Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Developments and applications of the OPTIMADE API for materials discovery, design, and data exchange†
Matthew L.
Evans
 ab,
Johan
Bergsma‡
ab,
Johan
Bergsma‡
 c,
Andrius
Merkys
c,
Andrius
Merkys
 d,
Casper W.
Andersen
d,
Casper W.
Andersen
 e,
Oskar B.
Andersson
e,
Oskar B.
Andersson
 f,
Daniel
Beltrán
g,
Evgeny
Blokhin
f,
Daniel
Beltrán
g,
Evgeny
Blokhin
 hi,
Tara M.
Boland
hi,
Tara M.
Boland
 j,
Rubén
Castañeda Balderas
j,
Rubén
Castañeda Balderas
 k,
Kamal
Choudhary
k,
Kamal
Choudhary
 l,
Alberto
Díaz Díaz
l,
Alberto
Díaz Díaz
 k,
Rodrigo
Domínguez García
k,
Rodrigo
Domínguez García
 k,
Hagen
Eckert
k,
Hagen
Eckert
 mn,
Kristjan
Eimre
mn,
Kristjan
Eimre
 o,
María Elena
Fuentes Montero
o,
María Elena
Fuentes Montero
 p,
Adam M.
Krajewski
p,
Adam M.
Krajewski
 q,
Jens Jørgen
Mortensen
q,
Jens Jørgen
Mortensen
 j,
José Manuel
Nápoles Duarte
j,
José Manuel
Nápoles Duarte
 p,
Jacob
Pietryga
p,
Jacob
Pietryga
 r,
Ji
Qi
s,
Felipe de Jesús
Trejo Carrillo
r,
Ji
Qi
s,
Felipe de Jesús
Trejo Carrillo
 k,
Antanas
Vaitkus
k,
Antanas
Vaitkus
 d,
Jusong
Yu
oah,
Adam
Zettel
d,
Jusong
Yu
oah,
Adam
Zettel
 mn,
Pedro Baptista
de Castro
mn,
Pedro Baptista
de Castro
 t,
Johan
Carlsson
t,
Johan
Carlsson
 u,
Tiago F. T.
Cerqueira
u,
Tiago F. T.
Cerqueira
 v,
Simon
Divilov
v,
Simon
Divilov
 mn,
Hamidreza
Hajiyani§
u,
Felix
Hanke
mn,
Hamidreza
Hajiyani§
u,
Felix
Hanke
 w,
Kevin
Jose
x,
Corey
Oses
w,
Kevin
Jose
x,
Corey
Oses
 y,
Janosh
Riebesell
y,
Janosh
Riebesell
 xz,
Jonathan
Schmidt
xz,
Jonathan
Schmidt
 aa,
Donald
Winston
aa,
Donald
Winston
 ab,
Christen
Xie
s,
Xiaoyu
Yang
ab,
Christen
Xie
s,
Xiaoyu
Yang
 acadae,
Sara
Bonella
acadae,
Sara
Bonella
 c,
Silvana
Botti
c,
Silvana
Botti
 af,
Stefano
Curtarolo
af,
Stefano
Curtarolo
 mn,
Claudia
Draxl
mn,
Claudia
Draxl
 ag,
Luis Edmundo
Fuentes Cobas
ag,
Luis Edmundo
Fuentes Cobas
 k,
Adam
Hospital
k,
Adam
Hospital
 g,
Zi-Kui
Liu
g,
Zi-Kui
Liu
 q,
Miguel A. L.
Marques
q,
Miguel A. L.
Marques
 af,
Nicola
Marzari
af,
Nicola
Marzari
 oah,
Andrew J.
Morris
oah,
Andrew J.
Morris
 ai,
Shyue Ping
Ong
ai,
Shyue Ping
Ong
 s,
Modesto
Orozco
s,
Modesto
Orozco
 g,
Kristin A.
Persson
g,
Kristin A.
Persson
 zaj,
Kristian S.
Thygesen
zaj,
Kristian S.
Thygesen
 j,
Chris
Wolverton
j,
Chris
Wolverton
 r,
Markus
Scheidgen
r,
Markus
Scheidgen
 ag,
Cormac
Toher
ag,
Cormac
Toher
 nak,
Gareth J.
Conduit
nak,
Gareth J.
Conduit
 xan,
Giovanni
Pizzi
xan,
Giovanni
Pizzi
 oah,
Saulius
Gražulis
oah,
Saulius
Gražulis
 dal,
Gian-Marco
Rignanese
dal,
Gian-Marco
Rignanese
 *abam and
Rickard
Armiento
*abam and
Rickard
Armiento
 *f
*f
aUCLouvain, Institut de la Matière Condensée et des Nanosciences (IMCN), Chemin des Étoiles 8, Louvain-la-Neuve 1348, Belgium. E-mail: gian-marco.rignanese@uclouvain.be
bMatgenix SRL, 185 Rue Armand Bury, 6534 Gozée, Belgium
cCentre Européen de Calcul Atomique et Moléculaire (CECAM), École Polytechnique Fédérale de Lausanne, Avenue de Forel 3, 1015 Lausanne, Switzerland
dInstitute of Biotechnology, Life Sciences Center, Vilnius University, Saulėtekio av. 7, LT-10257 Vilnius, Lithuania
eSINTEF, P.O. Box 4760 Torgarden, NO-7465 Trondheim, Norway
fMaterials Design and Informatics Unit, Department of Physics, Chemistry and Biology, Linköping University, Sweden. E-mail: rickard.armiento@liu.se
gInstitute for Research in Biomedicine (IRB Barcelona), Baldiri i Reixac 10-12, 08028 Barcelona, Spain
hTilde Materials Informatics, Straßmannstraße 25, 10249, Berlin, Germany
iMaterials Platform for Data Science, Sepapaja 6, 15551, Tallinn, Estonia
jComputational Atomic-Scale Materials Design, Technical University of Denmark, Kgs. Lyngby, Denmark
kCentro de Investigación en Materiales Avanzados, S.C. (CIMAV), Av. Miguel de Cervantes 120, Complejo Industrial Chihuahua, 31136, Chihuahua, Chih., Mexico
lMaterial Measurement Laboratory, National Institute of Standards and Technology, Gaithersburg, MD 20899, USA
mDepartment of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
nCenter for Extreme Materials, Duke University, Durham, NC 27708, USA
oTheory and Simulation of Materials (THEOS), and National Centre for Computational Design and Discovery of Novel Materials (MARVEL), École Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland
pUniversidad Autónoma de Chihuahua, Facultad de Ciencias Quimicas, 31125 Chihuahua, Mexico
qDepartment of Materials Science and Engineering, The Pennsylvania State University, University Park, PA 16802, USA
rDepartment of Materials Science and Engineering, Northwestern University, Evanston, IL 60208, USA
sDepartment of NanoEngineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0448, USA
tNational Institute for Materials Science, 1-2-1 Sengen, Tsukuba, Ibaraki 305-0047, Japan
uDassault Systèmes Germany GmbH, Am Kabellager 11-13, 51063 Cologne, Germany
vCFisUC, Department of Physics, University of Coimbra, Rua Larga, 3004-516 Coimbra, Portugal
wDassault Systèmes, 22 Science Park, CB4 0FJ, UK
xTheory of Condensed Matter, Cavendish Laboratory, Cambridge, UK
yDepartment of Materials Science and Engineering, Johns Hopkins University, Baltimore, MD 21218, USA
zLawrence Berkeley National Lab, Berkeley, CA, USA
aaMaterials Theory, ETH Zürich, Wolfgang-Pauli-Strasse 27, 8093 Zurich, Switzerland
abPolyneme LLC, New York, NY 10038, USA
acComputer Network Information Center, Chinese Academy of Sciences, Beijing, 100083, China
adUniversity of Chinese Academy of Sciences, Beijing, 101408, China
aeBeijing MaiGao MatCloud Technology Co. Ltd, Beijing, 100149, China
afResearch Center Future Energy Materials and Systems of the University Alliance Ruhr and Interdisciplinary Centre for Advanced Materials Simulation, Ruhr University Bochum, Universitätsstraße 150, D-44801 Bochum, Germany
agHumboldt-Universität zu Berlin, Institut für Physik and IRIS Adlershof, 12489 Berlin, Germany
ahLaboratory for Materials Simulations (LMS), Paul Scherrer Institute (PSI), 5232 Villigen PSI, Switzerland
aiSchool of Metallurgy and Materials, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK
ajDepartment of Materials Science and Engineering, UC Berkeley, Hearst Mining Memorial Building, Berkeley, 94720 CA, USA
akDepartment of Materials Science and Engineering and Department of Chemistry and Biochemistry, The University of Texas at Dallas, Richardson, TX 75080, USA
alInstitute of Computer Science, Faculty of Mathematics and Informatics, Vilnius University, Naugarduko g. 24, LT-03225 Vilnius, Lithuania
amSchool of Materials Science and Engineering, Northwestern Polytechnical University, Xi'an, Shaanxi 710072, China
anIntellegens Ltd, French's Rd, Cambridge, CB4 3NP, UK
First published on 18th April 2024
Abstract
The Open Databases Integration for Materials Design (OPTIMADE) application programming interface (API) empowers users with holistic access to a growing federation of databases, enhancing the accessibility and discoverability of materials and chemical data. Since the first release of the OPTIMADE specification (v1.0), the API has undergone significant development, leading to the v1.2 release, and has underpinned multiple scientific studies. In this work, we highlight the latest features of the API format, accompanying software tools, and provide an update on the implementation of OPTIMADE in contributing materials databases. We end by providing several use cases that demonstrate the utility of the OPTIMADE API in materials research that continue to drive its ongoing development.
1 Introduction
Industrial chemicals and materials underpin the global economy: for example, chemicals alone contribute $6.4 trillion24 annually to the global economy. Industrial chemicals and materials companies are under significant pressure to improve the environmental, social, and governance impact of their business,10 with a particular focus on reducing the carbon footprint. Unfortunately, the discovery of chemicals and drugs is traditionally a time-consuming and expensive process driven by experiment-led trial-and-improvement, delaying the response to the climate crisis. However, in the last few years, high-throughput calculations have led to an explosion in the volume of available materials data.41,57,77 Machine learning has emerged as a pivotal tool to exploit this data,125 accelerating the discovery of chemicals and drugs that meet the challenges faced today.A core requirement for the data and machine-learning revolution is data availability and interoperability. Therefore, the Open Databases Integration for Materials Design (OPTIMADE) universal application programming interface (API) was created to empower users with programmatic access to many leading materials databases. By organising under an open federation, and emphasising the interoperability of search as well as access, OPTIMADE improves the discoverability of materials data, especially from smaller, less known databases. As we move into the era of autonomous laboratories (both computational and experimental), the technical approach taken renders all OPTIMADE APIs machine actionable, allowing for automated serendipitous discovery of newly added data entries in a given materials space without needing to specify which databases to access. Such an extended data availability requires explicit clarification of data permissions and ownership, which can be different for each database.
Since the first release of the OPTIMADE specification (v1.0)2 with accompanying article,3 the OPTIMADE API format has enjoyed significant adoption, with 22 registered providers130,131 of 25 interoperable databases serving over 22 million crystal structures with associated properties. In the v1.2 release,5 the specification has undergone significant extensions and enhancements that enable novel use cases whilst making the format accessible to both users and developers. This gives users access to data from both large and well-known sources, and many specialist datasets focused on a family of materials of particular interest. The combination of a general overview of all possible materials and detailed knowledge of particular materials enables novel discovery and deep insights with for example machine learning.
In this paper, we highlight recent developments to and uses of the OPTIMADE API. First, in Section 2, we provide an overview of the OPTIMADE API format, and of the latest features. Next, in Section 3, we highlight the efforts of leading materials databases to provide access through the OPTIMADE API format. In Section 4, we show how the OPTIMADE API has been used in computational screening and for the creation of machine learning datasets. Section 5 discusses future plans for OPTIMADE, both in terms of new technical frontiers, and of the sustainability of the ecosystem and community. Finally, in Section 6, we summarise and look ahead to the future of materials databases.
2 Overview of the OPTIMADE API
The OPTIMADE API is well-positioned to set the standard to enable search, retrieval and annotation in a common way for all databases. Crystal structure data has benefited from decades of standardisation work in the form of the Crystallographic Information File (CIF)13,51 and related initiatives, which heavily inspired the crystal structure representation employed by the OPTIMADE API format. By building a standardised, open format, both proprietary and open data can be aggregated and used on the same footing.Building on these seminal standardisation efforts, OPTIMADE goes considerably further than standardising the representation of crystal structure data, by including: the means for filtering entries (via the OPTIMADE filter grammar), a standard for laying out resources on the web (by providing rules and expectations of URL formats), a means for introspectively defining additional properties and entry types per-database, and the creation of a decentralised federation of compatible databases. These additional aspects are what maximises the impact of the OPTIMADE API and enable new scientific applications. The OPTIMADE API is registered with FAIRsharing.org as a data standard,38 and releases are archived on Zenodo,2 with ongoing development occurring openly under the Materials-Consortia banner on GitHub (https://github.com/Materials-Consortia).129 The initial motivation for OPTIMADE, and a discussion of the previously existing materials API formats and filter mechanisms can be found in ref. 3 which described the first release.
The process for a user to access OPTIMADE compliant data is as follows. The user starts with the base URL defined by the database provider (available from the federated OPTIMADE provider list130,131), and then appends a common string describing the entry type to query, plus any filter or implementation parameters, which is submitted as an HTTP GET request. For example, to probe the Materials Project66 for materials containing SiO2, we make a GET request to the following URL:
This delivers the response in Box 1 that contains entries where oxygen and silicon occur in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The power of the OPTIMADE API is that the same generic request can be appended to the base URL of any other database, and its matching entries will be returned. This is a non-trivial step for database providers, who must convert the OPTIMADE filter grammar into the corresponding query for their own database engine. The benefit is that client code can then be written to unify the results from multiple databases, allowing users to receive the most comprehensive results for the query. Such an extended data availability requires explicit clarification of data permissions and ownership, which can be different for each database or even each entry.
1 ratio. The power of the OPTIMADE API is that the same generic request can be appended to the base URL of any other database, and its matching entries will be returned. This is a non-trivial step for database providers, who must convert the OPTIMADE filter grammar into the corresponding query for their own database engine. The benefit is that client code can then be written to unify the results from multiple databases, allowing users to receive the most comprehensive results for the query. Such an extended data availability requires explicit clarification of data permissions and ownership, which can be different for each database or even each entry.
Box 1: An excerpt of the JSON response showing the material attributes for one of the returned entries.
2.1 OPTIMADE core design principles
A materials database provider that implements the OPTIMADE API will have a database backend and one or more interfaces available to clients. These interfaces include the OPTIMADE API, but can also provide access by other means, e.g., a database-specific API or web-based graphical user interface. Fig. 1 serves as a schematic illustration of this point.The providers that presently support the OPTIMADE API represent a wide range of underlying backends. The primary backend component is, usually, a database engine, but the backend covers all parts of the system that manages the stored data. Relevant backends for OPTIMADE implementations range from simple flat files in a filesystem, to sophisticated setups with load-balanced distributed cloud hosting of relational database engines (e.g., Structured Query Language, SQL-based) or non-relational key-value, document or graph database engines (so-called NoSQL).
There exists a multitude of APIs for data access and storage (both for materials databases and more generally) that have been designed for a specific database backend. These APIs are typically designed around specific features the backend provides in terms of, e.g., browse, search, and retrieval, and, crucially, how these features are implemented by the backend. The API then typically becomes a thin wrapper for these features, which exposes the functionality of the backend implementation. There is thus a crucial need for a generic database interface, which should be based on the central principles:
• A core feature set that any reasonable materials database backend can implement via cheap, single-pass, on-the-fly translations in the API layer. Furthermore, the translations should be possible without modifying the underlying backend – i.e., participating databases should not be required to reformat or amend the stored data, software features, etc. to provide these core OPTIMADE features.
• Extended features beyond the core features that are shared among multiple (but not all) backends should be standardised as optional features. It may appear that this design works against interoperability, as it may lead to query Q working for databases A and B, but not for database C. However, if the database backend for C cannot support the type of query that Q represents (for example, a database containing molecular dynamics calculations of proteins cannot sensibly be searched on the chemical formula in the simulation cell), it means that there exists no way to make that query interoperable across A, B, and C (without altering the backend of C). Hence, the highest level of interoperability is achieved by standardising Q as an optional feature.
• For standardised optional features, multiple overlapping representations of the same features and/or data should be avoided. The reason is that, for two different ways of providing feature Q as Q1 and Q2, we may end up with database A supporting only Q1 and database B supporting only Q2. Hence, the client either has to tailor the query differently depending on the destination or provide multiple versions of the query in the request, which is an undesirable burden to put on clients and works against an interoperable design. What is strictly standardised is how a database that does not implement a certain feature should respond should that feature be requested.
• Those features and data that only occur in a particular database should be provided in isolated database-specific namespaces that other databases can recognise and handle appropriately. The OPTIMADE specification describes how references to such fields should be announced by the particular database and also how clients making multi-provider queries should handle in the response in various situations for optimal interoperability. The latest version of OPTIMADE even outlines a mechanism for creating sub-specifications that allow multiple database providers to collaborate on custom communal definitions, as shall be discussed later.
2.2 Recent OPTIMADE improvements
Since the initial v1.0 release2 of the OPTIMADE API in July 2020, many features have been added, driven by user feedback and use cases. The major enhancements to the specification introduced in versions 1.1 (ref. 4) and 1.2 are discussed below, with the full changelog (https://github.com/Materials-Consortia/OPTIMADE/blob/master/CHANGELOG.md) and specification text available online on GitHub (https://github.com/Materials-Consortia/OPTIMADE).129OPTIMADE v1.2 includes a full schema format, based on – and compatible with – JSONSchema,99 capable of fully describing in a machine-readable way what a property is (including definition of sophisticated data structures with multiple layers of lists and dictionary subfields). All properties are given clear versioned stable identifiers (URIs) that can be used to identify that multiple databases refer to the exact same property. A simple example is provided in Box 2, which displays the definition of the  field. Furthermore, a browsable interface to these definitions for the OPTIMADE standard properties is available at https://schemas.optimade.org/.
field. Furthermore, a browsable interface to these definitions for the OPTIMADE standard properties is available at https://schemas.optimade.org/.
There is related work outside of the specification on providing an enlarged set of shared definitions that can be incrementally adopted by the subsections of the community. This will allow databases to point to and share the same property definition without requiring the slow consensus-building step for the relevant fields to be promoted to the main specification. This is especially useful in cases where this shared information is the defining feature of a given database; for example, the first target namespace is that of stability predictions arising from density-functional theory calculations, the key property required to truly enable materials discovery applications with OPTIMADE (see Section 4.1.2 later for more information).
 or similar encodings), significantly increasing the size of the data to be transferred over the network. Adding support for a format like BSON, which does support binary data, or JSON variants which do support streamed processing (such as JSON Lines) could therefore improve the data transfer rates for OPTIMADE requests, and allow for immediate visualisation of partial results.
or similar encodings), significantly increasing the size of the data to be transferred over the network. Adding support for a format like BSON, which does support binary data, or JSON variants which do support streamed processing (such as JSON Lines) could therefore improve the data transfer rates for OPTIMADE requests, and allow for immediate visualisation of partial results.
Box 2: An example OPTIMADE property definition for the  property.
property.
In previous versions of OPTIMADE, the size of property data was limited so that a single database entry could fit in a single HTTP response. In the most recent version of OPTIMADE, the implementation can decide to defer large properties to be communicated over a separate simplified streamable protocol, which in practice can be implemented by serving large static files over HTTP. This addition was driven by the future application of serving trajectory data with OPTIMADE (see Section 5 for more information) where individual “properties”, such as atomic positions in each frame of a trajectory, can individually be prohibitively large.
Symmetry information for structures. The latest OPTIMADE release includes a set of standardised and comprehensive descriptions of structural symmetry. This has been achieved with 5 fields that can be variously used for filtering by symmetry and for reconstructing atomic site positions, building on existing standards laid out by the IUCr and others:
 endpoint.
The OPTIMADE specification has been extended to enable the description of files and their precise relationships to other OPTIMADE entries. To implement this, a new
endpoint.
The OPTIMADE specification has been extended to enable the description of files and their precise relationships to other OPTIMADE entries. To implement this, a new  entry endpoint has been defined. This addition allows, for example, linking
entry endpoint has been defined. This addition allows, for example, linking  entries with their representations in widely used structural data formats such as CIF, POSCAR, and SDF, or linking directly to the raw input or output files associated with a calculation involving a structure.
entries with their representations in widely used structural data formats such as CIF, POSCAR, and SDF, or linking directly to the raw input or output files associated with a calculation involving a structure.
Per property and per entry metadata. We plan to create a mechanism to provide metadata per property for each individual entry. This metadata could, for example, be confidence intervals, or information about how a property was calculated.
Request delay. In order not to overload a particular OPTIMADE server, the metadata property
 appears in the latest release to allow an implementation to suggest a specific back-off time delay between subsequent requests. It is up to the given server implementation to decide what to do with clients violating the requested delay: refuse to serve, intentionally delay the response, or ignore.
appears in the latest release to allow an implementation to suggest a specific back-off time delay between subsequent requests. It is up to the given server implementation to decide what to do with clients violating the requested delay: refuse to serve, intentionally delay the response, or ignore.
Licensing. The possibility of unsupervised database harvesting raises a need for machine-readable definition of data licenses. Whilst OPTIMADE is an open format, it can be used to serve proprietary or otherwise restricted data, the usage terms of which must be described. The latest OPTIMADE release addresses this issue by introducing the metadata fields
 ,
,  , and
, and  . The property
. The property  is intended for databases to link to human-readable licensing terms, while
is intended for databases to link to human-readable licensing terms, while  and
and  allow specifying machine-readable license identifiers following the Software Package Data Exchange (SPDX) standard, making interpreting requirements easier for automated clients and web crawlers. This standardised way to announce that either the whole database, or the individual entries, are explicitly available under certain licenses enables machine-actionable licensing decisions, e.g. for commercial re-use and republishing of data.
allow specifying machine-readable license identifiers following the Software Package Data Exchange (SPDX) standard, making interpreting requirements easier for automated clients and web crawlers. This standardised way to announce that either the whole database, or the individual entries, are explicitly available under certain licenses enables machine-actionable licensing decisions, e.g. for commercial re-use and republishing of data.
Substring comparisons on list elements. The OPTIMADE filter grammar defines substring comparison operators to match either the start, end or any part of a property value. However, until version 1.2, such comparisons could not be carried out on list elements. The latest version of OPTIMADE now explicitly supports such substring queries on elements of list properties.
Boolean values. Although there are no Boolean fields in the main OPTIMADE specification, the latest version of the OPTIMADE specification includes support in the filter grammar for defining and filtering on such custom fields defined by data providers.
2.3 Associated software tools
 is an open-source software (MIT license) package that provides tooling for serving, validating and consuming OPTIMADE APIs in Python,36 available on GitHub (https://github.com/Materials-Consortia/optimade-python-tools). Now in version 1.0, it provides a highly extensible reference server implementation of an OPTIMADE API, with support for different database backends. This server is provided as a Docker container for easy deployment and can be configured to use an existing database, or generate one from scratch in the OPTIMADE format. Existing databases wanting to make use of the library need to provide mappings to and from their existing data format and query mechanisms. The package contains isolated modules for various OPTIMADE-related functionalities, for example, a grammar and parser for the OPTIMADE filter language, mappers for querying different database backends, and a fuzzy validator that can dynamically generate requests to an OPTIMADE server to assess its compliance with the standard (which is also used to generate the OPTIMADE provider dashboard: https://www.optimade.org/providers-dashboard/).130 In addition to server-focused functionalities, the package includes reusable code that can help OPTIMADE consumers and clients, including adapters for converting OPTIMADE entries into common formats used in the community, such as ASE
is an open-source software (MIT license) package that provides tooling for serving, validating and consuming OPTIMADE APIs in Python,36 available on GitHub (https://github.com/Materials-Consortia/optimade-python-tools). Now in version 1.0, it provides a highly extensible reference server implementation of an OPTIMADE API, with support for different database backends. This server is provided as a Docker container for easy deployment and can be configured to use an existing database, or generate one from scratch in the OPTIMADE format. Existing databases wanting to make use of the library need to provide mappings to and from their existing data format and query mechanisms. The package contains isolated modules for various OPTIMADE-related functionalities, for example, a grammar and parser for the OPTIMADE filter language, mappers for querying different database backends, and a fuzzy validator that can dynamically generate requests to an OPTIMADE server to assess its compliance with the standard (which is also used to generate the OPTIMADE provider dashboard: https://www.optimade.org/providers-dashboard/).130 In addition to server-focused functionalities, the package includes reusable code that can help OPTIMADE consumers and clients, including adapters for converting OPTIMADE entries into common formats used in the community, such as ASE  ,72 pymatgen
,72 pymatgen  90 and AiiDA nodes.60
90 and AiiDA nodes.60
The package also contains an advanced asynchronous HTTP client that can be used within Python code or at the command line to concurrently filter multiple OPTIMADE databases with multiple queries, paginating and validating the results, as well as searching across databases for supported properties. Users can provide callbacks to store the results in local secondary databases for re-use in other projects.
 is fully documented online at https://www.optimade.org/optimade-python-tools/latest/ with guides for setting up and validating an API, deploying a server and using the client. In this way,
is fully documented online at https://www.optimade.org/optimade-python-tools/latest/ with guides for setting up and validating an API, deploying a server and using the client. In this way,  significantly lowers the barrier to retrieving data from OPTIMADE APIs, and to the development of new server implementations. Future developments will focus on extending the use cases of
significantly lowers the barrier to retrieving data from OPTIMADE APIs, and to the development of new server implementations. Future developments will focus on extending the use cases of  to operating on static data, so that it can be embedded within archival infrastructure, such as Materials Cloud,127 to serve user data without any additional input.
to operating on static data, so that it can be embedded within archival infrastructure, such as Materials Cloud,127 to serve user data without any additional input.
 is an open-source software (MIT license) package implementing a RESTful API in Python for querying OPTIMADE databases, available on GitHub at the Materials-Consortia gateway (https://github.com/Materials-Consortia/optimade-gateway). This provides a powerful yet straightforward tool to allow users of the Python programming language (that is widespread in machine learning and other branches of data science) to access material database results.
is an open-source software (MIT license) package implementing a RESTful API in Python for querying OPTIMADE databases, available on GitHub at the Materials-Consortia gateway (https://github.com/Materials-Consortia/optimade-gateway). This provides a powerful yet straightforward tool to allow users of the Python programming language (that is widespread in machine learning and other branches of data science) to access material database results.  supports both synchronous and asynchronous searches via HTTP GET and POST requests, respectively. It can return search results in the standard OPTIMADE data format, as well as a custom OPTIMADE-inspired data format. The main purpose and goal of the deployed service is to be a client backend. A gateway to version 0.4 is running at https://mmp-optimade-gateway.materialscloud.io/redoc as well as at https://optimade-gateway.fly.dev/redoc. Both services utilise a MongoDB database for time-limited caching of the query results, increasing response speeds for common queries.
supports both synchronous and asynchronous searches via HTTP GET and POST requests, respectively. It can return search results in the standard OPTIMADE data format, as well as a custom OPTIMADE-inspired data format. The main purpose and goal of the deployed service is to be a client backend. A gateway to version 0.4 is running at https://mmp-optimade-gateway.materialscloud.io/redoc as well as at https://optimade-gateway.fly.dev/redoc. Both services utilise a MongoDB database for time-limited caching of the query results, increasing response speeds for common queries.
 and is thus highly-portable, can be opened from anywhere, on any environment (e.g., on a smartphone or locally from a USB stick). To increase ease-of-use, a simple pattern-matching library was developed (https://github.com/mpds-io/optimade-mpds-nlp), transforming the free-text user input into a standard OPTIMADE query (e.g., a keyword
and is thus highly-portable, can be opened from anywhere, on any environment (e.g., on a smartphone or locally from a USB stick). To increase ease-of-use, a simple pattern-matching library was developed (https://github.com/mpds-io/optimade-mpds-nlp), transforming the free-text user input into a standard OPTIMADE query (e.g., a keyword  is transformed into
is transformed into  ). A separate Svelte user interface kit (https://github.com/basf/svelte-spectre) was developed offering a range of the modular GUI components, willingly accepted by the frontend community and already re-used in many other web-projects, including commercial ones. A standalone OPTIMADE client written in TypeScript was employed, being fully isomorphic (that is, the same code can be used inside the web browser and on the web server).
). A separate Svelte user interface kit (https://github.com/basf/svelte-spectre) was developed offering a range of the modular GUI components, willingly accepted by the frontend community and already re-used in many other web-projects, including commercial ones. A standalone OPTIMADE client written in TypeScript was employed, being fully isomorphic (that is, the same code can be used inside the web browser and on the web server).
3 Contributing databases
The burgeoning community of materials databases are the core that underpins the OPTIMADE consortium. Therefore, below we discuss the key features of the major materials databases that make data available through the OPTIMADE API. We first briefly introduce each database and its offering, and we then discuss its particular OPTIMADE implementation. Finally, we provide an updated table from our previous work3 that compares the amount of compliant data available in different databases.3.1 AFLOW
 prefix. A full list of keywords available to use with OPTIMADE to query AFLOW are available at the info endpoint
prefix. A full list of keywords available to use with OPTIMADE to query AFLOW are available at the info endpoint  .
.
Base URL: https://aflow.org/API/optimade
3.2 Alexandria
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062
062![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 521 entries, spanning nearly the entire periodic table with 89 elements. The database was primarily generated by scanning binary, ternary, and quaternary prototypes to identify stable compounds. This process employed crystal graph attention networks113,114,117 to predict the stability of all potential compositions for each prototype. Compounds that were found to be close to stability were subsequently confirmed using DFT. Additionally, the database includes compounds obtained from traditional high-throughput searches conducted previously.112,115,137
521 entries, spanning nearly the entire periodic table with 89 elements. The database was primarily generated by scanning binary, ternary, and quaternary prototypes to identify stable compounds. This process employed crystal graph attention networks113,114,117 to predict the stability of all potential compositions for each prototype. Compounds that were found to be close to stability were subsequently confirmed using DFT. Additionally, the database includes compounds obtained from traditional high-throughput searches conducted previously.112,115,137
Most entries were calculated using the PBE functional with parameters mostly consistent with those of the Materials Project,66 and includes 3D (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 489
489![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 295 entries), 2D (137
295 entries), 2D (137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833) and 1D (13
833) and 1D (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 295 entries) compounds. Additionally, a total of 422
295 entries) compounds. Additionally, a total of 422![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 098 materials were computed using the PBEsol and SCAN functionals to yield more precise geometries, formation energies, and bandgaps.116 The PBE version of the Alexandria database, which comprises 115
098 materials were computed using the PBEsol and SCAN functionals to yield more precise geometries, formation energies, and bandgaps.116 The PBE version of the Alexandria database, which comprises 115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 535 potentially stable materials, represents the most extensive publicly available DFT convex hull of thermodynamic stability in our knowledge. Furthermore, 771
535 potentially stable materials, represents the most extensive publicly available DFT convex hull of thermodynamic stability in our knowledge. Furthermore, 771![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 696 materials lie within a distance of less than 50 meV per atom from the convex hull. The database encompasses various properties, including structure (lattice and atomic positions), energy distance to the convex hull, formation energy and direct as well as indirect bandgaps. Continuous expansion of the database is underway through further ongoing high-throughput searches.
696 materials lie within a distance of less than 50 meV per atom from the convex hull. The database encompasses various properties, including structure (lattice and atomic positions), energy distance to the convex hull, formation energy and direct as well as indirect bandgaps. Continuous expansion of the database is underway through further ongoing high-throughput searches.
 36 reference implementation and provides a list of extra properties with the
36 reference implementation and provides a list of extra properties with the  prefix. Prior to this development, the Alexandria database was made available solely as a static archive that users had to download in its entirety to explore, but now the OPTIMADE format supports filtering on both composition and the predicted stability of database entries.
prefix. Prior to this development, the Alexandria database was made available solely as a static archive that users had to download in its entirety to explore, but now the OPTIMADE format supports filtering on both composition and the predicted stability of database entries.
Base URL: https://alexandria.icams.rub.de
3.3 BioExcel COVID-19
 has been set up, which provides the trajectory data at the
has been set up, which provides the trajectory data at the  endpoint. This server also provides protein specific properties and metadata under database specific fields with the
endpoint. This server also provides protein specific properties and metadata under database specific fields with the  prefix. Querying has been partially implemented, but is not yet available for all fields. No atomistic structures are shared, so the
prefix. Querying has been partially implemented, but is not yet available for all fields. No atomistic structures are shared, so the  endpoint is not available.
endpoint is not available.
Base URL: https://bioexcel-cv19.bsc.es/optimade/
3.4 Computational Materials Repository (CMR)
The flagship database in CMR is the Computational Two-Dimensional Materials Database (C2DB)44,50,88 that contains structural, thermodynamic, elastic, electronic, magnetic, and optical properties of more than 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 two-dimensional (2D) monolayer materials computed using the GPAW34,86 package. The core set of materials in C2DB have been obtained by extracting monolayers from experimentally known layered van der Waals crystals. Subsequently, new monolayers have been generated by systematic atom-substitution applied to the core materials, or using deep generative AI models.78 Recently, the C2DB has been complemented by the BiDB database containing homobilayers formed by stacking 1000 of the most stable monolayers from the C2DB in all possible commensurate configurations.94
000 two-dimensional (2D) monolayer materials computed using the GPAW34,86 package. The core set of materials in C2DB have been obtained by extracting monolayers from experimentally known layered van der Waals crystals. Subsequently, new monolayers have been generated by systematic atom-substitution applied to the core materials, or using deep generative AI models.78 Recently, the C2DB has been complemented by the BiDB database containing homobilayers formed by stacking 1000 of the most stable monolayers from the C2DB in all possible commensurate configurations.94
 library. At present only C2DB is available via an OPTIMADE API, but in the future other CMR databases will also be available via OPTIMADE.
library. At present only C2DB is available via an OPTIMADE API, but in the future other CMR databases will also be available via OPTIMADE.
Base URL: https://cmr-optimade.fysik.dtu.dk
3.5 Crystallography Open Database (COD)
 prefix. This allows OPTIMADE to include experimental position and composition disorder information following the Crystallographic Information Framework.51
prefix. This allows OPTIMADE to include experimental position and composition disorder information following the Crystallographic Information Framework.51
Base URL: https://www.crystallography.net/cod/optimade
3.6 Joint Automated Repository for Various Integrated Simulations (JARVIS)
 prefix.
prefix.
Base URL: https://jarvis.nist.gov/optimade/jarvisdft
3.7 Materials Cloud
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 3D crystals and 3000 2D crystals, respectively, providing properties of experimentally-known inorganic compounds obtained via DFT simulations. The contributed databases include 2D topological insulators, pyrene-based metal organic frameworks, high-throughput Wannierisation, SrTiO3–CeO2 interfaces, tail-corrections in the molecular simulations of porous materials, hidden spontaneous polarisation in the chalcohalide photovoltaic Sn2SbS2I3, and the CURATED covalent organic frameworks database. The data has been used to investigate transport properties such as mobility,122,123 to search for
000 3D crystals and 3000 2D crystals, respectively, providing properties of experimentally-known inorganic compounds obtained via DFT simulations. The contributed databases include 2D topological insulators, pyrene-based metal organic frameworks, high-throughput Wannierisation, SrTiO3–CeO2 interfaces, tail-corrections in the molecular simulations of porous materials, hidden spontaneous polarisation in the chalcohalide photovoltaic Sn2SbS2I3, and the CURATED covalent organic frameworks database. The data has been used to investigate transport properties such as mobility,122,123 to search for  topological order,80 to screen and discover quantum spin-Hall insulators47,79 and Weyl semimetals,48 to obtain tight-binding-like Wannier Hamiltonians in a fully automated fashion,103 and to develop machine-learning methods for fast identification of low-dimensional materials.134
topological order,80 to screen and discover quantum spin-Hall insulators47,79 and Weyl semimetals,48 to obtain tight-binding-like Wannier Hamiltonians in a fully automated fashion,103 and to develop machine-learning methods for fast identification of low-dimensional materials.134
Current developments are focusing on a second “Materials Cloud Archive” provider, allowing submissions to Materials Cloud Archive to specify whether and how data contributed by users should be served via an OPTIMADE API, to enable advanced federated search over archived data. The first proof-of-concept of this integration is a recently published dataset of novel electride materials.139,140 The full list of OPTIMADE databases served by the Materials Cloud can be explored at the  endpoint, with a similar list for the Materials Cloud Archive available at
endpoint, with a similar list for the Materials Cloud Archive available at  . A landing page for both OPTIMADE providers is available at https://www.materialscloud.org/optimade.
. A landing page for both OPTIMADE providers is available at https://www.materialscloud.org/optimade.
 36 package. In addition to the server implementations, Materials Cloud also offers users several web applications that include clients of the OPTIMADE API, as we describe in more detail in Section 4.2.1.
36 package. In addition to the server implementations, Materials Cloud also offers users several web applications that include clients of the OPTIMADE API, as we describe in more detail in Section 4.2.1.
Index base URL: https://www.materialscloud.org/optimade/main
Index base URL: https://www.materialscloud.org/optimade/archive
3.8 Materials Platform for Data Science
 endpoint), physical properties (
endpoint), physical properties ( endpoint), and phase diagrams (
endpoint), and phase diagrams ( endpoint). These three data types are inter-linked into about 200 thousand distinct phases (
endpoint). These three data types are inter-linked into about 200 thousand distinct phases ( endpoint). Any distinct phase is uniquely determined by the chemical formula, space group, and Pearson symbol. Furthermore, each distinct phase has the permanent integer identifier
endpoint). Any distinct phase is uniquely determined by the chemical formula, space group, and Pearson symbol. Furthermore, each distinct phase has the permanent integer identifier  , e.g., see brookite (https://mpds.io/#phase_id/27712). The MPDS OPTIMADE implementation is specifically designed for the low response time and high retrieval speed, therefore some expensive operators (
, e.g., see brookite (https://mpds.io/#phase_id/27712). The MPDS OPTIMADE implementation is specifically designed for the low response time and high retrieval speed, therefore some expensive operators ( ,
,  ) are currently not supported (cf.Table 1†).
) are currently not supported (cf.Table 1†).
| Provider | N1 | N2 | N3 | Ntot |
|---|---|---|---|---|
| a The final column indicates the total number of structures served by each OPTIMADE API. Providers that serve multiple databases are indicated with *. Results for Materials Cloud and Materials Cloud Archive have been aggregated under the same title. The corresponding values from the 2021 paper3 are provided in brackets, where appropriate. | ||||
| AFLOW | 704![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302 (700 302 (700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 192) 192) |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 017 (62 017 (62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 293) 293) |
413![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 (382 797 (382![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 554) 554) |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 330 330 |
| Alexandria* | 939![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 084 084 |
48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 510 |
437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 768 768 |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 055 055![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 842 842 |
| COD | 458![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249 (416 249 (416![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 314) 314) |
4082 (3896) | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 739 (32 739 (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 420) 420) |
512![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282 282 |
| CMR | 2811 | 386 | 0 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 789 789 |
| JARVIS-DFT | 9017 | 1426 | 8084 | 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 096 |
| Materials cloud* | 961![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 564 564 |
4218 | 136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 176 |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 515 515![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 120 |
| Materials project | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 (27 424 (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309) 309) |
3750 (3545) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 (10 861 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 501) 501) |
154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 387 387 |
| MPDD | 811![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 136 136 |
80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 195 195 |
490![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 975 975![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666 666 |
| MPOD | 91 | 8 | 16 | 401 |
| MPDS | — | — | — | 507![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
| NOMAD | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 451 451![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056 (3 056 (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 359 359![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 594) 594) |
587![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 (532 923 (532![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 123) 123) |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 092![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 989 (1 989 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 611 611![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302) 302) |
12,116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021 021 |
| odbx* | 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648 (55) 648 (55) |
3179 (54) | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 009 (0) 009 (0) |
523![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 216 216 |
| omdb | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 718 (58 718 (58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 718) 718) |
690 (690) | 7428 (7428) | 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 566 566 |
| OQMD | 261![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 (153 400 (153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 113) 113) |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375 (11 375 (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011) 011) |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 673 (70 673 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252) 252) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226 226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 781 781 |
| TCOD | 7161 (2631) | 296 (296) | 662 (660) | 7452 |
| 2DMatpedia | 1172 | 739 | 255 | 6351 |
Base URL: https://api.mpds.io
3.9 Materials Project
 .36 Since May 2022, Materials Project has been serving formation energy data via its OPTIMADE API. In June 2023, MP started exposing additional thermodynamic stability in the form of energy distance to the convex hull via OPTIMADE for all 154k materials in its core database. The convex hull distance to the Materials Project is one of the most important properties of theoretical structures for experimentalists and simulators alike, as it indicates whether a postulated material is potentially synthesisable.
.36 Since May 2022, Materials Project has been serving formation energy data via its OPTIMADE API. In June 2023, MP started exposing additional thermodynamic stability in the form of energy distance to the convex hull via OPTIMADE for all 154k materials in its core database. The convex hull distance to the Materials Project is one of the most important properties of theoretical structures for experimentalists and simulators alike, as it indicates whether a postulated material is potentially synthesisable.
The MP OPTIMADE integration is complemented by a convenient open-source pymatgen90 interface in the form of the  class (https://github.com/materialsproject/pymatgen/blob/ec750ca15d02cdd51b0c0a7a4408af8e0d259223/pymatgen/ext/optimade.py#L31), designed to streamline access to these resources for existing users of pymatgen and the Materials Project. Additionally, efforts are underway to further expose the full set of MP
class (https://github.com/materialsproject/pymatgen/blob/ec750ca15d02cdd51b0c0a7a4408af8e0d259223/pymatgen/ext/optimade.py#L31), designed to streamline access to these resources for existing users of pymatgen and the Materials Project. Additionally, efforts are underway to further expose the full set of MP  data via the OPTIMADE endpoint under the
data via the OPTIMADE endpoint under the  namespace, mirroring the complete set of data recorded in the
namespace, mirroring the complete set of data recorded in the  SummaryDoc (https://github.com/materialsproject/emmet/blob/bf8a4ef09a0d9f91bb6e9fe3e2fca0acd3582306/emmet-core/emmet/core/summary.py#L137).
SummaryDoc (https://github.com/materialsproject/emmet/blob/bf8a4ef09a0d9f91bb6e9fe3e2fca0acd3582306/emmet-core/emmet/core/summary.py#L137).
Base URL: https://optimade.materialsproject.org
3.10 Material-Property-Descriptor Database
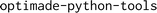 .36 Making the MPDD available via OPTIMADE was initially challenging, as MPDD stores and exchanges data in a way that prioritises high throughput and low storage requirements, including binary data, making it difficult or slow to make MPDD queryable as an OPTIMADE API on-the-fly. However, issues have been resolved by establishing a self-updating mirror of the dataset where structures are made OPTIMADE-compliant during transfer and with most associated MPDD-specific data available under the
.36 Making the MPDD available via OPTIMADE was initially challenging, as MPDD stores and exchanges data in a way that prioritises high throughput and low storage requirements, including binary data, making it difficult or slow to make MPDD queryable as an OPTIMADE API on-the-fly. However, issues have been resolved by establishing a self-updating mirror of the dataset where structures are made OPTIMADE-compliant during transfer and with most associated MPDD-specific data available under the  namespace, including dictionaries of metadata, properties, and descriptors.
namespace, including dictionaries of metadata, properties, and descriptors.
Base URL: http://optimade.mpdd.org
3.11 Materials Properties Open Database
 .36 In this context, the prefix
.36 In this context, the prefix  was employed to delineate specific database fields.
was employed to delineate specific database fields.
Base URL: http://mpod_optimade.cimav.edu.mx
3.12 Materials Resource Registry
3.13 Matterverse
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664
664![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858 hypothetical materials. The current structures were generated by combinatorial isovalent ionic substitutions on 5283 binary, ternary, and quaternary structural prototypes from the 2019 version of the ICSD database. A critical enabler for this database is the Materials 3-body Graph Network (M3GNet) universal interatomic potential encompassing 89 elements of the periodic table.18 Along with the information of lattice parameters, atom coordinates and Ehull, matterverse.ai (https://matterverse.ai/) also provides the predicted formation energies, bandgaps (of multiple fidelities, including PBE, HSE and experimental), and bulk and shear moduli. As an ongoing effort, matterverse.ai is growing in two directions, (i) increasing the number of hypothetical materials via various structure generation strategies, and (ii) increasing the number of ML-predicted properties.
858 hypothetical materials. The current structures were generated by combinatorial isovalent ionic substitutions on 5283 binary, ternary, and quaternary structural prototypes from the 2019 version of the ICSD database. A critical enabler for this database is the Materials 3-body Graph Network (M3GNet) universal interatomic potential encompassing 89 elements of the periodic table.18 Along with the information of lattice parameters, atom coordinates and Ehull, matterverse.ai (https://matterverse.ai/) also provides the predicted formation energies, bandgaps (of multiple fidelities, including PBE, HSE and experimental), and bulk and shear moduli. As an ongoing effort, matterverse.ai is growing in two directions, (i) increasing the number of hypothetical materials via various structure generation strategies, and (ii) increasing the number of ML-predicted properties.
 .36
.36
Base URL: https://optimade.matterverse.ai
3.14 NOMAD
NOMAD enables individual researchers to make their data available to a wide range of possible clients and applications. All data is formally described through rich metadata schema41 and can be analyzed with build in containerised tools and notebooks.109 Data in NOMAD is provided through the OPTIMADE API, NOMAD specific APIs, and a rich graphical user interface with faceted search, (meta)data explorer, and visualisations for material properties. API access is particularly important for re-use of the data, e.g. with artificial-intelligence (AI) methods. A collection of AI tools is available in the NOMAD AI Toolkit.109 Besides supporting the community with the central data infrastructure, NOMAD offers the same software111 for local installation through NOMAD Oasis, which allows research groups to manage and provide their own research data individually and customise the software accordingly.
Base URL: https://nomad-lab.eu/prod/rae/optimade
3.15 Open Database of Xtals (odbx)
 36 packages. As well as serving the standard OPTIMADE properties, odbx also serves stability data (hull distances, formation energies) and the DFT parameters used to relax the structures under the
36 packages. As well as serving the standard OPTIMADE properties, odbx also serves stability data (hull distances, formation energies) and the DFT parameters used to relax the structures under the  namespace. odbx serves multiple distinct datasets; the
namespace. odbx serves multiple distinct datasets; the  endpoint of the index base URL below can be used to retrieve them.
endpoint of the index base URL below can be used to retrieve them.
Index base URL: https://optimade-index.odbx.science
3.16 Open Materials Database (omdb)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264 structures for access via programmatic interaction using this toolkit. The structures are also accessible via a web interface. Recently, it is being integrated in the broader database effort Anyterial (https://www.anyterial.se/), which also includes the ADAQ database of point defects.27
264 structures for access via programmatic interaction using this toolkit. The structures are also accessible via a web interface. Recently, it is being integrated in the broader database effort Anyterial (https://www.anyterial.se/), which also includes the ADAQ database of point defects.27
Base URL: https://optimade.openmaterialsdb.se
3.17 Open Quantum Materials Database
 prefix, including formation energies, bandgap, and stabilities of compounds.
prefix, including formation energies, bandgap, and stabilities of compounds.
Base URL: https://oqmd.org/optimade
3.18 Comparison of data available
An important benefit of a universal API format such as OPTIMADE is the ability to simultaneously request and unify results from different databases. While the key features of the databases are highlighted under the subsections dedicated to the respective providers in Section 3, a summarizing list of the implementations tested and confirmed to support the OPTIMADE API is shown in Table 1.† These databases are all openly accessible and provide users with a broad range of materials classes, applications and modalities.The table shows the return from three requests that explore materials that contain at least one element from Group 14 (N1), and then constrains the search to cover only binary materials (N2), and only ternary materials without toxic lead (N3):
These queries directly duplicate the three detailed in the 2021 OPTIMADE paper.3 The ability to repeat the query attests to how the OPTIMADE API helps with reproducibility in research. In addition, we provide the total number of structures served by each OPTIMADE API in the Ntot column.
Comparison of this table to that in ref. 3 bears witness to the growth and impact of OPTIMADE. We see several additional providers (Alexandria, BioExcel, CMR, JARVIS, MPOD, MPDD and 2DMatpedia) that now support OPTIMADE, and several new databases hosted by pre-existing providers. Furthermore, among the databases that did support OPTIMADE in 2021, there has been an impressive growth in the volume of returned data, reflecting their continued efforts to assimilate further data.
4 Application of OPTIMADE to real-life problems
A key goal for the OPTIMADE API is for it to act as an enabling technology for materials discovery, design and other new research avenues. Feedback from users is crucial to motivate the future development of the API. Therefore, in the following sections, we spotlight several use cases of the application of the OPTIMADE API to real-life systems, firstly in Section 4.1 by supplying data for machine learning, and secondly in Section 4.2 by providing data for screening and other studies. We highlight examples which benefit from access to the wealth of data available in large databases (e.g., the hard-coating alloys database discussed immediately below), and examples that benefit from access to specialist data available only in the small and focused databases (e.g., Section 4.1.1).Furthermore, there has been additional use of OPTIMADE in the literature: firstly how OPTIMADE has been a central tool to access materials data for materials discovery, secondly as a template for materials data curation and access, and thirdly through online web-based interfaces:
4.1 Discovery
The hard-coating alloys database (HADB)74 exploited the OPTIMADE API to rapidly and easily provide the browser-based graphical web interface as well as a RESTful API. A second application of the OPTIMADE API was to query and retrieve an unprecedented volume of data to train an attention-based crystal graph convolutional neural network to accurately predict the formation energy, total energy, bandgap, and Fermi energy of a broad range of crystals.136 Finally, OPTIMADE has found application in materials discovery, where it was used in ref. 54 to assess the novelty of predicted structures in a high-throughput study on quaternary mixed metal chalcohalide perovskites using the client.36 As these structures were ingested into an OPTIMADE-compliant database, in this case NOMAD,111 any future OPTIMADE queries for novelty in this chemical space will yield the results of this study.
client.36 As these structures were ingested into an OPTIMADE-compliant database, in this case NOMAD,111 any future OPTIMADE queries for novelty in this chemical space will yield the results of this study.
4.2 Template
Development of the OPTIMADE API has motivated and guided data access in other ongoing projects. For example, firstly OPTIMADE API collaborates with, and is being used in, the development of the FAIRmat metadata, dictionaries, and materials ontology.110 The inclusion in other community efforts reflects the maturity and uptake of the OPTIMADE API. A second example is the BIG-MAP project, where the consortium plans to use the OPTIMADE API to guide the access of the data gathered in the Battery Interface Genome.164.3 Interfaces and integrations
The MarketPlace Project128 has integrated the OPTIMADE Gateway (Section 2.3.2) into the platform, which will make it possible to perform OPTIMADE queries through its global search functionality. OntoTrans91 has developed the Open Translation Environment to perform ontology-driven data pipelines to retrieve, parse, map, and transform data. As part of the project, an OPTIMADE plugin has been developed for the system, making it possible to request and digest OPTIMADE resources. The next steps include semantic mappings for the OPTIMADE data models for true semantic data interoperability.4.4 Machine learning
Machine learning is a promising tool that is already having a significant impact in the materials sciences. Machine learning starts from already computed data about a system and trains a model to capture trends. The machine learning model can then make predictions and design materials quicker and more cost effectively than performing additional experiments.Machine learning relies on having a pool of historical data available. This is where the OPTIMADE API offers a significant boost, by opening access to a wide range of materials databases that hold complementary data. We highlight the help offered by the OPTIMADE API to machine learning with two case studies.
We use the OPTIMADE API to retrieve high-entropy alloy materials using the filter query
The complete dataset obtained using OPTIMADE is split into training and testing sets (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) while ensuring that ratio of entries from each provider is the same in both training and testing sets. The training set is used to train the “combined” model M-C. Data points from the providers P1, P2, and P3 that appear in the training set are used to train the models M-P1, M-P2, M-P3, respectively. The predictive power of the models is assessed by calculating the R2 on the same test set.
1 ratio) while ensuring that ratio of entries from each provider is the same in both training and testing sets. The training set is used to train the “combined” model M-C. Data points from the providers P1, P2, and P3 that appear in the training set are used to train the models M-P1, M-P2, M-P3, respectively. The predictive power of the models is assessed by calculating the R2 on the same test set.
We choose Random Forest Regressor (with default parameters, from the  1.2 package in Python96) to construct the machine learning models for our example. Standard structural entries of the OPTIMADE specification,
1.2 package in Python96) to construct the machine learning models for our example. Standard structural entries of the OPTIMADE specification,  and
and  , are used to construct vectors codifying the composition of each material and also to calculate the density of each material. The ‘composition vectors’, described above, are used as input to machine learning models that are trained to predict the densities (output). Models that are trained on data from only one provider (M-P1, M-P2, and M-P3) perform poorly when tested on data from all the providers (R2 = 0.316, 0.104, −2.79 for M-P1, M-P2, and M-P3 respectively). Meanwhile, the “combined” model that is trained on data from all providers (M-C) performs very well (R2 = 0.995). A comparison of the R2 values is shown in the top-left of Fig. 2.
, are used to construct vectors codifying the composition of each material and also to calculate the density of each material. The ‘composition vectors’, described above, are used as input to machine learning models that are trained to predict the densities (output). Models that are trained on data from only one provider (M-P1, M-P2, and M-P3) perform poorly when tested on data from all the providers (R2 = 0.316, 0.104, −2.79 for M-P1, M-P2, and M-P3 respectively). Meanwhile, the “combined” model that is trained on data from all providers (M-C) performs very well (R2 = 0.995). A comparison of the R2 values is shown in the top-left of Fig. 2.
We can get a better insight into the benefits of leveraging data from multiple providers by looking at comparison of actual density values and those predicted by the machine learning models for small random sampling of materials (scatter plot in Fig. 2). For models trained only on data from a single provider (M-P1, M-P2, and M-P3), the prediction is quite accurate when tested on data from the same provider (indicted by ‘cross’ markers). However, most of the predictive power is lost when tested on data from another provider (‘dot’ markers). This explains their poor R2 values. Meanwhile, the model which is trained on data from all providers (M-C) retains its predictive power when tested on data from all the providers. The number of materials returned by each provider is shown in the bar-graph on the bottom right of Fig. 2. A Venn diagram of unique elements that appear in the materials from each provider are shown in the centre right in Fig. 2. Therefore, the OPTIMADE API offers the significant benefit to merge the information from the datasets together.
Another possible application is to repeat previous high-throughput materials design campaigns on the wider set of structures now available through OPTIMADE. As each structure is time-stamped, active or ongoing workflows can be implemented to constantly monitor and screen new crystal structures against the search criteria of the campaign, to avoid having to redo such searches from scratch. Structure-based property prediction models, e.g., MODNet (for small structure–property datasets)28,29,76 or graph-based models (where larger structure–property datasets are available),19,20,143 can be leveraged to sift through huge swathes of available crystal structures, with the results being used to prioritise future calculations, attempts at synthesis, characterisation experiments or model retraining, by selecting for structures with combinations of particular target properties.
4.5 Data provision
The OPTIMADE API has also been used to provide data for a rich variety of other scientific analysis approaches, below we highlight three projects that take advantage of the comprehensive range of data offered by OPTIMADE.To facilitate this goal, Materials Cloud127 provides the OPTIMADE client, a web application to perform the structure search task via a unified and user-friendly GUI, empowering users to not only generate and execute complex OPTIMADE queries, but also to provide immediate graphical access and visualisation of the resulting structures. The OPTIMADE client can be embedded in other applications or used as a standalone tool, which is hosted on Materials Cloud at https://optimadeclient.materialscloud.io/. The filtering section of the GUI is shown in Fig. 3. A dropdown (at the top) is provided to select any of the known and automatically discovered OPTIMADE database providers. A periodic table widget allows users to select which elements need to be included (green) or excluded (red) in the compounds; additional filtering tools are also provided, such as for the number of elements and of sites, and for the dimensionality. An OPTIMADE query string is then produced (which can be optionally manually modified). After the search button is clicked, the OPTIMADE query is sent to the selected database provider. The results are curated and shown in the results widget. Here, the structures are visualised and can be downloaded.
 | ||
| Fig. 3 The search interface of the standalone OPTIMADE client. The user can select any OPTIMADE-compliant database and make a query based on filters specified through GUI elements. | ||
Materials Cloud also provides various tools that leverage the power of the OPTIMADE client. One example is the Quantum ESPRESSO input generator, available as a tool at https://www.materialscloud.org/work/tools/qeinputgenerator. A structure can be sent directly to this tool using a button in the OPTIMADE client (shown at the bottom of Fig. 3). The Quantum ESPRESSO input generator enables any user to obtain a working input file for the Quantum ESPRESSO DFT code,42,43 including an automated selection of all numerical parameters, by just specifying a crystal structure (either by uploading it, or by selecting it from OPTIMADE). A second example is the Quantum ESPRESSO app (https://aiidalab-qe.readthedocs.io) developed within the AiiDAlab platform.144 It allows users to run complex computational workflows from the web browser, using straightforward graphical user interfaces for structure selection (including via OPTIMADE), parameter selection and inspection of the results.
Xerus uses a straightforward OPTIMADE interface to connect to the multiple databases hosted by members of the OPTIMADE consortia. The dynamic OPTIMADE providers list allows new databases to be automatically included in Xerus search results. Additional filtering parameters can be used to refine the searches towards materials stable (or predicted to be stable) at the experimental conditions (e.g., low temperature or high pressure).
Results are shown in Fig. 4(b), where we plot the intercalation voltage of Li–Ni–O materials from the extracted VASP70 data in Materials Project and NOMAD and compare predictions to those from CASTEP. The comparability of results illustrates the functionality of the workflow. Since databases contain different structures, the OPTIMADE API facilitates the process of materials investigations by aggregating the query of all of them.
MatCloud81 is a cloud-based integrated high-throughput computational materials infrastructure, which is directly connected to computing clusters and material property databases.145,146 Users worldwide can visually design structures, create and run simulation jobs through workflows, and retrieve crystal structures from multiple databases using OPTIMADE; all the user needs is a web browser. MatCloud provides a Graphical User Interface (GUI)-based environment for users to intuitively create, enact and monitor a workflow. The MatCloud workflow system includes a front-end workflow designer and a back-end workflow engine, and supports the creation of workflows by a drag-and-drop approach.
Fig. 5(a) shows a workflow that retrieves a Si8 crystal structure from the MPDS database through OPTIMADE, and replaces two Si atoms with Ge to produce structures in the Si–Ge chemical space (Fig. 5(b)). The band structure and density of states (DOS) are then simulated respectively over the chemical space in a high-throughput manner. Fig. 5(c) and (d) show the visualisation results for the band structure and total DOS of the structures.
5 Future of OPTIMADE
The rapid expansion of materials databases, the use cases presented, and the adoption of machine learning motivate the continued work on the OPTIMADE API. We describe below firstly the ongoing tutorials that introduce new users to OPTIMADE, secondly the workshops that enable the development and promulgation of the OPTIMADE API, and finally the features currently under development coming out of those recent workshops.5.1 Workshops & tutorials
The OPTIMADE consortium originated from the workshop “Open Databases Integration for Materials Design”, held at the Lorentz Center in Leiden, Netherlands in October 2016. There were follow-up workshops held at CECAM in Lausanne, Switzerland annually from 2018 to 2023, with events since 2020 also supporting remote attendees. The workshops in 2022 and 2023 were accompanied by on-site tutorials during the first two days. There was also a partner workshop, “Ontologies for machine learning driven materials design” held at Linköping University, Sweden in 2021. Going forward, the OPTIMADE API will undergo continued development through further annual workshops and publicly advertised monthly video calls. There are continual efforts to reach out to new databases to help accelerate their adoption of the format.Several tutorial exercises have been developed and delivered at a variety of conferences and topical workshops, outside of the OPTIMADE annual meetings. Eight different exercises developed by the community are now hosted on the Materials-Consortia GitHub repository (https://github.com/Materials-Consortia/optimade-tutorial-exercises), ranging from the basics of the OPTIMADE filter syntax and URL structure, to machine-learning pipelines operating on database-specific properties, all the way up to hosting an OPTIMADE API for a new database.
5.2 Upcoming features for future OPTIMADE releases
The ongoing real-life use cases of the OPTIMADE API have identified a series of opportunities to extend the API and make it more applicable to a wide range of materials systems. The features currently under development include: endpoint.
So far, OPTIMADE can only be used to describe static structures. We are, however, working to expand the OPTIMADE specification, so that OPTIMADE can be used for sharing trajectory data as well. Such data can originate from structural optimisations, or from Monte Carlo and molecular dynamics simulations. These trajectories could be used as a starting point for new simulations, to train machine learning potentials or to extract dynamical properties.
endpoint.
So far, OPTIMADE can only be used to describe static structures. We are, however, working to expand the OPTIMADE specification, so that OPTIMADE can be used for sharing trajectory data as well. Such data can originate from structural optimisations, or from Monte Carlo and molecular dynamics simulations. These trajectories could be used as a starting point for new simulations, to train machine learning potentials or to extract dynamical properties.
 endpoint.
It has been suggested several times whether it would be possible to have a method to create groups of entries. For example, all the structures that were used to generate a certain machine-learning potential, or all the structures that pertain to a certain research project. We therefore plan to introduce a
endpoint.
It has been suggested several times whether it would be possible to have a method to create groups of entries. For example, all the structures that were used to generate a certain machine-learning potential, or all the structures that pertain to a certain research project. We therefore plan to introduce a  endpoint, which would contain metadata for the collection as a whole, as well as references to all the individual entries that belong to this collection.
endpoint, which would contain metadata for the collection as a whole, as well as references to all the individual entries that belong to this collection.
 endpoint is to handle biomolecular structures, which can only be described statistically as a trajectory with multiple configurations. The OPTIMADE specification will therefore be extended to standardise the various dynamical fields and order parameters that are used to describe biomolecular structures. This covers fields that are typically stored in PDB files such as insertion codes, Chain IDs and sequence information of proteins, DNA and RNA.
endpoint is to handle biomolecular structures, which can only be described statistically as a trajectory with multiple configurations. The OPTIMADE specification will therefore be extended to standardise the various dynamical fields and order parameters that are used to describe biomolecular structures. This covers fields that are typically stored in PDB files such as insertion codes, Chain IDs and sequence information of proteins, DNA and RNA.
6 Conclusion
The OPTIMADE API provides users with easy access to many of the world leading materials databases. Since the initial release, the OPTIMADE API has not only been adopted by scientists as a tool to drive innovation, but furthermore served as a template for data curation. In this paper, we have provided use cases for how the breadth of data made available through OPTIMADE enables discovery in both academia and industry. The development of the OPTIMADE API has continued apace. Major new support for enhanced property definitions and partial data formats have recently been added and will underpin future work on trajectories and biomolecular data. The concept that sub-consortia of databases are responsible for the definition of new sets of shared properties will accelerate the extension of the OPTIMADE API to other disciplines and fields.Through monthly meetings, and with the continuing support of CECAM, the developers are continuing to extend the range of properties accessible via OPTIMADE APIs. Plans to both expand the format to cover challenges arising from dealing with molecular dynamics data, and continued outreach to support the adoption of the API by additional databases, will further expand the range of scientific use cases that OPTIMADE enables. Increasingly, use cases will take advantage of the unique advantages OPTIMADE has to offer; namely the robust and straightforward aggregation and data unification from the multitude of growing and federated data sources.
Author contributions
These authors were active developers and reviewers of the specification: Johan Bergsma, Matthew L. Evans, and Andrius Merkys. These authors were active developers of implementations for database providers and/or made contributions to specification: Johan Bergsma, Matthew L. Evans, Andrius Merkys, Oskar B. Andersson, Casper W. Andersen, Daniel Beltrán, Evgeny Blokhin, Tara Boland, Rubén Castañeda Balderas, Kamal Choudary, Alberto Díaz Díaz, Rodrigo Domínguez García, Hagen Eckert, Kristjan Eimre, María Elena Fuentes Montero, Adam M. Krajewski, Jens Jørgen Mortensen, José Manuel Nápoles Duarte, Jacob Pietryga, Ji Qi, Felipe de Jesús Trejo Carrillo, Antanas Vaitkus, Jusong Yu, Adam Zettel. These authors primarily contributed to the paper (along with all other authors): Pedro Baptista de Castro, Johan Carlsson, Tiago F. T. Cerqueira, Simon Divilov, Hamidreza Hajiyani, Felix Hanke, Kevin Jose, Corey Oses, Janosh Riebesell, Jonathan Schmidt, Donald Winston, Christen Xie, Xiaoyu Yang. These authors managed individual databases that have implemented the OPTIMADE API: Sara Bonella, Silvana Botti, Stefano Curtarolo, Claudia Draxl, Luis Edmundo Fuentes Cobas, Adam Hospital, Zi-Kui Liu, Miguel Marques, Nicola Marzari, Andrew Morris, Shyue Ping Ong, Modesto Orozco, Kristin A. Persson, Kristian Thygesen, Chris Wolverton. These authors are organisers of the OPTIMADE API and are also senior developers who contributed to code and/or to the specification: Rickard Armiento, Gareth J. Conduit, Saulius Gražulis, Giovanni Pizzi, Gian-Marco Rignanese, Markus Scheidgen, Cormac Toher.Conflicts of interest
G. J. C. is a shareholder and Director of Intellegens Ltd. G.-M. R. is a shareholder and Chief Innovation Officer of Matgenix SRL. E. B. is a shareholder and Director of Materials Platform for Data Science OÜ.Acknowledgements
The authors acknowledge support from CECAM in Lausanne (Switzerland) and the Lorentz Center in Leiden (Netherlands) for hosting OPTIMADE workshops; and for the partner workshop at Linköping University in 2021, support from Psi-k, NCCR MARVEL (a National Centre of Competence in Research, funded by the Swiss National Science Foundation, grant no. 205602), and the Swedish e-Science Research Centre (SeRC). G. J. C. would like to acknowledge financial support from the Royal Society. M. L. E. thanks the BEWARE scheme of the Wallonia-Brussels Federation for funding under the European Commission's Marie Curie-Skłodowska Action (Co-fund 847587). CT thanks NSF (DMR-2219788). M. S. and C. D. acknowledge support from the German Research Foundation (DFG) through the NFDI consortium FAIRmat (project 460197019). K. E., J. Y., S. B., N. M., and G. P. acknowledge funding by the NCCR MARVEL (a National Centre of Competence in Research, funded by the Swiss National Science Foundation, grant no. 205602). G. P. acknowledge funding by the Open Research Data Program of the ETH Board (project “PREMISE”: Open and Reproducible Materials Science Research, https://open-research-data-portal.ch/projects/open-and-reproducible-materials-science-research/). R. A. and O. B. A. acknowledges funding from Vetenskapsrådet grant no. 2020-05402 and the Swedish e-Science Research Centre. A. M., A. V. and S. G. acknowledge funding under the Programme “University Excellence Initiatives” of the Ministry of Education, Science and Sports of the Republic of Lithuania (Measure No. 12-001-01-01-01 “Improving the Research and Study Environment”). A. J. M. acknowledges support from EPSRC via CCP-NC (EP/T026642/1), CCP9 (EP/T026375/1), UKCP (EP/P022561/1) and Baskeville (EP/T022221/1). The work by J. S., D. W., J. Q., S. P. O., and K. A. P. were supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under contract no. DE-AC02-05-CH11231; the Materials Project program (KC23MP). D. B, A. H., and M. O. acknowledge funding by the European Union's Horizon 2020 programme under grant agreements No. 675728 (BioExcel), No. 823830 (BioExcel-2), No. 720270 (Human Brain Project SGA1), No. 785907 (Human Brain Project SGA2), No. 945539 (Human Brain Project SGA3) and by the Horizon Europe Programme under No. 101093290 (BioExcel-3) and No. 101094651 (Molecular Dynamics Data Bank). K. C. acknowledges funding support from the CHIPS Metrology Program, part of CHIPS for America, National Institute of Standards and Technology, U.S. Department of Commerce. X. Y. acknowledges the funding support from National Natural Science Foundation of China (NSFC) under the grant no. 62376258.References
- A. Calzolari, C. Oses, C. Toher, M. Esters, X. Campilongo, S. P. Stepanoff, D. E. Wolfe and S. Curtarolo, Plasmonic high-entropy carbides, Nat. Commun., 2022, 13, 5993, DOI:10.1038/s41467-022-33497-1.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, A. Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshøj, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu andX. Yang, The OPTIMADE Specification, version 1.0.0, Zenodo, 2020, p. 1, DOI:10.5281/zenodo.4195051.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, A. Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshøj, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu and X. Yang, OPTIMADE: an API for exchanging materials data, Sci. Data, 2021, 8, 217, DOI:10.1038/s41597-021-00974-z.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, A. Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshøj, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu andX. Yang, The OPTIMADE Specification, version 1.1.0, Zenodo, 2021, p. 1, DOI:10.5281/zenodo.4251947.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, A. Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshøj, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu andX. Yang, The OPTIMADE Specification, version 1.2.0, 2024, https://github.com/Materials-Consortia/OPTIMADE/blob/v1.2.0/optimade.rst Search PubMed.
- P. Andrio, A. Hospital, J. Conejero, L. Jordà, M. del Pino, L. Codo, S. Soiland-Reyes, C. Goble, D. Lezzi, R. Badia, M. Orozco and J. L. Gelpi, BioExcel Building Blocks, a software library for interoperable biomolecular simulation workflows, Sci. Data, 2019, 6, 169–179, DOI:10.1038/s41597-019-0177-4.
- R. Armiento. Database-Driven High-Throughput Calculations and Machine Learning Models for Materials Design, in Machine Learning Meets Quantum Physics, Lecture Notes in Physics, ed. K. T. Schütt, S. Chmiela, O. A. von Lilienfeld, A. Tkatchenko, K. Tsuda, and K.-R. Müller, Springer International Publishing, Cham, 2020, pp. 377–395, DOI:10.1007/978-3-030-40245-7_17, ISBN 978-3-030-40245-7.
- M. Aykol, S. Kirklin and C. Wolverton, Thermodynamic aspects of cathode coatings for lithium-ion batteries, Adv. Energy Mater., 2014, 4(17), 1400690, DOI:10.1002/aenm.201400690.
- P. Baptista de Castro, K. Terashima, M. G. Esparza Echevarria, H. Takeya and Y. Takano, XERUS: An Open-Source Tool for Quick XRD Phase Identification and Refinement Automation, Adv. Theory Simul., 2022, 5(5), 2100588, DOI:10.1002/adts.202100588.
- A. Baratta, A. Cimino, F. Longo, V. Solina and S. Verteramo, The Impact of ESG Practices in Industry with a Focus on Carbon Emissions: Insights and Future Perspectives, Sustainability, 2023, 15, 6685, DOI:10.3390/su15086685.
- M. Barjasteh, M. Vossoughi, M. Bagherzadeh and K. Pooshang Bagheri, MIL-100(Fe) a potent adsorbent of Dacarbazine: Experimental and molecular docking simulation, Chem. Eng. J., 2023, 452, 138987, DOI:10.1016/j.cej.2022.138987.
- D. Beltrán, A. Hospital, J. L. Gelpí and M. Orozco, A new paradigm for molecular dynamics databases: the COVID-19 database, the legacy of a titanic community effort, Nucleic Acids Res., 2023, 52(D1), D393–D403, DOI:10.1093/nar/gkad991.
- H. J. Bernstein, J. C. Bollinger, I. D. Brown, S. Gražulis, J. R. Hester, B. McMahon, N. Spadaccini, J. D. Westbrook and S. P. Westrip, Specification of the crystallographic information file format, version 2.0, J. Appl. Crystallogr., 2016, 49(1), 277–284, DOI:10.1107/S1600576715021871.
- T. B. Brown, B. Mann, N. Ryder, M. Subbiah, J. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. M. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever, and D. Amodei, Language Models are Few-Shot Learners, arXiv, 2020, preprint, arXiv:2005.14165, DOI:10.48550/arXiv.2005.14165.
- D. Campi, N. Mounet, M. Gibertini, G. Pizzi and N. Marzari, Expansion of the Materials Cloud 2D Database, ACS Nano, 2023, 17(12), 11268–11278, DOI:10.1021/acsnano.2c11510.
- I. E. Castelli, D. J. Arismendi-Arrieta, A. Bhowmik, I. Cekic-Laskovic, S. Clark, R. Dominko, E. Flores, J. Flowers, K. U. Frederiksen, J. Friis, A. Grimaud, K. V. Hansen, L. J. Hardwick, K. Hermansson, L. Königer, H. Lauritzen, F. L. Cras, H. Li, S. Lyonnard, H. Lorrmann, N. Marzari, L. Niedzicki, G. Pizzi, F. Rahmanian, H. Stein, M. Uhrin, W. Wenzel, M. Winter, C. Wölke and T. Vegge, Data management plans: the importance of data management in the BIG-MAP project, Batteries Supercaps, 2021, 4, 1803–1812, DOI:10.1002/batt.202100117.
- L. Chan, G. R. Hutchison and G. M. Morris, Understanding ring puckering in small molecules and cyclic peptides, J. Chem. Inf. Model., 2021, 61(2), 743–755, DOI:10.1021/acs.jcim.0c01144.
- C. Chen and S. P. Ong, A universal graph deep learning interatomic potential for the periodic table, Nat. Comput. Sci., 2022, 2(11), 718–728, DOI:10.1038/s43588-022-00349-3.
- C. Chen, W. Ye, Y. Zuo, C. Zheng and S. P. Ong, Graph Networks as a Universal Machine Learning Framework for Molecules and Crystals, Chem. Mater., 2019, 31(9), 3564–3572, DOI:10.1021/acs.chemmater.9b01294.
- K. Choudhary and B. DeCost, Atomistic Line Graph Neural Network for improved materials property predictions, npj Comput. Mater., 2021, 7(1), 1–8, DOI:10.1038/s41524-021-00650-1.
- K. Choudhary and K. Garrity, Designing high-Tc superconductors with BCS-inspired screening, density functional theory, and deep-learning, npj Comput. Mater., 2022, 8(1), 244, DOI:10.1038/s41524-022-00933-1.
- K. Choudhary, K. F. Garrity, A. C. Reid, B. DeCost, A. J. Biacchi, A. R. Hight Walker, Z. Trautt, J. Hattrick-Simpers, A. G. Kusne, A. Centrone, A. Davydov, J. Jiang, R. Pachter, G. Cheon, E. Reed, A. Agrawal, X. Qian, V. Sharma, H. Zhuang, S. V. Kalinin, B. G. Sumpter, G. Pilania, P. Acar, S. Mandal, K. Haule, D. Vanderbilt, K. Rabe and F. Tavazza, The joint automated repository for various integrated simulations (jarvis) for data-driven materials design, npj Comput. Mater., 2020, 6(1), 173, DOI:10.1038/s41524-020-00440-1.
- S. J. Clark, M. D. Segall, C. J. Pickard, P. J. Hasnip, M. J. Probert, K. Refson and M. C. Payne, First principles methods using CASTEP, Z. Kristallogr., 2005, 220(5–6), 567–570, DOI:10.1524/zkri.220.5.567.65075.
- T. B. R. Company, Chemicals global market report 2022, 2022, https://www.researchandmarkets.com/reports/5598260/chemicals-global-market-report-2022.
- D. S. A. Corporation, BIOVIA Pipeline Pilot, 2022, https://www.3ds.com/products-services/biovia/products/data-science/pipeline-pilot/.
- Crystallography Domain Ontology, https://github.com/emmo-repo/domain-crystallography, 2019.
- J. Davidsson, V. Ivády, R. Armiento and I. A. Abrikosov, ADAQ: automatic workflows for magneto-optical properties of point defects in semiconductors, Comput. Phys. Commun., 2021, 269, 108091, DOI:10.1016/j.cpc.2021.108091.
- P.-P. De Breuck, M. L. Evans and G.-M. Rignanese, Robust model benchmarking and bias-imbalance in data-driven materials science: a case study on MODNet, J. Phys.: Condens. Matter, 2021, 33(40), 404002, DOI:10.1088/1361-648x/ac1280.
- P.-P. De Breuck, G. Hautier and G.-M. Rignanese, Materials property prediction for limited datasets enabled by feature selection and joint learning with MODNet, npj Comput. Mater., 2021, 7(1), 1–8, DOI:10.1038/s41524-021-00552-2.
- S. Divilov, H. Eckert, D. Hicks, C. Oses, C. Toher, R. Friedrich, M. Esters, M. J. Mehl, A. C. Zettel, Y. Lederer, E. Zurek, J.-P. Maria, D. W. Brenner, X. Campilongo, S. Filipovic, W. G. Fahrenholtz, C. J. Ryan, C. M. DeSalle, R. J. Crealese, D. E. Wolfe, A. Calzolari and S. Curtarolo, Disordered enthalpy-entropy descriptor for high-entropy ceramics discovery, Nature, 2024, 625, 66–73, DOI:10.1038/s41586-023-06786-y.
- C. Draxl and M. Scheffler, NOMAD: The FAIR concept for big data-driven materials, MRS Bull., 2018, 43, 676–682, DOI:10.1557/mrs.2018.208.
- C. Draxl and M. Scheffler, The NOMAD laboratory: from data sharing to artificial intelligence, JPhys Mater., 2019, 2(3), 036001, DOI:10.1088/2515-7639/ab13bb.
- Elementary Multiperspective Material Ontology (EMMO), 2019, https://github.com/emmo-repo/EMMO.
- J. Enkovaara, C. Rostgaard, J. J. Mortensen, J. Chen, M. Dułak, L. Ferrighi, J. Gavnholt, C. Glinsvad, V. Haikola, H. A. Hansen, H. H. Kristoffersen, M. Kuisma, A. H. Larsen, L. Lehtovaara, M. Ljungberg, O. Lopez-Acevedo, P. G. Moses, J. Ojanen, T. Olsen, V. Petzold, N. A. Romero, J. Stausholm-Møller, M. Strange, G. A. Tritsaris, M. Vanin, M. Walter, B. Hammer, H. Häkkinen, G. K. H. Madsen, R. M. Nieminen, J. K. Nørskov, M. Puska, T. T. Rantala, J. Schiøtz, K. S. Thygesen and K. W. Jacobsen, Electronic structure calculations with GPAW: a real-space implementation of the projector augmented-wave method, J. Phys.: Condens. Matter, 2010, 22(25), 253202, DOI:10.1088/0953-8984/22/25/253202.
- M. Esters, C. Oses, S. Divilov, H. Eckert, R. Friedrich, D. Hicks, M. J. Mehl, F. Rose, A. Smolyanyuk, A. Calzolari, X. Campilongo, C. Toher and S. Curtarolo, aflow.org: A web ecosystem of databases, software and tools, Comput. Mater. Sci., 2023, 216, 111808, DOI:10.1016/j.commatsci.2022.111808.
- M. L. Evans, C. W. Andersen, S. Dwaraknath, M. Scheidgen, Á. Fekete and D. Winston, optimade-python-tools: a Python library for serving and consuming materials data via OPTIMADE APIs, J. Open Source Softw., 2021, 6(65), 3458, DOI:10.21105/joss.03458.
- M. L. Evans and A. J. Morris, matador: a Python library for analysing, curating and performing high-throughput density-functional theory calculations, J. Open Source Softw., 2020, 5(54), 2563, DOI:10.21105/joss.02563.
- FAIRsharing.org: OPTIMADE; Open Databases Integration for Materials Design, 2020, DOI:10.25504/FAIRsharing.xvfqAC.
- D. C. Ford, D. Hicks, C. Oses, C. Toher and S. Curtarolo, Metallic glasses for biodegradable implants, Acta Mater., 2019, 176, 297–305, DOI:10.1016/j.actamat.2019.07.008.
- L. Fuentes-Cobas, D. Chateigner, M. Fuentes-Montero, G. Pepponi and S. Grazulis, The representation of coupling interactions in the Material Properties Open Database (MPOD), Adv. Appl. Ceram., 2017, 116(8), 428–433, DOI:10.1080/17436753.2017.1343782.
- L. M. Ghiringhelli, C. Baldauf, T. Bereau, S. Brockhauser, C. Carbogno, J. Chamanara, S. Cozzini, S. Curtarolo, C. Draxl, S. Dwaraknath, Á. Fekete, J. Kermode, C. T. Koch, M. Kühbach, A. N. Ladines, P. Lambrix, M.-O. Himmer, S. V. Levchenko, M. Oliveira, A. Michalchuk, R. E. Miller, B. Onat, P. Pavone, G. Pizzi, B. Regler, G.-M. Rignanese, J. Schaarschmidt, M. Scheidgen, A. Schneidewind, T. Sheveleva, C. Su, D. Usvyat, O. Valsson, C. Wöll and M. Scheffler, Shared metadata for data-centric materials science, Sci. Data, 2023, 10(1), 626, DOI:10.1038/s41597-023-02501-8.
- P. Giannozzi, O. Andreussi, T. Brumme, O. Bunau, M. B. Nardelli, M. Calandra, R. Car, C. Cavazzoni, D. Ceresoli, M. Cococcioni, N. Colonna, I. Carnimeo, A. D. Corso, S. de Gironcoli, P. Delugas, R. A. DiStasio, A. Ferretti, A. Floris, G. Fratesi, G. Fugallo, R. Gebauer, U. Gerstmann, F. Giustino, T. Gorni, J. Jia, M. Kawamura, H.-Y. Ko, A. Kokalj, E. Küçükbenli, M. Lazzeri, M. Marsili, N. Marzari, F. Mauri, N. L. Nguyen, H.-V. Nguyen, A. O. de-la Roza, L. Paulatto, S. Poncé, D. Rocca, R. Sabatini, B. Santra, M. Schlipf, A. P. Seitsonen, A. Smogunov, I. Timrov, T. Thonhauser, P. Umari, N. Vast, X. Wu and S. Baroni, Advanced capabilities for materials modelling with Quantum ESPRESSO, J. Phys.: Condens. Matter, 2017, 29(46), 465901, DOI:10.1088/1361-648X/aa8f79.
- P. Giannozzi, S. Baroni, N. Bonini, M. Calandra, R. Car, C. Cavazzoni, D. Ceresoli, G. Chiarotti, M. Cococcioni, I. Dabo, A. Dal Corso, S. de Gironcoli, S. Fabris, G. Fratesi, R. Gebauer, U. Gerstmann, C. Gougoussis, A. Kokalj, M. Lazzeri, L. Martin-Samos, N. Marzari, F. Mauri, R. Mazzarello, S. Paolini, A. Pasquarello, L. Paulatto, C. Sbraccia, S. Scandolo, G. Sclauzero, A. Seitsonen, A. Smogunov, P. Umari and R. Wentzcovitch, Quantum ESPRESSO: a modular and open-source software project for quantum simulations of materials, J. Phys.: Condens. Matter, 2009, 21(39), 395502, DOI:10.1088/0953-8984/21/39/395502.
- M. N. Gjerding, A. Taghizadeh, A. Rasmussen, S. Ali, F. Bertoldo, T. Deilmann, N. R. Knøsgaard, M. Kruse, A. H. Larsen, S. Manti, T. G. Pedersen, U. Petralanda, T. Skovhus, M. K. Svendsen, J. J. Mortensen, T. Olsen and K. S. Thygesen, Recent progress of the computational 2D materials database (C2DB), 2D Materials, 2021, 8(4), 044002, DOI:10.1088/2053-1583/ac1059.
- V. Gomzi, I. M. Šapić and A. Vidak, ReaxFF force field development and application for toluene adsorption on MnMOx (M=Cu, Fe, Ni) catalysts, J. Phys. Chem. A, 2021, 125(50), 10649–10656, DOI:10.1021/acs.jpca.1c06939.
- R. E. A. Goodall, A. S. Parackal, F. A. Faber, R. Armiento and A. A. Lee, Rapid discovery of stable materials by coordinate-free coarse graining, Sci. Adv., 2022, 8(30), eabn4117, DOI:10.1126/sciadv.abn4117.
- D. Grassano, D. Campi, A. Marrazzo and N. Marzari, Complementary screening for quantum spin Hall insulators in two-dimensional exfoliable materials, Phys. Rev. Mater., 2023, 7, 094202, DOI:10.1103/PhysRevMaterials.7.094202.
- D. Grassano, N. Marzari and D. Campi, High-throughput screening of weyl semimetals, Phys. Rev. Mater., 2024, 8, 024201, DOI:10.1103/PhysRevMaterials.8.024201.
- S. Gražulis, A. Daškevič, A. Merkys, D. Chateigner, L. Lutterotti, M. Quirós, N. R. Serebryanaya, P. Moeck, R. T. Downs and A. L. Bail, Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration, Nucleic Acids Res., 2012, 40, D420–D427, DOI:10.1093/nar/gkr900.
- S. Haastrup, M. Strange, M. Pandey, T. Deilmann, P. S. Schmidt, N. F. Hinsche, M. N. Gjerding, D. Torelli, P. M. Larsen, A. C. Riis-Jensen, J. Gath, K. W. Jacobsen, J. J. Mortensen, T. Olsen and K. S. Thygesen, The Computational 2D Materials Database: high-throughput modeling and discovery of atomically thin crystals, 2D Materials, 2018, 5(4), 042002, DOI:10.1088/2053-1583/aacfc1.
- S. R. Hall, F. H. Allen and I. D. Brown, The crystallographic information file (CIF): a new standard archive file for crystallography, Acta Crystallogr., Sect. A: Found. Crystallogr., 1991, 47, 655–685, DOI:10.1107/S010876739101067X.
- A. F. Harper, M. L. Evans, J. P. Darby, B. Karasulu, C. P. Koçer, J. R. Nelson and A. J. Morris, Ab initio Structure Prediction Methods for Battery Materials: A review of recent computational efforts to predict the atomic level structure and bonding in materials for rechargeable batteries, Johnson Matthey Technol. Rev., 2020, 64(2), 103–118, DOI:10.1595/205651320x15742491027978.
- J. He, Z. Yao, V. I. Hegde, S. S. Naghavi, J. Shen, K. M. Bushick and C. Wolverton, Computational discovery of stable heteroanionic oxychalcogenides ABXO (A, B=metals; X=S, Se, and Te) and their potential applications, Chem. Mater., 2020, 32(19), 8229–8242, DOI:10.1021/acs.chemmater.0c01902.
- P. Henkel, J. Li, G. K. Grandhi, P. Vivo and P. Rinke, Screening Mixed-Metal Sn2M(III)Ch2X3 Chalcohalides for Photovoltaic Applications, Chem. Mater., 2023, 35(18), 7761–7769, DOI:10.1021/acs.chemmater.3c01629.
- D. Hicks, M. J. Mehl, M. Esters, C. Oses, O. Levy, G. L. W. Hart, C. Toher and S. Curtarolo, The AFLOW Library of Crystallographic Prototypes: Part 3, Comput. Mater. Sci., 2021, 199, 110450, DOI:10.1016/j.commatsci.2021.110450.
- D. Hicks, M. J. Mehl, E. Gossett, C. Toher, O. Levy, R. M. Hanson, G. L. W. Hart and S. Curtarolo, The AFLOW Library of Crystallographic Prototypes: Part 2, Comput. Mater. Sci., 2019, 161, S1–S1011, DOI:10.1016/j.commatsci.2018.10.043.
- L. Himanen, A. Geurts, A. S. Foster and P. Rinke, Data-Driven Materials Science: Status, Challenges, and Perspectives, Adv. Sci., 2019, 6(21), 1900808, DOI:10.1002/advs.201900808.
- A. Hospital, P. Andrio, C. Cugnasco, L. Codo, Y. Becerra, P. D. Dans, F. Battistini, J. Torrers, R. Goñi, M. Orozco and J. L. Gelpi, BIGNASim: a NoSQL database structure and analysis portal for nucleic acids simulation data, Nucleic Acids Res., 2016, 44, D272–D278, DOI:10.1093/nar/gkv1301.
- A. Hospital, F. Battistini, R. Soliva, J. L. Gelpi and M. Orozco, Surviving the deluge of biosimulation data, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2020, 10, e1449, DOI:10.1002/wcms.1449.
- S. P. Huber, S. Zoupanos, M. Uhrin, L. Talirz, L. Kahle, R. Häuselmann, D. Gresch, T. Müller, A. V. Yakutovich, C. W. Andersen, F. F. Ramirez, C. S. Adorf, F. Gargiulo, S. Kumbhar, E. Passaro, C. Johnston, A. Merkys, A. Cepellotti, N. Mounet, N. Marzari, B. Kozinsky and G. Pizzi, AiiDA 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance, Sci. Data, 2020, 7(1), 300, DOI:10.1038/s41597-020-00638-4.
- S. Im, S. L. Shang, N. D. Smith, A. M. Krajewski, T. Lichtenstein, H. Sun, B. J. Bocklund, Z. K. Liu and H. Kim, Thermodynamic properties of the Nd-Bi system via emf measurements, DFT calculations, machine learning, and CALPHAD modeling, Acta Mater., 2022, 223, 117448, DOI:10.1016/J.ACTAMAT.2021.117448.
- O. Isayev, D. Fourches, E. N. Muratov, C. Oses, K. Rasch, A. Tropsha and S. Curtarolo, Materials cartography: Representing and mining materials space using structural and electronic fingerprints, Chem. Mater., 2015, 27(3), 735–743, DOI:10.1021/cm503507h.
- IUCr, Data requirements for structures, 2023, https://journals.iucr.org/c/services/cif/reqdata.html.
- Y. Iwasaki, A. G. Kusne and I. Takeuchi, Comparison of dissimilarity measures for cluster analysis of X-ray diffraction data from combinatorial libraries, npj Comput. Mater., 2017, 3(1), 1–9, DOI:10.1038/s41524-017-0006-2.
- K. M. Jablonka, Q. Ai, A. Al-Feghali, S. Badhwar, J. D. Bocarsly, A. M. Bran, S. Bringuier, L. C. Brinson, K. Choudhary, D. Circi, S. Cox, W. A. de Jong, M. L. Evans, N. Gastellu, J. Genzling, M. V. Gil, A. K. Gupta, Z. Hong, A. Imran, S. Kruschwitz, A. Labarre, J. Lála, T. Liu, S. Ma, S. Majumdar, G. W. Merz, N. Moitessier, E. Moubarak, B. Mouriño, B. Pelkie, M. Pieler, M. C. Ramos, B. Ranković, S. G. Rodriques, J. N. Sanders, P. Schwaller, M. Schwarting, J. Shi, B. Smit, B. E. Smith, J. V. Herck, C. Völker, L. Ward, S. Warren, B. Weiser, S. Zhang, X. Zhang, G. A. Zia, A. Scourtas, K. J. Schmidt, I. Foster, A. D. White and B. Blaiszik, 14 examples of how LLMs can transform materials science and chemistry: a reflection on a large language model hackathon, Digital Discovery, 2023, 2(5), 1233–1250, 10.1039/d3dd00113j.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. A. Persson, Commentary: The Materials Project: A materials genome approach to accelerating materials innovation, APL Mater., 2013, 1(1), 011002, DOI:10.1063/1.4812323.
- S. Kirklin, J. E. Saal, V. I. Hegde and C. Wolverton, High-throughput computational search for strengthening precipitates in alloys, Acta Mater., 2016, 102, 125–135, DOI:10.1016/j.actamat.2015.09.016.
- A. M. Krajewski, J. W. Siegel, and Z.-K. Liu. Efficient structure-informed featurization and property prediction of ordered, dilute, and random atomic structures, arXiv, 2024, preprint, arXiv:2404.02849, DOI:10.48550/arXiv.2404.02849.
- A. M. Krajewski, J. W. Siegel, J. Xu and Z. K. Liu, Extensible Structure-Informed Prediction of Formation Energy with improved accuracy and usability employing neural networks, Comput. Mater. Sci., 2022, 208, 111254, DOI:10.1016/J.COMMATSCI.2022.111254.
- G. Kresse and J. Furthmüller, Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54(16), 11169–11186, DOI:10.1103/physrevb.54.11169.
- A. G. Kusne, H. Yu, C. Wu, H. Zhang, J. Hattrick-Simpers, B. DeCost, S. Sarker, C. Oses, C. Toher, S. Curtarolo, A. V. Davydov, R. Agarwal, L. A. Bendersky, M. Li, A. Mehta and I. Takeuchi, On-the-fly closed-loop materials discovery via bayesian active learning, Nat. Commun., 2020, 11, 5966, DOI:10.1038/s41467-020-19597-w.
- A. H. Larsen, J. J. Mortensen, J. Blomqvist, I. E. Castelli, R. Christensen, M. Dułak, J. Friis, M. N. Groves, B. Hammer, C. Hargus, E. D. Hermes, P. C. Jennings, P. B. Jensen, J. Kermode, J. R. Kitchin, E. L. Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J. B. Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K. S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng and K. W. Jacobsen, The atomic simulation environment—a Python library for working with atoms, J. Phys.: Condens. Matter, 2017, 29(27), 273002, DOI:10.1088/1361-648x/aa680e.
- J. N. Law, S. Pandey, P. Gorai and P. C. St. John, Upper-Bound Energy Minimization to Search for Stable Functional Materials with Graph Neural Networks, JACS Au, 2023, 3(1), 113–123, DOI:10.1021/jacsau.2c00540.
- H. Levämäki, F. Bock, D. G. Sangiovanni, L. J. S. Johnson, F. Tasnádi, R. Armiento and I. A. Abrikosov, HADB: A materials-property database for hard-coating alloys, Thin Solid Films, 2023, 766, 139627, DOI:10.1016/j.tsf.2022.139627.
- H. Li, O. Hartig, R. Armiento and P. Lambrix, Ontology-based GraphQL server generation for data access and data integration, Semant. Web, 2024, 1–37, DOI:10.3233/sw-233550.
- X. Liu, P.-P. De Breuck, L. Wang and G.-M. Rignanese, A simple denoising approach to exploit multi-fidelity data for machine learning materials properties, npj Comput. Mater., 2022, 8, 233, DOI:10.1038/s41524-022-00925-1.
- Z. Liu, Perspective on materials genome®, Chin. Sci. Bull., 2014, 59, 1619, DOI:10.1007/s11434-013-0072-x.
- P. Lyngby and K. S. Thygesen, Data-driven discovery of 2D materials by deep generative models, npj Comput. Mater., 2022, 8(1), 232, DOI:10.1038/s41524-022-00923-3.
- A. Marrazzo, M. Gibertini, D. Campi, N. Mounet and N. Marzari, Prediction of a large-gap and switchable Kane-Mele quantum spin Hall insulator, Phys. Rev. Lett., 2018, 120, 117701, DOI:10.1103/PhysRevLett.120.117701.
- A. Marrazzo, M. Gibertini, D. Campi, N. Mounet and N. Marzari, Relative abundance of Z2 topological order in exfoliable two-dimensional insulators, Nano Lett., 2019, 19(12), 8431–8440, DOI:10.1021/acs.nanolett.9b02689.
- Matcloud, http://matcloud.com.cn, accessed 2024.
- A. Medina-Smith, C. A. Becker, R. L. Plante, L. M. Bartolo, A. Dima, J. A. Warren and R. J. Hanisch, A Controlled Vocabulary and Metadata Schema for Materials Science Data Discovery, Data Sci. J., 2021, 20(1), 18, DOI:10.5334/dsj-2021-018.
- M. J. Mehl, D. Hicks, C. Toher, O. Levy, R. M. Hanson, G. L. W. Hart and S. Curtarolo, The AFLOW Library of Crystallographic Prototypes: Part 1, Comput. Mater. Sci., 2017, 136, S1–S828, DOI:10.1016/j.commatsci.2017.01.017.
- J. Mendenhall, B. P. Brown, S. Kothiwale and J. M. BCL, Conf: Improved Open-Source Knowledge-Based Conformation Sampling Using the Crystallography Open Database, J. Chem. Inf. Model., 2020, 61(1), 189–201, DOI:10.1021/acs.jcim.0c01140.
- A. Merchant, S. Batzner, S. S. Schoenholz, M. Aykol, G. Cheon and E. D. Cubuk, Scaling deep learning for materials discovery, Nature, 2023, 624(7990), 80–85, DOI:10.1038/s41586-023-06735-9.
- J. J. Mortensen, A. H. Larsen, M. Kuisma, A. V. Ivanov, A. Taghizadeh, A. Peterson, A. Haldar, A. O. Dohn, C. Schäfer, E. Ö. Jónsson, E. D. Hermes, F. A. Nilsson, G. Kastlunger, G. Levi, H. Jónsson, H. Häkkinen, J. Fojt, J. Kangsabanik, J. Sødequist, J. Lehtomäki, J. Heske, J. Enkovaara, K. T. Winther, M. Dulak, M. M. Melander, M. Ovesen, M. Louhivuori, M. Walter, M. Gjerding, O. Lopez-Acevedo, P. Erhart, R. Warmbier, R. Würdemann, S. Kaappa, S. Latini, T. M. Boland, T. Bligaard, T. Skovhus, T. Susi, T. Maxson, T. Rossi, X. Chen, Y. L. A. Schmerwitz, J. Schiøtz, T. Olsen, K. W. Jacobsen and K. S. Thygesen, GPAW: An open Python package for electronic structure calculations, J. Chem. Phys., 2024, 160(9), 092503, DOI:10.1063/5.0182685.
- N. Mounet, M. Gibertini, P. Schwaller, D. Campi, A. Merkys, A. Marrazzo, T. Sohier, I. E. Castelli, A. Cepellotti, G. Pizzi and N. Marzari, Two-dimensional materials from high-throughput computational exfoliation of experimentally known compounds, Nat. Nanotechnol., 2018, 13(3), 246–252, DOI:10.1038/s41565-017-0035-5.
- H. Moustafa, P. M. Larsen, M. N. Gjerding, J. J. Mortensen, K. S. Thygesen and K. W. Jacobsen, Computational exfoliation of atomically thin one-dimensional materials with application to Majorana bound states, Phys. Rev. Mater., 2022, 6(6), 064202, DOI:10.1103/PhysRevMaterials.6.064202.
- C. Nyshadham, C. Oses, J. E. Hansen, I. Takeuchi, S. Curtarolo and G. L. W. Hart, A computational high-throughput search for new ternary superalloys, Acta Mater., 2017, 122, 438–447, DOI:10.1016/j.actamat.2016.09.017.
- S. P. Ong, W. D. Richards, A. Jain, G. Hautier, M. Kocher, S. Cholia, D. Gunter, V. L. Chevrier, K. A. Persson and G. Ceder, Python Materials Genomics (pymatgen): A robust, open-source Python library for materials analysis, Comput. Mater. Sci., 2013, 68, 314–319, DOI:10.1016/j.commatsci.2012.10.028.
- Ontology driven Open Translation Environment (OntoTrans), https://ontotrans.eu, 2020, accessed 2023.
- C. Oses, M. Esters, D. Hicks, S. Divilov, H. Eckert, R. Friedrich, M. J. Mehl, A. Smolyanyuk, X. Campilongo, A. van de Walle, J. Schroers, A. G. Kusne, I. Takeuchi, E. Zurek, M. Buongiorno Nardelli, M. Fornari, Y. Lederer, O. Levy, C. Toher and S. Curtarolo, aflow++: A C++ framework for autonomous materials design, Comput. Mater. Sci., 2023, 217, 111889, DOI:10.1016/j.commatsci.2022.111889.
- Y. Ozaki, Y. Suzuki, T. Hawai, K. Saito, M. Onishi and K. Ono, Automated crystal structure analysis based on blackbox optimisation, npj Comput. Mater., 2020, 6(1), 1–7, DOI:10.1038/s41524-020-0330-9.
- S. Pakdel, A. Rasmussen, A. Taghizadeh, M. Kruse, T. Olsen and K. S. Thygesen, High-throughput computational stacking reveals emergent properties in natural van der Waals bilayers, Nat. Commun., 2024, 15(1), 932, DOI:10.1038/s41467-024-45003-w.
- Pauling File, 2024, https://paulingfile.com.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, Scikit-learn: Machine learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- G. Pepponi, S. Gražulis and D. Chateigner, MPOD: A Material Property Open Database linked to structural information, Nucl. Instrum. Methods Phys. Res., Sect. B, 2012, 284, 10–14, DOI:10.1016/j.nimb.2011.08.070.
- E. Perim, D. Lee, Y. Liu, C. Toher, P. Gong, Y. Li, W. N. Simmons, O. Levy, J. J. Vlassak, J. Schroers and S. Curtarolo, Spectral descriptors for bulk metallic glasses based on the thermodynamics of competing crystalline phases, Nat. Commun., 2016, 7, 12315, DOI:10.1038/ncomms12315.
- F. Pezoa, J. L. Reutter, F. Suarez, M. Ugarte, and D. Vrgoč, Foundations of JSON schema, in Proceedings of the 25th International Conference on World Wide Web, International World Wide Web Conferences Steering Committee, 2016, pp. 263–273, DOI:10.1145/2872427.2883029.
- G. Pizzi, A. Cepellotti, R. Sabatini, N. Marzari and B. Kozinsky, AiiDA: automated interactive infrastructure and database for computational science, Comput. Mater. Sci., 2016, 111, 218–230, DOI:10.1016/j.commatsci.2015.09.013.
- G. Pizzi, S. Milana, A. C. Ferrari, N. Marzari and M. Gibertini, Shear and breathing modes of layered materials, ACS Nano, 2021, 15(8), 12509–12534, DOI:10.1021/acsnano.0c10672.
- R. L. Plante, C. A. Becker, A. Medina-Smith, K. Brady, A. Dima, B. Long, L. M. Bartolo, J. A. Warren and R. J. Hanisch, Implementing a Registry Federation for Materials Science Data Discovery, Data Sci. J., 2021, 20(1), 15, DOI:10.5334/dsj-2021-015.
- J. Qiao, G. Pizzi and N. Marzari, Projectability disentanglement for accurate and automated electronic-structure Hamiltonians, npj Comput. Mater., 2023, 9(1), 208, DOI:10.1038/s41524-023-01146-w.
- F. L. Reyes Tirado, J. Perrin Toinin and D. C. Dunand, γ + γ′ microstructures in the Co-Ta-V and Co-Nb-V ternary systems, Acta Mater., 2018, 151, 137–148, DOI:10.1016/j.actamat.2018.03.057.
- F. Rose, C. Toher, E. Gossett, C. Oses, M. Buongiorno Nardelli, M. Fornari and S. Curtarolo, AFLUX: The LUX materials search API for the AFLOW data repositories, Comput. Mater. Sci., 2017, 137, 362–370, DOI:10.1016/j.commatsci.2017.04.036.
- J. E. Saal, S. Kirklin, M. Aykol, B. Meredig and C. Wolverton, Materials Design and Discovery with High-Throughput Density Functional Theory: The Open Quantum Materials Database (OQMD), JOM, 2013, 65(11), 1501–1509, DOI:10.1007/s11837-013-0755-4.
- S. Sanvito, C. Oses, J. Xue, A. Tiwari, M. Žic, T. Archer, P. Tozman, M. Venkatesan, J. M. D. Coey and S. Curtarolo, Accelerated discovery of new magnets in the Heusler alloy family, Sci. Adv., 2017, 3(4), e1602241, DOI:10.1126/sciadv.1602241.
- P. Sarker, T. Harrington, C. Toher, C. Oses, M. Samiee, J.-P. Maria, D. W. Brenner, K. S. Vecchio and S. Curtarolo, High-entropy high-hardness metal carbides discovered by entropy descriptors, Nat. Commun., 2018, 9(1), 4980, DOI:10.1038/s41467-018-07160-7.
- L. Sbailò, Á. Fekete, L. M. Ghiringhelli and M. Scheffler, The NOMAD artificial-intelligence toolkit: turning materials-science data into knowledge and understanding, npj Comput. Mater., 2022, 8(1), 250, DOI:10.1038/s41524-022-00935-z.
- M. Scheffler, M. Aeschlimann, M. Albrecht, T. Bereau, H.-J. Bungartz, C. Felser, M. Greiner, A. Groß, C. T. Koch, K. Kremer, W. E. Nagel, M. Scheidgen, C. Wöll and C. Draxl, FAIR data enabling new horizons for materials research, Nature, 2022, 604(7907), 635–642, DOI:10.1038/s41586-022-04501-x.
- M. Scheidgen, L. Himanen, A. N. Ladines, D. Sikter, M. Nakhaee, Á. Fekete, T. Chang, A. Golparvar, J. A. Márquez, S. Brockhauser, S. Brückner, L. M. Ghiringhelli, F. Dietrich, D. Lehmberg, T. Denell, A. Albino, H. Näsström, S. Shabih, F. Dobener, M. Kühbach, R. Mozumder, J. F. Rudzinski, N. Daelman, J. M. Pizarro, M. Kuban, C. Salazar, P. Ondračka, H.-J. Bungartz and C. Draxl, NOMAD: A distributed web-based platform for managing materials science research data, J. Open Source Softw., 2023, 8(90), 5388, DOI:10.21105/joss.05388.
- J. Schmidt, L. Chen, S. Botti and M. A. L. Marques, Predicting the stability of ternary intermetallics with density functional theory and machine learning, J. Chem. Phys., 2018, 148(24), 241728, DOI:10.1063/1.5020223.
- J. Schmidt, N. Hoffmann, H.-C. Wang, P. Borlido, P. J. M. A. Carriço, T. F. T. Cerqueira, S. Botti and M. A. L. Marques, Machine-learning-assisted determination of the global zero-temperature phase diagram of materials, Adv. Mater., 2023, 35(22), 2210788, DOI:10.1002/adma.202210788.
- J. Schmidt, L. Pettersson, C. Verdozzi, S. Botti and M. A. L. Marques, Crystal graph attention networks for the prediction of stable materials, Sci. Adv., 2021, 7(49), eabi7948, DOI:10.1126/sciadv.abi7948.
- J. Schmidt, J. Shi, P. Borlido, L. Chen, S. Botti and M. A. L. Marques, Predicting the thermodynamic stability of solids combining density functional theory and machine learning, Chem. Mater., 2017, 29(12), 5090–5103, DOI:10.1021/acs.chemmater.7b00156.
- J. Schmidt, H.-C. Wang, T. F. T. Cerqueira, S. Botti and M. A. L. Marques, A new dataset of 175k stable and metastable materials calculated with the PBEsol and SCAN functionals, Sci. Data, 2021, 12(1), 64, DOI:10.1038/s41597-022-01177-w.
- J. Schmidt, H.-C. Wang, G. Schmidt and M. A. Marques, Machine learning guided high-throughput search of non-oxide garnets, npj Comput. Mater., 2023, 9(1), 63, DOI:10.1038/s41524-023-01009-4.
- H. Schwarz, T. Uhlig, T. Lindner, T. Lampke, G. Wagner and T. Seyller, Hardness Enhancement in CoCrFeNi1-x(WC)x High-Entropy Alloy Thin Films Synthesised by Magnetron Co-Sputtering, Coatings, 2022, 12(2), 269, DOI:10.3390/coatings12020269.
- S.-L. Shang, H. Sun, B. Pan, Y. Wang, A. M. Krajewski, M. Banu, J. Li and Z.-K. Liu, Forming Mechanism of Equilibrium and Non-equilibrium Metallurgical Phases in Dissimilar Materials: Illustrated With Aluminum/steel (Al-Fe) Joints, Sci. Rep., 2021, 11, 24251, DOI:10.1038/s41598-021-03578-0.
- J. Shen, S. D. Griesemer, A. Gopakumar, B. Baldassarri, J. E. Saal, M. Aykol, V. I. Hegde and C. Wolverton, Reflections on one million compounds in the open quantum materials database (OQMD), JPhys Mater., 2022, 5(3), 031001, DOI:10.1088/2515-7639/ac7ba9.
- SMARTS – A Language for Describing Molecular Patterns, https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, accessed 2024.
- T. Sohier, D. Campi, N. Marzari and M. Gibertini, Mobility of two-dimensional materials from first principles in an accurate and automated framework, Phys. Rev. Mater., 2018, 2, 114010, DOI:10.1103/PhysRevMaterials.2.114010.
- T. Sohier, M. Gibertini, D. Campi, G. Pizzi and N. Marzari, Valley-engineering mobilities in two-dimensional materials, Nano Lett., 2019, 19(6), 3723–3729, DOI:10.1021/acs.nanolett.9b00865.
- V. Stanev, C. Oses, A. G. Kusne, E. Rodriguez, J. Paglione, S. Curtarolo and I. Takeuchi, Machine learning modeling of superconducting critical temperature, npj Comput. Mater., 2018, 4, 29, DOI:10.1038/s41524-018-0085-8.
- C. Suh, C. Fare, J. Warren and E. Pyzer-Knapp, Evolving the materials genome: How machine learning is fueling the next generation of materials discovery, Annu. Rev. Mater. Res., 2020, 50, 1–25, DOI:10.1146/annurev-matsci-082019-105100.
- SVELTE, 2024, https://svelte.dev.
- L. Talirz, S. Kumbhar, E. Passaro, A. V. Yakutovich, V. Granata, F. Gargiulo, M. Borelli, M. Uhrin, S. P. Huber, S. Zoupanos, C. S. Adorf, C. W. Andersen, O. Schütt, C. A. Pignedoli, D. Passerone, J. VandeVondele, T. C. Schulthess, B. Smit, G. Pizzi and N. Marzari, Materials Cloud, a platform for open computational science, Sci. Data, 2020, 7(1), 299, DOI:10.1038/s41597-020-00637-5.
- The MarketPlace Project, https://materials-marketplace.eu, 2018, accessed 2023.
- The OPTIMADE Developers, Open Database Integration for Materials Design (OPTIMADE), https://github.com/Materials-Consortia/OPTIMADE, accessed 2023.
- The OPTIMADE Developers, OPTIMADE Providers Dashboard, https://www.optimade.org/providers-dashboard/, accessed 2023.
- The OPTIMADE Developers, OPTIMADE Providers List, https://providers.optimade.org, accessed 2023.
- B. H. Toby and R. B. Von Dreele, GSAS-II: the genesis of a modern open-source all purpose crystallography software package, J. Appl. Crystallogr., 2013, 46(2), 544–549, DOI:10.1107/S0021889813003531.
- M. Uhrin, S. P. Huber, J. Yu, N. Marzari and G. Pizzi, Workflows in AiiDA: Engineering a high-throughput, event-based engine for robust and modular computational workflows, Comput. Mater. Sci., 2021, 187, 110086, DOI:10.1016/j.commatsci.2020.110086.
- M. T. Vahdat, K. V. Agrawal and G. Pizzi, Machine-learning accelerated identification of exfoliable two-dimensional materials, Mach. Learn.: Sci. Technol., 2022, 3(4), 045014, DOI:10.1088/2632-2153/ac9bca.
- A. van Roekeghem, J. Carrete, C. Oses, S. Curtarolo and N. Mingo, High-throughput computation of thermal conductivity of high-temperature solid phases: The case of oxide and fluoride perovskites, Phys. Rev. X, 2016, 6, 041061, DOI:10.1103/PhysRevX.6.041061.
- B. Wang, Q. Fan and Y. Yue, Study of crystal properties based on attention mechanism and crystal graph convolutional neural network, J. Phys.: Condens. Matter, 2022, 34, 195901, DOI:10.1088/1361-648X/ac5705.
- H.-C. Wang, S. Botti and M. A. L. Marques, Predicting stable crystalline compounds using chemical similarity, npj Comput. Mater., 2021, 7(1), 1–9, DOI:10.1038/s41524-020-00481-6.
- T. Wang, K. Zhang, J. Thé and H. Yu, Accurate prediction of band gap of materials using stacking machine learning model, Comput. Mater. Sci., 2022, 201, 110899, DOI:10.1016/j.commatsci.2021.110899.
- Z. Wang, Y. Gong, M. L. Evans, Y. Yan, S. Wang, N. Miao, R. Zheng, G.-M. Rignanese and J. Wang, Machine Learning-Accelerated Discovery of A2BC2 Ternary Electrides with Diverse Anionic Electron Densities, J. Am. Chem. Soc., 2023, 145(48), 26412–26424, DOI:10.1021/jacs.3c10538.
- Z. Wang, Y. Gong, M. L. Evans, Y. Yan, S. Wang, N. Miao, R. Zheng, G.-M. Rignanese and J. Wang, Machine learning-accelerated discovery of A2BC2 ternary electrides with diverse anionic electron densities, Materials Cloud Archive, 2023, 181, 26412–26424, DOI:10.24435/materialscloud:c8-gy.
- D. Weininger, SMILES, a chemical languge and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Comput. Sci., 1988, 28(1), 31–36, DOI:10.1021/ci00057a005.
- D. Wines, R. Gurunathan, K. F. Garrity, B. DeCost, A. J. Biacchi, F. Tavazza and K. Choudhary, Recent progress in the JARVIS infrastructure for next-generation data-driven materials design, Applied Physics Reviews, 2023, 10(4), 041302, DOI:10.1063/5.0159299.
- T. Xie and J. C. Grossman, Crystal Graph Convolutional Neural Networks for Accurate and Interpretable Prediction of Material Properties, Phys. Rev. Lett., 2018, 120(14), 145301, DOI:10.1103/PhysRevLett.120.145301.
- A. V. Yakutovich, K. Eimre, O. Schütt, L. Talirz, C. S. Adorf, C. W. Andersen, E. Ditler, D. Du, D. Passerone, B. Smit, N. Marzari, G. Pizzi and C. A. Pignedoli, AiiDAlab – an ecosystem for developing, executing, and sharing scientific workflows, Comput. Mater. Sci., 2021, 188, 110165, DOI:10.1016/j.commatsci.2020.110165.
- X. Yang, Z. Wang, X. Zhao, J. Song, C. Yu, J. Zhou and K. Li, A high-throughput computational materials infrastructure: Present, future visions and challenges, Chin. Phys. B, 2018, 27(11), 110301, DOI:10.1088/1674-1056/27/11/110301.
- X. Yang, Z. Wang, X. Zhao, J. Song, M. Zhang and H. Liu, Matcloud: A high-throughput computational infrastructure for integrated management of materials simulation, data and resources, Comput. Mater. Sci., 2018, 146, 319–333, DOI:10.1016/j.commatsci.2018.01.039.
- W. Ye, X. Lei, M. Aykol and J. H. Montoya, Novel inorganic crystal structures predicted using autonomous simulation agents, Sci. Data, 2022, 9(1), 302, DOI:10.1038/s41597-022-01438-8.
- S. Zivanovic, G. Bayarri, F. Colizzi, D. Moreno, J. L. Gelpi, R. Soliva, A. Hospital and M. Orozco, Bioactive conformational ensemble server and database. a public framework to speed up in silico drug discovery, J. Chem. Theory Comput., 2020, 16, 6586–6597, DOI:10.1021/acs.jctc.0c00305.
Footnotes |
| † Electronic supplementary information (ESI) available: Copy of Table 1 with web links. See DOI: https://doi.org/10.1039/d4dd00039k |
| ‡ Present address: Van't Hoff Institute for Molecular Sciences, University of Amsterdam, PO Box 94157, 1090 GD Amsterdam, Netherlands. |
| § Present address: Henkel, AID/Digital Twins & Data Analytics, 40589 Düsseldorf, Germany. |
| ¶ The double awarded Nobel laureate Linus Pauling personally endorsed this project and gave an explicit written permission to use his name. In 2019, the Pauling File's founder Pierre Villars was acknowledged with the NIMS Award (Tsukuba, Japan) for the fundamental research for data-driven materials development. |
| This journal is © The Royal Society of Chemistry 2024 |