451-03-47/2023-01/200111).References
- B. Rosenberg, L. Van Camp and T. Krigas, Nature, 1965, 205, 698–699 CrossRef CAS PubMed.
- S. Laufer, U. Holzgrabe and D. Steinhilber, Angew. Chem., Int. Ed., 2013, 52, 4072–4076 CrossRef CAS PubMed.
- T. Rau and R. van Eldik, Metal Ions in Biological Systems, Marcel Dekker, New York, 1996 Search PubMed.
- N. N. Stone and R. G. Stock, Eur. Urol., 2002, 41, 427–433 CrossRef CAS PubMed.
- L. Potters, Y. Cao, E. Calugaru, T. Torre, P. Fearn and X. H. Wang, Int. J. Radiat. Oncol., Biol., Phys., 2001, 50, 605–614 CrossRef CAS PubMed.
- J. Ruiz, J. Lorenzo, L. Sanglas, N. Cutillas, C. Vicente, M. D. Villa, F. X. Avilés, G. López, V. Moreno, J. Pérez and D. Bautista, Inorg. Chem., 2006, 45, 6347–6360 CrossRef CAS PubMed.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- F. Arnesano and G. Natile, Coord. Chem. Rev., 2009, 253, 2070–2081 CrossRef CAS.
- B. T. Khan, J. Bhatt, K. Najmuddin, S. Shamsuddin and K. Annapoorna, J. Inorg. Biochem., 1991, 44, 55–63 CrossRef CAS PubMed.
- H. Mansuri-Torshizi, T. S. Srivastava, H. K. Parekh and M. P. Chitnis, J. Inorg. Biochem., 1992, 45, 135–148 CrossRef CAS PubMed.
- Đ. S. Petrović, S. S. Jovičić Milić, M. B. Đukić, I. D. Radojević, M. M. Jurišević, N. M. Gajović, A. Petrović, N. N. Arsenijević, I. P. Jovanović, E. Avdović, D. Lj. Stojković and V. V. Jevtić, J. Inorg. Biochem., 2023, 246, 112283 CrossRef PubMed.
- L. N. Schoenberg, D. W. Cooke and C. F. Liu, Inorg. Chem., 1968, 7, 2386–2393 CrossRef CAS.
- A. Savić, S. Misirlić-Denčić, M. Dulović, L. E. Mihajlović-Lalić, M. Jovanović, S. Grgurić-Šipka, I. Marković and T. J. Sabo, Bioorg. Chem., 2014, 54, 73–80 CrossRef PubMed.
- A. Savić, L. Filipović, S. Aranđelović, B. Dojčinović, S. Radulović, T. J. Sabo and S. Grgurić-Šipka, Eur. J. Med. Chem., 2014, 82, 372–384 CrossRef PubMed.
- H. Liu, X. Shi, M. Xu, Z. Li, L. Huang, D. Bai and Z. Zeng, Eur. J. Med. Chem., 2011, 46, 1638–1647 CrossRef CAS PubMed.
- G. Fanali, A. Di Masi, V. Trezza, M. Marino, M. Fasano and P. Ascenzi, Mol. Aspects Med., 2012, 33, 209–290 CrossRef CAS PubMed.
- A. Sułkowska, J. Mol. Struct., 2002, 614, 227–232 CrossRef.
- L. Moriggi, M. A. Yaseen, L. Helm and P. Caravan, Chem. – Eur. J., 2012, 18, 3675–3686 CrossRef CAS PubMed.
- C. X. Zhang and S. J. Lippard, Curr. Opin. Chem. Biol., 2003, 7, 481–489 CrossRef CAS PubMed.
- Q. L. Zhang, J. G. Liu, H. Chao, G. Q. Xue and L. N. Ji, J. Inorg. Biochem., 2001, 83, 49–55 CrossRef CAS PubMed.
- K. C. Skyrianou, C. P. Raptopoulou, V. Psycharis, D. P. Kessissoglou and G. Psomas, Polyhedron, 2009, 28, 3265–3271 CrossRef CAS.
- A. Tarushi, G. Psomas, C. P. Raptopoulou and D. P. Kessissoglou, J. Inorg. Biochem., 2009, 103, 898–905 CrossRef CAS PubMed.
- L. P. Fioravanço, J. B. Pôrto, F. M. Martins, J. D. Siqueira, B. A. Iglesias, B. M. Rodrigues, O. A. Chaves and D. F. Back, J. Inorg. Biochem., 2023, 239, 112070 CrossRef PubMed.
- B. A. Iglesias, N. P. Peranzoni, S. I. Faria, L. B. Trentin, A. P. Schuch, O. A. Chaves, R. R. Bertoloni, S. Nikolaou and K. T. de Oliveira, Molecules, 2023, 28, 5217 CrossRef CAS PubMed.
- D. Lj. Stojković, V. V. Jevtić, G. P. Radić, D. V. Todorović, M. Petrović, M. Zarić, I. Nikolić, D. Baskić and S. R. Trifunović, J. Inorg. Biochem., 2015, 143, 111–116 CrossRef PubMed.
- G. C. Cavalcante, A. P. Schaan, G. F. Cabral, M. N. Santana-Da-Silva, P. Pinto, A. F. Vidal and Â. Ribeiro-Dos-Santos, Int. J. Mol. Sci., 2019, 20, 4133 CrossRef PubMed.
- R. Kumar, A. Saneja and A. K. Panda, in Lung Cancer: Methods and Protocols, Springer US, New York, 2021 Search PubMed.
- M. Zhou, J. C. Boulos, E. A. Omer, H. A. Rudbari, T. Schirmeister, N. Micale and T. Efferth, Eur. J. Pharmacol., 2023, 956, 175980 CrossRef CAS PubMed.
- R. Jan and G. E. S. Chaudhry, Adv. Pharm. Bull., 2019, 9, 205–218 CrossRef CAS PubMed.
- S. Qian, Z. Wei, W. Yang, J. Huang, Y. Yang and J. Wang, Front. Oncol., 2022, 12, 985363 CrossRef CAS PubMed.
- S. S. Jovičić Milić, V. V. Jevtić, S. R. Radisavljević, B. V. Petrović, I. D. Radojević, I. R. Raković, Đ. S. Petrović, D. Lj. Stojković, M. Jurišević, N. Gajović, A. Petrović, N. Arsenijević, I. Jovanović, O. R. Klisurić, N. L. Vuković, M. Vukić and M. Kačániová, J. Inorg. Biochem., 2022, 233, 111857 CrossRef PubMed.
- J. Espino, E. Fernández-Delgado, S. Estirado, F. de la Cruz-Martinez, S. Villa-Carballar, E. Viñuelas-Zahínos, F. Luna-Giles and J. A. Pariente, Sci. Rep., 2020, 10, 16745 CrossRef CAS PubMed.
- S. S. Menon, C. Guruvayoorappan, K. M. Sakthivel and R. R. Rasmi, Clin. Chim. Acta, 2019, 491, 39–45 CrossRef CAS PubMed.
- A. A. Franich, M. D. Živković, D. Ćoćić, B. Petrović, M. Milovanović, A. Arsenijević, J. Milovanović, D. Arsenijević, B. Stojanović, M. I. Đuran and S. Rajković, J. Biol. Inorg. Chem., 2019, 24, 1009–1022 CrossRef CAS PubMed.
- L. Ding, J. Cao, W. Lin, H. Chen, X. Xiong, H. Ao, M. Yu, J. Lin and Q. Cui, Int. J. Mol. Sci., 2020, 21, 1960 CrossRef CAS PubMed.
- Y. Chi, S. Huang, M. Liu, L. Guo, X. Shen and J. Wu, Cancer Cell Int., 2015, 26, 15–89 Search PubMed.
- Đ. S. Petrović, S. S. J. Milić, M. B. Đukić, I. D. Radojević, R. M. Jelić, M. M. Jurišević, G. P. Radić, N. M. Gajović, N. N. Arsenijević, I. P. Jovanović, N. V. Marković, D. Lj. Stojković and V. V. Jevtić, Inorg. Chim. Acta, 2021, 528, 120601 CrossRef.
- D. Lj. Stojković, V. V. Jevtić, G. P. Radić, M. B. Đukić, R. M. Jelić, M. M. Zarić, M. V. Anđelković, M. S. Mišić, D. D. Baskić and S. R. Trifunović, New J. Chem., 2018, 42, 3924–3935 RSC.
- G. P. Radić, V. V. Glođović, I. D. Radojević, O. D. Stefanović, Lj. R. Čomić, V. M. Đinović and S. R. Trifunović, Inorg. Chim. Acta, 2012, 391, 44–49 CrossRef.
- S. S. Jovičić Milić, V. V. Jevtić, D. Lj. Stojković, Đ. S. Petrović, E. H. Avdović, Z. S. Marković, I. D. Radojević, Lj. Čomić and V. S. Mladenović, Inorg. Chim. Acta, 2020, 510, 119743 CrossRef.
- G. P. Vasić, V. V. Glođović, I. D. Radojević, O. D. Stefanović, Lj. R. Čomić, V. M. Đinović and S. R. Trifunović, Inorg. Chim. Acta, 2010, 363, 3606–3610 CrossRef.
- I. Radojevic, O. Stefanovic, Lj. Čomić, V. Jevtić, G. Radić and S. Trifunović, Oxid. Commun., 2017, 40, 1070–1080 Search PubMed.
- J. Marmur, J. Mol. Biol., 1961, 3, 208–218 CrossRef CAS.
- M. E. Reichmann, S. A. Rice, C. A. Thomas and P. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef CAS.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, USA, 2006 Search PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- M. Jurišević, A. Arsenijević, J. Pantić, N. Gajović, J. Milovanović, M. Milovanović, J. Poljarević, T. Sabo, D. Vojvodić, G. D. Radosavljević and N. Arsenijević, Oncotarget, 2018, 9, 28195–28212 CrossRef PubMed.
- A. Popović, M. Nikolić, M. Mijajlović, Z. Ratković, V. Jevtić, S. R. Trifunović, G. Radić, M. Zarić, P. Čanović, M. Milovanović, S. Radisavljević, M. Međedović, B. Petrović and I. Jovanović, Transition Met. Chem., 2019, 44, 219–228 CrossRef.
- B. Konovalov, A. A. Franich, M. Jovanović, M. Jurišević, N. Gajović, M. Jovanović, N. Arsenijević, V. Marić, I. Jovanović, M. D. Živković and S. Rajković, Appl. Organomet. Chem., 2021, 35, 1–15 CrossRef.
- J. M. Andrews, J. Antimicrob. Chemother., 2005, 56, 60–76 CrossRef CAS PubMed.
- S. D. Sarker, L. Nahar and Y. Kumarasamy, Methods, 2007, 42, 321–324 CrossRef CAS PubMed.
- G. P. Radić, V. V. Glođović, I. D. Radojević, O. D. Stefanović, Lj. R. Čomić, Z. R. Ratković, A. Valkonen, K. Rissanen and S. R. Trifunović, Polyhedron, 2012, 31, 69–76 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt00659c |
| This journal is © The Royal Society of Chemistry 2024 |