Open Access Article
Open Access ArticleChemspyd: an open-source python interface for Chemspeed robotic chemistry and materials platforms†
Martin
Seifrid‡
 abc,
Felix
Strieth-Kalthoff‡
abc,
Felix
Strieth-Kalthoff‡
 bc,
Mohammad
Haddadnia
bc,
Mohammad
Haddadnia
 bd,
Tony C.
Wu
bc,
Emre
Alca§
b,
Leticia
Bodo
b,
Sebastian
Arellano-Rubach
e,
Naruki
Yoshikawa
bd,
Tony C.
Wu
bc,
Emre
Alca§
b,
Leticia
Bodo
b,
Sebastian
Arellano-Rubach
e,
Naruki
Yoshikawa
 cd,
Marta
Skreta
cd,
Rachel
Keunen
bf and
Alán
Aspuru-Guzik
*bcdfghi
cd,
Marta
Skreta
cd,
Rachel
Keunen
bf and
Alán
Aspuru-Guzik
*bcdfghi
aDepartment of Materials Science and Engineering, North Carolina State University, Raleigh, NC, USA
bDepartment of Chemistry, University of Toronto, Toronto, Ontario M5S 3H6, Canada. E-mail: alan@aspuru.com
cDepartment of Computer Science, University of Toronto, Toronto, ON M5S 3H6, Canada
dVector Institute for Artificial Intelligence, Toronto, ON M5S 1M1, Canada
eUniversity of Toronto Schools, Toronto, ON M5S 2R7, Canada
fAcceleration Consortium, University of Toronto, Toronto, Ontario M5S 3H6, Canada
gDepartment of Chemical Engineering & Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada
hDepartment of Materials Science, University of Toronto, Toronto, Ontario M5S 3E4, Canada
iLebovic Fellow, Canadian Institute for Advanced Research, Toronto, ON M5S 1M1, Canada
First published on 22nd May 2024
Abstract
We introduce Chemspyd, a lightweight, open-source Python package for operating the popular laboratory robotic platforms from Chemspeed Technologies. As an add-on to the existing proprietary software suite, Chemspyd enables dynamic communication with the automated platform, laying the foundation for its modular integration into customizable, higher-level laboratory workflows. We show the applicability of Chemspyd in a set of case studies from chemistry and materials science. We demonstrate how the package can be used with large language models to provide a natural language interface. By providing an open-source software interface for a commercial robotic platform, we hope to inspire the development of open interfaces that facilitate the flexible, adaptive integration of existing laboratory equipment into automated laboratories.
Introduction
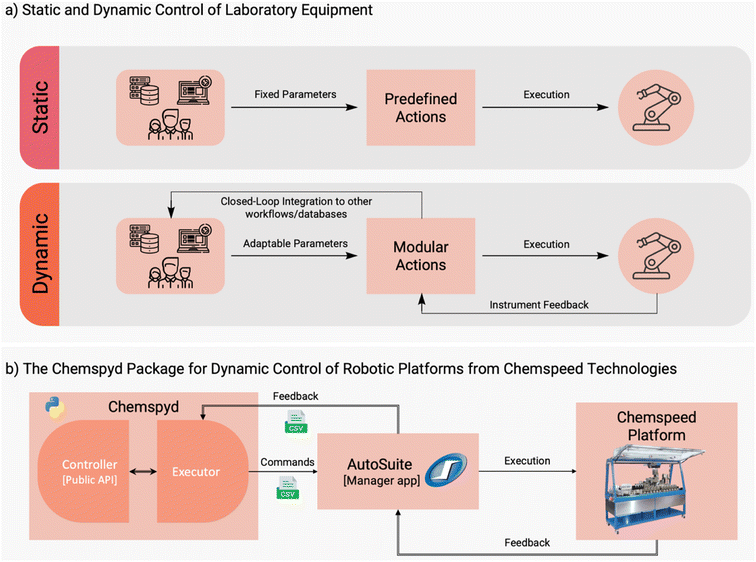
Laboratory automation has been identified as a key strategy for increasing the rate at which new discoveries are made in chemistry and materials science.1–6 Automation serves two central purposes: (1) to increase the experimental throughput via continuous and/or parallel execution of otherwise repetitive, manual tasks, and (2) to foster more standardized and reproducible results. While the history of automation in chemistry traces back to the mid-20th century,1 recent years have seen a “renaissance” of automation in both academic and industrial laboratories. Advances in robotics and engineering have enabled the automation of increasingly challenging laboratory operations such as thin-film fabrication,7 sample handling under inert gas,8,9 or dosing of powders, gels and slurries.10,11 Integrating such automated modules into larger workflows has demonstrated the potential to tackle increasingly complex scientific challenges in an automated fashion.12 This surge in automated experimentation has produced a growing market of instruments, particularly platform solutions consisting of multiple experimental modules. Arguably, the most prominent such systems have come from companies such as Chemspeed Technologies and Unchained Labs, and have shown the enormous potential to enable highly complex discovery workflows across various fields in chemistry and materials science. Examples include the discovery of battery electrolytes,13 new catalysts,14–16 organic laser materials,17,18 polymer formulations,19–21 or stereoselective synthesis.8The current phase in the evolution of automated laboratories involves the transition from static, pre-defined automation workflows to modular and flexible labs where decisions about the next experimental steps are adaptively made in real time (Fig. 1a).22–24 Particularly with recent strides in data-driven design and machine learning,25 this has the potential to optimize the use of automated resources, and thereby accelerate scientific discoveries. Especially against the background of modularity and adaptive decision making, the availability of open software interfaces (application programming interfaces, APIs) for automated platforms are essential for the seamless incorporation into flexible, customizable workflows.26,27 At the same time, such dynamic APIs are often not provided by instrument manufacturers, whose software tends to follow a workflow- and instrument-centric philosophy. In fact, available APIs are often constrained to the configuration and post-run evaluation of static workflows. This presents a major barrier to integrating further instruments into the workflow, or to making adaptive data-driven decisions in real time.
To address these gaps, we introduce Chemspyd, an open-source Python API specifically designed for Chemspeed platforms. Python has long been a foundational element of scientific computing and of automated laboratories, in particular. This API enables real-time, adaptive control of Chemspeed instruments by providing a simple interface through which Chemspeed instruments can be integrated with the rest of the scientific Python ecosystem. Users are able to integrate with other open source tools such as packages for optimization or design-of-experiment, other lab automation, online analysis, and in silico predictions. Ultimately, this empowers researchers to seamlessly integrate Chemspeed robots into custom workflows and automated or self-driving laboratories (SDLs). We use three experimental case studies to demonstrate how Chemspyd can be used for experiments in the chemical and materials sciences. Most importantly, Chemspyd is designed as a modular and expandable open-source project,28 and can therefore serve as a blueprint for the development of similar interfaces that meet the evolving demands of modern, flexible, and customizable automated laboratories.
“Under the hood”: the design of Chemspyd
Chemspyd's architecture is guided by three core design principles: (1) dynamic and fine-grained control over the robot's actions; (2) easy installation and usage with existing Chemspeed setups; (3) modular, extendable open-source architecture, facilitating continuous development by the community, and enabling effortless integration with experiment planning and scheduling workflows. Because of (2) and (3), Chemspyd comes as a lightweight Python package (i.e., easy to install and lacking heavy dependencies) that dynamically interacts with Chemspeed's proprietary AutoSuite software.Chemspyd is organized following object-oriented design principles and is structured into two main classes: the Controller and the Executor. Whereas the Executor handles the communication with the instrument's control software (for details, vide infra), the Controller provides a standardized, public API for users to develop customizable, adaptive workflows in Python. For this purpose, it houses an extensive catalog of elementary actions that encompass a wide range of the functionalities that the Chemspeed robotic platforms offer. These elementary actions enable dynamic and fine-grained control over the action space. A full list of elementary actions is provided in the ESI,† as well as the detailed package documentation.29
Chemspyd communicates with AutoSuite through the Executor, which reads and writes shared CSV files, providing a standardized means of communication that is human-readable and supported by both Python and AutoSuite (Fig. 1b). This enables bidirectional communication between AutoSuite (and thereby, the Chemspeed robotic platform) and Chemspyd, containing the instrument status, execution commands and parameters, instrument return values, and general metadata. A full description of the communication protocol is provided in the ESI.†
To enable dynamic control on the Chemspeed side, we created a dedicated AutoSuite application file, referred to as the Manager (see installation guide in the package documentation for file location), that listens for command files, and executes actions based on the provided keywords and parameters. Each elementary Controller method has an execution counterpart in the Manager. As a result, Chemspyd allows users to perform individual actions (helpful during development and troubleshooting) or perform different routines without needing to restart the platform.
Beyond the fine-grained control over elementary actions, we developed Chemspyd to contain a series of optional tools to assist with operation safety, accurate resource management, and standardized data collection. These safety checks include a simulation mode within Chemspyd that can be used to verify that the code will execute without generating internal Chemspyd errors prior to testing the operations in AutoSuite's own simulation mode, which validates other operational parameters. Together, these form a “digital twin” that enables users to simulate processes without the need to access a Chemspeed instrument. Chemspyd's resource management features also allow users to validate operations of workflows prior to execution to ensure that liquids or solids can be added or removed from the specified wells, and that the wells will not be overfilled or depleted. The required attributes of each well (type, volume, etc.) are automatically extracted from the instrument configuration, avoiding manual input by the user (see section “Installation and usage” for further details).
To streamline workflow development and enhance convenience for the user, we have organized common experimental routines within the routines sub-package. Examples of such routines include the priming of syringe pumps, evacuate-backfill cycles (i.e., “Schlenk cycles”), filtration and collection steps, and injection to on-deck HPLC ports. Notably, the routines sub-package provides a framework for implementing further custom experimental routines, highlighting the modular, open-source nature of Chemspyd, and fostering continued active development by the community.
Installation and usage
The Chemspyd Python package can be installed from the PyPI repository (Fig. 2a).30 The source code repository can be accessed at its GitLab page31 under the Apache 2.0 license, and provides extensive documentation,29 including installation instructions, usage guides and tutorial examples. Once installed, Chemspyd code can be written entirely in Python (versions ≥3.9), and, thus, enables users to developed and test their code on any platform.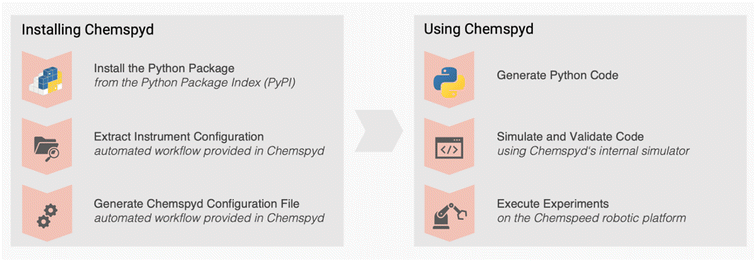 | ||
| Fig. 2 Installing and using the Chemspyd Python library. During initial installation, Chemspyd can automatically read the platform's configuration and convert it into the necessary configuration file. After installation, Chemspyd workflows can be coded and validated in Python before executing the code on the actual robotic platform. Code examples for this workflow can be found in Fig. S1.† | ||
The process of setting up Chemspyd on any local platform involves two stages: (1) creating a custom, local manager and (2) extracting the platform's hardware configuration. In the first stage, users should create a new manager application file in AutoSuite whose instrument configuration matches that of their platform. All pre-defined commands, which are provided as part of Chemspyd (see package documentation for further details) should be copied into this application file. Second, for extracting the hardware configuration from the manager and making it accessible to Chemspyd, we provide an automated solution to ease the installation process. For this purpose, Chemspyd interacts with AutoSuite's.NET API. For user convenience, this process is fully wrapped in the 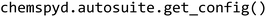 function (Fig. 2a, see package documentation for further details). As a result, the installation of Chemspyd is largely automated, and does not require a tedious configuration procedure, but is designed for the seamless integration with existing robotic setups. Should the API not be accessible, the resulting configuration file can also be created manually.
function (Fig. 2a, see package documentation for further details). As a result, the installation of Chemspyd is largely automated, and does not require a tedious configuration procedure, but is designed for the seamless integration with existing robotic setups. Should the API not be accessible, the resulting configuration file can also be created manually.
Once the Python package and the corresponding AutoSuite Manager have been properly set up, executing Chemspyd code on a Chemspeed platform requires the following two steps: (1) start the manager in AutoSuite, (2) execute one or multiple Chemspyd scripts, an example of which is shown in Fig. 2b.
Experimental use cases
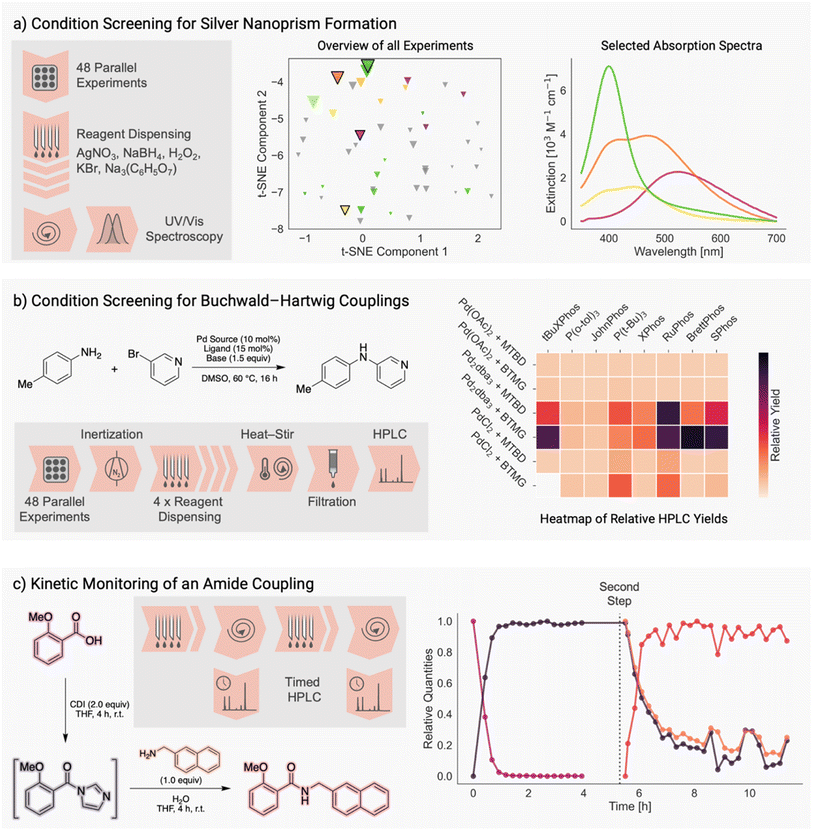
In order to showcase the different features of Chemspyd, we demonstrate a set of experiments from inorganic and organic chemistry as possible use cases of the software in automated laboratories. All experiments were performed on the Chemspeed SWING XL robot available in our laboratory at the Matter Lab at the University of Toronto.As a first use case, we performed a systematic evaluation of reaction conditions for the formation of silver nanoprisms.32–34 The size distribution of the nanoprisms – and thereby, their absorption properties – are determined by the stoichiometric ratios of the silver source (AgNO3), the reductive component (NaBH4), the oxidative component (H2O2), the buffer (sodium citrate) and the silver concentration mediator (KBr). We selected a representative set of conditions from this five-dimensional continuous parameter space through Latin hypercube sampling. Using Chemspyd, we were able to quickly write the execution code, simply looping over all hypercube samples, and the required liquid transfer and stirring operations were performed automatically. Optical absorption measurements were carried out on our spectroscopic characterization platform.17 The resulting dataset of spectroscopic properties of the obtained nanoprisms is shown in Fig. 3a.
Our second use case targeted the screening of experimental conditions for the Buchwald–Hartwig coupling reaction, one of the most prominent reaction classes in organic and medicinal chemistry.35 Specifically, we created a combinatorial dataset by varying three categorical parameters, namely the palladium precursor, ligand, and base.36 Exploiting our platform's capacity to perform reactions under an inert gas atmosphere, all synthesis (inertization, reagent addition, temperature control, vortex stirring), workup (filtration) and analysis (injection to an HPLC) were encoded in Chemspyd, and run without manual intervention. Notably, the modular design of Chemspyd was crucial for the software-level integration with our group's HPLC-MS instrument and its Python control code. Relative yields (with respect to an internal standard) for each reaction are visualized in the heatmap in Fig. 3b.
The dynamic nature of the communication between Chemspyd and the instrument is emphasized in a third experiment, in which we perform a two-step amide coupling with continuous reaction monitoring.37 Here, a Python script dynamically orchestrates the execution of the individual reaction steps, the timed preparation and derivatization of aliquots, and their injection to our HPLC system. At the same time, the script interacts with the HPLC instrument to ensure synchronization of both instrument operations. The kinetic traces of both reagents, the proposed intermediate, and the reaction product are shown in Fig. 3c, and are in good agreement with the traces obtained by Liu et al. in their dedicated reaction monitoring platform.37
Simplified adoption through a natural language interface
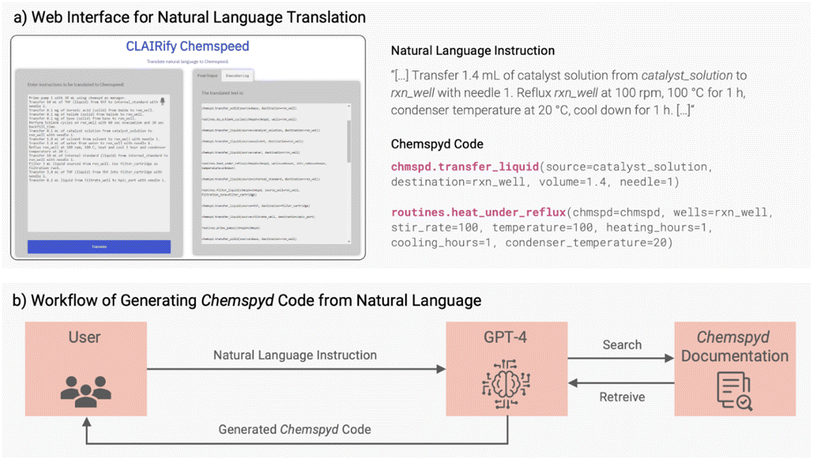
To further facilitate the adoption of Chemspyd and its rapid implementation into new laboratory routines, we provide a natural language interface for generating Chemspyd code based on iterative prompting of GPT-4. Analogous methods have recently proven to be powerful enabling technologies for automated or self-driving laboratories.38,39 For instance, ChemCrow, reported by Schwaller, White and co-workers,38 uses iterative large language model (LLM) prompting and tool integration to propose synthetic routes for organic molecules. Coscientist, recently described by Boiko et al.,39 uses a related approach and adds the ability to generate instrument operation code for automated systems. Notably, Chemspyd could readily be integrated into these types of systems. However, our natural language interface targets a slightly different use case: namely, facilitating adoption of automation tools by users who are not familiar with programming in Python, which is characteristic of a significant portion of the chemistry and materials science communities at the moment.Similar to our recent work,40 we provide a web interface that uses a LLM to convert the natural language inputs into structured Chemspyd output.41 In our implementation, all Chemspyd functions, along with their natural language documentation and all parameters, are organized in an associative array. We use a similar approach as Boiko et al.,39 demonstrating that it generalizes well to a domain-specific language unknown to GPT-4. Incoming natural language instructions are segmented into structured commands, which are then matched to the classes and functions in the associative array based on cosine similarity. Subsequently, OpenAI's GPT-4 (ref. 42) is employed to translate the instructions into the corresponding code. Command-by-command, each section of the generated Chemspyd code is sent back to the user for feedback and validation. This match-translate cycle is repeated iteratively until satisfactory Chemspyd code is reached (Fig. 4b). We maintain user feedback in the match-translate cycle because there are many cases where semantic errors in the generated code cannot be detected in simulation. As an example, if the natural language input specifies “add ethanol” and the generated code incorrectly adds methanol, the generated code does not reflect the intent of the user but the simulation may not yield errors. This kind of semantic error detection requires a high-level natural language understanding ability beyond simulation, and forms part of our future work.
Eventually, the outcome is a responsive interface that effectively bridges the gap between user intent and the correct Chemspyd code, showcasing the power of NLP in user–system interactions, and providing a useful tool for non-expert programmers to generate experiments with Chemspyd.
Summary and outlook
We have introduced Chemspyd as a simple, lightweight and easy-to-use Python API for Chemspeed platforms. In contrast to the existing graphical user interface control software (AutoSuite), Chemspyd allows for fine-grained, dynamic instrument control through Python, thereby facilitating integration of Chemspeed instruments in custom workflows and SDLs. With the rapid spread of Chemspeed platforms across academic and industrial laboratories across the world, we envision widespread adoption of this package, particularly in those scenarios where dynamic control and flexible integration with third-party software or hardware is required. Importantly, Chemspyd is an open-source project. Therefore, we encourage feedback and contributions from the community, and hope to inspire development of further functionality based on the needs of users outside our laboratory.Beyond extending the package's functionality, the next critical steps will be to integrate Chemspyd with open-source standards for laboratory instrumentation, such as the XDL standard for encoding synthesis procedures,43 the SiLA2 standard for inter-device communication,44 and operating frameworks for orchestrating self-driving laboratories.27 We are convinced that such open, community-driven standards will be key for reusable, open-source software development.28 Eventually, we believe that Chemspyd can serve as an inspiration and blueprint for instrument manufacturers to provide the open APIs necessary for operating experimental modules in self-driving labs.
Data availability
The code for the Chemspyd Python package can be found at https://gitlab.com/aspuru-guzik-group/self-driving-lab/instruments/chemspyd. Code for the experimental demonstrations can be found at https://gitlab.com/aspuru-guzik-group/self-driving-lab/instruments/chemspyd/-/tree/main/demos. Chemspyd version 1.0.0 was used for this study.Author contributions
Conceptualization: M. S., F. S.-K., A. A.-G. Data curation: M. S., F. S.-K., L. B. Formal analysis: M. S., F. S.-K. Funding acquisition: A. A.-G. Investigation: M. S., F. S.-K., M. H., T. C. W., E. A., L. B., R. K. Methodology: M. S., F. S.-K., M. H., T. C. W., L. B., N. Y., M. Sk., R. K. Project administration: M. S., F. S.-K. Resources: A. A.-G. Software: M. S., F. S.-K., M. H., T. C. W., E. A., S. A.-R., N. Y., M. Sk. Supervision: M. S., F. S.-K., A. A.-G. Validation: M. S., F. S.-K., M. H., L. B., R. K. Visualization: F. S.-K., M. H. Writing (original draft): M. S., F. S.-K., M. H., N. Y. Writing (review and editing): M. S., F. S.-K., M. H., N. Y., M. Sk., A. A.-G.Conflicts of interest
A. A.-G. is chief visionary officer and board member of Kebotix Inc., a company that carries out closed-loop molecular materials discovery.Acknowledgements
The authors acknowledge the Defense Advanced Research Projects Agency (DARPA) under the Accelerated Molecular Discovery Program under Cooperative Agreement No. HR00111920027 dated August 1, 2019. The content of the information presented in this work does not necessarily reflect the position or the policy of the Government. F. S.-K. is a postdoctoral fellow in the Eric and Wendy Schmidt AI in Science Postdoctoral Fellowship Program, a program by Schmidt Futures. A. A.-G. thanks Anders G. Frøseth for his generous support. A. A.-G. also acknowledges funding by Natural Resources Canada and the Canada 150 Research Chairs program.References
- R. J. Spinrad, Science, 1967, 158, 55–60 CrossRef CAS PubMed.
- J. Boyd, Science, 2002, 295, 517–518 CrossRef CAS PubMed.
- R. D. King, J. Rowland, S. G. Oliver, M. Young, W. Aubrey, E. Byrne, M. Liakata, M. Markham, P. Pir, L. N. Soldatova, A. Sparkes, K. E. Whelan and A. Clare, Science, 2009, 324, 85–89 CrossRef CAS PubMed.
- M. Christensen, L. P. E. Yunker, P. Shiri, T. Zepel, P. L. Prieto, S. Grunert, F. Bork and J. E. Hein, Chem. Sci., 2021, 12, 15473–15490 RSC.
- J. Bai, L. Cao, S. Mosbach, J. Akroyd, A. A. Lapkin and M. Kraft, JACS Au, 2022, 2, 292–309 CrossRef CAS PubMed.
- R. L. Greenaway, K. E. Jelfs, A. C. Spivey and S. N. Yaliraki, Nat. Rev. Chem., 2023, 7, 527–528 CrossRef PubMed.
- B. P. MacLeod, F. G. L. Parlane, T. D. Morrissey, F. Häse, L. M. Roch, K. E. Dettelbach, R. Moreira, L. P. E. Yunker, M. B. Rooney, J. R. Deeth, V. Lai, G. J. Ng, H. Situ, R. H. Zhang, M. S. Elliott, T. H. Haley, D. J. Dvorak, A. Aspuru-Guzik, J. E. Hein and C. P. Berlinguette, Sci. Adv., 2020, 6, eaaz8867 CrossRef CAS PubMed.
- V. Fasano, R. C. Mykura, J. M. Fordham, J. J. Rogers, B. Banecki, A. Noble and V. K. Aggarwal, Nat. Synth., 2022, 1, 902–907 CrossRef.
- N. L. Bell, F. Boser, A. Bubliauskas, D. R. Willcox, V. S. Luna and L. Cronin, Nat. Chem. Eng., 2024, 1, 180–189 CrossRef.
- A. M. Lunt, H. Fakhruldeen, G. Pizzuto, L. Longley, A. White, N. Rankin, R. Clowes, B. Alston, L. Gigli, G. M. Day, A. I. Cooper and S. Y. Chong, Chem. Sci., 2024, 15, 2456–2463 RSC.
- N. J. Szymanski, B. Rendy, Y. Fei, R. E. Kumar, T. He, D. Milsted, M. J. McDermott, M. Gallant, E. D. Cubuk, A. Merchant, H. Kim, A. Jain, C. J. Bartel, K. Persson, Y. Zeng and G. Ceder, Nature, 2023, 624, 86–91 CrossRef CAS PubMed.
- G. Tom, S. P. Schmid, S. G. Baird, Y. Cao, K. Darvish, H. Hao, S. Lo, S. Pablo-García, E. M. Rajaonson, M. Skreta, N. Yoshikawa, S. Corapi, G. D. Akkoc, F. Strieth-Kalthoff, M. Seifrid and A. Aspuru-Guzik, ChemRxiv, 2024, preprint, DOI:10.26434/chemrxiv-2024-rj946.
- F. Rahmanian, M. Vogler, C. Wölke, P. Yan, S. Fuchs, M. Winter, I. Cekic-Laskovic and H. S. Stein, Sci. Data, 2023, 10, 43 CrossRef CAS PubMed.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai, X. Wang, X. Li, B. M. Alston, B. Li, R. Clowes, N. Rankin, B. Harris, R. S. Sprick and A. I. Cooper, Nature, 2020, 583, 237–241 CrossRef CAS PubMed.
- A. Ramirez, E. Lam, D. Pacheco, Y. Hou, H. Tribukait, L. Roch, C. Copéret and P. Laveille, Chem Catal., 2024, 4(2), 100888 CrossRef CAS.
- P. Laveille, P. Miéville, S. Chatterjee, E. Clerc, J.-C. Cousty, F. de Nanteuil, E. Lam, E. Mariano, A. Ramirez, U. Randrianarisoa, K. Villat, C. Copéret and N. Cramer, Chimia, 2023, 77, 154–158 CrossRef CAS PubMed.
- T. C. Wu, A. Aguilar-Granda, K. Hotta, S. A. Yazdani, R. Pollice, J. Vestfrid, H. Hao, C. Lavigne, M. Seifrid, N. Angello, F. Bencheikh, J. E. Hein, M. Burke, C. Adachi and A. Aspuru-Guzik, Adv. Mater., 2023, 35, 2207070 CrossRef CAS PubMed.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wolos, R. Roszak, C.-T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Gránda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Science, 2024, 384, eadk9227 CrossRef PubMed.
- C. Guerrero-Sanchez, R. Yañez-Macias, M. Rosales-Guzmán, M. A. De Jesus-Tellez, C. Piñon-Balderrama, J. J. Haven, G. Moad, T. Junkers and U. S. Schubert, in RAFT Polymerization, John Wiley & Sons, Ltd, 2021, pp. 1051–1076 Search PubMed.
- T. Schuett, J. Kimmig, S. Zechel and U. S. Schubert, Polymers, 2022, 14, 292 CrossRef CAS PubMed.
- A. Vriza, H. Chan and J. Xu, Chem. Mater., 2023, 35, 3046–3056 CrossRef CAS.
- F. Häse, L. M. Roch and A. Aspuru-Guzik, Trends Chem., 2019, 1, 282–291 CrossRef.
- B. P. MacLeod, F. G. L. Parlane, A. K. Brown, J. E. Hein and C. P. Berlinguette, Nat. Mater., 2022, 21, 722–726 CrossRef CAS PubMed.
- R. El-khawaldeh and J. E. Hein, Trends Chem., 2024, 6, 1–4 CrossRef.
- H. Wang, T. Fu, Y. Du, W. Gao, K. Huang, Z. Liu, P. Chandak, S. Liu, P. Van Katwyk, A. Deac, A. Anandkumar, K. Bergen, C. P. Gomes, S. Ho, P. Kohli, J. Lasenby, J. Leskovec, T.-Y. Liu, A. Manrai, D. Marks, B. Ramsundar, L. Song, J. Sun, J. Tang, P. Veličković, M. Welling, L. Zhang, C. W. Coley, Y. Bengio and M. Zitnik, Nature, 2023, 620, 47–60 CrossRef CAS PubMed.
- L. M. Roch, F. Häse, C. Kreisbeck, T. Tamayo-Mendoza, L. P. E. Yunker, J. E. Hein and A. Aspuru-Guzik, PLOS One, 2020, 15, e0229862 CrossRef CAS PubMed.
- M. Sim, M. G. Vakili, F. Strieth-Kalthoff, H. Hao, R. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, Matter, 2024 DOI:10.1016/j.matt.2024.04.022.
- S. Lehtola, J. Chem. Phys., 2023, 159, 180901 CrossRef CAS PubMed.
- Chemspyd Package Documentation, https://aspuru-guzik-group.gitlab.io/self-driving-lab/instruments/chemspyd/.
- Chemspyd (on the Python Package Index), https://pypi.org/project/chemspyd/.
- Chemspyd: An Open-Source Python Interface for Chemspeed Robotic Platforms, https://gitlab.com/aspuru-guzik-group/self-driving-lab/instruments/chemspyd.
- G. S. Métraux and C. A. Mirkin, Adv. Mater., 2005, 17, 412–415 CrossRef.
- J. E. Millstone, S. J. Hurst, G. S. Métraux, J. I. Cutler and C. A. Mirkin, Small, 2009, 5, 646–664 CrossRef CAS PubMed.
- A. J. Frank, N. Cathcart, K. E. Maly and V. Kitaev, J. Chem. Educ., 2010, 87, 1098–1101 CrossRef CAS.
- D. G. Brown and J. Boström, J. Med. Chem., 2016, 59, 4443–4458 CrossRef CAS PubMed.
- D. T. Ahneman, J. G. Estrada, S. Lin, S. D. Dreher and A. G. Doyle, Science, 2018, 360, 186–190 CrossRef CAS PubMed.
- J. Liu, Y. Sato, F. Yang, A. J. Kukor and J. E. Hein, Chem.: Methods, 2022, 2, e202200009 CAS.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Nat. Mach. Intell., 2024 DOI:10.1038/s42256-024-00832-8.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Nature, 2023, 624, 570–578 CrossRef CAS PubMed.
- N. Yoshikawa, M. Skreta, K. Darvish, S. Arellano-Rubach, Z. Ji, L. Bjørn Kristensen, A. Z. Li, Y. Zhao, H. Xu, A. Kuramshin, A. Aspuru-Guzik, F. Shkurti and A. Garg, Auton. Robots, 2023, 47, 1057–1086 CrossRef.
- CLAIRIFY-Chemspeed: A natural language interface for Chemspyd code generation, https://github.com/ac-rad/clairify-chemspeed/.
- J. Achiam, S. Adler, S. Agarwal, L. Ahmad, I. Akkaya, F. L. Aleman, D. Almeida, J. Altenschmidt, S. Altman, S. Anadkat, R. Avila, I. Babuschkin, S. Balaji, V. Balcom, P. Baltescu, H. Bao, M. Bavarian, J. Belgum, I. Bello, J. Berdine, G. Bernadett-Shapiro, C. Berner, L. Bogdonoff, O. Boiko, M. Boyd, A.-L. Brakman, G. Brockman, T. Brooks, M. Brundage, K. Button, T. Cai, R. Campbell, A. Cann, B. Carey, C. Carlson, R. Carmichael, B. Chan, C. Chang, F. Chantzis, D. Chen, S. Chen, R. Chen, J. Chen, M. Chen, B. Chess, C. Cho, C. Chu, H. W. Chung, D. Cummings, J. Currier, Y. Dai, C. Decareaux, T. Degry, N. Deutsch, D. Deville, A. Dhar, D. Dohan, S. Dowling, S. Dunning, A. Ecoffet, A. Eleti, T. Eloundou, D. Farhi, L. Fedus, N. Felix, S. P. Fishman, J. Forte, I. Fulford, L. Gao, E. Georges, C. Gibson, V. Goel, T. Gogineni, G. Goh, R. Gontijo-Lopes, J. Gordon, M. Grafstein, S. Gray, R. Greene, J. Gross, S. S. Gu, Y. Guo, C. Hallacy, J. Han, J. Harris, Y. He, M. Heaton, J. Heidecke, C. Hesse, A. Hickey, W. Hickey, P. Hoeschele, B. Houghton, K. Hsu, S. Hu, X. Hu, J. Huizinga, S. Jain, S. Jain, J. Jang, A. Jiang, R. Jiang, H. Jin, D. Jin, S. Jomoto, B. Jonn, H. Jun, T. Kaftan, Ł. Kaiser, A. Kamali, I. Kanitscheider, N. S. Keskar, T. Khan, L. Kilpatrick, J. W. Kim, C. Kim, Y. Kim, H. Kirchner, J. Kiros, M. Knight, D. Kokotajlo, Ł. Kondraciuk, A. Kondrich, A. Konstantinidis, K. Kosic, G. Krueger, V. Kuo, M. Lampe, I. Lan, T. Lee, J. Leike, J. Leung, D. Levy, C. M. Li, R. Lim, M. Lin, S. Lin, M. Litwin, T. Lopez, R. Lowe, P. Lue, A. Makanju, K. Malfacini, S. Manning, T. Markov, Y. Markovski, B. Martin, K. Mayer, A. Mayne, B. McGrew, S. M. McKinney, C. McLeavey, P. McMillan, J. McNeil, D. Medina, A. Mehta, J. Menick, L. Metz, A. Mishchenko, P. Mishkin, V. Monaco, E. Morikawa, D. Mossing, T. Mu, M. Murati, O. Murk, D. Mély, A. Nair, R. Nakano, R. Nayak, A. Neelakantan, R. Ngo, H. Noh, L. Ouyang, C. O'Keefe, J. Pachocki, A. Paino, J. Palermo, A. Pantuliano, G. Parascandolo, J. Parish, E. Parparita, A. Passos, M. Pavlov, A. Peng, A. Perelman, F. d A. B. Peres, M. Petrov, H. P. d O. Pinto, M. Pokorny, M. Pokrass, V. Pong, T. Powell, A. Power, B. Power, E. Proehl, R. Puri, A. Radford, J. Rae, A. Ramesh, C. Raymond, F. Real, K. Rimbach, C. Ross, B. Rotsted, H. Roussez, N. Ryder, M. Saltarelli, T. Sanders, S. Santurkar, G. Sastry, H. Schmidt, D. Schnurr, J. Schulman, D. Selsam, K. Sheppard, T. Sherbakov, J. Shieh, S. Shoker, P. Shyam, S. Sidor, E. Sigler, M. Simens, J. Sitkin, K. Slama, I. Sohl, B. Sokolowsky, Y. Song, N. Staudacher, F. P. Such, N. Summers, I. Sutskever, J. Tang, N. Tezak, M. Thompson, P. Tillet, A. Tootoonchian, E. Tseng, P. Tuggle, N. Turley, J. Tworek, J. F. C. Uribe, A. Vallone, A. Vijayvergiya, C. Voss, C. Wainwright, J. J. Wang, A. Wang, B. Wang, J. Ward, J. Wei, C. J. Weinmann, A. Welihinda, P. Welinder, J. Weng, L. Weng, M. Wiethoff, D. Willner, C. Winter, S. Wolrich, H. Wong, L. Workman, S. Wu, J. Wu, M. Wu, K. Xiao, T. Xu, S. Yoo, K. Yu, Q. Yuan, W. Zaremba, R. Zellers, C. Zhang, M. Zhang, S. Zhao, T. Zheng, J. Zhuang, W. Zhuk and B. Zoph, arXiv, 2023, perprint, DOI:10.48550/arXiv.2303.08774.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, Science, 2020, 370, 101–108 CrossRef CAS PubMed.
- L. Bromig, D. Leiter, A.-V. Mardale, N. von den Eichen, E. Bieringer and D. Weuster-Botz, SoftwareX, 2022, DOI:10.1016/j.softx.2022.100991.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00046c |
| ‡ Authors contributed equally. |
| § Present address: Department of Systems Biology, Harvard Medical School, Boston, MA, USA. |
| This journal is © The Royal Society of Chemistry 2024 |