Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
FSL-CP: a benchmark for small molecule activity few-shot prediction using cell microscopy images†
Son V.
Ha
,
Lucas
Leuschner
and
Paul
Czodrowski
 *
*
JGU Mainz, Germany. E-mail: czodpaul@uni-mainz.de
First published on 26th February 2024
Abstract
Predicting small molecule activities using information from high-throughput microscopy images has been shown to tremendously increase hit rates and chemical diversity of the hits in previous drug discovery projects. However, due to high cost of acquiring data or ethical reasons, data sparsity remains a big challenge in drug discovery. This opens up the opportunity for few-shot prediction: fine-tuning a model on a low-data assay of interest after pretraining on other more populated assays. Previous efforts have been made to establish a benchmark for few-shot learning of molecules based on molecular structures. With cell images as a molecular representation, methods in the computer vision domain are also applicable for activity prediction. In this paper, we make two contributions: (a) a public data set for few-shot learning with cell microscopy images for the scientific community and (b) a range of baseline models encompassing different existing single-task, multi-task and meta-learning approaches.
1 Introduction
High-throughput imaging (HTI) has been a powerful tool in drug discovery, having yielded many biological discoveries.1–3 It often involves capturing the morphological changes of cells induced by chemical compounds and quantifying these changes into a large set of numerical features4 such as staining intensity, texture, shape and spatial correlations. They act as ‘fingerprints’ that can be used to characterise compounds in a relatively unbiased way. This technique, known as morphological profiling, has proven to be useful for a variety of applications, such as optimizing the diversity of compound libraries,5 determining the mechanism of action of compounds,6–8 and clustering genes based on their biological functions.9,10Cell painting is a morphological profiling method in which cells are perturbed with a compound, have their different compartments stained using six dyes, and have their images of the five fluorescence channels captured.4 Cell painting data have been used for a range of applications, from predicting mitochondrial toxicity,3,11in vitro toxicity,12 hit identification,13 and more. In addition, cell painting can be used in combination with other modalities to enhance prediction such as chemical structure14 and gene expression data.15
1.1 Small molecule activity prediction
Prediction of small molecule activity against a drug target is an important task in drug discovery. It helps identify and optimise compounds for a desired activity, as well as recognise and avoid off-target activities. This leads to the identification of compounds with the highest potential in the early drug discovery pipeline.HTI data have been used in bioactivity prediction by Simm et al.16 in two drug discovery projects, which led to a tremendous increase in hit rates by 50- to 250-fold, while increasing the chemical structure diversity of the hits. In these projects, only 1.6% of the label matrix is filled, for over 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds and 1200 prediction tasks. This reflects the need for a modeling paradigm which can not only adapt to new tasks quickly with little data, but also leverage the availability of many low-data related tasks. We find that this setting is ideal to form a few-shot learning challenge.
000 compounds and 1200 prediction tasks. This reflects the need for a modeling paradigm which can not only adapt to new tasks quickly with little data, but also leverage the availability of many low-data related tasks. We find that this setting is ideal to form a few-shot learning challenge.
1.2 Few-shot learning
In the few-shot learning setting, there is not one big dataset D to learn from, but instead many small datasets we called tasks, denoted T. The aim of few-shot methods is to generalise over new tasks {Tu}Uu=1 ∈ Dtest efficiently with only a small number of available datapoints. Each task Tu consists of a support set S for learning and a query set Q for evaluation, Tu = 〈S, Q〉. Typically the size of support set S is very small to reflect the low-data setting.Few-shot models adapt efficiently to low-data tasks by using an advantage initialisation of their parameters, normally through some sort of pretraining on a large data corpus such as a set of auxiliary tasks {Tv}Vv=1 ∈ Dtrain. We expect that knowledge gained from pretraining can be transferred effectively to new unseen tasks, so that models can quickly learn these new tasks using only little data. This can be compared to, for example, a person who already has prior knowledge of music picking up a new musical instrument relativelyquickly with little demonstration.
Most state-of-the-art few-shot methods come from computer vision and natural language processing domains.17–21 Drug discovery is another field where there is growing interest in few-shot learning,22 since data scarcity is a common setting for many prediction tasks. In this paper, we propose to expand another challenge in drug discovery to the few-shot learning area: the aforementioned small molecule activity prediction with cell imaging data. We find that this is a real-world scientific problem with an ideal few-shot setting: there are many related low-data tasks convenient for knowledge transfer between each other. Furthermore, with cell images as a molecular representation, methods in the computer vision domain can be adapted for activity prediction. We believe that the field of cell imaging/analysis would greatly benefit from these algorithmic innovations.
In this paper, we make two contributions:
• A data set for few-shot prediction of small molecule activity using cell microscopy images, which we named FSL-CP. The dataset is curated so that it is easy for future researchers to experiment with their few-shot methods.
• A benchmark of models encompassing different existing single-task, multi-task and meta-learning approaches on the dataset. This acts as both a diverse baseline for future algorithms and a means to study the strengths and weaknesses of different modelling paradigms.
2 Methods
2.1 FSL-CP: few-shot learning data set with cell microscopy images
The FSL-CP dataset comprises compounds at the intersection of ChEMBL23 version 31 and the cell painting4 public dataset. We provide an overview of the data construction process below (Fig. 1), and the exact reproducible source code is available on GitHub, at https://github.com/czodrowskilab/FSL_CP_DataPrep.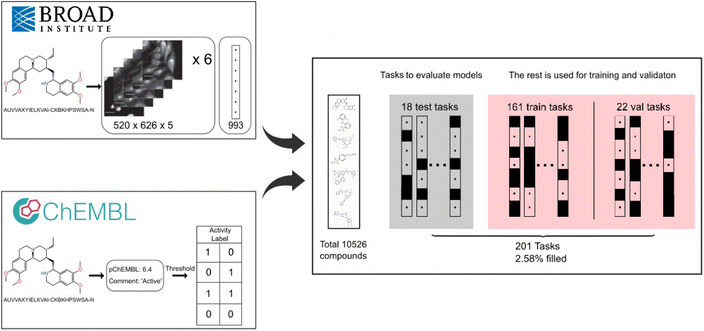 | ||
Fig. 1 FSL-CP data curation and processing. Cell painting images and features from CellProfiler24 come from Bray et al.4 Each well is represented by six 520 × 696 × 5 images and a feature vector of length 993. Small molecule activity labels are retrieved from assays in ChEMBL31.23 Then a threshold procedure is applied to binarise the labels, producing different tasks from assays. The intersection of the two sources results in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526 unique compounds and 201 prediction tasks. 18 tasks are chosen for model evaluation based on a set of criteria, and the rest are for training and validation (referred to as auxiliary tasks). 526 unique compounds and 201 prediction tasks. 18 tasks are chosen for model evaluation based on a set of criteria, and the rest are for training and validation (referred to as auxiliary tasks). | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 265 five-channel views, representing 30
265 five-channel views, representing 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 616 compounds. In this cell painting protocol, U2OS cells have 8 major organelles and sub-compartments stained using a mixture of 6 fluorescent dyes, resulting in 5 different image channels. A CellProfiler24 pipeline is then used to extract 1783 single-cell morphological features from those images.
616 compounds. In this cell painting protocol, U2OS cells have 8 major organelles and sub-compartments stained using a mixture of 6 fluorescent dyes, resulting in 5 different image channels. A CellProfiler24 pipeline is then used to extract 1783 single-cell morphological features from those images.
• Tasks in Dtest do not share the same targets as those in the Dtrain and Dval, unless a target is unknown (denoted as ‘unchecked’ on ChEMBL). This is to avoid the overlap of very similar tasks during training and inference.
• Test tasks must have over 96 datapoints, to enable model comparison for a range of support set sizes.
• Test tasks must have a ratio of active compounds between 0.3 and 0.7, to avoid strongly imbalanced data affecting the model comparison. Some methods might be better because they overcome the data imbalance problem, not the low data problem which is what we focus on.
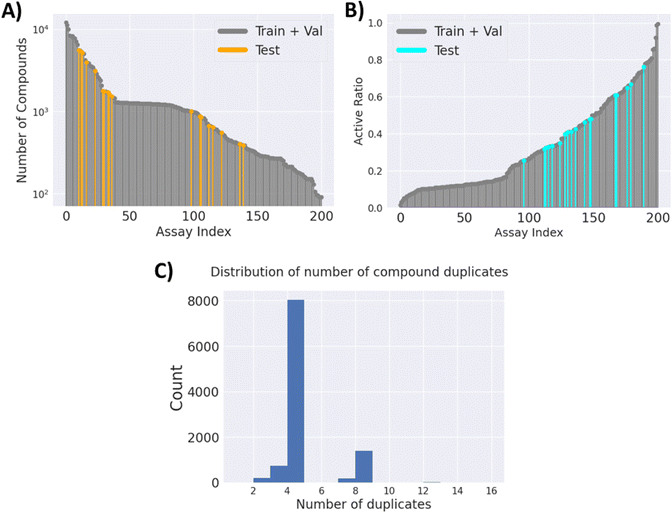
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526 unique compounds, with only 2.58% of the label matrix filled. The number of compounds for each task and their active ratio are visualised in (Fig. 2A) and (Fig. 2B), respectively. It is worth noting that the majority of the compounds have 4 replicates, as per the design of the cell painting assay. However, there are cases where there are fewer or more replicates (Fig. 2C), potentially due to the omission of low-quality images and repeated purchases of some compounds.
526 unique compounds, with only 2.58% of the label matrix filled. The number of compounds for each task and their active ratio are visualised in (Fig. 2A) and (Fig. 2B), respectively. It is worth noting that the majority of the compounds have 4 replicates, as per the design of the cell painting assay. However, there are cases where there are fewer or more replicates (Fig. 2C), potentially due to the omission of low-quality images and repeated purchases of some compounds.
2.2 Evaluation
For every test task, we report model performances averaged over 100 episodes in order to eliminate variations from sampling. Additionally, the results are recorded over a range of support set sizes M: 8, 16, 32, 64, 96, to monitor how well models perform as the size of available data increases.
In addition, the results reported in terms of F1 score, balanced accuracy, Cohen's kappa, and ΔAUPRC22 can also be found in the ESI.†
2.3 Benchmark models
In this section, we provide a detailed description of different modelling paradigms for this particular few-shot problem. The code for all of the models and the training/inference scripts can be found at https://github.com/czodrowskilab/FSL_CP. As a naming convention, models with _img are trained directly on the images, _cp means that they are trained on the original CP features, and _cp+ means that they are trained on the enhanced CP features.For our benchmark, the same FNN model as that in the single-task case is used, but with a head of length 183 instead. Pretraining and validation are performed using 183 auxiliary tasks in Dtrain and Dval in a multi-task manner. Then the weights are frozen, and the head is replaced with a new one of length 1 for fine-tuning. During evaluation, for each episode, the same frozen model has its last layer fine-tuned using the support set, evaluated on the query set, and reverted back to the state before fine-tuning. We tried training on both sets of CP features but neither led to drastic improvements over the other. We decided to report the result for the model trained on the original CP to reflect the methods by Simm et al. This model is denoted as multitask_cp.
One subclass of meta-learning is a metric-based method, which tries to learn a distance function over data samples. For example, the prototypical network17 uses a backbone model to generate an embedding. Then classification is made using k-means clustering based on the Euclidean distance from the embedding to the cluster prototypes. Since the backbone for a prototypical network can be any kind of embedding generator, we try 2 versions: a ResNet50 and an FNN backbone, which generate embeddings from CP images and CP features, respectively. These models are named protonet_img, protonet_cp and protonet_cp+.
The optimisation-based method is another subclass of meta-learning. This approach intends to make gradient-based optimisation converge within a small number of optimisation steps. MAML18 (model-agnostic meta-learning) achieves this by obtaining good weight initialisation through pretraining, so that fine-tuning to unseen tasks can be more efficient. Thanks to MAML working with any algorithm that uses gradient descent, we provide the results of ResNet50 and an FNN after being trained by MAML (denoted as maml_img and maml_cp+).
It is important to mention that, unlike feature-based models, image-based models are highly computationally expensive to train. Hence to train these models and tune the hyperparameters in a reasonable time frame, only one out of six available views is used, plus random cropping and down-sizing of images are performed. With an 11GiB NVIDIA GeForce RTX 2080 Ti, the training and inference for one hyperparameter configuration take between 1 and 2 weeks, depending on the model.
3 Results
In order to compare performances of different methods benchmarked on FSL-CP, we plot the mean AUROC across 18 test tasks of each method at different support set sizes (Fig. 3A). In addition, a paired Wilcoxon sign-rank test is performed for each pair of models as demonstrated in Table 1, with the alternative hypothesis that the method in the left column outperforms the method in the upper row. These figures also provide insight into how the performance of each model changes as the amount of available data increases.| Protonet_cp | Multitask_cp | Singletask_cp | Maml_cp+ | Logistic_cp+ | Maml_img | Protonet_img | |
|---|---|---|---|---|---|---|---|
| a The test compares mean AUROC (over 100 episodes) of models in 18 test tasks. Marked in bold are significant p-values at α = 0.01. | |||||||
| (a) Support set size 8 | |||||||
| Protonet_cp+ | 3.60 × 10−1 | 7.89 × 10−1 | 1.56 × 10−1 | 3.79 × 10−1 | 1.05 × 10 −3 | 3.09 × 10 −3 | 5.38 × 10 −4 |
| Protonet_cp | 8.90 × 10−1 | 2.74 × 10−1 | 6.12 × 10−1 | 1.79 × 10 −3 | 5.23 × 10 −3 | 4.52 × 10 −4 | |
| Multitask_cp | 3.96 × 10−2 | 3.04 × 10−1 | 2.69 × 10 −4 | 1.11 × 10 −3 | 1.43 × 10 −4 | ||
| Singletask_cp | 6.03 × 10−1 | 3.09 × 10 −3 | 8.05 × 10−2 | 1.21 × 10 −3 | |||
| Maml_cp+ | 8.84 × 10−2 | 1.92 × 10−2 | 2.74 × 10−2 | ||||
| Logistic_cp+ | 6.42 × 10−1 | 9.16 × 10−3 | |||||
| Maml_img | 3.47 × 10−2 | ||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| (b) Support set size 16 | |||||||
| Protonet_cp+ | 1.45 × 10−2 | 5.42 × 10−2 | 9.36 × 10 −4 | 2.77 × 10 −3 | 2.49 × 10 −4 | 5.34 × 10 −5 | 7.63 × 10 −6 |
| Protonet_cp | 5.41 × 10−1 | 1.08 × 10−2 | 2.21 × 10−2 | 8.53 × 10 −4 | 5.35 × 10 −4 | 2.63 × 10 −4 | |
| Multitask_cp | 5.22 × 10 −4 | 1.13 × 10−2 | 2.08 × 10 −4 | 9.54 × 10 −5 | 1.46 × 10 −4 | ||
| Singletask_cp | 6.51 × 10−1 | 1.68 × 10 −3 | 3.49 × 10−2 | 3.43 × 10 −4 | |||
| Maml_cp+ | 2.01 × 10−2 | 2.98 × 10 −3 | 1.03 × 10 −3 | ||||
| Logistic_cp+ | 2.48 × 10−1 | 1.50 × 10 −3 | |||||
| Maml_img | 1.27 × 10−1 | ||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| (c) Support set size 32 | |||||||
| Protonet_cp+ | 8.63 × 10−2 | 7.97 × 10 −3 | 3.83 × 10 −4 | 1.42 × 10 −4 | 1.46 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 |
| Protonet_cp | 6.49 × 10−2 | 1.46 × 10 −3 | 2.67 × 10 −4 | 2.67 × 10 −5 | 1.74 × 10 −4 | 1.14 × 10 −5 | |
| Multitask_cp | 4.03 × 10 −4 | 4.56 × 10 −3 | 3.81 × 10 −6 | 3.81 × 10 −6 | 3.81 × 10 −6 | ||
| Singletask_cp | 3.61 × 10−1 | 2.80 × 10 −3 | 1.22 × 10 −3 | 2.56 × 10 −4 | |||
| Maml_cp+ | 3.69 × 10 −2 | 1.65 × 10 −3 | 5.23 × 10 −4 | ||||
| Logistic_cp+ | 7.97 × 10 −3 | 1.36 × 10 −4 | |||||
| Maml_img | 2.55 × 10−1 | ||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| (d) Support set size 64 | |||||||
| Protonet_cp+ | 2.62 × 10−1 | 3.43 × 10−1 | 6.97 × 10 −3 | 4.40 × 10 −4 | 7.63 × 10 −6 | 3.81 × 10 −6 | 3.81 × 10 −6 |
| Protonet_cp | 3.88 × 10−1 | 1.46 × 10−2 | 6.05 × 10 −4 | 1.91 × 10 −5 | 1.46 × 10 −4 | 7.63 × 10 −6 | |
| Multitask_cp | 2.07 × 10 −4 | 2.47 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 | 3.81 × 10 −6 | ||
| Singletask_cp | 9.31 × 10−2 | 1.43 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 | |||
| Maml_cp+ | 4.10 × 10 −3 | 1.14 × 10 −5 | 1.14 × 10 −5 | ||||
| Logistic_cp+ | 2.02 × 10 −3 | 2.11 × 10 −4 | |||||
| Maml_img | 4.18 × 10−1 | ||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| (e) Support set size 96 | |||||||
| Protonet_cp+ | 6.14 × 10−1 | 2.84 × 10−1 | 7.73 × 10−2 | 7.25 × 10 −5 | 3.81 × 10 −6 | 3.81 × 10 −6 | 3.81 × 10 −6 |
| Protonet_cp | 2.09 × 10−1 | 6.44 × 10−2 | 2.29 × 10 −4 | 2.16 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 | |
| Multitask_cp | 4.00 × 10−2 | 1.45 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 | 3.81 × 10 −6 | ||
| Singletask_cp | 3.36 × 10 −4 | 1.44 × 10 −4 | 3.81 × 10 −6 | 3.81 × 10 −6 | |||
| Maml_cp+ | 1.85 × 10−1 | 1.14 × 10 −5 | 4.44 × 10 −4 | ||||
| Logistic_cp+ | 3.21 × 10 −4 | 3.81 × 10 −6 | |||||
| Maml_img | 2.33 × 10−1 | ||||||
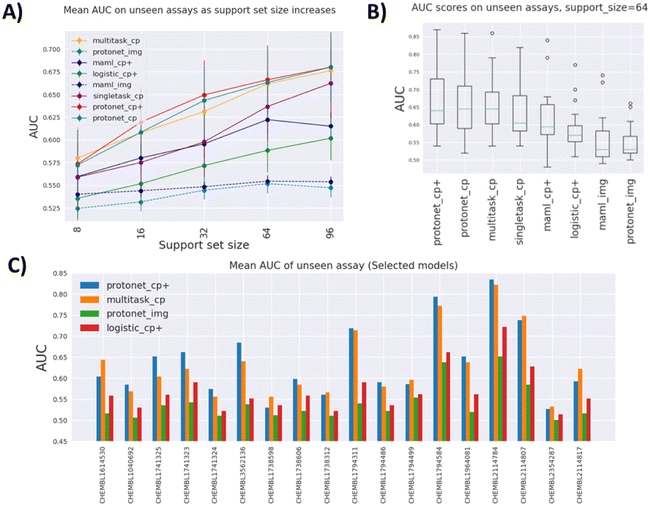
Fig. 3A indicates that the best performing models overall are variants of prototypical networks protonet_cp+ and protonet_cp, followed closely by multitask_cp, although according to the Wilcoxon signed-rank test, protonet_cp+ only outperforms multitask_cp for medium-sized datasets (support set size 32). There is no sufficient evidence to reject the null hypothesis that they perform equally well at other support set sizes with α = 0.01. We note that multitask_cp even slightly outperforms protonet_cp+ and protonet_cp at support set size 8.
The best single-task model singletask_cp is surprisingly powerful, being able to catch up with maml_cp+ at lower support set size, and outperforms it by a wide margin at large support set size. For these single-task models with no pretraining, the availability of more data in the support set can lead to dramatic improvements in performance. It is highly likely that their performances will keep improving and eventually might overtake other methods beyond support set size 96. In contrast, improvements in AUROC scores of meta-learning methods slow down at higher support set size and even drop as in the case of maml_cp+. However, it is worth noting that at support set size 96, some test tasks are excluded from the evaluation process due to insufficient data points. Plus, fewer tasks in Dtrain and Dval are included in the pretraining for meta-learning models at high support set size, due to the fact that there may not be enough datapoints to sample for episodic training. All of these factors can affect meta-learning methods' performance in higher data setting.
Image-based models such as protonet_img and maml_img substantially under-perform compared to other feature-based methods, likely because only one view out of six is used and down-sizing of images of fairly small cells leads to drastic information loss.
We also try to leverage deep-learning-based features by concatenating the original CP features with a ResNet50 embedding of size 1000. While in some cases it leads to higher AUROC, as evidenced by the fact that many models use the enhanced feature, the improvements are somewhat minute. For example, when comparing protonet_cp+ against protonet_cp, Table 1 shows insufficient evidence of improvement across tasks, and as shown in Fig. 3A, the additional features lead to only small improvements at support set sizes 16, 32 and 64. However, this still poses an interesting question for future research: how meaningful embedding from cell images can be produced using deep learning methods.
Better performing models have a larger spread of AUROC across test tasks, as shown in Fig. 3B, indicating that model performances are fairly dependent on tasks. This is further demonstrated in Fig. 3C, where some tasks (e.g. CHEMBL2114784) consistently show high AUROC across models and some (e.g. CHEMBL2354287) are not predictive at all. Additionally, for some tasks protonet_cp+ is the best method, but in a few other cases multitask_cp or singletask_cp is the better method.
Fig. 3C also gives insight on how much pretraining on auxiliary tasks benefits prediction. Again, this is highly task-dependent. Some tasks benefit greatly from pretraining (CHEMBL3562136 and CHEMBL2114807), as seen from the improvements of the two pretraining models over the single-task models. However, pretraining can offer no improvement, or even be detrimental in tasks such as CHEMBL1738598 and CHEMBL1738312.
4 Discussion
We have presented FSL-CP, a dataset for small molecule activity few-shot prediction using cell microscopy images. This few-shot challenge mimics a screening process in early stage drug discovery, where the aim is to identify potent compounds targeting a specific protein from high-content cell images with little data. Previous efforts have been made to benchmark few-shot methods on molecules as graph-structured data.22 But in machine learning, the primary focus of few-shot learning has been in the computer vision and natural language processing domains. The fact that our dataset uses cell images as a molecular representation opens up opportunities to adapt state-of-the-art ideas from computer vision to enhance modeling.This dataset allows us to establish benchmarks that compare the performances of different few-shot learning paradigms. Our result indicates that the feature-based prototypical network and multitask FNN pretrained on auxiliary tasks generally perform well across all support set sizes. We also observe improvement in performance slowing down for meta-learning methods at high support set size, in contrast to single-task methods, which greatly benefit from the availability of more available data. However, more labelled compounds and their cell painting data are needed in order to accurately point out whether eventually single-task models outperform pretrained models, and if yes, at what support set size.
Image-based models underperform on our benchmark, and the fact that each datapoint consists of six high-definition five-channel images makes it tremendously computationally expensive to train. We had to use only one randomly cropped, downsized image to train the models in a reasonable time-frame with our infrastructure, and this leads to high information loss. Training on full-resolution cell images has been shown to offer better performance than that on CP features in some settings.25 However, in realistic drug discovery projects, larger-size images are used, and there are typically more compounds and more prediction tasks. These make pretraining on full-resolution images difficult, especially if the model needs to be regularly retrained.
A less expensive way to leverage the power of computer vision is to enhance the CP feature with the embedding of an image using a pretrained model such as ResNet or Inception. We tried a simple approach with ResNet50 as an embedding generator, which yielded small improvements. Since most vision models are pretrained on ImageNet, this suggests that there is some transferable knowledge obtained from training on a large unrelated image database, but not enough to make a significant improvement. We expect that a more informative embedding can be achieved by pretraining the embedding generator end-to-end on cell images with a more relevant pretraining task, such as multi-task or contrastive learning.39,40
The benchmark also provides insight on how effective transferring knowledge from pretraining models on auxiliary tasks is to new tasks. Mostly, new tasks benefit from such a pretraining scheme, but the degree to which different tasks improve varies. To what degree a new task benefits from pretraining is still an open question for research. As a general observation, it seems that already predicted tasks tend to benefit more from pretraining. Companies aiming to pretrain their models on auxiliary tasks can use a combination of tasks from public sources as well as their own databases to benefit from as much data as possible.
FSL-CP offers a promising and interesting research question, though not one without unique challenges of its own. Through this study, we hope to encourage further research in few-shot and computer vision methods in the domain of cell imaging. The code for generating the dataset, model training, inference, and tutorials is publicly available on GitHub.
Data availability
The dataset, model codes, plots and results are all publicly available on Github: https://github.com/czodrowskilab/FSL_CP. In addition, since the FSL-CP dataset is curated from two larger public databases: ChEMBL and Broad Institute, the code for data processing and curation is also available on Github https://github.com/czodrowskilab/FSL_CP_DataPrep.Author contributions
S. V. H. and P. C. formulated the project. S. V. H. curated the data. L. L. coded and trained the single-task models. S. V. H. coded and trained the multi-task and the meta-learning models. S. V. H. wrote the manuscript, and all authors contributed to revising the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The first author's research position was funded by the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie Innovative Training Network – European Industrial Doctorate grant agreement no. 956832, “Advanced machine learning for Innovative Drug Discovery”. The authors thank Steffen Jaensch, Lorena G. A. Freitas for proofreading the manuscript, and others from Janssen Pharmaceutica for providing feedback for the project. In addition, we thank Axel Pahl and Jose Manuel-Gally from the Max-Planck Institute in Dortmund and other members of the Czodrowski lab for the fruitful discussions.Notes and references
- J. G. Moffat, J. Rudolph and D. Bailey, Nat. Rev. Drug Discovery, 2014, 13, 588–602 CrossRef CAS PubMed.
- C. M. Johannessen, P. A. Clemons and B. K. Wagner, Trends Genet., 2015, 31, 16–23 CrossRef CAS PubMed.
- D. Herman, M. M. Kańduła, L. G. A. Freitas, C. van Dongen, T. Le Van, N. Mesens, S. Jaensch, E. Gustin, L. Micholt, C.-H. Lardeau, C. Varsakelis, J. Reumers, S. Zoffmann, Y. Will, P. J. Peeters and H. Ceulemans, Chem. Res. Toxicol., 2023, 36, 1028–1036 Search PubMed.
- M.-A. Bray, S. M. Gustafsdottir, M. H. Rohban, S. Singh, V. Ljosa, K. L. Sokolnicki, J. A. Bittker, N. E. Bodycombe, V. Dančík, T. P. Hasaka, C. S. Hon, M. M. Kemp, K. Li, D. Walpita, M. J. Wawer, T. R. Golub, S. L. Schreiber, P. A. Clemons, A. F. Shamji and A. E. Carpenter, GigaScience, 2017, 6, 1–5 Search PubMed.
- J. C. Caicedo, S. Singh and A. E. Carpenter, Curr. Opin. Biotechnol., 2016, 39, 134–142 CrossRef CAS PubMed.
- F. Reisen, A. Sauty de Chalon, M. Pfeifer, X. Zhang, D. Gabriel and P. Selzer, Assay Drug Dev. Technol., 2015, 13, 415–427 CrossRef CAS PubMed.
- D. W. Young, A. Bender, J. Hoyt, E. McWhinnie, G.-W. Chirn, C. Y. Tao, J. A. Tallarico, M. Labow, J. L. Jenkins, T. J. Mitchison and Y. Feng, Nat. Chem. Biol., 2008, 4, 59–68 CrossRef CAS PubMed.
- V. Ljosa, P. D. Caie, R. Ter Horst, K. L. Sokolnicki, E. L. Jenkins, S. Daya, M. E. Roberts, T. R. Jones, S. Singh, A. Genovesio, P. A. Clemons, N. O. Carragher and A. E. Carpenter, J. Biomol. Screening, 2013, 18, 1321–1329 CrossRef CAS PubMed.
- C. Collinet, M. Stöter, C. R. Bradshaw, N. Samusik, J. C. Rink, D. Kenski, B. Habermann, F. Buchholz, R. Henschel, M. S. Mueller, W. E. Nagel, E. Fava, Y. Kalaidzidis and M. Zerial, Nature, 2010, 464, 243–249 CrossRef CAS PubMed.
- F. Fuchs, G. Pau, D. Kranz, O. Sklyar, C. Budjan, S. Steinbrink, T. Horn, A. Pedal, W. Huber and M. Boutros, Mol. Syst. Biol., 2010, 6, 370 CrossRef PubMed.
- M. Garcia de Lomana, P. A. Marin Zapata and F. Montanari, Chem. Res. Toxicol., 2023, 36, 1107–1120 Search PubMed.
- A. Liu, S. Seal, H. Yang and A. Bender, SLAS Discovery, 2023, 28, 53–64 Search PubMed.
- M. Akbarzadeh, I. Deipenwisch, B. Schoelermann, A. Pahl, S. Sievers, S. Ziegler and H. Waldmann, Cell Chem. Biol., 2022, 29, 1053–1064 Search PubMed.
- S. Seal, H. Yang, M.-A. Trapotsi, S. Singh, J. Carreras-Puigvert, O. Spjuth and A. Bender, J. Cheminf., 2023, 15, 56 Search PubMed.
- S. Seal, J. Carreras-Puigvert, M.-A. Trapotsi, H. Yang, O. Spjuth and A. Bender, Commun. Biol., 2022, 5, 858 CrossRef CAS PubMed.
- J. Simm, G. Klambauer, A. Arany, M. Steijaert, J. K. Wegner, E. Gustin, V. Chupakhin, Y. T. Chong, J. Vialard, P. Buijnsters, I. Velter, A. Vapirev, S. Singh, A. E. Carpenter, R. Wuyts, S. Hochreiter, Y. Moreau and H. Ceulemans, Cell Chem. Biol., 2018, 25, 611–618 CrossRef CAS PubMed.
- J. Snell, K. Swersky and R. S. Zemel, arXiv, 2017, arXiv:1703.05175 [cs.LG], DOI:10.48550/arXiv.1703.05175.
- C. Finn, P. Abbeel and S. Levine, arXiv, 2017, arXiv:1703.03400 [cs.LG], DOI:10.48550/arXiv.1703.03400.
- T. B. Brown, B. Mann, N. Ryder, M. Subbiah, J. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. M. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, arXiv, 2020, arXiv:2005.14165 [cs.CL], DOI:10.48550/arXiv.2005.14165.
- R. Geng, B. Li, Y. Li, X. Zhu, P. Jian and J. Sun, arXiv, 2019, arXiv:1902.10482 [cs.CL] , DOI:10.48550/arXiv.1902.10482.
- O. Vinyals, C. Blundell, T. Lillicrap, K. Kavukcuoglu and D. Wierstra, arXiv, 2017, arXiv:1606.04080 [cs.LG], DOI:10.48550/arXiv.1606.04080.
- M. Stanley, J. F. Bronskill, K. Maziarz, H. Misztela, J. Lanini, M. Segler, N. Schneider and M. Brockschmidt, NeurIPS 2021 Track Datasets and Benchmarks, 2021 Search PubMed.
- D. Mendez, A. Gaulton, A. P. Bento, J. Chambers, M. De Veij, E. Félix, M. Magariños, J. Mosquera, P. Mutowo, M. Nowotka, M. Gordillo-Marañón, F. Hunter, L. Junco, G. Mugumbate, M. Rodriguez-Lopez, F. Atkinson, N. Bosc, C. Radoux, A. Segura-Cabrera, A. Hersey and A. Leach, Nucleic Acids Res., 2018, 47, D930–D940 CrossRef PubMed.
- A. Carpenter, T. Jones, M. Lamprecht, C. Clarke, I. Kang, O. Friman, D. Guertin, J. Chang, R. Lindquist, J. Moffat, P. Golland and D. Sabatini, Genome Biol., 2006, 7, R100 CrossRef PubMed.
- M. Hofmarcher, E. Rumetshofer, D.-A. Clevert, S. Hochreiter and G. Klambauer, J. Chem. Inf. Model., 2019, 59, 1163–1171 CrossRef CAS PubMed.
- K. He, X. Zhang, S. Ren and J. Sun, arXiv, 2015, arXiv:1512.03385 [cs.CV], DOI:10.48550/arXiv.1512.03385.
- O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh, S. Ma, Z. Huang, A. Karpathy, A. Khosla, M. Bernstein, A. C. Berg and L. Fei-Fei, Int. J. Comput. Vis., 2015, 115, 211–252 CrossRef.
- L. Schiff, B. Migliori, Y. Chen, D. Carter, C. Bonilla, J. Hall, M. Fan, E. Tam, S. Ahadi, B. Fischbacher, A. Geraschenko, C. J. Hunter, S. Venugopalan, S. DesMarteau, A. Narayanaswamy, S. Jacob, Z. Armstrong, P. Ferrarotto, B. Williams, G. Buckley-Herd, J. Hazard, J. Goldberg, M. Coram, R. Otto, E. A. Baltz, L. Andres-Martin, O. Pritchard, A. Duren-Lubanski, A. Daigavane, K. Reggio, P. C. Nelson, M. Frumkin, S. L. Solomon, L. Bauer, R. S. Aiyar, E. Schwarzbach, S. A. Noggle, F. J. Monsma, D. Paull, M. Berndl, S. J. Yang, B. Johannesson and N. G. S. C. A. Team, Nat. Commun., 2022, 13, 1590 CrossRef CAS PubMed.
- C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke and A. Rabinovich, arXiv, 2014, arXiv:1409.4842 [cs.CV], DOI:10.48550/arXiv.1409.4842.
- C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna, arXiv, 2015, arXiv:1512.00567 [cs.CV], DOI:10.48550/arXiv.1512.00567.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742–754 CrossRef CAS PubMed.
- A. Mauri, V. Consonni, M. Pavan and R. Todeschini, MATCH Communications in Mathematical and in Computer Chemistry, 2006, 56, 237–248 CAS.
- O. Soufan, W. Ba-alawi, A. Magana-Mora, M. Essack and V. B. Bajic, Sci. Rep., 2018, 8, 9110 CrossRef PubMed.
- D. Butina, M. D. Segall and K. Frankcombe, Drug Discovery Today, 2002, 7, S83–S88 CrossRef CAS PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- A. Mayr, G. Klambauer, T. Unterthiner and S. Hochreiter, Front. Environ. Res., 2016, 3, 80 Search PubMed.
- W. Hu, B. Liu, J. Gomes, M. Zitnik, P. Liang, V. Pande and J. Leskovec, arXiv, 2020, arXiv:1905.12265 [cs.LG], DOI:10.48550/arXiv.1905.12265.
- L. Weng, https://www.lilianweng.github.io, 2018.
- K. Chaitanya, E. Erdil, N. Karani and E. Konukoglu, arXiv, 2020, arXiv:2006.10511 [cs.CV], DOI:10.48550/arXiv.2006.10511.
- A Radford, J. W. Kim, C. Hallacy, A. Ramesh, G. Goh, S. Agarwal, G. Sastry, A. Askell, P. Mishkin, J. Clark, G. Krueger and I. Sutskever, arXiv, 2021, arXiv:2103.00020 [cs.CV], DOI:10.48550/arXiv.2103.00020.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00205e |
| This journal is © The Royal Society of Chemistry 2024 |