Open Access Article
Open Access ArticleHybrid-LLM-GNN: integrating large language models and graph neural networks for enhanced materials property prediction†
Youjia
Li
 *a,
Vishu
Gupta
*a,
Vishu
Gupta
 abc,
Muhammed Nur Talha
Kilic
abc,
Muhammed Nur Talha
Kilic
 d,
Kamal
Choudhary
d,
Kamal
Choudhary
 ef,
Daniel
Wines
ef,
Daniel
Wines
 e,
Wei-keng
Liao
a,
Alok
Choudhary
a and
Ankit
Agrawal
*a
e,
Wei-keng
Liao
a,
Alok
Choudhary
a and
Ankit
Agrawal
*a
aDepartment of Electrical and Computer Engineering, Northwestern University, Evanston, IL, USA. E-mail: youjia@northwestern.edu; ankit-agrawal@northwestern.edu
bLewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA
cLudwig Institute for Cancer Research, Princeton University, Princeton, NJ, USA
dDepartment of Computer Science, Northwestern University, Evanston, IL, USA
eMaterial Measurement Laboratory, National Institute of Standards and Technology, 100 Bureau Dr, Gaithersburg, MD, USA
fDeepMaterials LLC, Silver Spring, MD 20906, USA
First published on 18th December 2024
Abstract
Graph-centric learning has attracted significant interest in materials informatics. Accordingly, a family of graph-based machine learning models, primarily utilizing Graph Neural Networks (GNN), has been developed to provide accurate prediction of material properties. In recent years, Large Language Models (LLM) have revolutionized existing scientific workflows that process text representations, thanks to their exceptional ability to utilize extensive common knowledge for understanding semantics. With the help of automated text representation tools, fine-tuned LLMs have demonstrated competitive prediction accuracy as standalone predictors. In this paper, we propose to integrate the insights from GNNs and LLMs to enhance both prediction accuracy and model interpretability. Inspired by the feature-extraction-based transfer learning study for the GNN model, we introduce a novel framework that extracts and combines GNN and LLM embeddings to predict material properties. In this study, we employed ALIGNN as the GNN model and utilized BERT and MatBERT as the LLM model. We evaluated the proposed framework in cross-property scenarios using 7 properties. We find that the combined feature extraction approach using GNN and LLM outperforms the GNN-only approach in the majority of the cases with up to 25% improvement in accuracy. We conducted model explanation analysis through text erasure to interpret the model predictions by examining the contribution of different parts of the text representation.
1 Introduction
The accurate prediction of material properties from crystal structures is a fundamental aspect of materials science. As artificial intelligence plays an increasingly critical role in the fourth, data-driven paradigm of science,1,2 advanced data mining techniques powered by deep learning algorithms have been employed to make accurate predictions for material properties.3–12 Recently, deep learning approaches have been extended to work on graph-structured crystal structure representation, giving rise to a family of graph neural network architectures13–19 that achieve state-of-art performance in material property prediction. These GNN models provide a distinct advantage in predicting material properties by effectively encoding and utilizing the geometric information in the connections of atoms with bonds as edges.13 On top of this, the Atomistic Line Graph Neural Network (ALIGNN) model18 extends to another layer of abstraction, which represents inter-bond relationships as edges of a line graph. This innovative approach has enabled ALIGNN to surpass several other contemporary models, demonstrating its superiority in material property prediction.Despite the unique strength of GNN architecture, its reliability and accuracy are dependent on the size of available datasets. Although material datasets are regularly growing in size,20–22 there are still several material properties that are expensive to compute. To tackle the performance degradation challenges due to limited dataset size, Gupta et al.23–25 proposed a series of transfer learning (TL) frameworks that capture cross-property learnings from a source model trained with source property to improve the predictive ability for target property model with small datasets. Work in (ref. 23) applied two cross-property transfer learning strategies to the GNN-based model to learn structure-aware representations from the source property model. Firstly, a fine-tuning approach was explored by leveraging a pre-trained ALIGNN model for parameter initialization. Secondly, a feature-extraction-based TL approach was investigated by extracting embeddings from the knowledge model as features. The predictive model performance collected from diverse materials datasets demonstrated that the latter approach is better suited for small datasets.
Meanwhile, Large Language Models (LLMs), with their generality and transferability, offer an alternative solution for materials science knowledge discovery.26–28 In recent years, the rapid development of LLMs has led to a surge of revolutions for numerous text-related tasks in various domains.29–32 Their exceptional performance has incentivized researchers to apply them in structure–property relationship discovery. Particularly, pre-trained domain-specific language models have been proven to be effective in encapsulating latent knowledge embedded within domain literature.33,34 The combination of fine-tuning-based transfer learning and pre-trained domain-specific language models exhibits state-of-art performances in both property prediction tasks26,33,34 and material generation tasks.35 In the era of LLMs, there has been a growing focus on investigating the potential of LLMs to enhance the generalization, transferability, and few-shot learning capabilities of Graph Learning.36–40 However, in the context of crystal property prediction, there is less visibility of works that attempt to combine textual information extracted from natural language and structural-aware learnings from the aforementioned GNN model.
In this work, we present a novel framework that combines contextual word embeddings extracted from pre-trained LLMs into the structure-aware embeddings extracted from GNNs. This integration aims to combine the strengths of these two models to enhance both the predictive accuracy and model interpretability. The workflow comparison of the original GNN-based transfer learning and the proposed approach is shown in Fig. 1a. In the proposed workflow, we first reproduce the GNN embeddings as extracted structure-aware feature vectors. On a separate working thread, we employ pre-trained LLM models to generate contextual embedding for string representation of the same data samples. Lastly, we concatenate the embeddings from two sources and feed them to the data mining model to predict target material properties. With this concatenation, we aim to combine unique insights from natural language contexts with structural learnings represented in GNN layers. LLM embeddings can provide a deep understanding of text sequences, including nuanced semantic relationships, syntactic structures, and commonsense reasoning. These complementary learnings are anticipated to refine data sample representations for the downstream predictive model. In addition, by harnessing human-readable text inputs, LLM embeddings enable a direct mapping between the model's predictions and the string representation it operates on. This approach facilitates model interpretability by enabling tracing the impact of specific text representations on the model's outputs.
2 Model architectures
2.1 Hybrid-LLM-GNN feature extractor
The general workflow of the hybrid feature extractor is presented in Fig. 1b. We first start by reproducing the GNN-based embeddings as proposed by previous transfer learning work23 based on ALIGNN model trained on the formation energy of Materials Project (MP) dataset. Alongside the GNN-based feature extractor, the input crystal structure samples are processed by NLP-based text generation library to produce materials chemistry text description. We utilize two sources of text generators in this work: Robocystallographer41 and ChemNLP.42 The generated string representations are piped into the general Bidirectional Encoder Representations from Transformers (BERT)43 or domain-specific MatBERT44 model. To extract LLM embeddings, we process the text through the model and focus on the last layer of hidden states, which contains the most refined representations of each token. By averaging the embeddings of all tokens in the final layer, we obtain a single vector that encapsulates the overall context and meaning. Lastly, the embeddings from two different sources are concatenated to a fully-connected deep neural network for prediction.3 Results and analysis
3.1 Datasets
We use two datasets of density functional theory (DFT)-computed properties for different purposes in this work: Materials Project (MP)22 and Joint Automated Repository for Various Integrated Simulations45,46 (JARVIS).For the source GNN knowledge model, we use Formation Energy from the MP dataset. For the target property prediction, we use multiple DFT-computed properties from the 2022.12.12 version of JARVIS-3D dataset, which consists of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 materials with properties including formation energies, energy above the hull, modified Becke Johnson potential (MBJ) bandgaps,47 spectroscopic limited maximum efficiency (SLME),48 magnetic moments, topological spin–orbit spillage49,50 and superconducting transition temperature.51 Across all properties, we use 80%
993 materials with properties including formation energies, energy above the hull, modified Becke Johnson potential (MBJ) bandgaps,47 spectroscopic limited maximum efficiency (SLME),48 magnetic moments, topological spin–orbit spillage49,50 and superconducting transition temperature.51 Across all properties, we use 80%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10%
10%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10% splits with random shuffling for training, validation and testing.
10% splits with random shuffling for training, validation and testing.
3.2 Model performance
Here, we present the performance of the proposed transfer learning model on 7 different target material properties within the JARVIS-3D dataset. Several factors can influence the performance of language models. First, the aforementioned sources of string representation create variations in text content and the level of understanding perceived by the LLM model. We compare and analyze the model performance difference between the two versions of string representations. Second, we can optionally select the domain-specific variant for the LLM encoder. We compare model performance results using BERT and MatBERT. The latter is trained using a large corpus of text from materials science scientific papers with Masked-language Modeling (MLM) objective.Table 1 indicates that the proposed transfer learning approach (ALIGNN-MatBERT-based TL) outperforms ALIGNN scratch models in 5/7 cases. When compared against ALIGNN embedding-only transfer learning approach, the proposed hybrid approach produces superior performance for all 7 properties. The results illustrate the advantage of using the proposed hybrid representation when the data size is small. We believe the combined feature representation has benefited from the information-dense embeddings from LLM model, which contains textual insights from the description. The comparison of pre-trained LLM models shows that MatBERT is leading in most cases by generating more informative word embeddings. This aligns with expectations because the generated text incorporates domain-specific knowledge, benefiting from MatBERT's specialization in better understanding materials science terminology and scientific reasoning. We further delve into the performance gain of combining GNN and LLM embeddings through a parity plot of DFT-calculated versus machine-learning-predicted bandgap property. As illustrated in Fig. 2, the successful elimination of extreme prediction errors in ALIGNN Scratch model contributes to the overall MAE performance gain.
| Property | Data size | XGBoost with matminer | ALIGNN scratch | ALIGNN-based TL | ALIGNN-BERT-based TL | ALIGNN-MatBERT-based TL | % error change | |||
|---|---|---|---|---|---|---|---|---|---|---|
| ChemNLP | Robo-crystallographer | ChemNLP | Robo-crystallographer | Against ALIGNN scratch | Against ALIGNN-based TL | |||||
| Formation energy (eV per atom) | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 799 799 |
0.0640 | 0.0297 | 0.0346 | 0.0347 | 0.0366 | 0.0345 | 0.0339 | 13.94 | −2.16 |
| E hull (eV per atom) | 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 926 926 |
0.0543 | 0.0332 | 0.0383 | 0.0365 | 0.0366 | 0.0359 | 0.0357 | 7.65 | −6.80 |
| Magout (μB) | 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212 212 |
0.5509 | 0.5246 | 0.4465 | 0.4115 | 0.4385 | 0.3932 | 0.4211 | −25.05 | −11.95 |
| BandgapmBJ (eV) | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556 556 |
0.304 | 0.2571 | 0.2771 | 0.2559 | 0.2523 | 0.2720 | 0.2516 | −2.13 | −9.21 |
| Spillage | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 301 301 |
0.3337 | 0.3344 | 0.3285 | 0.3210 | 0.3226 | 0.3215 | 0.3140 | −6.10 | −4.41 |
| SLME (%) | 9762 | 5.0917 | 4.7064 | 4.6516 | 4.8874 | 4.7128 | 4.6579 | 4.5634 | −3.04 | −1.90 |
| Tc_supercon (K) | 1054 | 2.7499 | 2.5144 | 2.2441 | 2.1469 | 2.0363 | 2.2055 | 2.1095 | −19.01 | −9.26 |
Text source comparison shows that Robocystallographer marginally outperforms ChemNLP in generating text descriptions as it leads in 10/14 cases. Here, we present the sample string representation of LiCeO2 in Fig. 4 for comparison of different text representations. At a high level, while both paragraphs from two different sources share a formal, technical style and neutral tone, the text from Robocystallographer is a more direct, conversational, and versatile language use, possibly making it more accessible and easier to comprehend. In contrast, the text from ChemNLP has a dense style and is more detailed and descriptive with a sheer amount of numerical property information. As a result, the concise and more natural language-like style of the former could possibly enhance the accuracy of prediction.
3.3 Model explanation
| Property | Robocrystallographer text | ChemNLP text | ||||
|---|---|---|---|---|---|---|
| ALIGNN-MatBERT-based TL | ALIGNN-based TL | MatBERT-based TL | ALIGNN-MatBERT-based TL | ALIGNN-based TL | MatBERT-based TL | |
| Formation energy (eV per atom) | 0.0339 | 0.0346 | 0.0871 | 0.0345 | 0.0346 | 0.1018 |
| E hull (eV per atom) | 0.0357 | 0.0383 | 0.0601 | 0.0359 | 0.0383 | 0.0683 |
| Magout (μB) | 0.4211 | 0.4465 | 0.5571 | 0.3932 | 0.4465 | 0.5712 |
| BandgapmBJ (eV) | 0.2516 | 0.2771 | 0.3598 | 0.2720 | 0.2771 | 0.4012 |
| Spillage | 0.3140 | 0.3285 | 0.3507 | 0.3215 | 0.3285 | 0.3523 |
| SLME (%) | 4.5634 | 4.6516 | 5.3658 | 4.6579 | 4.6516 | 6.0009 |
| Tc_supercon (K) | 2.1095 | 2.2441 | 2.5879 | 2.2055 | 2.2441 | 2.3506 |
The first text-based model explanation analysis we consider is the word-level rationale extracted from string representations. In the regression task setting, this can be done by masking the target word or token at the inference stage and measuring the significance of predicted property change. Fig. 3 illustrates this approach, showing that the bond length value (2.50) is the most influential word among all candidates for this particular crystal sample.
Despite these insights, the above model interpretation has clear limitations in both applicability and effectiveness. As the string representation varies in length and vocabulary across samples, word-level analysis is limited to individual samples and cannot be generalized to the entire dataset. Furthermore, in a regression setting, the impact of masking a particular word from one sample is not directional, making it unclear whether the model's performance is improved or degraded. Therefore, we also perform a second model interpretation analysis. At the sentence level, both text generations from two sources are well-organized across all samples, allowing for the systematic removal of specific descriptive sentences from the entire dataset to measure the impact on prediction performance.
To facilitate text-based removal, we tag sentences in generated descriptions based on the textual information contained as illustrated in Fig. 4. The text generated by Robocystallographer starts with an opening introduction sentence (tagged as [summary]) that states the material's crystallization and space group. Then, for each element, it describes the number of primary atomic sites for multi-site elements (only present in multi-site samples, tagged as [site info]). Then the description iterates through each atomic site and describes the bonding environment and geometric arrangement for each site ([tagged as structure coordination]). Following this, it includes the measurement of bond distances (tagged as [bond length]) and bond angles (present only in some samples, tagged as [bond angle]). On the other hand, the text sourced from ChemNLP begins with the chemical information (tagged as [chemical info]), which details chemical properties including formula and atomic fractions. Then it introduces the structure information (tagged as [structure info]), detailing the lattice parameters, space group, top X-ray diffraction (XRD) peaks, material density, crystallization system, point group and Wyckoff positions. Finally, the bond lengths (tagged as [bond length]) are included for every atomic pair present in the structure.
In the family of erasure-based explainable AI (XAI) techniques25,54–57 for NLP tasks, rationale comprehensiveness54 provides a theoretical framework for classification tasks by measuring the decrease in model confidence in the correct prediction when the tokens comprising the provided rationale are erased. Here, we extend the concept of rational comprehensiveness to the regression problem setting by measuring the MAE increase with the removal of the target subregion of text across all samples. We systematically categorize string descriptions into 5 tags based on the content across all samples. Then we can measure the comprehensiveness of each tag by constructing a contrast text representation dataset for original text data T, T/ti, which is the original text dataset T with tag ti removed from all samples. At the testing stage, two versions of the text dataset will be piped into the trained ALIGNN-MatBERT-based TL forward model. We can then calculate the comprehensiveness of tag ti as the MAE difference between the original full-text representation and the erased version using formula (1):
| Comprehensivenessi = MAE(T/ti) − MAE(T) | (1) |
A high comprehensiveness score here implies that the tagged text significantly influenced the prediction, whereas a low score suggests the opposite.
Results from model explanation analysis emphasize the importance of structure information, particularly structural coordination descriptions. During the analysis, the model is not retrained, and only the input text representation is adjusted at the inference stage. The MAE values after removal for each property are collected in Fig. 5. As shown in Fig. 4 and 5, for Robocrystallographer text, the removal of each tag causes a varied level of degradation in prediction performance. The most impactful tag observed is structure coordination across all properties. Bond lengths and summary tags are ranked in the second tier of impactful tags. Similarly, for ChemNLP text, structure information has the greatest impact on performance gain with a drastic MAE increase (114.59%) observed in the prediction of energy above hull (Ehull) property. One divergence observed in the two text representation sources is the significance of the bond length tag. For ChemNLP text, the bond distance information has a negligible impact on the MAE changes. This can be attributed to the different textual representations of bond information from the two text sources. The bond length description by Robocystallographer follows a more logical sequence and a more natural language-like style, which turns out to be favored by the downstream LLM.
4 Discussion
In this work, we explore the potential of combining GNN embeddings and contextual word embeddings extracted from LLM in enhancing material property predictions. The ablation test for embeddings indicates that LLM embedding alone is not enough to catch up with the performance level of ALIGNN-based TL. However, the concatenation of ALIGNN embeddings and MatBERT embeddings further enhances the ALIGNN-based TL performance. This concatenation paves an effective and efficient way for combining structural learnings and textual learnings in forward predictive models.Our model explanation analysis takes advantage of the natural-language interface provided by the text representation of crystal structures. The presented erasure-based analysis is an illustration of interpreting model predictions by relating performance to a human-readable text representation. Additionally, the results from the model explanation analysis emphasize the importance of structural information. This suggests that despite the dense structural learnings from the pre-trained GNN source model, there are still complementary structural learnings in the text format that remain untapped. When we compare the structural descriptions from the generated text with the ALIGNN model input features, we find overlapping structural properties, such as bond distances, which are directly encoded in the GNN model input, as well as other structural insights that are either missing or indirectly encoded in the GNN model input, such as crystal system. Therefore, the additional structural learnings can either be sourced from the enhanced representation of existing structural feature properties or new structural information that is uniquely present in text descriptions.
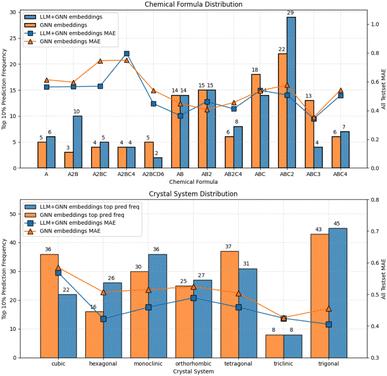
To further explore the impact of incorporating LLM embeddings into the transfer learning pipeline, we analyzed the test dataset performance categorized by crystal system and composition prototype. Using bandgap as a representative target property, we plotted the distribution of the top 10% accurate predictions and the overall MAE level in Fig. 6. The bar plot shows the distribution of the top 10% accurate predictions for each crystal system type or chemical composition prototype, while the line plot represents the overall MAE level across the entire test set. We compared predictions from ALIGNN-MatBERT-based embeddings against those from ALIGNN-embeddings-only. Looking at the MAE values grouped by composition prototypes, we find that LLM embeddings improved predictions for A2BC, AB and A2BCD6 prototypes. The top 10% accurate predictions highlights the samples for which the model performs well. The comparison of the composition prototype distribution reveals a shift in the model's predictive strengths, with an increased frequency for ABC2 and A2B prototypes and a decreased frequency for ABC and ABC3 prototypes. For crystal systems, hexagonal and monoclinic systems shows the most significant improvements by LLM embeddings, evidenced by both a higher frequency of top 10% predictions and a lower overall MAE.
5 Future works
By combining GNN and LLM for material property prediction, our research shows that feature-extraction-based transfer learning is a viable and effective approach for such an integration. Despite its efficacy and simplicity, this solution has some limitations. The concatenation of embeddings establishes a tenuous connection between graphs and text representations. The inserted LLM embeddings lack contextual adaptation to the structural patterns learned by the GNN. There remain unexplored modeling efforts that can potentially provide a more systematic integration framework of GNN and LLM for materials science knowledge discovery. One promising direction is the recently proposed iterative structure58,59 that co-trains LLM and GNN by generating pseudo labels for each other.On a different note, the performance gain achieved with the domain-specific MatBERT over the general BERT model underscores the unique value of a domain-specific tokenizer for knowledge discovery in materials science. A domain-specific tokenizer tailored to the materials science field enhances text processing by accurately recognizing and tokenizing specialized vocabulary, technical terms, chemical formulas, and abbreviations unique to the discipline. To better mine insights from materials science literature, one promising direction is to develop domain-specific tokenization for the pre-training phase of LLMs.
Code availability
The codebase used in this study to extract and merge LLM embeddings is available at https://github.com/Jonathanlyj/ALIGNN-BERT-TL-crystal.Data availability
The datasets used in this paper are publicly available from the corresponding websites- MP4 from https://materialsproject.org/, JARVIS from https://jarvis.nist.gov.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was carried out with the support of the following financial assistance award 70NANB19H005 from U.S. Department of Commerce, National Institute of Standards and Technology as part of the Center for Hierarchical Materials Design (CHiMaD). Partial support is also acknowledged from NSF awards CMMI-2053929 and OAC-2331329, DOE award DE-SC0021399, and Northwestern Center for Nanocombinatorics. Certain equipment, instruments, software, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement of any product or service by NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.Notes and references
- A. Agrawal and A. Choudhary, APL Mater., 2016, 4, 5 CrossRef.
- V. Gupta, W.-k. Liao, A. Choudhary and A. Agrawal, MRS Commun., 2023, 13, 754–763 CrossRef.
- V. Gupta, A. Peltekian, W.-k. Liao, A. Choudhary and A. Agrawal, Sci. Rep., 2023, 13, 9128 CrossRef PubMed.
- G. Pilania, Comput. Mater. Sci., 2021, 193, 110360 CrossRef.
- D. Jha, V. Gupta, W.-k. Liao, A. Choudhary and A. Agrawal, Sci. Rep., 2022, 12, 11953 CrossRef PubMed.
- J. Westermayr, M. Gastegger, K. T. Schütt and R. J. Maurer, J. Chem. Phys., 2021, 154, 230903 CrossRef PubMed.
- A. Mannodi-Kanakkithodi and M. K. Chan, Trends Chem., 2021, 3, 79–82 CrossRef.
- V. Gupta, Y. Li, A. Peltekian, M. N. T. Kilic, W.-k. Liao, A. Choudhary and A. Agrawal, J. Cheminf., 2024, 16, 1–13 Search PubMed.
- P. Friederich, F. Häse, J. Proppe and A. Aspuru-Guzik, Nat. Mater., 2021, 20, 750–761 CrossRef PubMed.
- V. Gupta, K. Choudhary, Y. Mao, K. Wang, F. Tavazza, C. Campbell, W.-k. Liao, A. Choudhary and A. Agrawal, J. Chem. Inf. Model., 2023, 63, 1865–1871 CrossRef.
- R. Pollice, G. dos Passos Gomes, M. Aldeghi, R. J. Hickman, M. Krenn, C. Lavigne, M. Lindner-D'Addario, A. Nigam, C. T. Ser and Z. Yao, et al. , Acc. Chem. Res., 2021, 54, 849–860 CrossRef.
- V. Gupta, W.-k. Liao, A. Choudhary and A. Agrawal, Proceedings of the 2022 SIAM international conference on data mining (SDM), 2022, pp. 343–351 Search PubMed.
- T. Xie and J. C. Grossman, Phys. Rev. Lett., 2018, 120, 145301 CrossRef PubMed.
- K. Schütt, P.-J. Kindermans, H. E. Sauceda Felix, S. Chmiela, A. Tkatchenko and K.-R. Müller, Advances in neural information processing systems, 2017, vol. 30 Search PubMed.
- C. Chen, W. Ye, Y. Zuo, C. Zheng and S. P. Ong, Chem. Mater., 2019, 31, 3564–3572 CrossRef.
- J. Gasteiger, S. Giri, J. T. Margraf and S. Günnemann, arXiv, 2020, preprint, arXiv:2011.14115, DOI:10.48550/arXiv.2011.14115.
- C. W. Park and C. Wolverton, Phys. Rev. Mater., 2020, 4, 063801 CrossRef.
- K. Choudhary and B. DeCost, npj Comput. Mater., 2021, 7, 185 CrossRef.
- Z. Qiao, M. Welborn, A. Anandkumar, F. R. Manby and T. F. Miller, J. Chem. Phys., 2020, 153, 124111 CrossRef.
- S. Curtarolo, G. L. Hart, M. B. Nardelli, N. Mingo, S. Sanvito and O. Levy, Nat. Mater., 2013, 12, 191–201 CrossRef PubMed.
- J. E. Saal, S. Kirklin, M. Aykol, B. Meredig and C. Wolverton, JOM, 2013, 65, 1501–1509 CrossRef.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner and G. Ceder, et al. , APL Mater., 2013, 011002 CrossRef.
- V. Gupta, K. Choudhary, B. DeCost, F. Tavazza, C. Campbell, W.-k. Liao, A. Choudhary and A. Agrawal, npj Comput. Mater., 2024, 10, 1 CrossRef.
- V. Gupta, K. Choudhary, F. Tavazza, C. Campbell, W.-k. Liao, A. Choudhary and A. Agrawal, Nat. Commun., 2021, 12, 6595 CrossRef PubMed.
- V. Gupta, W.-k. Liao, A. Choudhary and A. Agrawal, International Joint Conference on Neural Networks (IJCNN), 2023, pp. 1–8 Search PubMed.
- A. N. Rubungo, C. Arnold, B. P. Rand and A. B. Dieng, arXiv, 2023, preprint, arXiv:2310.14029, DOI:10.48550/arXiv.2310.14029.
- K. Choudhary, J. Phys. Chem. Lett., 2024, 15, 6909–6917 CrossRef.
- G. Lei, R. Docherty and S. J. Cooper, Digital Discovery, 2024, 3(7), 1249–1442 RSC.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Nature, 2023, 624, 570–578 CrossRef PubMed.
- N. J. Szymanski, B. Rendy, Y. Fei, R. E. Kumar, T. He, D. Milsted, M. J. McDermott, M. Gallant, E. D. Cubuk and A. Merchant, et al. , Nature, 2023, 624, 86–91 CrossRef.
- Z. Lin, H. Akin, R. Rao, B. Hie, Z. Zhu, W. Lu, N. Smetanin, R. Verkuil, O. Kabeli and Y. Shmueli, et al. , Science, 2023, 379, 1123–1130 CrossRef.
- R. Luo, L. Sun, Y. Xia, T. Qin, S. Zhang, H. Poon and T.-Y. Liu, Briefings Bioinf., 2022, 23, bbac409 CrossRef.
- V. Korolev and P. Protsenko, Patterns, 2023, 4, 100803 CrossRef.
- T. Xie, Y. Wan, K. Lu, W. Zhang, C. Kit and B. Hoex, AI for Accelerated Materials Design-NeurIPS 2023 Workshop, 2023 Search PubMed.
- N. Gruver, A. Sriram, A. Madotto, A. G. Wilson, C. L. Zitnick and Z. Ulissi, arXiv, 2024, preprint, arXiv:2402.04379, DOI:10.48550/arXiv.2402.04379.
- W. Fan, S. Wang, J. Huang, Z. Chen, Y. Song, W. Tang, H. Mao, H. Liu, X. Liu, D. Yin, et al., arXiv, 2024, preprint, arXiv:2404.14928, DOI:10.48550/arXiv.2404.14928.
- Y. Shi, A. Zhang, E. Zhang, Z. Liu and X. Wang, arXiv, 2023, preprint, arXiv:2310.13590, DOI:10.48550/arXiv.2310.13590.
- Z. Guo, L. Xia, Y. Yu, Y. Wang, Z. Yang, W. Wei, L. Pang, T.-S. Chua and C. Huang, arXiv, 2024, preprint, arXiv:2402.15183, DOI:10.48550/arXiv.2402.15183.
- Z. Chai, T. Zhang, L. Wu, K. Han, X. Hu, X. Huang and Y. Yang, arXiv, 2023, preprint, arXiv:2310.05845, DOI:10.48550/arXiv.2310.05845.
- Y. Liang, R. Zhang, L. Zhang and P. Xie, arXiv, 2023, preprint, arXiv:2309.03907, DOI:10.48550/arXiv.2309.03907.
- A. M. Ganose and A. Jain, MRS Commun., 2019, 9, 874–881 CrossRef.
- K. Choudhary and M. L. Kelley, J. Phys. Chem. C, 2023, 127, 17545–17555 CrossRef.
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, arXiv, 2018, preprint, arXiv:1810.04805, DOI:10.48550/arXiv.1810.04805.
- N. Walker, A. Trewartha, H. Huo, S. Lee, K. Cruse, J. Dagdelen, A. Dunn, K. Persson, G. Ceder and A. Jain, Patterns, 2022, 3, 100488 CrossRef PubMed.
- K. Choudhary, K. F. Garrity, A. C. Reid, B. DeCost, A. J. Biacchi, A. R. H. Walker, Z. Trautt, J. Hattrick-Simpers, A. G. Kusne, A. Centrone, et al., arXiv, 2020, preprint, arXiv:2007.01831, DOI:10.48550/arXiv.2007.01831.
- D. Wines, R. Gurunathan, K. F. Garrity, B. DeCost, A. J. Biacchi, F. Tavazza and K. Choudhary, Appl. Phys. Rev., 2023, 10, 041302 Search PubMed.
- K. Choudhary, Q. Zhang, A. C. Reid, S. Chowdhury, N. Van Nguyen, Z. Trautt, M. W. Newrock, F. Y. Congo and F. Tavazza, Sci. Data, 2018, 5, 1–12 CrossRef PubMed.
- K. Choudhary, M. Bercx, J. Jiang, R. Pachter, D. Lamoen and F. Tavazza, Chem. Mater., 2019, 31, 5900–5908 CrossRef PubMed.
- K. Choudhary, K. F. Garrity and F. Tavazza, Sci. Rep., 2019, 9, 8534 CrossRef PubMed.
- K. Choudhary, K. F. Garrity, N. J. Ghimire, N. Anand and F. Tavazza, Phys. Rev. B, 2021, 103, 155131 CrossRef CAS.
- K. Choudhary and K. Garrity, npj Comput. Mater., 2022, 8, 244 CrossRef CAS.
- H. Kaur, H. Nori, S. Jenkins, R. Caruana, H. Wallach and J. Wortman Vaughan, Proceedings of the 2020 CHI conference on human factors in computing systems, 2020, pp. 1–14 Search PubMed.
- D. S. Weld and G. Bansal, Commun. ACM, 2019, 62, 70–79 CrossRef.
- J. DeYoung, S. Jain, N. F. Rajani, E. Lehman, C. Xiong, R. Socher and B. C. Wallace, arXiv, 2019, preprint, arXiv:1911.03429, DOI:10.48550/arXiv.1911.03429.
- Z. Chen, C. Jiang and K. Tu, arXiv, 2024, preprint, arXiv:2404.02068, DOI:10.48550/arXiv.2404.02068.
- S. Kim, J. Yi, E. Kim and S. Yoon, arXiv, 2024, preprint, arXiv:2010.13984, DOI:10.48550/arXiv.2010.13984.
- S. Feng, E. Wallace, A. Grissom II, M. Iyyer, P. Rodriguez and J. Boyd-Graber, arXiv, 2018, preprint, arXiv:1804.07781, DOI:10.48550/arXiv.1804.07781.
- J. Yang, Z. Liu, S. Xiao, C. Li, D. Lian, S. Agrawal, A. Singh, G. Sun and X. Xie, Advances in Neural Information Processing Systems, 2021, vol. 34, pp. 28798–28810 Search PubMed.
- J. Zhao, M. Qu, C. Li, H. Yan, Q. Liu, R. Li, X. Xie and J. Tang, arXiv, 2022, preprint, arXiv:2210.14709, DOI:10.48550/arXiv.2210.14709.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00199k |
| This journal is © The Royal Society of Chemistry 2025 |