 Open Access Article
Open Access ArticleAnalyses of the electronic structures of FeFe-cofactors compared with those of FeMo- and FeV-cofactors and their P-clusters†
Zhen-Lang
Xie
 a,
Wan-Ting
Jin
b and
Zhao-Hui
Zhou
a,
Wan-Ting
Jin
b and
Zhao-Hui
Zhou
 a
a
aState Key Laboratory of Physical Chemistry of Solid Surfaces and College of Chemistry and Chemical Engineering, Xiamen University, Xiamen, 361005, China. E-mail: zhzhou@xmu.edu.cn; Fax: +86-592-2183047; Tel: +86 -592-2184531
bCollege of Chemical and Material Engineering, Quzhou University, Quzhou, 324000, China
First published on 23rd January 2024
Abstract
The electronic structures of FeFe-cofactors (FeFe-cos) in resting and turnover states, together with their PN clusters from iron-only nitrogenases, have been calculated using the bond valence method, and their crystallographic data were reported recently and deposited in the Protein Data Bank (PDB codes: 8BOQ and 8OIE). The calculated results have also been compared with those of their homologous Mo- and V-nitrogenases. For FeFe-cos in the resting state, Fe1/2/4/5/6/7/8 atoms are prone to Fe3+, while the Fe3 atom shows different degrees of mixed valences. The results support that the Fe8 atom at the terminal positions of FeFe-cos possesses the same oxidation states as the Mo3+/V3+ atoms of FeMo-/FeV-cos. In the turnover state, the overall oxidation state of FeFe-co is slightly reduced than those in the resting species, and its electronic configuration is rearranged after the substitution of S2B with OH, compatible with those found in CO-bound FeV-co. Moreover, the calculations give the formal oxidation states of 6Fe2+–2Fe3+ for the electronic structures of PN clusters in Fe-nitrogenases. By the comparison of Mo-, V- and Fe-nitrogenases, the overall oxidation levels of 7Fe atoms (Fe1–Fe7) for both FeFe- and FeMo-cos in resting states are found to be higher than that of FeV-co. For the PN clusters in MoFe-, VFe- and FeFe-proteins, they all exhibit a strong reductive character.
1 Introduction
Nitrogenases uniquely reduce atmospheric inert dinitrogen (N2) to bioavailable ammonia (NH3) under ambient conditions, providing nearly two-thirds of the worldwide nitrogen resource.1 A catalytic reaction occurs at the dinitrogenase component, while a dinitrogenase reductase provides the required electrons and is also the site of adenosine triphosphate (ATP) hydrolysis to drive the reaction.2 Three homologous nitrogenase isoforms, Mo-, V- and Fe-nitrogenases, are known to date, which primarily differ in the architecture of their catalytically active sites, named M-clusters or FeMo-/FeV-/FeFe-cofactors (FeMo-/FeV-/FeFe-cos), respectively.3–9 Their N2-reducing activities decrease from MoFe, VFe and FeFe proteins sequentially, and the latter two are thus generally regarded as backup systems.10,11 Three nitrogenases catalyze the reactions as follows:12,13Mo-nitrogenase:
| N2 + 8e− + 8H+ + 16MgATP → 2NH3 + H2 + 16MgADP + 16Pi |
V-nitrogenase:
| N2 + 18e− + 18H+ + 36MgATP → 2NH3 + 6H2 + 36MgDP + 36Pi |
Fe-nitrogenase:
| N2 + 20e− + 20H+ + 40MgATP → 2NH3 + 7H2 + 40MgADP + 40Pi |
There are three recognized reversible states of the P-cluster in nitrogenase: the reduced PN state, the one electron-oxidized P1+ state, and the two electron-oxidized P2+ state.8 It has been proposed that electrons for N2 reduction are transferred from the P-cluster to FeMo/FeV/FeFe-cos.14–16 The formal electronic configuration of FeMo-co in the resting state has been assigned to Mo3+–3Fe2+–4Fe3+ by spectroscopic studies and theoretical calculations,17–19 while a reduced oxidation state of FeV-co is suggested as V3+–3Fe2+–4Fe3+.20 Although Fe-only nitrogenase has been studied extensively,21–25 the first crystallographic structure was isolated recently from Azotobacter vinelandii (PDB code 8BOQ) and determined by X-ray diffraction.7 The cryogenic electron microscopy of Fe-only nitrogenase from Rhodobacter capsulatus was also presented (PDB code 8OIE).26 FeFe-co was therefore identified as Fe8S9C[R-(H)homocitrate],7,26 which carries an organic homocitrate ligand that is strikingly similar to those observed in FeMo- and FeV-cos.4,27,28 Intriguingly, Fe-only nitrogenase can retain its reactivity towards N2 without the exchange of the apical iron of the M-cluster with heterometals Mo or V.7,26 In particular, recent work has shown that Fe-only nitrogenase has the unique ability to produce significant quantities of methane (CH4) by CO2 reduction, not shared by the other two homologous nitrogenases.29
However, the electronic structures of FeFe-co and the P-cluster in Fe-nitrogenases are still uncertain, and their oxidation states are crucial for understanding the potential electron transfer sites and pathways. This prompts us to apply a classical bond-valence method for analysing the oxidation states of FeFe-cos and their P-clusters, where the data are taken from the crystal structures of Fe-nitrogenases in the Protein Data Bank (PDB codes 8BOQ and 8OIE). Moreover, the resolution of 8BOQ is high at 1.55 Å, which indicates good reliability of the bond distances with a high weight scheme in calculations.
The bond valence method is a simple but powerful approach for determining the oxidation states of metal ions based on the bond distances of the metal–ligand from their crystal structures.30,31 It was initially used to analyse inorganic crystal structures,32–34 and was gradually applied to other fields, especially in the field of high-resolution protein structures.19,35–39 In contrast to density functional theory (DFT) calculations, whose reliability in nitrogenase models has been questioned by different results,40 the BVS method is much more convenient for studying metalloenzyme systems without considering the spin state of metal ions. Herein, we have employed the BVS method for comparing the resting and turnover states of FeFe-cos in Fe-nitrogenases, trying to figure out the electron distributions of different metal atoms in iron-only nitrogenases and assess the rationality of their structures with the P-cluster. Only PN states of P-clusters are found in FeFe protein in the PDB codes 8BOQ and 8OIE. Therefore, all the following calculations involve PN clusters alone. To further investigate the nitrogen fixation mechanism with different nitrogenases, we have also performed comprehensive comparisons among FeMo-, FeV- and FeFe-cos, together with their PN clusters in nitrogenases.
2. Calculation method
2.1 Criteria for valence assignment
Bond valence sums (BVS) were calculated according to eqn (1)–(3): | (1) |
| d = Si − n | (2) |
 | (3) |
In the FeFe-co and PN cluster of Fe-nitrogenases, rij is the bond length between metal i (Fe) and coordinated atom j (S/O/N/C), measured by Pymol from the crystal structures of FeFe proteins deposited in the Protein Data Bank (PDB). B is considered a constant and is equal to 0.37 Å.32 Here, R0 values used for the calculations of FeFe-co and the PN cluster in Fe-nitrogenases are listed in Table 1. They were determined empirically so that the BVS is generally quite close to the oxidation states of the metal ions.35 Least-squares fitting procedures were used to determine the initial set of R0 values by minimizing the differences between the BVS and the known oxidation states for crystallographically characterized materials. Most R0 values can be found on the web and relevant studies.42Si refers to the calculated bond valence sum of each Fe ion. The d value allows us to assess the deviation between calculated valence Si and assumed valence n. The absolute deviation |d| can serve as an evaluation index of the calculated result. Some valences calculated from either R0 (+2) and R0 (+3) might give a similar |d| value due to the electron delocalization in FeFe-cos and the PN cluster.11 In this case, we assume the existence of mixed valences instead of integral valences. If the |d| values calculated from R0 (+2) and R0 (+3) differ significantly, the assignment of the oxidation state of the Fe atom can be associated with the smallest |d| value. Although the Thorp group has stated that BVS values calculated from Brown's lengths were trustworthy within ±0.25 units,43 electron delocalizations in FeFe-cos and the PN cluster require a wider error-tolerance range than those in inorganic crystals. Therefore, we established an acceptable D value of 0.30 as a valence assignment criterion to distinguish integral or mixed valences.39,44 If the absolute deviation |d| is less than the D value, then iron is assumed to be Fe2+ or Fe3+; otherwise the valence is designated as indeterminate mixed valence Fen+ (2 < n < 3).
S t in eqn (3) represents the calculated valence sum of all eight Fe atoms Fe1–Fe8 in FeFe-co and the PN cluster (abbreviated as 8Fe). The valences of 8Fe are compared with the values of 16 and 24, which correspond to the valences of eight all-ferrous and all-ferric iron atoms, respectively. The average value of eight Fe atoms Fe1–Fe8 is denoted as 8Feav. The detailed bond valence calculations of FeFe-co and the PN cluster (PDB codes: 8BOQ and 8OIE) are given in Tables S1–S10.†
2.2 Weight schemes for the different resolutions of FeFe proteins
The bond valence method requires highly accurate bond distance data.36 To account for variations in the resolutions of crystallographic data, we have used a weighting scheme based on the d-spacing resolutions of structures in the PDB to calculate the bond valence of each iron Si in all calculated FeFe-co or the PN cluster. The smaller the value of the d-spacing resolution limit Ai, the larger the weight wi. As a result, we have revised the inverse distance weighted (IDW) interpolation method for assigning multiple weights: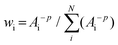 | (4) |
 | (5) |
Eqn (4) is of the same general form as the IDW principle.47 The bond valence Si calculated from a higher resolution Ai (e.g. 8BOQ at 1.55 Å) has greater effects on weighted average valences than that from a lower resolution (e.g. 8OIE at 2.35 Å).  in eqn (5) is obtained from the weighted average of calculated valences Si of different Fe atoms from all analysed FeFe-co or the PN cluster. N is the number of samples containing FeFe-co or the PN cluster of two FeFe proteins.
in eqn (5) is obtained from the weighted average of calculated valences Si of different Fe atoms from all analysed FeFe-co or the PN cluster. N is the number of samples containing FeFe-co or the PN cluster of two FeFe proteins. 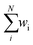 is actually equal to 1 in this equation. p is an index parameter set by the user.48,49 We have previously set p = 1,39,44 which would generate concordant wi with balanced distribution in the P-clusters of MoFe proteins. Here, we also use p = 1 to calculate FeFe proteins for further comparisons with MoFe and VFe proteins.
is actually equal to 1 in this equation. p is an index parameter set by the user.48,49 We have previously set p = 1,39,44 which would generate concordant wi with balanced distribution in the P-clusters of MoFe proteins. Here, we also use p = 1 to calculate FeFe proteins for further comparisons with MoFe and VFe proteins.
3. Results and discussion
3.1 Valence analyses of iron in FeFe-co
The Fo − Fc difference in the electron density map of FeFe proteins (PDB code: 8BOQ) revealed dual conformations, which are in an equilibrium of resting and turnover states.7 In the resting state conformation, Fe2 and Fe6 are bridged by S2B, and Gln176D points away from S2B, with its amide Nε2 weakly hydrogen-bonded to homocitrate. In the turnover state conformation, residue Gln176D rotates its side chain to accept a hydrogen bond from the Nε2 atom of the imidazole side chain of His180D to the Oε1 atom, forming the turnover state. That is, in the turnover state, S2B in FeFe-co is replaced by OH, which is suggested to represent a reaction intermediate, similar to the 6FEA structure with the OH group in the FeV-cofactor.50 Thus, in the PDB code 8BOQ, two FeFe-cos are in mixed states (resting and turnover states). In contrast, two FeFe-cos in the PDB code 8OIE are in resting states only.Fig. 2a and b show the absolute deviations |d| between the calculated and the expected valences of the weighted average value of eight Fe atoms (8Feav) and individual Fe atoms (Fe1–Fe8) in the resting and turnover states of FeFe-cos, respectively. Fig. 2c and d exhibit the valence distributions of Fe atoms in the resting and turnover states of FeFe-cos. Four FeFe-cos in the resting states from the PDB codes 8BOQ and 8OIE were calculated by weighted average values, and two FeFe-cos in the turnover state from the PDB code 8BOQ were calculated by an average value. Two groups of errors are generated from the parameters R0 (+2) (green) and R0 (+3) (orange), respectively. Table 2 lists the weighted average valences and absolute deviations of iron atoms calculated from R0 (+2) and R0 (+3) in FeFe-cos. In conjunction with Fig. 2a and Table 2, the two sets of |d| values calculated using R0 (+2) and R0 (+3) can be clearly distinguished for Fe1, Fe4, Fe5, Fe6 and Fe8 atoms in the resting state. The assignments of Fe3+ for these atoms are reliable with small errors (0.110 < |d| < 0.249), indicating a good fit to the parameters and credible calculated results. Similarly, Fe2 and Fe7 atoms may be favourable for Fe3+, due to their |d| values calculated from R0 (+3) below the valence assignment criteria D value of 0.30. The Fe8 atom is coordinated by R-(H)homocitrate and residue His423D, and the weighted average bond lengths of Fe8–O5 (2.217av Å), Fe8–O7 (2.160av Å) and Fe8–N (2.416av Å) are close than those of heterometals (Mo3+ and V3+) in Mo- and V-nitrogenases.4,6,7 The designation of Fe8 as Fe3+ with a small error (0.231) in the R0 (+3) group in the resting states is consistent with the oxidation states of Mo3+/V3+ in FeMo-/FeV-cos.19 This means that the types of metallic elements have no obvious effect on the oxidation states of the metal (Mo/V/Fe) at the end positions of the cofactors. Thus, the octahedral Fe8 atom probably serves a similar purpose as Mo3+ or V3+ in the other isoforms.7 Importantly, Fe6 is identified as the most oxidized site in the ground state with a calculated value of 2.955av from R0 (+3) in the PDB code 8BOQ, which is in accordance with spatially resolved anomalous dispersion analysis and BVS calculations of FeMo-co,18,19 and thus Fe6 is likely a site of reduction from the P-cluster.18,19 In contrast, the dots calculated from R0 (+2) and R0 (+3) for Fe3 are 0.492 and 0.321, respectively, which are above the valence assignment criteria D value of 0.30 (red dashed line in Fig. 2a). Thus, as depicted in Fig. 2c, the Fe3 atom may perform as mixed valence with strong electron delocalization. With respect to the total eight iron atoms in FeFe-co, the 8Feav values are 0.585 and 0.216 calculated from R0 (+2) and R0 (+3), respectively. The parameter R0 (+3) gives a more reliable result with a small error, suggesting more oxidative iron segments in the resting states of FeFe-co. The metal centers in the resting states could be formally equivalent to Fe2.5+–7Fe3+. Additionally, Fe1, Fe2, Fe4, Fe5, Fe6, Fe7 and Fe8 are more likely to have a higher oxidation state of +3, whereas Fe3 is more reduced and exhibits some degree of mixed valence. The calculations for the crystal structures of FeFe-co in the resting state seems not to be the same as the traditional view that FeFe-co is “4Fe2+–4Fe3+”, which is based on the analyses conducted in reductive solutions with excess dithionite-reduced agents.51
| PDB codes | States | Res (Å) | w i | R 0 (+2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe1 | Fe2 | Fe3 | Fe4 | Fe5 | Fe6 | Fe7 | Fe8 | 8Fe | 8Feav | ||||
| FeFe-co | |||||||||||||
| 8BOQ 7 | Resting | 1.55 | 0.301 | 2.627 | 2.532 | 2.390 | 2.534 | 2.594 | 2.763 | 2.466 | 2.721 | 20.627 | 2.578 |
| 8BOQ 7 | Resting | 1.55 | 0.301 | 2.598 | 2.525 | 2.425 | 2.478 | 2.521 | 2.680 | 2.508 | 2.415 | 20.150 | 2.519 |
| 8OIE 26 | Resting | 2.35 | 0.199 | 2.625 | 2.616 | 2.548 | 2.418 | 2.498 | 2.807 | 2.712 | 2.536 | 20.760 | 2.595 |
| 8OIE 26 | Resting | 2.35 | 0.199 | 3.358 | 2.469 | 2.693 | 2.772 | 2.646 | 2.430 | 2.569 | 2.547 | 21.484 | 2.686 |

|
2.763 | 2.534 | 2.492 | 2.541 | 2.563 | 2.681 | 2.548 | 2.557 | 20.679 | 2.585 | |||
| Weighted |d| | 0.763 | 0.534 | 0.492 | 0.541 | 0.563 | 0.681 | 0.548 | 0.557 | 4.632 | 0.585 | |||
| 8BOQ 7 | Turnover | 1.55 | 2.627 | 2.298 | 2.390 | 2.534 | 2.594 | 2.441 | 2.466 | 2.721 | 19.909 | 2.509 | |
| 8BOQ 7 | Turnover | 1.55 | 2.598 | 2.546 | 2.425 | 2.478 | 2.521 | 2.222 | 2.508 | 2.415 | 19.713 | 2.464 | |
| Average | 2.613 | 2.422 | 2.408 | 2.506 | 2.558 | 2.332 | 2.487 | 2.568 | 19.894 | 2.487 | |||
| |d| | 0.613 | 0.422 | 0.408 | 0.506 | 0.558 | 0.332 | 0.487 | 0.568 | 3.894 | 0.737 | |||
| PDB codes | States | Res (Å) | w i | R 0 (+3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe1 | Fe2 | Fe3 | Fe4 | Fe5 | Fe6 | Fe7 | Fe8 | 8Fe | 8Feav | ||||
| FeFe-co | |||||||||||||
| 8BOQ 7 | Resting | 1.55 | 0.301 | 2.841 | 2.751 | 2.596 | 2.753 | 2.817 | 2.999 | 2.678 | 2.949 | 22.384 | 2.798 |
| 8BOQ 7 | Resting | 1.55 | 0.301 | 2.810 | 2.742 | 2.634 | 2.692 | 2.737 | 2.910 | 2.724 | 2.620 | 21.869 | 2.734 |
| 8OIE 26 | Resting | 2.35 | 0.199 | 2.839 | 2.767 | 2.626 | 2.840 | 3.047 | 2.945 | 2.713 | 2.740 | 22.519 | 2.815 |
| 8OIE 26 | Resting | 2.35 | 0.199 | 2.799 | 2.682 | 2.925 | 3.010 | 2.873 | 2.640 | 2.791 | 2.752 | 22.471 | 2.809 |

|
2.823 | 2.738 | 2.679 | 2.803 | 2.850 | 2.890 | 2.721 | 2.769 | 22.273 | 2.784 | |||
| Weighted |d| | 0.177 | 0.262 | 0.321 | 0.197 | 0.150 | 0.110 | 0.279 | 0.231 | 1.779 | 0.216 | |||
| 8BOQ 7 | Turnover | 1.55 | 2.841 | 2.491 | 2.596 | 2.753 | 2.817 | 2.645 | 2.678 | 2.949 | 21.595 | 2.721 | |
| 8BOQ 7 | Turnover | 1.55 | 2.810 | 2.756 | 2.634 | 2.692 | 2.737 | 2.410 | 2.724 | 2.620 | 21.383 | 2.673 | |
| Average | 2.826 | 2.624 | 2.615 | 2.722 | 2.777 | 2.528 | 2.701 | 2.785 | 21.578 | 2.697 | |||
| |d| | 0.174 | 0.376 | 0.385 | 0.278 | 0.223 | 0.472 | 0.299 | 0.215 | 2.422 | 0.303 | |||
In the turnover state, the sulfide S2B was replaced by an OH group. As shown in Fig. 1b, calculations for Fe2 and Fe6 atoms in the turnover state show that the |d| values are 0.422 and 0.332 calculated from R0 (+2), and 0.376 and 0.472 calculated from R0 (+3), respectively. The two sets of errors for Fe2 and Fe6 atoms are larger than 0.30. Thus, neither the assumed valences +2 nor +3 can be reliably assigned for the oxidation states of these Fe atoms, indicating that some degree of mixed valences exists among these metal centres (Fig. 1d). For the total of eight iron atoms, the average values 8Feav of |d| are 0.487 and 0.303 calculated from R0 (+2) and R0 (+3), implying the presence of electron delocalization in the iron segments of FeFe-co in the turnover state. The total formal oxidation states of 8Fe in the turnover state of FeFe-co could be equal to 2Fe2+–6Fe3+ or Fe2+–7Fe3+, where St = 22 calculated from 2Fe2+–6Fe3+ is close to 21.578 calculated from R0 (+3).
 | ||
| Fig. 1 The resting states of (a) FeMo-co, (b) FeV-co and (c) FeFe-co (PDB codes: 3U7Q,45N6Y6 and 8BOQ7), respectively. Color code: Fe, brown; Mo, teal; V, violet; S yellow; O red; N blue; C grey; H, turquoise. Hydrogen atoms on homocitrates are added based on the vibrational circular dichroism (VCD) experiment for the FeMo-cofactor.41 | ||
 | ||
| Fig. 2 (a) The weighted average |d| values of 8Feav and Fe1–Fe8 atoms calculated from R0 (+2) and R0 (+3) for resting states in the FeFe-co (PDB codes: 8BOQ and 8OIE). (b) The average |d| values of 8Feav and Fe1–Fe8 atoms calculated from R0 (+2) and R0 (+3) for the turnover state in FeFe-co (PDB code: 8BOQ). The red dashed line represents D = 0.3. The valence distributions of Fe atoms in resting (c) and turnover (d) states in FeFe-co. Red, orange and green balls represent n values of 2.7 < n ≤ 3.0, 2.3 < n ≤ 2.7, and 2.0 < n ≤ 2.3 for Fen+, respectively. | ||
Overall, we have found that FeFe-co in the turnover state is more reduced than that in the resting state in he PDB code 8BOQ. The electron distributions in Fe2 and Fe6 atoms have been rearranged with the change of the structure. OH-bound sites Fe2 and Fe6 undergo reduction and exhibit mixed valence in the turnover state. Electron transfer from the Fe protein promotes FeFe-co from the resting state E0 to the E1 state, which leads to the reduction of Fe6, weakening its interaction with S2B, and then receives the proton from His180.7,52 Based on the above analyses, the electron configurations of FeFe-co in the turnover state seem to be more complicated and disturbed by the replacement of the bridging sulfur S2B.
3.2 Valence analyses of iron in the PN clusters in FeFe proteins
Fig. 3a presents the weighted average absolute deviations for 8Feav and individual iron atoms between calculated and expected valences in the PN cluster. Fig. 3b shows the valence distributions of Fe atoms in PN clusters. Table 3 shows the weighted average calculated valences of iron atoms using R0 (+2) and R0 (+3). As shown in Fig. 3a, it can be seen that Fe1, Fe2, Fe5 and Fe6 atoms can be clearly distinguished between the two groups of errors calculated from either R0 (+2) and R0 (+3), and the electrons are well localized in these iron atoms. The one-side match means that +2 valences are more suitable for Fe1/2/5/6. In contrast, the |d| values of the two sets of R0 (+2) and R0 (+3) for Fe3, Fe4, Fe7 and Fe8 are almost similar, which suggests that Fe3/4/7/8 atoms have higher oxidation states than Fe(II) with mixed valences. | ||
| Fig. 3 (a) The weighted average |d| values of 8Feav and Fe1–Fe8 atoms in PN clusters (PDB codes: 8BOQ and 8OIE) calculated from R0 (+2) and R0 (+3). Red dashed lines represent D = 0.3. (b) The valence distributions of Fe atoms in PN clusters of FeFe proteins. | ||
| PDB codes | Res (Å) | w i | R 0 (+2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe1 | Fe2 | Fe3 | Fe4 | Fe5 | Fe6 | Fe7 | Fe8 | 8Fe | 8Feav | |||
| P-cluster | ||||||||||||
| 8BOQ 7 | 1.55 | 0.301 | 2.279 | 2.088 | 2.634 | 2.452 | 2.146 | 1.881 | 2.414 | 2.460 | 18.345 | 2.294 |
| 8BOQ 7 | 1.55 | 0.301 | 2.067 | 2.208 | 2.530 | 2.344 | 2.274 | 1.873 | 2.430 | 2.532 | 18.259 | 2.282 |
| 8OIE 26 | 2.35 | 0.199 | 2.195 | 2.299 | 2.186 | 2.353 | 2.386 | 2.394 | 2.458 | 2.540 | 18.812 | 2.351 |
| 8OIE 26 | 2.35 | 0.199 | 2.189 | 2.228 | 2.291 | 2.616 | 2.450 | 2.468 | 2.347 | 2.276 | 18.864 | 2.358 |

|
2.181 | 2.194 | 2.445 | 2.433 | 2.293 | 2.097 | 2.415 | 2.461 | 18.519 | 2.315 | ||
| Weighted |d| | 0.181 | 0.194 | 0.445 | 0.433 | 0.293 | 0.097 | 0.415 | 0.461 | 2.519 | 0.315 | ||
| PDB codes | Res (Å) | w i | R 0 (+3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe1 | Fe2 | Fe3 | Fe4 | Fe5 | Fe6 | Fe7 | Fe8 | 8Fe | 8Feav | |||
| P-cluster | ||||||||||||
| 8BOQ 7 | 1.55 | 0.301 | 2.465 | 2.258 | 2.848 | 2.652 | 2.321 | 2.035 | 2.611 | 2.660 | 19.850 | 2.481 |
| 8BOQ 7 | 1.55 | 0.301 | 2.236 | 2.388 | 2.737 | 2.535 | 2.459 | 2.025 | 2.629 | 2.739 | 19.748 | 2.469 |
| 8OIE 26 | 2.35 | 0.199 | 2.374 | 2.486 | 2.364 | 2.545 | 2.581 | 2.593 | 2.659 | 2.747 | 20.349 | 2.544 |
| 8OIE 26 | 2.35 | 0.199 | 2.368 | 2.409 | 2.478 | 2.829 | 2.650 | 2.320 | 2.538 | 2.461 | 20.053 | 2.507 |

|
2.359 | 2.373 | 2.645 | 2.631 | 2.480 | 2.200 | 2.611 | 2.662 | 19.961 | 2.495 | ||
| Weighted |d| | 0.641 | 0.627 | 0.355 | 0.369 | 0.520 | 0.800 | 0.389 | 0.338 | 4.039 | 0.505 | ||
Regarding the total eight iron atoms in the PN cluster, the 8Feav value of |d| in the R0 (+2) group is 0.315, if we assume that the PN clusters are all-ferrous, while the corresponding |d| in the R0 (+3) group is 0.505 when the PN clusters are supposed to be all-ferric. The smaller error calculated from R0 (+2) suggests that the PN clusters in FeFe proteins may exhibit strong reductive character, similar to our previous calculations for the PN clusters in MoFe and VFe proteins.39 The weighted average valence sum of 8Fe calculated by R0 (+2) is 18.519 instead of 16, resembling the calculated value for the synthetic model compound of the PN cluster (CSD code DUGNEZ, St = 18.373).53 Therefore, we deduce that the oxidation states of the eight irons in the PN clusters of FeFe proteins are not all-ferrous, but contain four Fe2+ atoms and four mixed-valence irons. The formal oxidation states for 8Fe could be approximately equal to 4Fe2.5+–4Fe3+ or 6Fe2+–2Fe3+ with delocalized electrons.
As mentioned above, the Fe6 atom undergoes a reduction in the turnover state of FeFe-co, which may be due to the electron transfer from the PN cluster to FeFe-co. Judging from the spatial positions of the FeFe-co and PN cluster in the PDB code 8BOQ shown in Fig. 4a, terminal Fe3 is the closest atom among the eight iron atoms in the PN cluster to Fe6 of the FeFe-co, with a distance of 14.4 Å. Thus, we deduce that Fe3 might be responsible for electron transfer to FeFe-co. Similarly, Fe3 has the shortest distance (14.5 Å) in the eight iron atoms in the PN cluster to Fe6 in the M-cluster in the PDB code 8OIE. Besides, Fe6 is the nearest iron to the electron donor [Fe4S4], with a distance of 15.0 Å as shown in Fig. 4b.
 | ||
| Fig. 4 The spatial positions of FeFe-co and PN cluster in the PDB codes (a) 8BOQ and (b) 8OIE, respectively. The latter contains Fe and FeFe proteins simultaneously. | ||
3.3 Comparison of the metal oxidation states in FeMo-, FeV- and FeFe-cos and their PN clusters in nitrogenases
As depicted in Fig. 5a–c, FeMo-, FeV- and FeFe-cos in resting states show some similarities that Fe1, Fe2, Fe6 and Fe7 are more likely to be Fe3+. Fe3 atoms in FeMo-co and FeFe-co have a tendency to be mixed valence and Fe3+, respectively, while those in FeV-co prefers to be the Fe2+ state. Fe4 and Fe5 atoms (bound to a CO32− ligand) in FeV-co are more inclined toward Fe2+, which is different from those in FeMo-co and FeFe-co with higher oxidation states. The overall oxidation level of 7Fe atoms (Fe1–Fe7, St = 19.504) calculated from R0 (+3) for FeFe-co in the resting state is close to the average value of FeMo-co (St = 19.950), but obviously higher than that of FeV-co (St = 18.649).19 In the resting state, the designation of Fe8 as Fe3+ at the terminal positions of FeFe-co has the same oxidation states as the Mo3+/V3+ atoms of FeMo-/FeV-co, indicating that the types of metallic elements do not affect the oxidation of metal (Mo/V/Fe) states at the terminal positions of the cofactors. Therefore, we infer that the octahedral Fe8 atom in FeFe-co probably serves a similar purpose to Mo3+ or V3+ in the other isoforms. | ||
| Fig. 5 The weighted average |d| values of 7Fe and Fe1–Fe7 atoms in resting states (a–c) and turnover states (d–f) in the FeMo-, FeV- and FeFe-cos calculated from R0 (+2) and R0 (+3). (g–i) The |d| values of 8Fe and Fe1–Fe7 atoms in PN clusters of FeMo, FeV and FeFe proteins. The data for FeMo, FeV-cos and PN clusters come from our previous calculated results.19,38,39 | ||
Fig. 5d–f show the |d| values of CO-bound FeMo-co (PDB code: 4TKV), CO-bound FeV-co (PDB code 7ADR) and FeFe-co in turnover states (PDB code 8BOQ), respectively. The 7Fe values of |d| calculated from R0 (+2) and R0 (+3) are 3.940 and 1.494, respectively, for CO-bound FeMo-co, suggesting strong oxidative iron segments. The overall oxidation level of 7Fe atoms (St = 19.506) calculated from R0 (+3) for CO-bound FeMo-co is close to that of FeMo-co in the resting state (St = 19.950). This indicates that the dissociated S2B ligand (replaced by CO) takes two protons with it.54 For FeV-co, the 7Fe values of |d| produced from R0 (+2) and R0 (+3) are 0.973 and 3.451 for CO-bound FeV-co, and 2.292 and 2.391 for FeV-co in the resting state, respectively, which implies that FeV-co in the CO-bound state are more reduced than that in the resting state. Fe2 and Fe6 atoms undergo reduction and are assigned accurately as iron(II) after S2B substituted by the CO ligand. This may be helpful for the comprehension that V-nitrogenase holds a unique property for the conversion of CO to hydrocarbons with yields over 93% of C2H4.52,55 Similarly, the 7Fe values of |d| calculated from R0 (+2) and R0 (+3) are 3.326 and 2.207 for FeFe-co in the turnover state, and 4.122 and 1.548 for FeFe-co in the resting state, respectively. As mentioned above, FeFe-co in the turnover state is slightly reduced than that in the resting state, and Fe2 and Fe6 atoms are rearranged as mixed valence with the substitution of S2B by OH. The slightly reduced valences of Fe2 and Fe6 in FeFe-co in the turnover state are compatible with those found in CO-bound FeV-co. These observations in FeV- and FeFe-cos indicate the presence of a reduced and rearranged oxidation state in the substrate-binding state, especially at the bound sites of Fe2 and Fe6.
For the PN clusters of FeMo, FeV and FeFe proteins shown in Fig. 5g–i, the 8Feav value of |d| in the R0 (+2) group is below 0.30, indicating that these PN clusters exhibit strong reductive character. In particular, Fe1, Fe2, Fe5 and Fe6 tend to be Fe2+, while Fe8 exhibit some mixed valences. The electron distributions of Fe3, Fe4 and Fe7 atoms in the PN clusters of FeMo, FeV and FeFe proteins are somewhat different. From the point of view of the valence sum St of 8Fe by R0 (+2), the PN clusters in FeMo (St = 18.353), FeV (St = 17.960) and FeFe proteins (St = 18.515) may have a similar formal oxidation state with 6Fe2+–2Fe3+.
In general, the electron distributions of FeMo- and FeFe-cos are somewhat different from that of VFe-co. These distinctions could be attributed to the different structures of MoFe, VFe and FeFe proteins, which result in different electron transfer channels that induce the formation of the relevant iron oxidation states. Intriguingly, the oxidation states of 7Fe in FeV- and FeFe-cos in the turnover state are much reduced than those in the resting species, and Fe2 and Fe6 atoms undergo reduction in the substrate-binding state. MoFe, VFe and FeFe proteins possess the same PN cluster core structure [Fe8S7], and their formal oxidation states for 8Fe could be approximately equal to 6Fe2+–2Fe3+ with delocalized electrons.
4. Conclusions
In summary, we have calculated the valences of metal centres in the FeFe-cos and PN clusters of iron-only nitrogenases (PDB codes: 8BOQ and 8OIE) using the bond valence method with weight schemes. The electronic structures of FeMo-, FeV- and FeFe-cos and their P-clusters have been further compared. For FeFe-co in the resting state, Fe1/2/4/5/6/7/8 atoms ought to be designated as Fe3+, while Fe3 is more reduced and exhibits some degree of mixed valences. Moreover, the overall oxidation state of FeFe-co in the turnover state is slightly reduced than those in resting states. Interestingly, the electron distributions in Fe2 and Fe6 atoms are rearranged as mixed-valence states with the substitution of S2B by OH. In the PN clusters of FeFe proteins, the formal oxidation states of 8Fe could be approximately equal to 6Fe2+–2Fe3+ with delocalized electrons.A comparative study of the metal oxidation states for the FeMo-, FeV- and FeFe-cos suggests that the overall oxidation levels of 7Fe atoms (Fe1–Fe7) for both FeFe- and FeMo-cos in resting states are higher than that of FeV-co. Fe8 in the resting state of FeFe-co is calculated as the Fe3+ state, suggesting that the types of metallic elements have no obvious effect on the oxidation states of the apical Mo/V/Fe of the M-clusters. Moreover, Fe2 and Fe6 atoms undergo reduction in FeV- and FeFe-cos in the turnover state. Despite sharing the same core structure [Fe8S7] of the PN cluster in MoFe, VFe and FeFe proteins, it can be found that their electron distributions of the PN cluster exhibit some differences. These differences can be attributed to the unique structures of MoFe, VFe and FeFe proteins, which may lead to diverse channels of electron transfer and induce the formation of relevant iron oxidation states.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (22179110) for the generous financial support.References
- L. M. Rubio and P. W. Ludden, Annu. Rev. Microbiol., 2008, 62, 93–111 CrossRef CAS PubMed.
- F. A. Tezcan, J. T. Kaiser, D. Mustafi, M. Y. Walton, J. B. Howard and D. C. Rees, Science, 2005, 309, 1377–1380 CrossRef CAS PubMed.
- R. R. Eady, Chem. Rev., 1996, 96, 3013–3030 CrossRef CAS PubMed.
- T. Spatzal, M. Aksoyoglu, L. Zhang, S. L. A. Andrade, E. Schleicher, S. Weber, D. C. Rees and O. Einsle, Science, 2011, 334, 940–940 CrossRef CAS PubMed.
- O. Einsle, J. Biol. Inorg. Chem., 2014, 19, 737–745 CrossRef CAS PubMed.
- D. Sippel and O. Einsle, Nat. Chem. Biol., 2017, 13, 956–960 CrossRef CAS PubMed.
- C. Trncik, F. Detemple and O. Einsle, Nat. Catal., 2023, 6, 415–424 CrossRef CAS.
- A. J. Jasniewski, C. C. Lee, M. W. Ribbe and Y. L. Hu, Chem. Rev., 2020, 120, 5107–5157 CrossRef CAS PubMed.
- Y. L. Hu, A. W. Fay, C. C. Lee, J. A. Wiig and M. W. Ribbe, Dalton Trans., 2010, 39, 2964–2971 RSC.
- Y. L. Hu, C. C. Lee and M. W. Ribbe, Dalton Trans., 2012, 41, 1118–1127 RSC.
- O. Einsle and D. C. Rees, Chem. Rev., 2020, 120, 4969–5004 CrossRef CAS PubMed.
- D. F. Harris, D. A. Lukoyanov, S. Shaw, P. Compton, M. Tokmina-Lukaszewska, B. Bothner, N. Kelleher, D. R. Dean, B. M. Hoffman and L. C. Seefeldt, Biochemistry, 2018, 57, 701–710 CrossRef CAS PubMed.
- D. F. Harris, D. A. Lukoyanov, H. Kallas, C. Trncik, Z. Y. Yang, P. Compton, N. Kelleher, O. Einsle, D. R. Dean, B. M. Hoffman and L. C. Seefeldt, Biochemistry, 2019, 58, 3293–3301 CrossRef CAS PubMed.
- J. W. Peters, K. Fisher, W. E. Newton and D. R. Dean, J. Biol. Chem., 1995, 270, 27007–27013 CrossRef CAS PubMed.
- K. Danyal, D. R. Dean, B. M. Hoffman and L. C. Seefeldt, Biochemistry, 2011, 50, 9255–9263 CrossRef CAS PubMed.
- H. L. Rutledge and F. A. Tezcan, Chem. Rev., 2020, 120, 5158–5193 CrossRef CAS PubMed.
- R. Bjornsson, F. Neese and S. DeBeer, Inorg. Chem., 2017, 56, 1470–1477 CrossRef CAS PubMed.
- T. Spatzal, J. Schlesier, E. M. Burger, D. Sippel, L. Zhang, S. L. A. Andrade, D. C. Rees and O. Einsle, Nat. Commun., 2016, 7, 10902 CrossRef CAS PubMed.
- W. T. Jin, M. Yang, S. S. Zhu and Z. H. Zhou, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2020, 76, 428–437 CrossRef CAS PubMed.
- Z. Y. Yang, E. Jimenez-Vicente, H. Kallas, D. A. Lukoyanov, H. Yang, J. S. Martin del Campo, D. R. Dean, B. M. Hoffman and L. C. Seefeldt, Chem. Sci., 2021, 12, 6913–6922 RSC.
- J. R. Chisnell, R. Premakumar and P. E. Bishop, J. Bacteriol., 1988, 170, 27–33 CrossRef CAS PubMed.
- E. Krahn, B. Weiss, M. Kröckel, J. Groppe, G. Henkel, S. Cramer, A. Trautwein, K. Schneider and A. Müller, J. Biol. Inorg. Chem., 2002, 7, 37–45 CrossRef CAS PubMed.
- K. Schneider and A. Müller, in Catalysts for Nitrogen Fixation: Nitrogenases, Relevant Chemical Models and Commercial Processes, ed. B. E. Smith, R. L. Richards and W. E. Newton, Springer Netherlands, Dordrecht, 2004, pp. 281–307 Search PubMed.
- D. A. Lukoyanov, D. F. Harris, Z. Y. Yang, A. Pérez-González, D. R. Dean, L. C. Seefeldt and B. M. Hoffman, Inorg. Chem., 2022, 61, 5459–5464 CrossRef CAS PubMed.
- S. Greed, Nat. Rev. Chem., 2023, 7, 379–379 CrossRef PubMed.
- F. V. Schmidt, L. Schulz, J. Zarzycki, S. Prinz, N. N. Oehlmann, T. J. Erb and J. G. Rebelein, Nat. Struct. Mol. Biol., 2024, 31, 150–158 CrossRef CAS PubMed.
- K. M. Lancaster, M. Roemelt, P. Ettenhuber, Y. Hu, M. W. Ribbe, F. Neese, U. Bergmann and S. DeBeer, Science, 2011, 334, 974–977 CrossRef CAS PubMed.
- D. Sippel and O. Einsle, Nat. Chem. Biol., 2017, 13, 956–960 CrossRef CAS PubMed.
- Y. Zheng, D. F. Harris, Z. Yu, Y. Fu, S. Poudel, R. N. Ledbetter, K. R. Fixen, Z. Y. Yang, E. S. Boyd, M. E. Lidstrom, L. C. Seefeldt and C. S. Harwood, Nat. Microbiol., 2018, 3, 281–286 CrossRef CAS PubMed.
- I. D. Brown and R. D. Shannon, Acta Crystallogr., Sect. A: Cryst. Phys., Diffr., Theor. Gen. Crystallogr., 1973, 29, 266 CrossRef CAS.
- I. D. Brown, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1977, 33, 1305–1310 CrossRef.
- I. D. Brown and D. Altermatt, Acta Crystallogr., Sect. B: Struct. Sci., 1985, 41, 244–247 CrossRef.
- M. H. Chiang, M. R. Antonio, C. W. Williams and L. Soderholm, Dalton Trans., 2004, 801–806 RSC.
- S. Matsia, O. Tsave, A. Hatzidimitriou, C. Gabriel and A. Salifoglou, J. Inorg. Biochem., 2021, 222, 111469 CrossRef CAS PubMed.
- W. Liu and H. H. Thorp, Inorg. Chem., 1993, 32, 4102–4105 CrossRef CAS.
- P. Muller, S. Kopke and G. M. Sheldrick, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2003, 59, 32–37 CrossRef PubMed.
- I. D. Brown, Chem. Rev., 2009, 109, 6858–6919 CrossRef CAS PubMed.
- C. Yuan, W. T. Jin and Z. H. Zhou, New J. Chem., 2022, 46, 9519–9525 RSC.
- C. Yuan, W. T. Jin and Z. H. Zhou, RSC Adv., 2022, 12, 5214–5224 RSC.
- L. Cao and U. Ryde, Phys. Chem. Chem. Phys., 2019, 21, 2480–2488 RSC.
- L. Deng, H. Wang, C. H. Dapper, W. E. Newton, S. Shilov, S. Wang, S. P. Cramer and Z. H. Zhou, Commun. Chem., 2020, 3, 145 CrossRef CAS PubMed.
- I. Brown, Accumulated table of bond-valence parameters. https://www.iucr.org/__data/assets/file/0011/150779/bvparm2020.cif, 2020.
- H. H. Thorp, Inorg. Chem., 1992, 31, 1585–1588 CrossRef CAS.
- Z. L. Xie, C. Yuan and Z. H. Zhou, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2023, 79, 401–408 CrossRef CAS PubMed.
- I. D. Brown and D. Altermatt, Acta Crystallogr., Sect. B: Struct. Sci., 1985, 41, 244–247 CrossRef.
- W. Liu and H. H. Thorp, Inorg. Chem., 1993, 32, 4102–4105 CrossRef CAS.
- Y. Shi, W. He, J. Zhao, A. Hu, J. Pan, H. Wang and H. Zhu, J. Cleaner Prod., 2020, 253, 119965 CrossRef.
- D. Shepard , presented in part at the Proceedings of the 1968 23rd ACM national conference, 1968.
- P. M. Bartier and C. P. Keller, Comput. Geosci., 1996, 22, 795–799 CrossRef CAS.
- D. Sippel, M. Rohde, J. Netzer, C. Trncik, J. Gies, K. Grunau, I. Djurdjevic, L. Decamps, S. L. A. Andrade and O. Einsle, Science, 2018, 359, 1484–1489 CrossRef CAS PubMed.
- E. Krahn, B. J. R. Weiss, M. Kröckel, J. Groppe, G. Henkel, S. P. Cramer, A. X. Trautwein, K. Schneider and A. Müller, J. Biol. Inorg. Chem., 2002, 7, 37–45 CrossRef CAS PubMed.
- M. Rohde, K. Laun, I. Zebger, S. T. Stripp and O. Einsle, Sci. Adv., 2021, 7, eabg4474 CrossRef CAS PubMed.
- Y. Ohki, A. Murata, M. Imada and K. Tatsumi, Inorg. Chem., 2009, 48, 4271–4273 CrossRef CAS PubMed.
- J. Bergmann, E. Oksanen and U. Ryde, J. Inorg. Biochem., 2021, 219, 111426 CrossRef CAS PubMed.
- M. Rohde, K. Grunau and O. Einsle, Angew. Chem., Int. Ed., 2020, 59, 23626–23630 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of bond distances and valence calculations of PDB codes 8BOQ and 8OIE. See DOI: https://doi.org/10.1039/d3dt04126c |
| This journal is © The Royal Society of Chemistry 2024 |
