Open Access Article
Open Access ArticleSilver(I) complexes containing antifungal azoles: significant improvement of the anti-Candida potential of the azole drug after its coordination to the silver(I) ion†
Mia
Stanković
 a,
Jakob
Kljun
a,
Jakob
Kljun
 b,
Nevena Lj.
Stevanović
b,
Nevena Lj.
Stevanović
 a,
Jelena
Lazic
a,
Jelena
Lazic
 c,
Sanja
Skaro Bogojevic
c,
Sanja
Skaro Bogojevic
 c,
Sandra
Vojnovic
c,
Sandra
Vojnovic
 c,
Matija
Zlatar
c,
Matija
Zlatar
 d,
Jasmina
Nikodinovic-Runic
d,
Jasmina
Nikodinovic-Runic
 c,
Iztok
Turel
c,
Iztok
Turel
 *b,
Miloš I.
Djuran
*b,
Miloš I.
Djuran
 *e and
Biljana Đ.
Glišić
*e and
Biljana Đ.
Glišić
 *a
*a
aUniversity of Kragujevac, Faculty of Science, Department of Chemistry, R. Domanovića 12, 34000 Kragujevac, Serbia. E-mail: biljana.glisic@pmf.kg.ac.rs
bUniversity of Ljubljana, Faculty of Chemistry and Chemical Technology, Večna pot 113, SI-1000, Ljubljana, Slovenia. E-mail: Iztok.Turel@fkkt.uni-lj.si
cUniversity of Belgrade, Institute of Molecular Genetics and Genetic Engineering, Vojvode Stepe 444a, 11042 Belgrade, Serbia
dUniversity of Belgrade-Institute of Chemistry, Technology and Metallurgy, Department of Chemistry, Njegoševa 12, 11000 Belgrade, Serbia
eSerbian Academy of Sciences and Arts, Knez Mihailova 35, 11000 Belgrade, Serbia. E-mail: milos.djuran@pmf.kg.ac.rs
First published on 2nd January 2024
Abstract
Inspired by the emergence of resistance to currently available antifungal therapy and by the great potential of metal complexes for the treatment of various diseases, we synthesized three new silver(I) complexes containing clinically used antifungal azoles as ligands, [Ag(ecz)2]SbF6 (1, ecz is econazole), {[Ag(vcz)2]SbF6}n (2, vcz is voriconazole), and [Ag(ctz)2]SbF6 (3, ctz is clotrimazole), and investigated their antimicrobial properties. The synthesized complexes were characterized by mass spectrometry, IR, UV-vis and 1H NMR spectroscopy, cyclic voltammetry, and single-crystal X-ray diffraction analysis. In the mononuclear complexes 1 and 3 with ecz and ctz, respectively, the silver(I) ion has the expected linear geometry, in which the azoles are monodentately coordinated to this metal center through the N3 imidazole nitrogen atom. In contrast, the vcz-containing complex 2 has a polymeric structure in the solid state in which the silver(I) ions are coordinated by four nitrogen atoms in a distorted tetrahedral geometry. DFT calculations were done to predict the most favorable structures of the studied complexes in DMSO solution. All the studied silver(I) complexes have shown excellent antifungal and good to moderate antibacterial activities with minimal inhibitory concentration (MIC) values in the ranges of 0.01–27.1 and 2.61–47.9 μM on the selected panel of fungi and bacteria, respectively. Importantly, the complexes 1–3 have exhibited a significantly improved antifungal activity compared to the free azoles, with the most pronounced effect observed in the case of complex 2 compared to the parent vcz against Candida glabrata with an increase of activity by five orders of magnitude. Moreover, the silver(I)-azole complexes 2 and 3 significantly inhibited the formation of C. albicans hyphae and biofilms at the subinhibitory concentration of 50% MIC. To investigate the impact of the complex 3 more thoroughly on Candida pathogenesis, its effect on the adherence of C. albicans to A549 cells (human adenocarcinoma alveolar basal epithelial cells), as an initial step of the invasion of host cells, was studied.
Introduction
Fungal infections are a worldwide global health problem, affecting millions of patients per year. Many fungal species are responsible for these invasive infections, which kill about one and a half million people every year. These high mortality rates, which can exceed 50%, have been attributed to the difficulty of diagnosis, and shortcomings in the current antifungal arsenal.1–3 More than 90% of all reported fungal-related deaths result from species that belong to one of the four genera: Cryptococcus, Candida, Aspergillus, and Pneumocystis, although diseases caused by rarer fungi are becoming more common.4 Furthermore, it has been anticipated that global warming will bring new fungal diseases to mammals.5Although fungi were recognized as pathogens prior to bacteria, progress in the development of effective antifungals has lagged in antibacterial research. The reason for this lies in the fact that the eukaryotic nature of fungi makes them metabolically and physiologically similar to mammalian cells, and therefore offers limited pathogen-specific targets.6 The key to the successful development of antifungal agents is based on their specificity to arrest or kill pathogenic fungal cells in preference to host cells. The variance between fungal and host cells, as well as the variance among different fungal cells, provide the basis for the identification of druggable targets for new antifungal agents.
Up to now, only five main antifungal agent classes have been approved for human consumption: azoles, polyenes, echinocandins, allylamines, and antimetabolites.2 The antifungal azoles are a principal class of small molecules that consist of an imidazole or a triazole ring attached to a carbon atom. They are used to treat fungal infections by targeting the cytochrome P450-dependent enzyme, sterol 14α-demethylase (P450DM), preventing the synthesis of ergosterol, a major component of fungal plasma membranes.7 However, the main drawbacks of currently used antifungal azoles include fungistatic rather than fungicidal activity, narrow spectrum of antifungal activity, non-optimal pharmacokinetics, and hepatotoxicity.8 Also, a common and serious problem of these, but also other, antifungal agents is the emergence of severe drug resistance, which has significantly reduced their therapeutic efficacy.9–13 The limited efficacy of azoles in the treatment of invasive fungal infections highlights the urgent need for the development of new agents, particularly new structures that act upon novel targets or that could overcome currently used antifungal drug side effects.
Metal ions can play an important role in modifying the pharmacological properties of known organic-based drugs, most of which can be coordinated with a metal ion. The resulting coordination compounds have different physical and pharmacological properties, and their mechanism of activity fundamentally differs from those of clinically used organic drugs. This can be explained by the fact that metal complexes of such organic drugs can have different geometries and generally possess a higher 3D character,14,15 which is one of the crucial factors in determining the biological activity of metal-based compounds.16–18 It is thus not surprising that research of metal complexes as antifungals is quite popular nowadays.19,20
In light of the abovementioned facts, the complexation reactions between clinically used antifungal azoles and different metal ions (ruthenium(II), copper(II), zinc(II), silver(I) and gold(III)) have been recently investigated in our laboratories.21–24 The antimicrobial potential of metal-azole complexes obtained from these reactions was investigated by comparing their antimicrobial activity with that of the corresponding antifungal azoles. For instance, nine organoruthenium complexes with clotrimazole, tioconazole and miconazole have been evaluated against the fungus Culvularia lunata, showing significant antifungal activity, which decreased with the increasing number of ligands.21 Further, the complexation reaction of fluconazole (fcz) with copper(II) and zinc(II) ions led to the formation of polymeric {[CuCl2(fcz)2]·5H2O}n and {[ZnCl2(fcz)2]·2C2H5OH}n complexes of octahedral geometry, which have shown higher in vitro antifungal activity against C. krusei and C. parapsilosis than fluconazole.22 Moreover, these two complexes have shown strong inhibition of C. albicans filamentation and biofilm formation at subinhibitory concentrations. A high binding affinity of silver(I) ion for itraconazole (icz) was observed in the reaction between AgNO3 and this antifungal drug. The spectroscopic and crystallographic results for the [Ag(icz)2]NO3·H2O complex revealed that two icz ligands are monodentately coordinated to the silver(I) ion via the triazole nitrogen atom forming a cationic [Ag(icz)2]+ part, which is neutralized by the nitrate anion.23 The investigated silver(I) complex showed enhanced in vitro antifungal activity than icz, being 2.3- and 4.5-fold more active against C. albicans and C. glabrata, respectively. This complex was also more efficient in inhibiting yeast to hyphae transition process in C. albicans, as an important step in its pathogenesis. The toxicity evaluation in the zebrafish model (Danio rerio) suggests that the [Ag(icz)2]NO3·H2O complex has a better therapeutic profile and improved antifungal efficacy with respect to the parent drug. More recently, the complexation reactions of four antifungal azoles (clotrimazole, econazole, tioconazole and voriconazole) with gold(III) ion have been performed, leading to the formation of the mononuclear gold(III) complexes of the general formula [AuCl3(azole)].24 These complexes demonstrated enhanced antimicrobial effectiveness than the corresponding azoles in most cases, with the best improvement obtained after the auration of tioconazole and voriconazole.
In the course of the above investigations, inspired by the differences in both potency and selectivity between the different antifungal azoles, herein we report the synthesis and structural characterization of new silver(I) complexes with clinically used antifungal azoles, econazole (ecz), voriconazole (vcz) and clotrimazole (ctz) (Scheme 1), namely [Ag(ecz)2]SbF6 (1), {[Ag(vcz)2]SbF6}n (2) and [Ag(ctz)2]SbF6 (3). These complexes along with the azoles used for their synthesis were evaluated for their in vitro antimicrobial activity against the panel of bacterial and Candida species, while their cytotoxicity was tested against the human normal lung fibroblast cell line (MRC-5). Moreover, the influence of complexes 2 and 3 on the formation of C. albicans hyphae and biofilms was studied at subinhibitory concentrations. The adherence of C. albicans to A549 cells (human adenocarcinoma alveolar basal epithelial cells) was investigated in the presence of the complex 3 to study its effect on Candida pathogenesis more thoroughly.
 | ||
| Scheme 1 Structural formula of the antifungal azoles, econazole (ecz), clotrimazole (ctz) and voriconazole (vcz), used as ligands for the synthesis of silver(I) complexes in the present study. | ||
Results and discussion
Synthesis and structural characterization of the silver(I) complexes 1–3
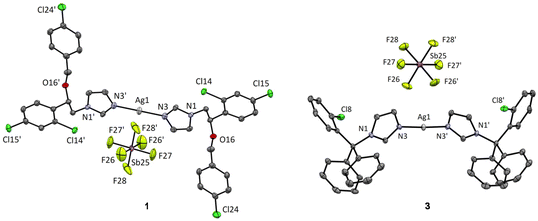
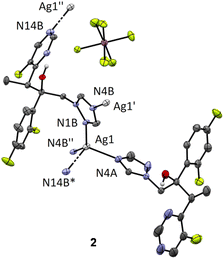
Silver(I) complexes with the antifungal agents econazole (ecz), voriconazole (vcz) and clotrimazole (ctz), [Ag(ecz)2]SbF6 (1), {[Ag(vcz)2]SbF6}n (2) and [Ag(ctz)2]SbF6 (3), were synthesized according to the procedure described in the Experimental section (vide infra). Silver(I) hexafluoroantimonate(V) was mixed with an equimolar amount of the corresponding azole in ethanol at ambient temperature. The crystals of the silver(I) complex 1 suitable for single-crystal X-ray diffraction analysis were obtained after the white precipitate from the reaction was recrystallized in acetonitrile, while the crystals of 2 and 3 were formed by recrystallization of the resulting precipitates in acetonitrile/ethanol (v/v 1/1) at ambient temperature. The composition of the synthesized complexes was elucidated from 1H NMR, IR, UV-vis and mass spectra, elemental analysis measurement, DFT calculations and single-crystal X-ray diffraction analysis.On the other hand, voriconazole as a ligand offers much greater potential for structural diversity because its structure includes four aromatic nitrogen atoms and an alcohol oxygen atom with coordination ability. The CSD25 reports three known structures of silver(I)-vcz complexes synthesized with different salts as silver(I) source, namely, triflate, nitrate and perchlorate.27,28 We report here the crystal structure of a fourth complex containing hexafluoroantimonate(V) counter-anion (Fig. 2). These four structures can be divided into two groups. In the structure of 2, vcz ligands bind to silver(I) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal
2 metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand ratio. We can observe the same coordination pattern as in the triflate28 and perchlorate27 salts, which form polymeric structures in which silver(I) ions are coordinated by four nitrogen atoms and adopt a distorted tetrahedral geometry (Fig. 2). However, the two ligands are crystallographically different – molecule A acts as a terminal ligand, binding to only one Ag(I) ion through its N4A triazole nitrogen, and molecule B acts as a bridging ligand binding to three different silver(I) ions through the N1B and N4B triazole nitrogen atoms and the N14B pyrimidine nitrogen atom (Fig. 2). Each silver(I) ion is thus coordinated by four vcz ligand molecules. The previously reported silver(I)-vcz complex with nitrate counter-anion, on the other hand, has a much more cross-linked structure in which we can observe that the asymmetric unit contains two different silver(I) ions (one tetracoordinated and one pentacoordinated) and four bridging ligands.27
ligand ratio. We can observe the same coordination pattern as in the triflate28 and perchlorate27 salts, which form polymeric structures in which silver(I) ions are coordinated by four nitrogen atoms and adopt a distorted tetrahedral geometry (Fig. 2). However, the two ligands are crystallographically different – molecule A acts as a terminal ligand, binding to only one Ag(I) ion through its N4A triazole nitrogen, and molecule B acts as a bridging ligand binding to three different silver(I) ions through the N1B and N4B triazole nitrogen atoms and the N14B pyrimidine nitrogen atom (Fig. 2). Each silver(I) ion is thus coordinated by four vcz ligand molecules. The previously reported silver(I)-vcz complex with nitrate counter-anion, on the other hand, has a much more cross-linked structure in which we can observe that the asymmetric unit contains two different silver(I) ions (one tetracoordinated and one pentacoordinated) and four bridging ligands.27
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(C
Car) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N))) in the expected region (1618–1406 cm−1).24 As far as complex 2 is concerned, a broad band at 3415 cm−1 in its IR spectra, due to the O–H stretching vibration of voriconazole, is observed.31
N))) in the expected region (1618–1406 cm−1).24 As far as complex 2 is concerned, a broad band at 3415 cm−1 in its IR spectra, due to the O–H stretching vibration of voriconazole, is observed.31
The ambient temperature UV-vis spectra of silver(I) complexes 1–3 and the corresponding azoles were recorded in DMSO (Fig. S1†). Due to the similarity in the shape of the UV-vis spectra of the silver(I) complex and the respective azole ligand, it can be proposed that the absorbance peaks for the complexes 1–3 at λmax = 271, 292 and 260 nm, respectively, originate from the intra-ligand charge transfer transitions. This is in accordance with the UV-vis spectroscopic features of the previously reported silver(I) complexes with various nitrogen- and sulfur-donor ligands.32,33 The absorbance peaks of complexes 1–3 show bathochromic red shifts compared to those of the corresponding azoles (λmax ∼255 nm; see ESI†). The UV-vis spectra were also recorded at 24 and 48 h after dissolution of the complexes 1–3 in DMSO and in DMSO/PBS (PBS is phosphate buffered saline) (Fig. S1 and S2†). No significant changes were seen between the initial UV-vis spectra and those recorded after 48 h (only a slight decrease in the intensity of the absorption maxima of the studied complexes was observed during this period; up to 5% for 1), being in accordance with the fact that the corresponding azole ligands remains coordinated to the silver(I) ion in solution. Despite this fact, it could be assumed that the silver(I) ion will be released from [Ag(azole)2]+ moiety under the physiological conditions in the presence of proteins and other cellular biomolecules, since the interactions between silver(I) ion and these biomolecules are responsible for its antimicrobial activity.34
The 1H NMR spectra of silver(I) complexes 1–3 and the respective azoles were recorded in DMSO-d6 (Fig. S3–S5†) and their assignment was performed based on the NMR data reported in the literature for the studied azoles, whose 1H NMR spectra were recorded in different solvents.24,26,35 As can be seen in Fig. S3–S5,† most of the 1H resonances for the coordinated azoles are downfield shifted concerning those for the uncoordinated ligand. In some cases, the Δ(1H)coord coordination shifts (determined with respect to the azole in DMSO-d6) are not significant, what can be expected for NMR spectroscopic properties of silver(I) complexes in DMSO-d6 solution.36,37 In all cases, the largest chemical shift was observed for the proton which is adjacent to the nitrogen binding center, i.e. C4H for ecz (+0.18 ppm), C5H for vcz (+0.08 ppm) and C2H for ctz (+0.31 ppm).
The positive ion ESI mass spectra for silver(I) complexes 1–3 (vide infra; Fig. S6–S8†) were consistent with the patterns simulated for the corresponding molecular cations, and additionally confirmed the coordination of the corresponding azole to the silver(I) ion in solution.
Computational results
DFT calculations have been carried out to gain a better understanding of the structures of the complexes in DMSO solutions. We have investigated the thermodynamics of possible reactions starting from the X-ray determined structures of the complexes 1–3. The free energy changes, ΔrG (298 K), for the formation of likely coordination structures in solution have been studied (Table 1).38,39 ΔrG (298 K) was calculated from the difference between the Gibbs free energies of the products and the reactants. A geometry optimization at the ZORA-BP86-D4/TZP-COSMO(DMSO) level of theory was performed. Electronic energies used for the calculation of the Gibbs free energies of studied species were evaluated at two levels of theory – ZORA-MN15/TZ2P-COSMO(DMSO) and ZORA-TPSSh-D4/TZ2P-COSMO(DMSO), and consistent results were obtained (Table 1). Complexes 1 and 3 are stable in DMSO solution, maintaining their linear coordination with two azole ligands coordinated in a monodentate fashion (Fig. 3). In the case of the polymeric structure 2, the tetracoordinated complex [Ag(vcz)4]+ was the starting point for the computational analysis. It was found that the dissociation of two vcz ligands is thermodynamically favored, leading to the linear [Ag(vcz)2]+ (Fig. 3 and Table 1). The [Ag(vcz)2]+ species is stable in DMSO solution, similar to complexes 1 and 3 (Table 1). Thus, based on our calculations, all three complexes are ascertained as [Ag(azole)2]+ in DMSO solution (Fig. 3), with two azole ligands linearly coordinated to the silver(I) center, corroborating spectroscopic results.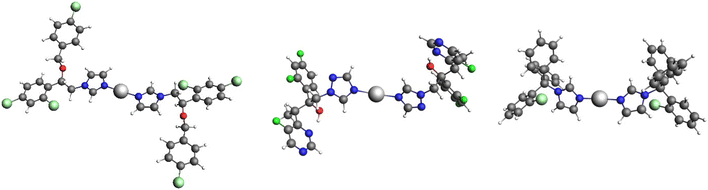 | ||
| Fig. 3 The structures of [Ag(ecz)2]+ (left), [Ag(vcz)2]+ (middle) and [Ag(ctz)2]+ (right) optimized at the ZORA-BP86-D4/TZP-COSMO(DMSO) level of theory. | ||
| Reaction | ΔrG (298 K) | |
|---|---|---|
| MN15/TZ2P | TPSSh-D4/TZ2P | |
| [Ag(ecz)2]+ + 2DMSO ⇌ [Ag(DMSO)2]+ + 2ecz | +9.5 | +12.9 |
| [Ag(ctz)2]+ + 2DMSO ⇌ [Ag(DMSO)2]+ + 2ctz | +11.2 | +14.4 |
| [Ag(vcz)4]+ ⇌ [Ag(vcz)2]+ + 2vcz | −2.3 | −2.6 |
| [Ag(vcz)2]+ + 2DMSO ⇌ [Ag(DMSO)2]+ + 2vcz | +3.1 | +7.0 |
Electrochemical behavior
To better understand the stability and biological activity of a redox active compound both in vitro and in vivo, it is very important to investigate its redox properties. Cyclic voltammetry determines the potential at which the compound can be reduced and/or oxidized.40 Thus, the cyclic voltammograms of the presently synthesized complexes were recorded at the glassy carbon (GC) electrode in DMSO and 0.1 M tetrabutylammonium hexafluorophosphate (TBAHP) as a supporting electrolyte under the following conditions, Ebegin = −2.0 V, Eend = 2.0 V, scan rate = 50 mV s−1 (Fig. S9†). In the voltammograms of the complexes 1–3, one oxidation peak Ia at ∼1.27, 1.48 and 1.47 V, respectively, was observed, which was attributed to the oxidation of silver(I) to silver(II) in accordance with the previous reports for the redox active silver(I) complexes.41–43 In cathodic direction, two reduction peaks Ic,1 and Ic,2 (at ∼−0.28 and −0.92 V for 1; at ∼−0.40 and −1.04 V for 2 and 3; Fig. S9†) are visible in the cyclic voltammograms of the complexes and are due to the reduction processes silver(II) → silver(I) and silver(I) → silver(0), respectively.41–43 It is important to note that silver can exert biological activity in all these oxidation states.44–46Evaluation of biological activity of silver(I) complexes 1–3
To determine the influence of the coordinated silver(I) ion on the antimicrobial activity of complexes 1–3, the biological properties of these complexes were compared with those of the corresponding antifungal azoles (Table 2). The investigated silver(I)-azole complexes 1–3 have shown excellent antifungal and good to moderate antibacterial activities. In comparison to the uncoordinated vcz and ctz azoles, complexes 2 and 3, respectively, exhibited significantly better antifungal activity, whereby 2 showed increased activity of 30-fold (C. parapsilosis), 75-fold (C. albicans), 140-fold (C. krusei) and 9533-fold (C. glabrata) in comparison to vcz, while 3 showed increased activity of 9-fold (C. glabrata), 22-fold (C. albicans), 47-fold (C. krusei) and 1020-fold (C. parapsilosis) in comparison to ctz. An improved antifungal activity was also observed for complex 1 in comparison to ecz against all tested strains except C. krusei, but this effect was less significant (1.7- to 5.0-fold).
| 1 | 2 | 3 | ecz | vcz | ctz | AgSbF6 | |
|---|---|---|---|---|---|---|---|
| a IC50 is the concentration of a compound, which inhibits 50% of cell growth after treatment of 48 h. | |||||||
| C. albicans ATCC 10231 | 2.25 ± 0.10 | 0.48 ± 0.02 | 0.12 ± 0.03 | 7.00 ± 0.14 | 35.80 ± 0.29 | 2.60 ± 0.15 | 3.64 ± 0.20 |
| C. parapsilosis ATCC 22019 | 2.25 ± 0.11 | 0.01 ± 0.001 | 0.01 ± 0.001 | 3.90 ± 0.19 | 0.30 ± 0.09 | 10.20 ± 0.28 | 0.09 ± 0.01 |
| C. krusei ATCC 6258 | 27.10 ± 0.15 | 0.01 ± 0.001 | 0.03 ± 0.01 | 14.10 ± 0.23 | 1.40 ± 0.08 | 1.40 ± 0.09 | 4.66 ± 0.19 |
| C. glabrata ATCC 2001 | 11.29 ± 0.25 | 0.06 ± 0.01 | 0.97 ± 0.06 | 56.20 ± 0.13 | 572.00 ± 0.01 | 9.10 ± 0.19 | 2.33 ± 0.07 |
| P. aeruginosa NCTC 10332 | 22.6 ± 0.22 | 11.9 ± 0.21 | 12.1 ± 0.22 | >500 | >500 | >500 | 36.38 ± 0.11 |
| E. coli NCTC 2001 | 11.3 ± 0.15 | 23.9 ± 0.25 | 12.1 ± 0.17 | >500 | >500 | >500 | 18.19 ± 0.15 |
| S. aureus ATCC 25923 | 2.71 ± 0 14 | 47.9 ± 0.25 | 2.61 ± 0.20 | 225.00 ± 0.01 | >500 | 290.00 ± 0.02 | 36.38 ± 0.07 |
| L. monocytogenes NCTC 11994 | 22.6 ± 0.15 | 47.9 ± 0.16 | 12.1 ± 0.27 | >500 | >500 | >500 | 36.38 ± 0.22 |
| MRC-5 cells | 10.0 ± 1.0 | 36.1 ± 1.5 | 16.0 ± 0.9 | 10.3 ± 1.0 | 859.0 ± 5.0 | 8.7 ± 0.4 | 18.3 ± 0.3 |
The obtained results are in line with those previously achieved by complexation of icz with silver(I) ion, whereby [Ag(icz)2]NO3·H2O complex is 2.3- and 4.5-fold more active than icz against C. albicans and C. glabrata.23 Moreover, the previously reported silver(I) complexes with vcz, [Ag(vcz)2]n(ClO4)n and [Ag2(vcz)4]n(NO3)2n,27 have shown an improvement in the activity against the panel of Candida spp., including vcz resistant isolates. The MIC values (μM) for the corresponding salt used for their synthesis, AgSbF6, are also given in Table 2 and although these values are comparable to the effect of the analyzed antifungal drugs, as well as the corresponding silver(I) complexes, its potential application is limited because this salt undergoes fast dissociation leading to the formation of AgCl precipitate under physiological conditions, as was mentioned above.34
The complexation of different organic ligands with silver(I) ion led to the formation of complexes, which also possess significant antibacterial activity.34,49,50 In comparison to the uncoordinated azoles, silver(I)-azoles complexes 1–3 exhibited significantly better antibacterial activity against all tested strains, ranging from 2.61–47.9 μM (Table 2). All investigated silver(I)-azole complexes had antibacterial activity, with the best activity observed for 1 and 3 against Gram-positive S. aureus, at 2.71 and 2.61 μM, respectively. Interestingly, when tested alone, two of the azole ligands which are in use for antifungal treatment in clinical practice (ecz and ctz), also had an effect against the same bacterial strain, while vcz was inactive against S. aureus at tested concentrations (among the complexes, 2 exhibited the lowest antibacterial activity). None of the uncoordinated azoles has shown antibacterial activity against other tested bacterial strains.
When cytotoxicity of the novel silver(I)-azole complexes against healthy human fibroblasts (MRC-5) was analyzed, it was observed that complex 1 exhibited a comparable effect to ecz, 2 was 24-fold more cytotoxic compared to vcz, while only 3 had a more favorable toxicity profile compared to ctz (1.8-fold lower) (Table 2). The selectivity index (SI) was determined for silver(I) complexes 1–3 and the corresponding azoles by dividing the obtained IC50 with MIC value (Table S1†). The SI values for the complexes on the tested Candida strains are in the range from exceptionally high at 3610 (2 for C. parapsilosis and C. krusei) to 0.4 (1 for C. krusei). For instance, clinically used silver(I) sulfadiazine, AgSD, has the highest SI of 4 against C. parapsilosis and C. krusei.51
Considering that clinical azoles are known to interfere with ergosterol biosynthesis by inhibiting the cytochrome P450,52 we have investigated the effect of complexes 2 and 3, the corresponding azoles vcz and ctz, and AgSbF6 on this process (Fig. S10†). In accordance with this, the ergosterol levels in C. albicans ATCC 10231 treated with these compounds were assessed. As can be seen from Fig. S10,† complexes 2 and 3 significantly reduced the ergosterol levels at 0.5 × MIC, while vcz and ctz, as well as AgSbF6, did not reduce or slightly reduced the ergosterol levels at the same concentration, compared to the untreated control (C. albicans ATCC 10231). These results reveal that the inhibition of ergosterol biosynthesis in the presence of complexes 2 and 3 is in correlation with the results of their antimicrobial activity.
Experimental
Materials and measurements
Silver(I) hexafluoroantimonate(V) (AgSbF6), econazole (ecz), voriconazole (vcz), clotrimazole (ctz), ethanol, acetonitrile, dimethyl sulfoxide (DMSO) and deuterated dimethyl sulfoxide (DMSO-d6) were obtained from the commercial suppliers (Sigma-Aldrich and Acros Organics). All chemicals were of analytical grade and used without further purification. Solvents were used as received.Elemental analyses of the synthesized silver(I)-azole complexes 1–3 for carbon, hydrogen and nitrogen were performed using a PerkinElmer 2400 Series II instrument (CHN). The ESI-HRMS spectra in the positive mode were recorded after dissolving the corresponding compound in CH3CN with an Agilent 62224 accurate mass spectrometer, using time of flight liquid chromatography/mass spectrometry. The IR spectra were recorded on a PerkinElmer Spectrum 2 spectrometer by KBr pellet technique over the wavenumber range of 4000–450 cm−1 (abbreviations used: br for broad, vs for very strong, s for strong, m for medium, w for weak). The UV-vis spectra were recorded over the wavelength range of 1100–200 nm on a Shimadzu UV-1800 spectrophotometer at ambient temperature. The concentration of DMSO solutions for UV-vis measurements was 6.4 × 10−4 M (1), 7.4 × 10−4 M (2) and 4.8 × 10−4 M (3). Additionally, the measurement of the UV-vis spectra for 1–3 was repeated after 24 and 48 h standing in the dark at ambient temperature. Moreover, the solution behavior of complexes 1–3 was studied by UV-vis measurements in DMSO/PBS (v/v is 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) during 48 h at ambient temperature. The 1H NMR spectra of silver(I) complexes 1–3 and the corresponding azoles were recorded on a Varian Gemini 2000 spectrometer at 200 MHz at ambient temperature. 5.0 mg of each compound was dissolved in 0.6 mL of DMSO-d6 and transferred into a 5 mm NMR tube. Chemical shifts, σ, are reported in ppm (parts per million) and scalar couplings, J, are given in Hz (Hertz). The splitting of protons is designated as: s, singlet; d, doublet; dd, doublet of doublets; dt, doublet of triplets; t, triplet; td, triplet of doublets; q, quartet and m, multiplet. The cyclic voltammetry (CV) measurements were performed using a potentiostat/galvanostat AutoLab PGSTAT204 (Utrecht, Netherlands). The three-electrode system consisted of a glassy carbon (GC) electrode as a working electrode, Ag/AgCl (saturated KCl) as a reference electrode and a platinum wire as a counter electrode. The electrode surface was renewed before each measurement by polishing with Al2O3 micro-powder and with a piece of cotton due to the strong adsorption of the complex. The concentration of the solution of the complexes in DMSO used for these measurements was 1.0 × 10−3 M.
3) during 48 h at ambient temperature. The 1H NMR spectra of silver(I) complexes 1–3 and the corresponding azoles were recorded on a Varian Gemini 2000 spectrometer at 200 MHz at ambient temperature. 5.0 mg of each compound was dissolved in 0.6 mL of DMSO-d6 and transferred into a 5 mm NMR tube. Chemical shifts, σ, are reported in ppm (parts per million) and scalar couplings, J, are given in Hz (Hertz). The splitting of protons is designated as: s, singlet; d, doublet; dd, doublet of doublets; dt, doublet of triplets; t, triplet; td, triplet of doublets; q, quartet and m, multiplet. The cyclic voltammetry (CV) measurements were performed using a potentiostat/galvanostat AutoLab PGSTAT204 (Utrecht, Netherlands). The three-electrode system consisted of a glassy carbon (GC) electrode as a working electrode, Ag/AgCl (saturated KCl) as a reference electrode and a platinum wire as a counter electrode. The electrode surface was renewed before each measurement by polishing with Al2O3 micro-powder and with a piece of cotton due to the strong adsorption of the complex. The concentration of the solution of the complexes in DMSO used for these measurements was 1.0 × 10−3 M.
Data from single crystals of 1–3 (Fig. S11 and S12†) were collected at 150 K on a SuperNova diffractometer with Atlas detector using CrysAlis software with monochromated Mo Kα (0.71073 Å).61 The initial structural models were solved with direct methods implemented in SHELXT using the Olex2 graphical user interface.62 A full-matrix least-squares refinement on F2 magnitudes with anisotropic displacement parameters for all non-hydrogen atoms using Olex2 or SHELXL-2018/3 was performed.62,63 All non-hydrogen atoms were refined anisotropically, while hydrogen atoms were placed at calculated positions and treated as riding on their parent atoms. Further details on the crystal data, data acquisition and refinement are given in Table S3.† Figures were prepared with Mercury.64
Synthesis of silver(I) complexes 1–3
Silver(I) complexes 1–3 were synthesized by modification of the previously reported procedure for the synthesis of silver(I) complex with itraconazole.23 The solution of 0.5 mmol of the corresponding azole (190.8 mg of ecz for 1, 174.6 mg of vcz for 2 and 172.4 mg of ctz for 3) in 10.0 mL of ethanol was added slowly under stirring to the solution containing an equimolar amount of AgSbF6 (171.8 mg) dissolved in 5.0 mL of ethanol. The obtained solutions were mixed at ambient temperature for 3 h in the absence of light. The crystals of complex 1 were obtained after the white precipitate from the reaction was recrystallized in acetonitrile, while the crystals of 2 and 3 were formed by recrystallization of the resulting white precipitates in acetonitrile/ethanol (v/v 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and after evaporation at ambient temperature for 3–5 days. The obtained crystals were filtered off and dried in the dark at room temperature. Yield: 210.3 mg (76%) for 1, 185.0 mg (71%) for 2 and 188.6 mg (73%) for 3.
1) and after evaporation at ambient temperature for 3–5 days. The obtained crystals were filtered off and dried in the dark at room temperature. Yield: 210.3 mg (76%) for 1, 185.0 mg (71%) for 2 and 188.6 mg (73%) for 3.
Anal. calcd for 1 (C36H30AgCl6F6N4O2Sb; MW = 1106.96 g mol−1): C, 39.06; H, 2.73; N, 5.06. Found: C, 39.35; H, 2.79; N, 5.43%. HRMS-ESI (CH3CN): m/z calcd for [Ag(ecz)2]+ [C36H30AgCl6N4O2]+: 866.9545; found 866.9583. IR (KBr, ν, cm−1): 3146m, 3101w, 3090w, 3072w, 3051m (ν(Cdiazole–H) and ν(Car–H)), 2939w, 2899w, 2871w, 2861w (ν(C–H)), 1599m, 1589m, 1561m, 1524s, 1492s, 1470s, 1439m, 1407m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(C
Car) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1246m, 1225m (β(Car–H) and β(Cdiazole–H)), 1109s (ν(C–O)), 1094vs (ν(Car–Cl)), 838m, 830m, 819m, 810m, 788m, 761m, 720w, 696w (γ(Car–H)) and (γ(Cdiazole–H)), 656vs (ν(SbF6)). 1H NMR (200 MHz, DMSO-d6): δ 7.80 (s, 1H, C2H), 7.68 (d, J = 2.0 Hz, 1H, C4H), 7.48 (dd, J = 8.4, 2.0 Hz, 1H, C10H), 7.37 (dd, J = 7.3, 5.2 Hz, 2H, C5H, C13H), 7.33 (s, 1H, C12H), 7.22 (s, 1H, C17H), 7.20 (s, 1H, C19H), 7.16 (s, 1H, C16H), 7.01 (s, 1H, C20H), 5.01 (t, J = 5.3 Hz, 1H, C7H), 4.40 (m, 2H, C6H), 4.29 ppm (d, J = 12.4 Hz, 2H, C14H). UV-vis (DMSO, λmax, nm): 271 (ε = 1.2 × 103 M−1 cm−1).
N)), 1246m, 1225m (β(Car–H) and β(Cdiazole–H)), 1109s (ν(C–O)), 1094vs (ν(Car–Cl)), 838m, 830m, 819m, 810m, 788m, 761m, 720w, 696w (γ(Car–H)) and (γ(Cdiazole–H)), 656vs (ν(SbF6)). 1H NMR (200 MHz, DMSO-d6): δ 7.80 (s, 1H, C2H), 7.68 (d, J = 2.0 Hz, 1H, C4H), 7.48 (dd, J = 8.4, 2.0 Hz, 1H, C10H), 7.37 (dd, J = 7.3, 5.2 Hz, 2H, C5H, C13H), 7.33 (s, 1H, C12H), 7.22 (s, 1H, C17H), 7.20 (s, 1H, C19H), 7.16 (s, 1H, C16H), 7.01 (s, 1H, C20H), 5.01 (t, J = 5.3 Hz, 1H, C7H), 4.40 (m, 2H, C6H), 4.29 ppm (d, J = 12.4 Hz, 2H, C14H). UV-vis (DMSO, λmax, nm): 271 (ε = 1.2 × 103 M−1 cm−1).
Anal. calcd for 2 (C32H28AgF12N10O2Sb; MW = 1042.26 g mol−1): C, 36.88; H, 2.71; N, 13.44. Found: C, 36.79; H, 2.68; N, 13.38%. HRMS-ESI (CH3CN): m/z calcd for [Ag(vcz)(MeCN)]+ [C18H17AgF3N6O]+: 497.0467; found 497.0447. IR (KBr, ν, cm−1): 3415br (ν(O–H)), 3192w (ν(Car–H)), 2992w, 2943w, 2976w (ν(C–H)), 1618s, 1596s, 1524m, 1499s, 1457m, 1423m, 1406vs (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(C
Car) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1291m (δ(O–H)), 1242w, 1218w (β(Car–H) and β(Ctriazole–H)), 1132m (ν(C–F)), 966m (ν(C–O)), 852m, 827w, 804w, 781w, 756w, 737w (γ(Car–H)) and (γ(Ctriazole–H)), 660vs (ν(SbF6)), 622m (β(Car–F)). 1H NMR (200 MHz, DMSO-d6): δ = 9.06 (d, J = 2.9 Hz, 1H, C18H), 8.87 (d, J = 2.1 Hz, 1H, C20H), 8.32 (s, 1H, C5H), 7.68 (s, 1H, C3H), 7.27 (m, 1H, C9H), 7.18 (m, 1H, C12H), 6.93 (td, J = 8.5, 2.5 Hz, 1H, C10H), 6.01 (s, 1H, OH), 4.83 (d, J = 14.3 Hz, 1H, C6H), 4.36 (d, J = 14.2 Hz, 1H, C6H), 3.94 (q, J = 7.0 Hz, 1H, C14H), 1.12 ppm (d, J = 7.0 Hz, 3H, C15H). UV-vis (DMSO, λmax, nm): 292 (ε = 1.1 × 103 M−1cm−1).
N)), 1291m (δ(O–H)), 1242w, 1218w (β(Car–H) and β(Ctriazole–H)), 1132m (ν(C–F)), 966m (ν(C–O)), 852m, 827w, 804w, 781w, 756w, 737w (γ(Car–H)) and (γ(Ctriazole–H)), 660vs (ν(SbF6)), 622m (β(Car–F)). 1H NMR (200 MHz, DMSO-d6): δ = 9.06 (d, J = 2.9 Hz, 1H, C18H), 8.87 (d, J = 2.1 Hz, 1H, C20H), 8.32 (s, 1H, C5H), 7.68 (s, 1H, C3H), 7.27 (m, 1H, C9H), 7.18 (m, 1H, C12H), 6.93 (td, J = 8.5, 2.5 Hz, 1H, C10H), 6.01 (s, 1H, OH), 4.83 (d, J = 14.3 Hz, 1H, C6H), 4.36 (d, J = 14.2 Hz, 1H, C6H), 3.94 (q, J = 7.0 Hz, 1H, C14H), 1.12 ppm (d, J = 7.0 Hz, 3H, C15H). UV-vis (DMSO, λmax, nm): 292 (ε = 1.1 × 103 M−1cm−1).
Anal. calcd for 3 (C44H34AgCl2F6N4Sb; MW = 1033.27 g mol−1): C, 51.14; H, 3.32; N, 5.42. Found: C, 51.07; H, 3.35; N, 5.35%. HRMS-ESI (CH3CN): m/z calcd for [Ag(ctz)2]+ [C44H34AgCl2N4]+: 795.1206; found 795.1209. IR (KBr, ν, cm−1): 3177w, 3162w, 3140w, 3108w, 3089w, 3037w (ν(Ctriazole–H) and ν(Car–H)), 2927w (ν(C–H)), 1603w, 1568w, 1530w, 1505m, 1495 m, 1465w, 1447m, 1433m (ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and ν(Car
Car) and ν(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1276m, 1225m (β(Car–H)) and β(Cdiazole–H), 1130m (ν(C–O)), 1093s (ν(Car–Cl)), 765s, 753s, 748s (γ(Car–H)), 655vs (ν(SbF6)), 633m (γ(Car–H)). 1H NMR (200 MHz, DMSO-d6): δ = 7.83 (s, 1H, C2H), 7.57 (dd, J = 7.7, 2.0 Hz, 1H, C8H), 7.51 (d, J = 1.7 Hz, 1H, C10H), 7.46 (m, 2H, C14H, C15H), 7.42 (m, 5H, C9H, C16H, C20H, C21H, C22H), 7.15 (m, 1H, C13H), 7.09 (m, 3H, C17H, C19H, C23H), 7.06 (d, J = 1.7 Hz, 2H, C4H, C5H), 6.94 ppm (dd, J = 7.7, 1.5 Hz, 1H, C11H). UV-vis (DMSO, λmax, nm): 260 (ε = 1.7 × 103 M−1cm−1).
N)), 1276m, 1225m (β(Car–H)) and β(Cdiazole–H), 1130m (ν(C–O)), 1093s (ν(Car–Cl)), 765s, 753s, 748s (γ(Car–H)), 655vs (ν(SbF6)), 633m (γ(Car–H)). 1H NMR (200 MHz, DMSO-d6): δ = 7.83 (s, 1H, C2H), 7.57 (dd, J = 7.7, 2.0 Hz, 1H, C8H), 7.51 (d, J = 1.7 Hz, 1H, C10H), 7.46 (m, 2H, C14H, C15H), 7.42 (m, 5H, C9H, C16H, C20H, C21H, C22H), 7.15 (m, 1H, C13H), 7.09 (m, 3H, C17H, C19H, C23H), 7.06 (d, J = 1.7 Hz, 2H, C4H, C5H), 6.94 ppm (dd, J = 7.7, 1.5 Hz, 1H, C11H). UV-vis (DMSO, λmax, nm): 260 (ε = 1.7 × 103 M−1cm−1).
For comparative purposes, the corresponding azoles were characterized by 1H NMR, IR and UV-vis spectroscopy and these data are given in ESI.†
Computational details
All DFT calculations were done with the ADF65 engine in the Amsterdam Modeling Suite (version 2023.104).66 The scalar-relativistic Zeroth Order Regular Approximation (ZORA) was used to account for relativistic effects.67–69 The COSMO solvation model,70,71 as implemented in ADF,72 with DMSO as solvent was used in all calculations. Geometry optimizations were performed with the BP86 exchange–correlation functional,73–75 with Grimme's fourth generation dispersion energy corrections,76i.e., BP86-D4. The all-electron triple-zeta Slater-type orbitals plus one polarization function (TZP) basis set was used for all atoms. The harmonic frequencies were calculated at the same level of theory. Vibrational analysis with low vibrational frequency free rotor interpolation corrections, as proposed by Li/Head-Gordon77 and Grimme78 has been used to evaluate internal energies and entropic effects to the Gibbs free energy at 298 K (frequency cut-off 100 cm−1). Correction due to the standard state conversion (1 atm. to 1 mol dm−3 solution standard state) is applied (1.89 kcal mol−1 to the free energies at 298 K). For DMSO, the free energy correction due to the conversion to the solvent standard state is made (3.46 kcal mol−1, at 298.15 K). Electronic energies used to calculate the Gibbs free energy were evaluated with MN1579 and TPSSh-D476,80,81meta-hybrid functionals at ZORA-BP86-D4/TZP-COSMO(DMSO) geometries. The all-electron triple-zeta Slater-type orbitals with two polarization functions (TZ2P) basis set were used for all atoms to calculate electronic energies. The LibXC library82 was used for all calculations employing MN15 functional.Antimicrobial activity
Minimum inhibitory concentration (MIC) values of the complexes 1–3, AgSbF6, and the corresponding azoles were determined according to the standard broth microdilution assays, recommended by the National Committee for Clinical Laboratory Standards (M07-A8) for bacteria and Standards of the European Committee on Antimicrobial Susceptibility Testing (v 7.3.1: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts) for Candida spp.83 The tested compounds were dissolved in DMSO at a concentration of 50 mg mL−1. The highest concentration used for antifungal testing was 250 mg mL−1, and for antibacterial testing was 200 mg mL−1. The tested microorganisms included four fungal Candida strains obtained from the American Type Culture Collection (ATCC): C. albicans ATCC 10231, C. parapsilosis ATCC 22019, C. krusei ATCC 6258 and C. glabrata ATCC 2001, and four bacterial strains: Pseudomonas aeruginosa PAO1 NCTC 10332, Escherichia coli NCTC 2001, Staphylococcus aureus ATCC 25923 and Listeria monocytogenes NCTC 11994. The inoculums were 1 × 105 colony forming units (cfu) mL−1 for Candida species and 5 × 105 cfu mL−1 for bacteria. The MIC value was recorded as the lowest concentration that inhibited the growth after 24 h at 37 °C, using a plate reader (Epoch Microplate Spectrophotometer, BioTek Instruments, Inc., USA).Cell viability and proliferation assay
Antiproliferative activity was measured using the standard colorimetric MTT assay.84 Briefly, cells were plated in a 96-well flat-bottomed microtiter plate at a concentration of 1 × 104 cells per well, grown in a humidified atmosphere of 95% air with 5% CO2 at 37 °C and maintained as monolayer cultures in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. Serial dilutions of each tested compound were added to the cells, starting with 50 μM as the maximum concentration. In the case of vcz, the maximum tested concentration was 1000 μM. The treatment lasted for 48 h. The MTT assay was performed two times in four replicates and the results were presented as a percentage of the DMSO-treated control that was arbitrarily set to 100%. The extent of MTT reduction was measured spectrophotometrically at 540 nm using a plate reader (Epoch Microplate Spectrophotometer, BioTek Instruments, Inc., USA). Cytotoxicity was expressed as the concentration of compound inhibiting the cell growth by 50% (IC50) in comparison with the negative control (DMSO-treated cells).Ergosterol analysis
Ergosterol levels were measured in untreated cultures of C. albicans ATCC 10231 and cultures treated with 0.5 × MIC of AgSbF6, azoles vcz and ctz, and complexes 2 and 3 after 18 h incubation at 37 °C on the rotary shaker (180 rpm) following the procedure.85 Ultrospec 3300pro 573 (Amersham Biosciences, Amersham, UK), spectrophotometer was used for following the ergosterol levels (concentrations) by scanning absorbance between 240 and 300 nm.Filamentation test on C. albicans ATCC 10231
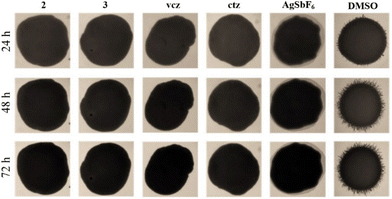
The effect of the tested compounds on C. albicans ATCC 10231 hyphae formation was assessed using solid composition.86 Briefly, 1 mL of C. albicans cells from an overnight culture grown in Sabouraud dextrose broth medium at 37 °C were centrifuged for 10 min at 16.873 rcf, and the pellet was washed several times with sterile PBS (phosphate-buffered saline; Sigma Aldrich, Munich, Germany). The pellet was resuspended in 200 μL PBS, and 2 μL of the cell suspension was poured over the solid composition with or without the tested compounds and incubated at 37 °C for 72 h. Morphological changes of C. albicans ATCC 10231 in the presence of subinhibitory concentration (0.5 × MIC) of compounds or without them were analyzed using bright field microscopy (SMZ143-N2GG, Motic, Germany). C. albicans ATCC 10231 treated with DMSO was used as a control.Anti-biofilm activity assessment on C. albicans ATCC 10231
The effect of the tested compounds on biofilm formation and disruption of preformed biofilms was determined on the C. albicans ATCC 10231 strain. Antibiofilm assays were conducted using previously reported methodologies.41,87 Briefly, the assays were carried out in 96-well round-bottom polystyrene microtiter plates. Cells were harvested from overnight grown cultures, washed twice with sterile PBS, and resuspended in RPMI 1640 medium (Sigma Aldrich, Munich, Germany) containing 2% glucose (w/v) at a concentration of 1 × 105 cfu mL−1. C. albicans suspension was incubated with 0.5 × MIC in 200 μL final volume per well for 48 h at 37 °C to allow biofilm formation. Biofilm growth was analyzed by crystal violet (CV) staining of adherent cells and the absorbance at 590 nm was read by a plate reader (Epoch Microplate Spectrophotometer, BioTek Instruments, Inc., USA). In the biofilm disruption assay, preformed biofilms (24 h at 37 °C) were incubated for 24 h with decreasing concentrations of the compound. Biofilm growth was quantified by CV staining of adherent cells and estimated as absorbance at 530 nm on an Epoch Microplate Spectrophotometer, BioTek Instruments, Inc.Adherence assay
The ability of red fluorescence protein (RFP)-labeled C. albicans SC5314 cells to infect human adenocarcinoma alveolar basal epithelial cells A549 (obtained from ATCC) was tested in an adherence assay, as described previously.88,89 A549 cells were grown on 22 mm glass coverslips in RPMI 1640 medium for two days. Overnight culture of C. albicans SC5314-RFP grown in Sabouraud dextrose broth medium at 37 °C was centrifuged for 10 min at 16.873 rcf. The pellet was washed several times with sterile PBS, 50 μL was inoculated into 5 mL RPMI 1640 medium without FBS or any antibiotic and grown at 37 °C to A530 = 0.2 (A530 = 0.3 corresponds to 1.0–2.5 × 106 cells per mL). A549 monolayers on glass coverslips were inoculated with ∼1 × 106C. albicans cells in the log growth phase (without centrifugation) and co-incubated with the selected compounds added at concentrations of 0.5 × MIC and 1 × MIC for 1.5 h in RPMI 1640 without FBS at 37 °C and 5% CO2. After co-incubation, non-adherent cells were removed by extensively rinsing three times with PBS and samples fixed with 4% p-formaldehyde. Next, A549 cells were stained with 1 μg mL−1 of 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, Sigma Aldrich, Munich, Germany) for 10 min in the dark. Both A549 and C. albicans SC5314-RFP cells were visualized using a fluorescence microscope (Olympus BX51, Applied Imaging Corp., San Jose, USA) at 20× magnification.Statistical analysis
The results are presented as mean ± standard deviation (SD). Statistical analysis was done by comparing means using t-test (Two-Sample Assuming Equal Variances) and one-way ANalysis of VAriance (ANOVA, Single Factor), with Fisher's Least Significant Difference (LSD) post-hoc test. The level of statistical significance is expressed as a p-value (probability value), and p ≤ 0.05 was considered statistically significant. Statistical analysis tests were performed in Microsoft Excel Spreadsheet Software by Data Analysis Tools add-in.Conclusions
The investigation of the reactions of silver(I) salt, AgSbF6, with econazole (ecz), voriconazole (vcz) and clotrimazole (ctz) has shown that silver(I) ion has good coordinating abilities for these clinically used antifungal azoles. The geometry of the silver(I)-azole complexes in the solid state depends on the structural features of the coordinating azole. Thus, the coordination of silver(I) ion with ecz and ctz, both containing imidazole ring, leads to the formation of mononuclear complexes of linear geometry, [Ag(ecz)2]SbF6 (1) and [Ag(ctz)2]SbF6 (3), respectively. However, the reaction of silver(I) ion with vcz, containing a triazole ring, results in formation of polymeric {[Ag(vcz)2]SbF6}n complex (2), in which silver(I) ions are coordinated by four nitrogen atoms and adopt a distorted tetrahedral geometry. Nevertheless, in the case of the latter complex, the formation of [Ag(vcz)2]+ species is favored in solution. The most important finding from the in vitro antimicrobial activity and toxicity evaluation is the fact that the synthesized complexes 1–3 have shown an improved anti-Candida activity and therapeutic profile in comparison to the corresponding clinically used antifungal azoles (with an increase of activity by up to five orders of magnitude). Moreover, the coordination of silver(I) ion to the azoles has endowed them also with antibacterial activity. Furthermore, the silver(I)-azole complexes have significantly contributed to the inhibition of the formation of C. albicans hyphae and biofilms at their subinhibitory concentration, which is a highly desirable property for a new antimicrobial agent. Taken all these facts together, it can be concluded that compounds obtained from the complexation reaction of the metal ion with antifungal azoles deserve further consideration as potential new antifungal agents with favorable pharmacological properties, being more effective and safer than the clinically used antifungal azoles in the treatment of lethal Candida infections.Author contributions
Conceptualization, M. S., J. N. R., I. T., M. I. Dj. and B. Đ. G.; methodology, M. S., J. K. and S. V.; software, J. K. and M. Z.; validation, J. N. R., I. T., M. I. Dj. and B. Đ. G.; investigation, M. S., J. K., N. Lj. S., M. Z., J. L., S. S. B. and S. V.; resources, J. N. R., M. Z., I. T., M. I. Dj. and B. Đ. G.; writing – original draft preparation, J. K., N. Lj. S., M. Z., J. L., S. S. B. and S. V.; writing – review and editing, J. N. R., I. T., M. I. Dj. and B. Đ. G.; visualization, M. S., J. K., N. Lj. S., M. Z., J. L. and S. S. B.; supervision, I. T., M. I. Dj. and B. Đ. G. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research has been financially supported by the Ministry of Science, Technological Development and Innovation of the Republic of Serbia (Agreements No. 451–03–47/2023–01/200026, 451-03-47/2023-01/200042 and 451-03-47/2023-01/200122) and by the Centre for Research Infrastructure at the University of Ljubljana, Faculty of Chemistry and Chemical Technology, which is part of the Network of Research and Infrastructural Centres UL (MRIC UL) financially supported by the Slovenian Research Agency (grant P1-0175 and Infrastructure programme No. I0-0022). The EN → FIST Centre of Excellence, Trg OF 13, SI-1000 Ljubljana, Slovenia, is acknowledged for the use of the SuperNova diffractometer. We thank Marta Počkaj for advice on modelling and refining the disordered atoms in crystal structures. This research has also received funding from the Serbian Academy of Sciences and Arts under project No. F128. N. Lj. S. is gratefully acknowledged to the Ministry of Education, Science and Sport of the Republic of Slovenia for awarding the nine months Mobility Grant (CMEPIUS) at the University of Ljubljana, Faculty of Chemistry and Chemical Technology. This article is also based upon work from COST Action EURESTOP, CA21145, supported by COST (European Cooperation in Science and Technology) and from Bilateral project No. 19 between University of Kragujevac, Faculty of Science, Serbia and University of Ljubljana, Faculty of Chemistry and Chemical Technology, Ljubljana, Slovenia. M. Z. acknowledges the support of the Science Fund of the Republic of Serbia for the computational part of this work (project #7750288, Tailoring Molecular Magnets and Catalysts Based on Transition Metal Complexes – TMMagCat).References
- G. D. Brown, D. W. Denning, N. A. R. Gow, S. M. Levitz, M. G. Netea and T. C. White, Sci. Transl. Med., 2012, 4, 165rv13 CrossRef PubMed.
- K. C. Howard, E. K. Dennis, D. S. Watt and S. Garneau-Tsodikova, Chem. Soc. Rev., 2020, 49, 2426–2480 RSC.
- Y. Lin, H. Betts, S. Keller, K. Cariou and G. Gasser, Chem. Soc. Rev., 2021, 50, 10346–10402 RSC.
- J. Jampilek, Expert Opin. Drug Discovery, 2016, 11, 1–9 CrossRef CAS PubMed.
- M. A. Garcia-Solache and A. Casadevall, mBio, 2010, 18, e00061–e00010 Search PubMed.
- N. A. R. Gow, J.-P. Latge and C. A. Munro, Microbiol. Spectrum, 2017, 5 DOI:10.1128/microbiolspec.funk-0035-2016.
- D. J. Sheehan, C. A. Hitchcock and C. M. Sibley, Clin. Microbiol. Rev., 1999, 12, 40–79 CrossRef CAS PubMed.
- G. Aperis and E. Mylonakis, Expert Opin. Invest. Drugs, 2006, 15, 579–602 CrossRef CAS PubMed.
- M. A. Pfaller, Am. J. Med., 2012, 125, S3–S13 CrossRef CAS PubMed.
- M. A. Pfaller, D. J. Diekema, M. G. Rinaldi, R. Barnes, B. Hu, A. V. Veselov, N. Tiraboschi, E. Nagy, D. L. Gibbs and on behalf of the Global Antifungal Surveillance Group, J. Clin. Microbiol., 2005, 43, 5848–5859 CrossRef CAS PubMed.
- S. Wu, Y. Wang, N. Liu, G. Dong and C. Sheng, J. Med. Chem., 2017, 60, 2193–2211 CrossRef CAS PubMed.
- M. C. Fisher, N. J. Hawkins, D. Sanglard and S. J. Gurr, Science, 2018, 360, 739–742 CrossRef CAS PubMed.
- Y. Lee, E. Puumala, N. Robbins and L. E. Cowen, Chem. Rev., 2021, 121, 3390–3411 CrossRef CAS PubMed.
- A. Frei, Antibiotics, 2020, 9, 90 CrossRef CAS PubMed.
- C. N. Morrison, K. E. Prosser, R. W. Stokes, A. Cordes, N. Metzler-Nolte and S. M. Cohen, Chem. Sci., 2020, 11, 1216–1225 RSC.
- A. W. Hung, A. Ramek, Y. Wang, T. Kaya, J. A. Wilson, P. A. Clemons and D. W. Young, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6799–6804 CrossRef CAS PubMed.
- W. H. B. Sauer and M. K. Schwarz, J. Chem. Inf. Comput. Sci., 2003, 43, 987–1003 CrossRef CAS PubMed.
- W. R. J. D. Galloway, A. Isidro-Llobet and D. R. Spring, Nat. Commun., 2010, 1, 80 CrossRef PubMed.
- A. Frei, A. G. Elliott, A. Kan, H. Dinh, S. Bräse, A. E. Bruce, M. R. Bruce, F. Chen, D. Humaidy, N. Jung, A. P. King, P. G. Lye, H. K. Maliszewska, A. M. Mansour, D. Matiadis, M. Paz Muñoz, T.-Y. Pai, S. Pokhrel, P. J. Sadler, M. Sagnou, M. Taylor, J. J. Wilson, D. Woods, J. Zuegg, W. Meyer, A. K. Cain, M. A. Cooper and M. A. T. Blaskovich, JACS Au, 2022, 2, 2277–2294 CrossRef CAS PubMed.
- R. Navarro-Peñaloza, B. Landeros-Rivera, H. López-Sandoval, R. Castro-Ramírez and N. Barba-Behrens, Coord. Chem. Rev., 2023, 494, 215360 CrossRef.
- J. Kljun, A. J. Scott, T. Lanišnik Rižner, J. Keiser and I. Turel, Organometallics, 2014, 33, 1594–1601 CrossRef CAS.
- N. Lj. Stevanović, I. Aleksic, J. Kljun, S. Skaro Bogojevic, A. Veselinovic, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Đ. Glišić, Pharmaceuticals, 2021, 14, 24 CrossRef PubMed.
- N. Lj. Stevanović, B. Đ. Glišić, S. Vojnovic, H. Wadepohl, T. P. Andrejević, S. Ž. Đurić, N. D. Savić, J. Nikodinovic-Runic, M. I. Djuran and A. Pavic, J. Mol. Struct., 2021, 1232, 130006 CrossRef.
- N. Lj. Stevanović, J. Kljun, I. Aleksic, S. Skaro Bogojevic, D. Milivojevic, A. Veselinovic, I. Turel, M. I. Djuran, J. Nikodinovic-Runic and B. Đ. Glišić, Dalton Trans., 2022, 51, 5322–5334 RSC.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS PubMed.
- Y. Cortat, M. Nedyalkova, K. Schindler, P. Kadakia, G. Demirci, S. Nasiri Sovari, A. Crochet, S. Salentinig, M. Lattuada, O. M. Steiner and F. Zobi, Antibiotics, 2023, 12, 619 CrossRef CAS PubMed.
- Y.-H. Ou, R.-K. Du, S.-P. Zhang, Y. Ling, S. Li, C.-J. Zhao, W.-Z. Zhang and L. Zhang, J. Mol. Struct., 2020, 1215, 128229 CrossRef CAS.
- Q. Li, T. Wu, J.-C. Lai, Z.-L. Fan, W.-Q. Zhang, G.-F. Zhang, D. Cui and Z.-W. Gao, Eur. J. Inorg. Chem., 2015, 2015, 5281–5290 CrossRef CAS.
- Q.-L. Ren, S.-S. Zhao, L.-X. Song, S.-S. Qian and J. Qin, J. Coord. Chem., 2016, 69, 227–237 CrossRef CAS.
- N. D. Savić, S. Vojnovic, B. Đ. Glišić, A. Crochet, A. Pavic, G. V. Janjić, M. Pekmezović, I. M. Opsenica, K. M. Fromm, J. Nikodinovic-Runic and M. I. Djuran, Eur. J. Med. Chem., 2018, 156, 760–773 CrossRef PubMed.
- A. B. D. Nandiyanto, R. Oktiani and R. Ragadhita, Indones. J. Sci. Technol., 2019, 4, 97–118 CrossRef.
- J.-A. Zhang, M. Pan, J.-Y. Zhang, H.-K. Zhang, Z.-J. Fan, B.-S. Kang and C.-Y. Su, Polyhedron, 2009, 28, 145–149 CrossRef CAS.
- Y. Jiang, C.-F. Zhu, Z. Zheng, J.-B. He and Y. Wang, Inorg. Chim. Acta, 2016, 451, 143–147 CrossRef CAS.
- S. Medici, M. Peana, V. M. Nurchi and M. A. Zoroddu, J. Med. Chem., 2019, 62, 5923–5943 CrossRef CAS PubMed.
- J. Kujawski, K. Czaja, E. Jodłowska, K. Dettlaff, M. Politańska, J. Żwawiak, R. Kujawski, T. Ratajczak, M. K. Chmielewski and M. K. Bernard, J. Mol. Struct., 2016, 1119, 250–258 CrossRef CAS.
- W. Liu, K. Bensdorf, A. Hagenbach, U. Abram, B. Niu, A. Mariappan and R. Gust, Eur. J. Med. Chem., 2011, 46, 5927–5934 CrossRef CAS PubMed.
- U. Kalinowska-Lis, A. Felczak, L. Chęcińska, M. Małecka, K. Lisowska and J. Ochocki, New J. Chem., 2016, 40, 694–704 RSC.
- N. Stevanović, M. Zlatar, I. Novaković, A. Pevec, D. Radanović, I. Z. Matić, M. Đorđić Crnogorac, T. Stanojković, M. Vujčić, M. Gruden, D. Sladić, K. Anđelković, I. Turel and B. Čobeljić, Dalton Trans., 2021, 51, 185–196 RSC.
- T. P. Andrejević, I. Aleksic, J. Kljun, M. Počkaj, M. Zlatar, S. Vojnovic, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Glišić, RSC Adv., 2023, 13, 4376–4393 RSC.
- M. Baranowska, K. Suliborska, W. Chrzanowski, B. Kusznierewicz, J. Namieśnik and A. Bartoszek, Redox Biol., 2018, 17, 355–366 CrossRef CAS PubMed.
- T. P. Andrejević, D. Milivojevic, B. Đ. Glišić, J. Kljun, N. Lj. Stevanović, S. Vojnovic, S. Medic, J. Nikodinovic-Runic, I. Turel and M. I. Djuran, Dalton Trans., 2020, 49, 6084–6096 RSC.
- J.-Y. Wu, Y.-L. Pan, X.-J. Zhang, T. Sun, Y.-P. Tian, J.-X. Yang and Z.-N. Chen, Inorg. Chim. Acta, 2007, 360, 2083–2091 CrossRef CAS.
- P. Połczyński, R. Jurczakowski and W. Grochala, J. Phys. Chem. C, 2013, 117, 20689–20696 CrossRef.
- K. D. Trotter, O. Owojaiye, S. P. Meredith, P. E. Keating, M. D. Spicer, J. Reglinski and C. M. Spickett, BioMetals, 2019, 32, 627–640 CrossRef CAS PubMed.
- J. H. B. Nunes, D. H. Nakahata, P. P. Corbi and R. E. F. de Paiva, Coord. Chem. Rev., 2023, 490, 215228 CrossRef.
- T. Bruna, F. Maldonado-Bravo, P. Jara and N. Caro, Int. J. Mol. Sci., 2021, 22, 7202 CrossRef CAS PubMed.
- D. Żyro, L. Radko, A. Śliwińska, L. Chęcińska, J. Kusz, I. Korona-Głowniak, A. Przekora, M. Wójcik, A. Posyniak and J. Ochocki, Cancers, 2022, 14, 900 CrossRef PubMed.
- N. A. Johnson, M. R. Southerland and W. J. Youngs, Molecules, 2017, 28, 1263 CrossRef PubMed.
- S. Medici, M. Peana, V. M. Nurchi, J. I. Lachowicz, G. Crisponi and M. A. Zoroddu, Coord. Chem. Rev., 2015, 284, 329–350 CrossRef CAS.
- S. Medici, M. Peana, G. Crisponi, V. M. Nurchi, J. I. Lachowicz, M. Remelli and M. A. Zoroddu, Coord. Chem. Rev., 2016, 327–328, 349–359 CrossRef CAS.
- N. D. Savić, B. B. Petković, S. Vojnovic, M. Mojicevic, H. Wadepohl, K. Olaifa, E. Marsili, J. Nikodinovic-Runic, M. I. Djuran and B. Đ. Glišić, Dalton Trans., 2020, 49, 10880–10894 RSC.
- A. G. Warrilow, J. E. Parker, D. E. Kelly and S. L. Kelly, Antimicrob. Agents Chemother., 2013, 57, 1352–1360 CrossRef CAS PubMed.
- J. A. Romo, C. G. Pierce, A. K. Chaturvedi, A. L. Lazzell, S. F. McHardy, S. P. Saville and J. L. Lopez-Ribot, mBio, 2017, 8, e01991–e01917 CrossRef CAS PubMed.
- T. P. Andrejević, I. Aleksic, J. Kljun, M. Počkaj, M. Zlatar, S. Vojnovic, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Đ. Glišić, RSC Adv., 2023, 13, 4376–4393 RSC.
- H. T. Taff, K. F. Mitchell, J. A. Edward and D. R. Andes, Future Microbiol., 2013, 8, 1325 CrossRef CAS PubMed.
- F. Saleh, A. Harb, N. Soudani and H. Zaraket, J. Infect. Public Health, 2020, 13, 1142–1147 CrossRef PubMed.
- A. J. Carterson, K. Höner zu Bentrup, C. M. Ott, M. S. Clarke, D. L. Pierson, C. R. Vanderburg, K. L. Buchanan, C. A. Nickerson and M. J. Schurr, Infect. Immun., 2005, 73, 1129–1140 CrossRef CAS PubMed.
- K. S. dos Santos, L. T. Oliveira, M. de Lima Fontes, K. F. Migliato, A. M. Fusco-Almeida, M. J. S. M. Giannini and A. Moroz, J. Fungi, 2023, 9, 634 CrossRef CAS PubMed.
- K. E. de Visser, A. Eichten and L. M. Coussens, Nat. Rev. Cancer, 2006, 6, 24–37 CrossRef CAS PubMed.
- J. Kljun, I. E. Leon, Š. Peršič, J. F. Cadavid-Vargas, S. B. Etcheverry, W. He, Y. Bai and I. Turel, J. Inorg. Biochem., 2018, 186, 187 CrossRef CAS PubMed.
- CrysAlis PRO, Oxford Diffraction Ltd, Yarnton, Oxfordshire, England, 2011 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- G. M. Sheldrick, Shelxl 2018/3, University of Göttingen, Germany, 2018 Search PubMed.
- C. F. Macrae, P. R. Edgington, P. McCabe, E. Pidcock, G. P. Shields, R. Taylor, M. Towler and J. Van De Streek, J. Appl. Crystallogr., 2006, 39, 453–457 CrossRef CAS.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. Fonseca Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- R. Rüger, M. Franchini, T. Trnka, A. Yakovlev, E. van Lenthe, P. Philipsen, T. van Vuren, B. Klumpers and T. Soini, AMS 2023.1, SCM, Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, https://www.scm.com.
- E. van Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1993, 99, 4597–4610 CrossRef CAS.
- E. van Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1994, 101, 9783–9792 CrossRef CAS.
- C. van Wüllen, J. Chem. Phys., 1998, 109, 392–399 CrossRef.
- A. Klamt and G. Schoermann, J. Chem. Soc., Perkin Trans. 2, 1993, 799–805 RSC.
- A. Klamt, J. Phys. Chem., 1995, 99, 2224–2235 CrossRef CAS.
- C. C. Pye and T. Ziegler, Theor. Chem. Acc., 1999, 101, 396–408 Search PubMed.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098–3100 CrossRef CAS PubMed.
- J. P. Perdew, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 33, 8822–8824 CrossRef PubMed.
- J. P. Perdew, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 34, 7406–7406 CrossRef PubMed.
- E. Caldeweyher, S. Ehlert, A. Hansen, H. Neugebauer, S. Spicher, C. Bannwarth and S. Grimme, J. Chem. Phys., 2019, 150, 154122 CrossRef PubMed.
- Y. P. Li, J. Gomes, S. M. Sharada, A. T. Bell and M. Head-Gordon, J. Phys. Chem. C, 2015, 119, 1840–1850 CrossRef CAS.
- S. Grimme, Chem. – Eur. J., 2012, 18, 9955–9964 CrossRef CAS PubMed.
- H. S. Yu, X. He, S. L. Li and D. G. Truhlar, Chem. Sci., 2016, 7, 5032–5051 RSC.
- J. Tao, J. Perdew, V. Staroverov and G. Scuseria, Phys. Rev. Lett., 2003, 91, 146401 CrossRef PubMed.
- V. N. Staroverov, G. E. Scuseria, J. Tao and J. P. Perdew, J. Chem. Phys., 2003, 119, 12129 CrossRef CAS.
- S. Lehtola, C. Steigemann, M. J. T. Oliveira and M. A. L. Marques, SoftwareX, 2018, 7, 1–5 CrossRef.
- M. C. Arendrup, M. Cuenca-Estrella, C. Lass-Flörl and W. Hope, EUCAST–AFST, Clin. Microbiol. Infect., 2012, 18, E246–E247 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- B. A. Arthington-Skaggs, H. Jradi, T. Desai and C. J. Morrison, J. Clin. Microbiol., 1999, 39, 3332–3337 CrossRef PubMed.
- H. Liu, J. Köhler and G. R. Fink, Science, 1994, 266, 1723–1726 CrossRef CAS PubMed.
- C. G. Pierce, P. Uppuluri, A. R. Tristan, F. L. Wormley Jr., E. Mowat, G. Ramage and J. L. Lopez-Ribot, Nat. Protoc., 2008, 3, 1494–1500 CrossRef CAS PubMed.
- A. Jakab, S. Mogavero, T. M. Forster, M. Pekmezovic, N. Jablonowski, V. Dombradi, I. Pocsi and B. Hube, Microbiology, 2016, 162, 2116–2125 CrossRef CAS PubMed.
- B. Wächtler, D. Wilson, K. Haedicke, F. Dalle and B. Hube, PLoS One, 2011, 6, e17046 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S12 and Tables S1–S3. CCDC 2277941–2277943. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3dt03010e |
| This journal is © The Royal Society of Chemistry 2024 |