DOI:
10.1039/D3CS00171G
(Review Article)
Chem. Soc. Rev., 2024, Advance Article
Fluorescent probes for targeting the Golgi apparatus: design strategies and applications
Received
31st January 2024
First published on 21st June 2024
Abstract
The Golgi apparatus is an essential organelle constructed by the stacking of flattened vesicles, that is widely distributed in eukaryotic cells and is dynamically regulated during cell cycles. It is a central station which is responsible for collecting, processing, sorting, transporting, and secreting some important proteins/enzymes from the endoplasmic reticulum to intra- and extra-cellular destinations. Golgi-specific fluorescent probes provide powerful non-invasive tools for the real-time and in situ visualization of the temporal and spatial fluctuations of bioactive species. Over recent years, more and more Golgi-targeting probes have been developed, which are essential for the evaluation of diseases including cancer. However, when compared with systems that target other important organelles (e.g. lysosomes and mitochondria), Golgi-targeting strategies are still in their infancy, therefore it is important to develop more Golgi-targeting probes. This review systematically summarizes the currently reported Golgi-specific fluorescent probes, and highlights the design strategies, mechanisms, and biological uses of these probes, we have structured the review based on the different targeting groups. In addition, we highlight the future challenges and opportunities in the development of Golgi-specific imaging agents and therapeutic systems.
 Silin Xu | Si-Lin Xu received his PhD degree in 2020 from Jiangxi Normal University, China. Now he is a lecturer at Xuchang University (Xuchang, China). His current research interests include development of novel synthetic methodologies for peptide and the design and synthesis of small molecule with structural diversity for chemical biology research. |
 Kai-Cheng Yan | Kai-Cheng Yan received his bachelor's degree in Industrial Engineering (2017) and Master's degree in Pharmaceutical Engineering from ECUST at the School of Chemistry and Molecular Engineering (2020). He is currently pursuing his PhD degree in Chemistry, at the University of Bath, UK, where his projects are related to chemical biology for pathogenic microorganism theranostics. |
 From left to right: Tony D. James, Zhi-Hong Xu and Yuan Wang | Tony D. James is a Professor at the University of Bath and Fellow of the Royal Society of Chemistry. He received his PhD from the University of Victoria (1991). He has an h-index of 90 and has been listed by Clarivate as a Highly Cited Researcher since 2022. Zhi-Hong Xu: received his PhD degree in 2008 from Lanzhou University, China. He is professor at the College of Chemical and Materials Engineering in Xuchang University. His current research interests include development of chemosensors and nanosensors for biological and environmental relevance. Yuan Wang: received his PhD degree in 2008 from Lanzhou University (Lanzhou, China). Now he is professor at Henan Polytechnic University (Jiaozuo, China). His main scientific interests are the design and synthesis of small molecule with structural diversity for chemical biology research. |
1. Introduction
Prokaryote organisms are different from eukaryotic cells in that they do not contain subcellular organelles, such as lysosomes/endosomes, mitochondria, lipid droplets (LDs), plasma membranes, nucleus, Golgi apparatus and endoplasmic reticulum (ER).1 These organelles exhibit distinctive properties and performs unique and/or essential biological functions in various cellular processes.2
The Golgi apparatus which is widely distributed in eukaryotic cells and is dynamically regulated during cell cycles, exhibits a slightly acidic environment (6.0–6.7) and expresses several specific enzymes or proteins such as cyclooxygenases and furin. The organelle is formed by the stacking of flattened vesicles and plays an important role in a variety of metabolic pathways and cell signaling mechanisms. Where one of the primary biological functions is collecting, processing, sorting, transporting and secreting important proteins/enzymes from the endoplasmic reticulum to intra- and extra-cellular destinations. The Golgi apparatus is not only involved in protein processing and transport, but also in many physiological processes such as ion exchange, oxidative stress and apoptosis, which helps maintain normal physiological function and cell survival.3–8
Redox homeostasis, the organelle microenviroment, metal ion homeostasis, and enzyme activity are intimately associated with the correct function of the Golgi. It has been established that Golgi-related functions are closely related to many important biological species (ions, reactive oxygen species, reactive nitrogen species, reactive sulfur species, enzymes etc.) and microenvironmental parameters (pH, viscosity, polarity etc.) present in the Golgi, and the deficiency or excess of such bioactive species can cause morphological changes and dysfunction of the Golgi apparatus, which is directly linked to a range of human disorders including cancer, neuronal, kidney, eye, and liver diseases.9,10 Therefore, the accurate in situ monitoring of bioactive species in the Golgi will provide a comprehensive appreciation of the biological functions and pathological processes linked with the Golgi apparatus, which can help facilitate the diagnosis of Golgi related diseases and monitor therapeutic agents during the treatment of diseases.11–15
Organelle-specific fluorescent probes provide powerful non-invasive tools for the real-time and in situ visualization of the temporal and spatial fluctuations of bioactive species in cells and in vivo with high selectivity and high sensitivity.16–35 However, compared with systems for other key organelles (e.g. lysosomes and mitochondria), Golgi-targeting strategies which are vital to monitor cellular health are still in their infancy.
With this review, we have summarized Golgi-specific fluorescent probes, which can respond to specific bioactive molecules, and highlighted the design strategies, mechanisms, and biological applications. We also provide insights and provide guidance for the future development of this area of research. It worth to mentioning that some Golgi-targeting markers that simply stain the Golgi were excluded from this review. We expect that this review will steer the development of probes targeting the Golgi and facilitate the evaluation of related biomedical areas.
2. Golgi targeting probes design
Visualization of the Golgi apparatus is of great significance for real-time monitoring of the functions of the Golgi apparatus. Over a hundred years ago, Camillo Golgi invented the silver staining method for the Golgi apparatus and instigated research into Golgi structure-function correlations.36 In the mid-20th century, Felix observed the basic shape of the Golgi apparatus using an electron microscopy. While in 1985, Lipsky et al. observed the Golgi apparatus for the first time in living cells using ceramide derivative staining.37 In the late 20th century and early 21st century, the discovery of fluorescent proteins (green fluorescent protein, yellow fluorescent protein, and so on) and Furin cleavage technology provided biological methods for Golgi imaging.38,39 However, these classical methods can only achieve intracellular Golgi imaging, but are unsuitable for the real-time detection of biomolecules in the Golgi apparatus. Significantly, the recent development of fluorescence imaging technologies has provided many probes for Golgi imaging and the monitoring of the concentration of bioactive molecules in the Golgi apparatus.40
Although Golgi apparatus targeting fluorescent probes have previously been developed, the targeting mechanism needs clarifying. When developing Golgi-specific fluorescent probes, the main obstacle is the lack of precise Golgi-targeting groups, as such determining appropriate systems that can improve localization efficiency of a targeting group is essential for the development of Golgi-targeting fluorescent probes.
Generally speaking, in order to obtain good Golgi-targeting efficiency, the Golgi localization groups should tightly and selectively bind to specific enzymes present in the Golgi apparatus, or match well with the weakly acidic environment of the Golgi or form non-convalent or covalent bonds with biomacromolecules present in the Golgi, or linked with Golgi-targeting peptides. In this review, we illustrate some representative targeting groups for the development of Golgi-specific fluorescent probes. For example, cyclooxygenase-2 (COX-2) inhibitors such as indomethacin and celecocoxib can bind tightly and selectively to the side pocket of COX-2 in the Golgi apparatus, and phenylsulfonamide also can bind to specific regions of COX-2, and 2-trifluoromethyl-7-aminoquinoline with a pyridyl moiety matches well with the slightly acidic microenvironment of the Golgi, while cysteine or cysteine-enriched BSA can form disulphide bonds by binding with thiol groups present in the Golgi. As such these are common targeting groups for evaluating the Golgi apparatus using non-covalent or covalent interactions with biomacromolecules present in the Golgi. Herein, we have classified the reported fluorescent probes based on their Golgi-targeting functional motif (Scheme 1).
 |
| | Scheme 1 Common Golgi-targeting groups. | |
3. Classification of Golgi-targeting probes based on the functional motif
3.1 Probes containing cyclooxygenase-2 (COX-2) inhibitors
Cyclooxygenases (COXs) control the synthesis of prostaglandins and are pivotal in tumor development and progression.41 The evolution of the Golgi apparatus and tumorigenesis, is associated with cyclooxygenase COX-2, which is an inducible enzyme accumulating in the Golgi apparatus. COX-2 is an attractive target for molecular imaging because it is upregulated in most cancer cells but less expressed in normal cells.42–49 COX-2 has been detected in premalignant lesions of the esophagus and colon, and its levels appear to increase during tumor progression, which indicates the importance of COX-2 in tumor progression.50,51 In addition, it has been confirmed that COX-2 inhibitors such as indomethacin (IMC) and Celecoxib (CXB) conjugates bind tightly and selectively to the side pocket of COX-2.42,52–54 Thus, IMC and CXB can be used as targeting groups for COX-2 and used for imaging the Golgi apparatus. Scheme 2 and Table 1 highlight Golgi-targeting probes using cyclooxygenase-2 (COX-2) inhibitors as the functional motif and their uses. Non-Golgi-targeting research using COX-2 inhibitors will not be covered in this section.
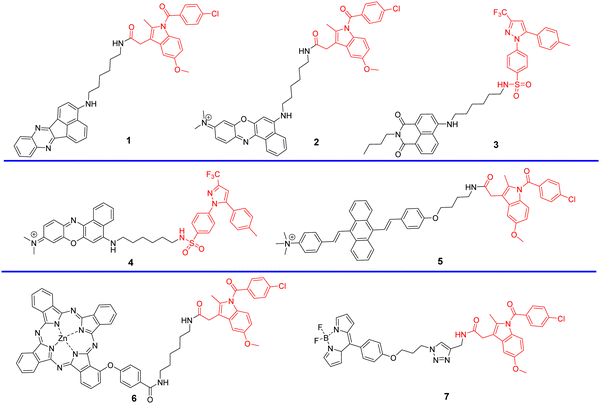 |
| | Scheme 2 Golgi-targeting probes containing cyclooxygenase-2 (COX-2) inhibitors as the functional motif. The Golgi-targeting units are highlighted in red. | |
Table 1 Golgi-targeting fluorescent probes with cyclooxygenase-2 (COX-2) inhibitors as functional motif
| Probe |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 1 |
OP 457/547 |
0.12–0.72 μg mL−1 |
0.11 μg mL−1 |
0.97 |
Differentiate cancer cells from normal cells; imaging of Golgi apparatus during cancer cell apoptosis |
55 |
| TP 800/— |
| 2 |
630/670 |
— |
— |
0.96, 0.97, 0.93 |
Differentiate cancer cells from normal cells; In vivo tumor optical imaging |
56 |
| 3 |
OP 488/— |
— |
— |
0.96 |
Differentiate cancer cells from normal cells; Tissue slice and tumor mouse imaging |
57 |
| TP 870/— |
| 4 |
630/700 |
— |
— |
0.94 |
Differentiate cancer cells from normal cells; Tissue slice and tumor mouse imaging |
58 |
| 5 |
420/574 |
0.14–0.8 μg mL−1 |
0.24 μg mL−1 |
— |
Differentiate cancer cells from normal cells; tumor-bearing mice imaging |
59 |
| 6 |
674/687 |
— |
— |
0.92 |
Photosensitizer for tumor cells |
60 |
| 7 |
488/515 |
1.8–950 cP |
— |
0.923, 0.930 |
Fluorescence lifetime imaging in live cells and zebrafish |
61 |
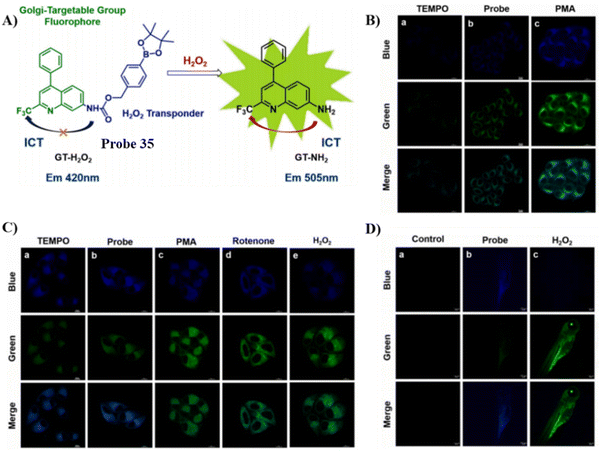
In 2013, the group of Peng developed a COX-2 turn-on fluorescent probe 1 (Scheme 2) for the Golgi apparatus of cancerous cells based on photoinduced electron transfer (PeT), which integrated a COX-2 inhibitor indomethacin (IMC) with the fluorophore acenaphtho[1,2-b]-quinoxaline (ANQ) using a hexanediamine linker.55 The free probe 1 exists in a folded conformation owing to the weak π-stacking interactions between ANQ and IMC, PeT from IMC to ANQ quenched the fluorescence of the probe (Fig. 1A). However, the fluorescence was activated (Fig. 1B) by attenuating the PeT process upon binding with COX-2, where probe 1 was forced to adopt an unfolded form. In addition, probe 1 exhibited significant two-photon properties (Fig. 1C) and was used for 3D depth imaging of living cancer tissues, probe 1 targeted the Golgi apparatus of cancer cells and was successfully used to monitor real-time changes of the Golgi apparatus during cancer cell apoptosis.
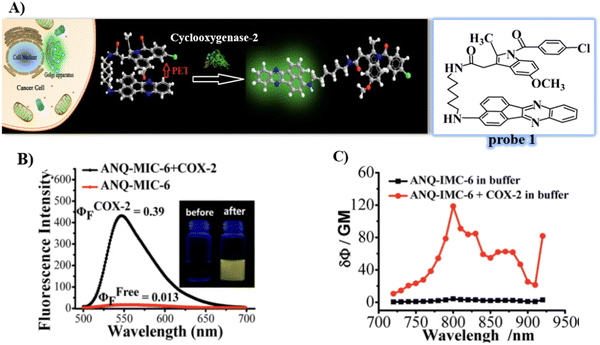 |
| | Fig. 1 (A) Detection mechanisms of probe 1 for COX-2. (B) Fluorescence emission spectra of probe 1 with and without addition of COX-2. (C) Two-photon spectra of probe 1 in the absence and presence of COX-2. Reproduced with permission from ref. 55. Copyright 2013, American Chemical Society. | |
In 2015, using the same strategy, and taking advantage of Near-infrared (NIR) fluorophores that exhibit minimum photodamage, minimal interference and deep tissue penetration, the group of Fan developed a Golgi-localized NIR fluorescent probe 2 (Scheme 2) for COX-2 (Fig. 2A) by replacing ANQ with Nile Blue, which was used to detect cancer cells.56 Probe 2 localized in the Golgi apparatus (Fig. 2B) and was used to image deep tissue tumors in a mouse model (Fig. 2C).
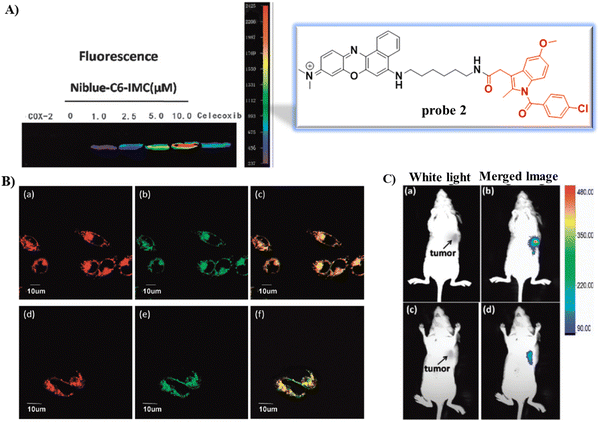 |
| | Fig. 2 (A) Fluorescence images of COX-2 with various concentration of probe 2 and in the presence of Celecoxib. (B) Fluorescence images of MCF-7 (a)–(c) and HeLa cells (d)–(f) stained with probe 2. (a) and (d) 2; (b) and (e) NBD C6-ceramide; (c) overlay of (a) and (b); (f) overlay of (d) and (e). (C) In vivo tumor optical imaging. (a) and (c) White light images of nude mice with tumor (b) treated with 2; (d) pre-dosed with Celecoxib. Reproduced with permission from ref. 56. Copyright 2015, The Royal Society of Chemistry. | |
Celecoxib (CXB) is an effective inhibitor of COX-2, which has been used for non-steroidal treatment of inflammation. In 2018 and 2020, the group of Peng developed two Golgi-targeting fluorescent probes 3 and 4 (Scheme 2) for COX-2, using CXB as a Golgi targeting group, which was then conjugated with naphthalamide and Nile Blue respectively via a flexible six-carbon linker.57,58 Probe 3 displays intense one- and two- photon fluorescence emission in cancer cells that express elevated levels of COX-2, and can be used for the fluorescence imaging of tumors (Fig. 3). Probe 4 exhibits strong NIR fluorescence in tumor cells/tissues with enhanced expression of COX-2, and can distinguish cancer cells from healthy cells, exhibits Golgi apparatus localization, and was used for the fluorescence visualization of tumors in mice and tissue slices (Fig. 4).
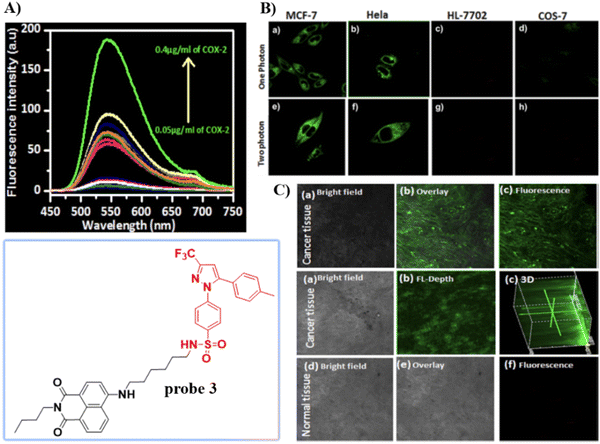 |
| | Fig. 3 (A) Fluorescence spectra of 3 upon the addition of COX-2. (B) One (a)–(d) and two photon (e)–(h) fluorescence images of cancer cells MCF-7 cells (a) and (e), HeLa cells (b) and (f) and normal cells HL-7702 cells (c) and (g), COS-7 cells (d) and (h) stained with 3. (C) Fluorescence imaging of cancerous and normal tissue. Reproduced with permission from ref. 57. Copyright 2018, American Chemical Society. | |
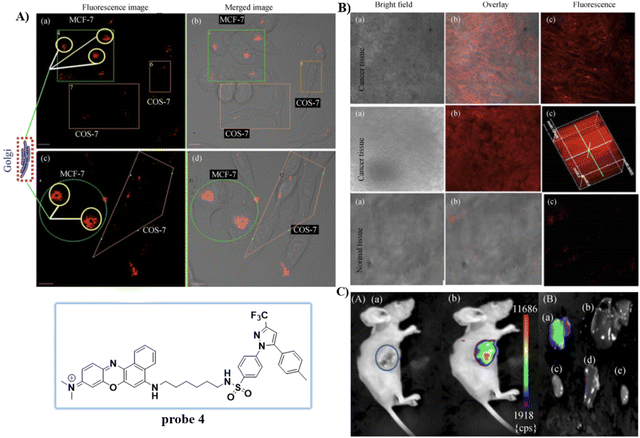 |
| | Fig. 4 (A) Normal cells (COS-7) and cancer cells (MCF-7) co-cultured and stained with 4. (a) and (c) Overlay fluorescence imaging of co-cultured cancer and normal cells (b) and (d). (B) Fluorescence imaging of cancerous and normal tissue stained with 4. (C) Fluorescence imaging of tumor mouse and organs. Reproduced with permission from ref. 58. Copyright 2020, Springer. | |
To overcome the aggregation-caused quenching found with traditional organic dyes including Nile blue, naphthalamide, and so on, in 2021, the group of Tian developed a “turn-on” aggregation-induced emission (AIE) based COX-2 fluorescent probe 5 (Scheme 2), using 9,10-distyrylanthracene as fluorophore and IMC as the targeting group. As such probe 5 could visualize sites that over express COX-2 in the Golgi apparatus of cancer cells, due to the restriction of intramolecular motion of the AIE-active probe 5 when bound to COX-2, enabling the discrimination of cancerous from normal cells and could be used to image tumor-bearing mice (Fig. 5).59
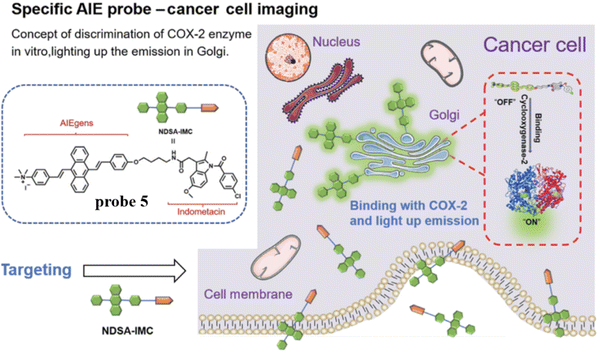 |
| | Fig. 5 Discrimination of COX-2 enzyme using probe 5. Reproduced with permission from ref. 59. Copyright 2021, Chinese Chemical Society. | |
Concerned by the poor tumor targeting and aggregation of photosensitizers, limited radius of activity and lifetime of reactive oxygen species that results in the limited therapeutic efficiency of photosensitizers in photodynamic therapy (PDT). The Chen group developed an ultra-efficient photosensitizer 6 by conjugating a COX-2 binding IMC with a zinc phthalocyanines (ZnPc) photosensitizing agent in order to improve therapeutic efficacy, the indomethacin moiety enhances the action of ZnPc by selectively targeting COX-2 expressing tumor cells, and targeting the Golgi apparatus. As such photosensitizer 6 is an encouraging candidate for improved therapeutic efficacy (Fig. 6).60
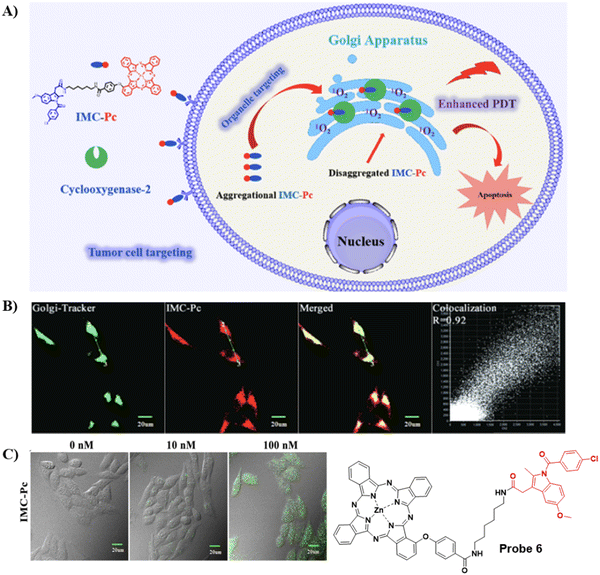 |
| | Fig. 6 (A) Illustration of 6 for highly-efficient anticancer therapy. (B) Co-localization fluorescence cell imaging of 6 with Golgi Tracker Green. (C) Fluorescence images of intracellular ROS production by 6 in HepG2 cells. Reproduced with permission from ref. 60. Copyright 2022, Elsevier. | |
To detect and diagnose the early stages of alcoholic liver injury, the Xiao group designed a Golgi-targeting fluorescent probe 7 for monitoring Golgi viscosity, which consists of a BODIPY fluorophore and a Golgi-targeting IMC moiety.61 Variations in viscosity of the Golgi during alcohol-induced liver injury were then evaluated quantitatively using fluorescence lifetime imaging in zebrafish and live cells, providing information on the link between viscosity of the Golgi and alcohol-induced liver injury (Fig. 7).
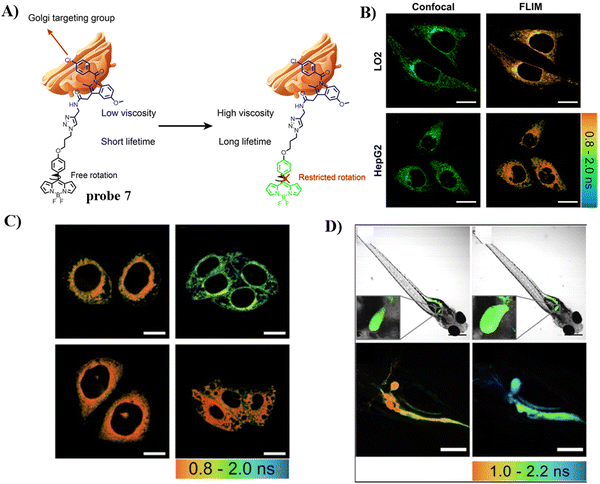 |
| | Fig. 7 (A) Structure and response mechanism of probe 7. (B) Fluorescence and FLIM imaging of LO2 and HepG2 cells with probe 7. (C) FLIM imaging in an alcohol-induced liver injury LO2 cell model of the Golgi with probe 7. (D) Fluorescence and FLIM imaging of zebrafish with the stimulation of ethanol. Reproduced with permission from ref. 61. Copyright 2022, The Royal Society of Chemistry. | |
3.2 Probes containing a phenylsulfonamide group
Given that COX-2 is upregulated in the Golgi apparatus of most cancer cells, fluorescent derivatives of COX-2 inhibitors represent an appropriate approach for Golgi-targeted imaging and monitoring. Research has shown that Golgi enzyme COX-2 inhibitor SC-558, derives selectivity from the phenylsulfonamide group which binds to specific regions of COX-2.62 therefore, the phenylsulfonamide unit is a promising Golgi targeting group for the Golgi apparatus.22,63 Scheme 3 and Table 2 highlight Golgi-targeting probes using phenylsulfonamide as the targeting group.
 |
| | Scheme 3 Golgi-targeting probes using phenylsulfonamide as the functional motif. Red moiety represents the Golgi-targeting unit. | |
Table 2 Golgi-targeting fluorescent probes with phenylsulfonamide as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 8 |
H2O2 |
405/470, 560 |
0–350 μM |
0.20 μM |
0.94 |
TP imaging in living cells and the kidney tissues of mice with hypertension |
63 |
| 9 |
H2S |
440/550 |
0–30 μM |
0.10 μM |
0.92 |
Imaging in living cells and zebrafish; the production of H2S under Golgi stress |
64 |
| 10 |
pH |
675/750, 810 |
pH 5.9–7.7 |
— |
0.9 |
Fluorescent images in cell; PAI in vivo; TMEM165 deficiencies influence the pH homeostasis |
65 |
| 11 |
Polarity |
675/405, 442 |
— |
— |
0.94 |
TP ratiometric imaging in cells; TP fluorescence imaging of kidney tissues |
66 |
| 12, 13 |
— |
— |
— |
— |
0.91, 0.92 |
Golgi apparatus localization |
67 |
| 14 |
FA |
440/546 |
0–100 μM |
0.35 μM |
0.806 |
In food; imaging in live cells |
68 |
| 15 |
CORM-3 |
430/520 |
0.5–80 μM |
0.35 μM |
0.93 |
Imaging in live cells and zebrafish |
69 |
| 16 |
ONOO− |
675/477, 650 |
— |
13 nM |
0.9 |
Imaging in live cells and DILI |
70 |
| 17 |
ONOO− |
420/600 |
8–40 μM |
250 nM |
0.9191 |
Imaging in live cells |
71 |
| 18 |
— |
366/612 |
— |
— |
0.92 |
In vivo/vitro imaging |
72 |
| 19 |
pH |
580/664 |
— |
— |
0.88 |
Imaging in live cells |
73 |
| 20 |
NTR |
440/534 |
0.1–5.0 μg mL−1 |
0.623 μM |
0.84 |
Imaging in live cells and in tumor tissue sections |
74 |
| 21 |
ClO− |
488/550 |
0–300 μM |
27 nM |
0.98 |
Imaging in live cells |
75 |
| 22 |
NO |
560/589 |
0–20 μM |
45 nM |
0.95 |
Imaging in live cells |
76 |
| 23 |
O2˙−, H2O2 |
633/740 |
0–35 μM |
62 nM |
0.827, 0.839 |
Imaging in live cells and mice |
77 |
At this point it should be noted that p-toluenesulfonamide is an ER targeting group which can localize within the ER through binding to sulfonylurea receptors in the ER, while a Golgi-targeting group is phenylsulfonamide, which can target Golgi apparatus through binding to COX-2 in the Golgi. While the p-toluenesulfonamide and phenylsulfonamide have very similar structures, they localize specifically in the ER and Golgi organelles respectively.
Using the Golgi-targeting phenylsulfonamide group, the Tang group developed a two-photon (TP) fluorescent probe 8, for evaluating oxidative stress in the Golgi apparatus during hypertension,63 where a phenylsulfonamide acts as the Golgi-targeting group and the boronic acid ester acts as a H2O2-recognition group. Probe 8 was used to ratiometrically image the generation of H2O2 in situ and revealed an increase of H2O2 levels in the Golgi of kidney tissues from mice with hypertension (Fig. 8). These outcomes indicate that phenylsulfonamide is an effective Golgi-targeting group. In addition, phenylsulfonamide is readily available, and can be chemically modified. Therefore, this targeting group can provide an ideal strategy for developing Golgi-targeting probes.
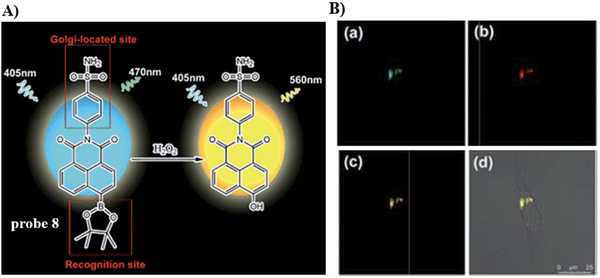 |
| | Fig. 8 (A) Probe 8 response mechanism. (B) Co-localization cell fluorescence imaging of 8 and Golgi Tracker Red. Reproduced with permission from ref. 63. Copyright 2019, The Royal Society of Chemistry. | |
Using the same Golgi-targeting group with a naphthalimide fluorophore as structural skeleton, the Zhu group developed a Golgi-targeting H2S fluorescent probe 9, for the visualization of endogenous and exogenous H2S in zebrafish and living cells (Fig. 9).64 The Golgi-targeting probe exhibited good sensitivity and biocompatibility and was used to monitor the production of H2S in the Golgi under stress.
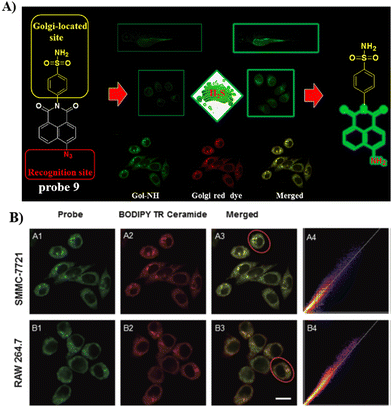 |
| | Fig. 9 (A) Probe 9 response mechanism. (B) Co-localization cell imaging of 9 and BODIPY TR ceramide. Reproduced with permission from ref. 64. Copyright 2020, The Royal Society of Chemistry. | |
In 2020, the Tang group developed a dual mode ratiometric photoacoustic (PA) and fluorescent imaging probe 10 (Fig. 10) for measuring the pH of Golgi in real-time with high sensitivity and excellent selectivity.65 The phenylsulfonamide moiety was selected as an effective Golgi-targeting anchor, and tricarbocyanine was selected as the fluorophore and PA reporter given that the tricarbocyanine fluorescence emission was in the NIR region78–83 and the tricarbocyanine unit also generates PA signals.84–89 The probe exhibited strong fluorescence emission at 810 nm under an acidic pH, and the emission of the probe shifts to 750 nm under basic conditions. As such acid–base changes of the system, resulted in the ratiometric fluorescence values (F750/F810) of the probe increasing by more than 100-fold. While at 800 nm the photoacoustic response of the probe was reduced with increasing pH, whilst at 690 nm the PA intensity was hardly affected. Significantly, the pH of the Golgi increases dramatically during monensin induced oxidative stress. Additional experiments indicated that transmembrane protein 165 (TMEM165) deficiency affected the proton equilibrium and resulted in Golgi acidification. This research not only provides confirmation that TMEM165 regulates pH homeostasis of the Golgi but also provided a potential Golgi pH indicator for various biological processes.
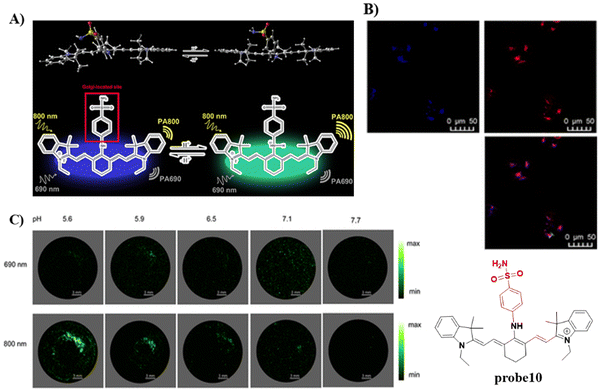 |
| | Fig. 10 (A) Probe 10 response mechanism. (B) Co-localization cell fluorescence imaging of 10 and Golgi Tracker Red. (C) pH monitoring by in vitro PA imaging. Reproduced with permission from ref. 65. Copyright 2020, American Chemical Society. | |
In 2021, by anchoring a Golgi-targeting moiety phenylsulfonamide onto a hemicyanine, the Tang group developed a TP ratiometric fluorescent probe 11 for Golgi polarity.66 Probe 11 was not only used for monitoring the changes of Golgi polarity in the kidneys of mice with acute kidney injury (AKI), but was used to image the variability of Golgi polarity during different physiological and biochemical processes (Fig. 11). This research confirms that Golgi polarity and AKI are correlated, and provides a potential high-throughput tool for AKI detection during drug development.
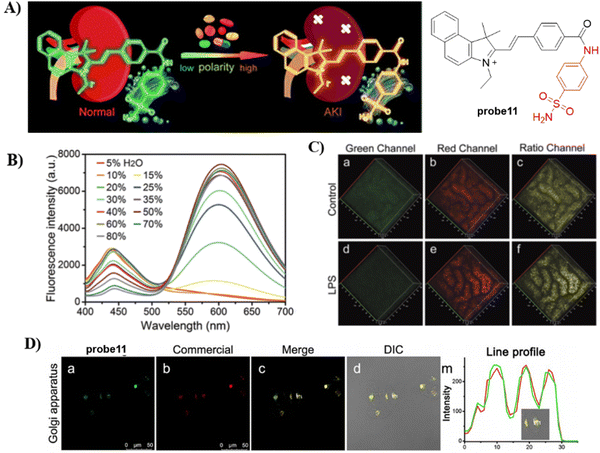 |
| | Fig. 11 (A) Probe 11 fluorescence and photoacoustic based dual-mode response mechanism. (B) Ratiometric fluorescence spectra of 11 towards polarity. (C) Two-photon fluorescence imaging in control mice and lipopolysaccharide (LPS) induced AKI mice of kidney tissues. (D) Co-localization cell imaging of 11 and Golgi Tracker Red. Reproduced with permission from ref. 66. Copyright 2021, The Royal Society of Chemistry. | |
The development of efficient and cost-effective endoplasmic reticulum (ER) and Golgi apparatus (GA) specific probes based on the AIE platform is of particular importance since it could provide a route to explore the interplay of organelles in living systems, but the development of such systems remains challenging. In 2021, the Tang group developed an AIE based fluorescence platform for GA and ER probes (12 to 13), which was achieved by changing the phenylsulfonamide to a glibenclamide unit attached to the fluorophore (Fig. 12A).67 For the design of organelle targeting probes, the phenylsulfonamide moiety was used to achieve GA-targeting and the glibenclamide unit binds to the ATP-sensitive potassium channel (KATP)55,62 to provide ER-targeting, and the triphenylamine (TPA) unit was used as rotor an electron donor (D). For Golgi localized probes, double bonds and cyano groups can be introduced to extend π-conjugation and enhance the D–A interaction, leading to transformation from ACQ-to-AIE (12 to 13) (Fig. 12B). Cellular imaging experiments indicated that these AIE-based probes which were simple to prepare exhibited good photostability, specificity, biocompatibility and high brightness when compared with commercially available probes.
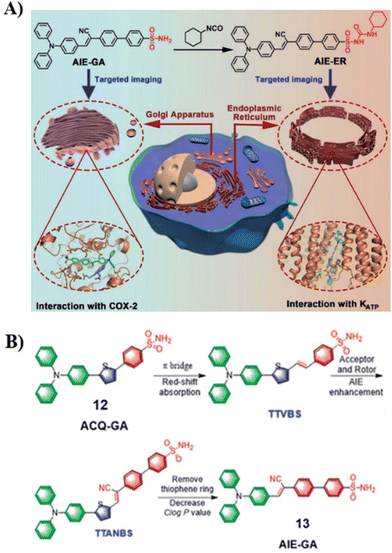 |
| | Fig. 12 (A) Schematic illustration of GA and ER targeting by AIEgens. (B) Rational design of GA probes. Reproduced with permission from ref. 67. Copyright 2021, The Royal Society of Chemistry. | |
In 2021, the Kim group prepared a fluorescent probe 14 for formaldehyde (FA) which contains hydrazine as the FA reaction site, integrated with a naphthalimide fluorophore and Golgi-targeting phenylsulfonamide moiety. Probe 14 exhibits a turn-on response due to the formation of a hydrazone and suppression of PeT and was suitable for the imaging of breast cancer cells (Fig. 13).68
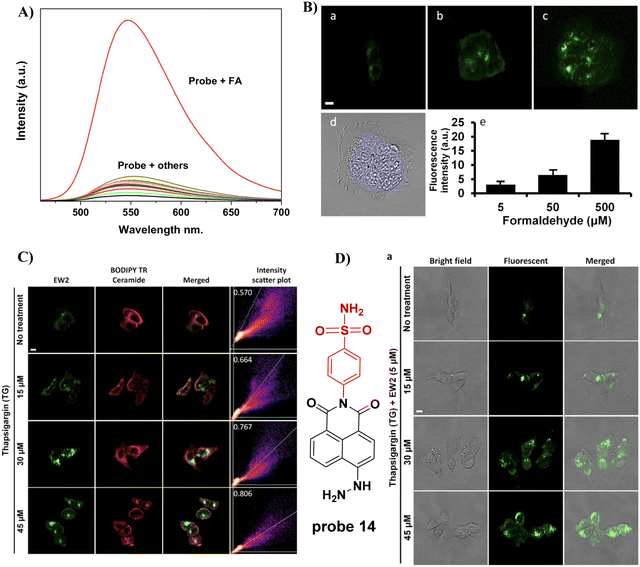 |
| | Fig. 13 (A) Fluorescence selectivity of probe 14. (B) The fluorescence imaging of FA in live MCF7 cells. (C) Co-localization of 14 and BODIPY TR ceramide tracker. (D) Imaging endogenous FA in Cells. Reproduced with permission from ref. 68. Copyright 2021, MDPI (Basel, Switzerland). | |
In 2022, the He group developed a Golgi-targeting fluorescent probe 15 for the detection of CO-releasing molecule-3 (CORM-3), with a naphthalimide fluorophore, and Golgi-targeting phenyl sulfonamide group, in addition a CORM-3-responsive nitro group was incorporated into the probe (Fig. 14).69 Probe 15 was used to monitor changes of CORM-3 in zebrafish, HepG2 and HeLa cells.
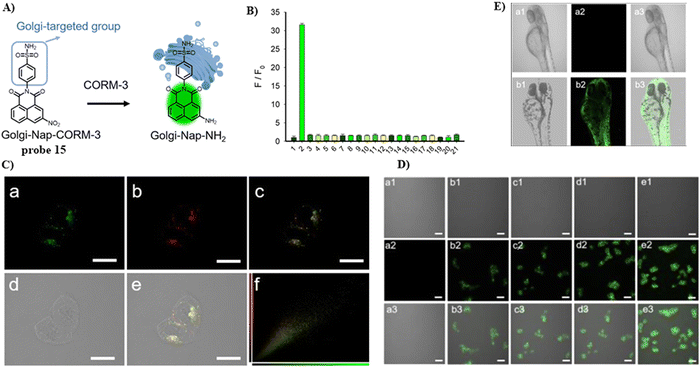 |
| | Fig. 14 (A) Probe 15 response mechanism. (B) Probe 15 fluorescence selectivity. (C) Co-localization of 15 and Golgi Tracker Red. (C) Confocal fluorescence images of HeLa cells. (E) Confocal fluorescence images of zebrafish. Reproduced with permission from ref. 69. Copyright 2022, American Chemical Society. | |
In 2022, the Feng group developed a Golgi-targeting fluorescent probe 16 for ONOO−, generated by linking a Golgi-targeting sulfanilamide group with a conjugated hemi-cyanine and coumarin.70 Probe 16 exhibited good sensitivity and selectivity rapid response for ONOO− and distinct ratiometric fluorescence changes and exhibited low cytotoxicity. Significantly, probe 16 can target the cell membrane and then the Golgi apparatus, making it suitable for detecting ONOO− in the Golgi and also cell membranes, the probe could be used to observe fluctuations of ONOO− during oxidative stress of the Golgi and in drug-induced liver injury (DILI) (Fig. 15).
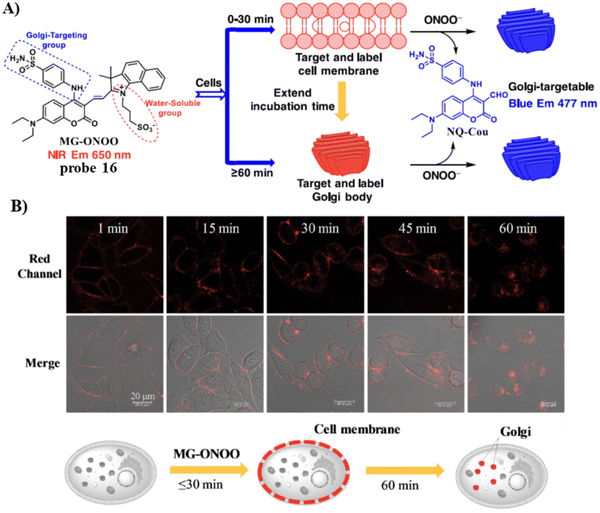 |
| | Fig. 15 (A) Membrane and Golgi-targeting fluorescent probe 16 for the ratiometric detection of ONOO−. (B) HeLa cell imaging with 16 at varying times. Reproduced with permission from ref. 70. Copyright 2022, Elsevier. | |
Using a quinolinemalononitrile based fluorophore, benzenesulfonamide targeting group, and a pinacol boronate ester as the recognition site the Ye group developed a AIE Golgi-targeting probe 17 to detect ONOO− in cells.71 Probe 17 exhibits a rapid response, good selectivity, large Stoke's shift and low detection limit. As such, the probe was used to detect endogenous and exogenous ONOO− in HeLa cells, and for monitoring the levels of ONOO− in the Golgi apparatus during monensin-induced oxidative stress (Fig. 16).
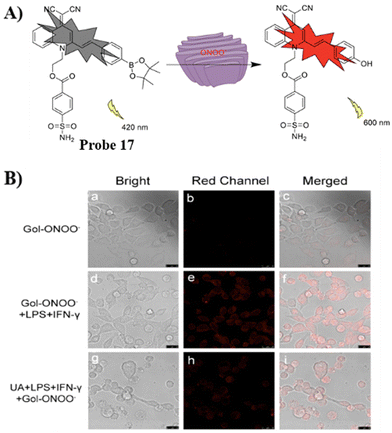 |
| | Fig. 16 (A) Possible reaction mechanism of 17 for detecting ONOO−. (B) Fluorescence imaging of probe 17 for endogenous ONOO−. Reproduced with permission from ref. 71. Copyright 2022, Elsevier. | |
Using hydrogen bonding between the COX-2 and benzenesulfonamide groups, the Yang group developed Golgi-targeting orange-emitting carbon dots (18) to image and monitor the Golgi apparatus. The carbon dots were prepared from p-phenylenediamine and benzenesulfonamide using one-step solvothermal methods. Probe 18 exhibits low toxicity and was suitable for imaging over 200 min (Fig. 17).72
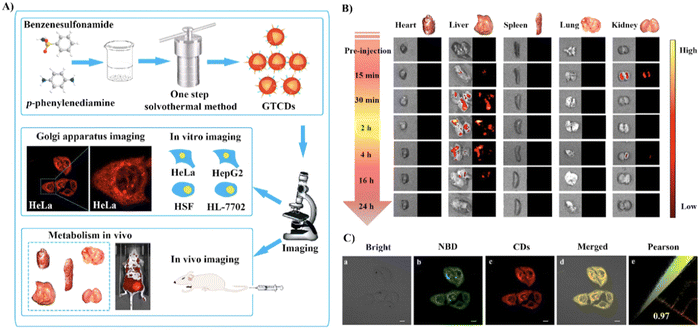 |
| | Fig. 17 (A) Synthesis and biological characterization of 18. (B) Optical and ex vivo fluorescence imaging of BALB/c mice organs at different treatment times with 18. (C) Co-localization cell imaging of 18 and NBD. Reproduced with permission from ref. 72. Copyright 2022, Elsevier. | |
In 2022, using a NIR rhodamine derivative as the fluorophore and p-methoxylbenzsulfamide as the targeting group, the Wang group developed a Golgi-targeting fluorescent probe 19 to detect changes in pH.73 The probe exhibits pH-dependent sensing behavior from pH 7.4–3.0 at 664 nm with a remarkable fluorescence “off–on” responses and a pKa value of 3.62. The probe selectively diffused into Golgi and was successfully used for visualizing and real-time tracking of Golgi pH variations in living cells (Fig. 18).
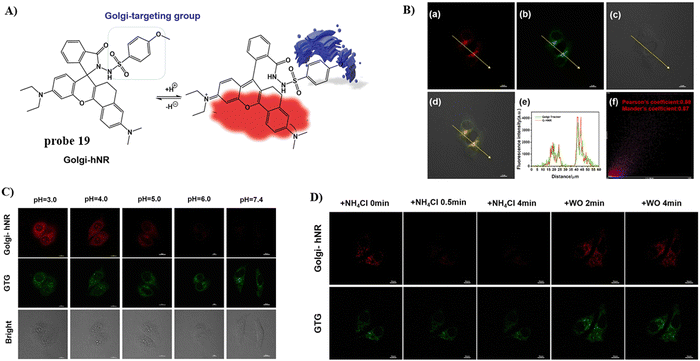 |
| | Fig. 18 (A) Possible reaction mechanism of 19 for pH. (B) Co-localization cell imaging of 19 and Golgi Tracker Green. (C) Fluorescence imaging using probe 19 for pH. (D) Visual monitoring of the pH changes upon different stimulations in the Golgi apparatus. Reproduced with permission from ref. 73. Copyright 2022, Elsevier. | |
Using naphthalimide as the fluorescent core and a nitro group as the response group, attached to a Golgi targeting sulfonamide group, the Lin group developed a Golgi-targeting fluorescent probe 20 (Gol-NTR) for detecting nitroreductase (NTR).74 The probe exhibits high biocompatibility, good photostability, high sensitivity and selectivity for NTR recognition, and was used to detect NTR in the Golgi apparatus. Consequently, the Golgi apparatus state could be monitored under different oxygen levels via the detection of NTR (Fig. 19).
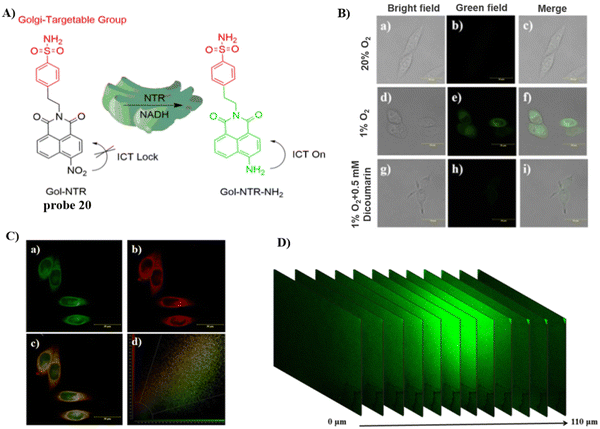 |
| | Fig. 19 (A) Structure and possible mechanism of 20 for monitoring NTR. (B) Fluorescence imaging of NTR using probe 20 in HeLa cells. (C) Co-localization cell imaging of 20 and Golgi Tracker Red. (D) Two-photon fluorescence imaging of NTR in live tumor tissue sections. Reproduced with permission from ref. 74. Copyright 2022, The Royal Society of Chemistry. | |
In 2023, the Song group developed a dual-functional, reversible, and ratiometric fluorescent probe 21 with coumarin fluorophore, benzenesulfonamide for Golgi-targeting, and selenium (Se) as the recognition site.75 Probe 21 could evaluate dynamic changes of Golgi oxidative stress by monitoring dynamic changes in the ClO−/GSH-mediated redox state in HeLa cells with rapid response, high selectivity, high sensitivity and low detection limit. More importantly, the probe facilitates the monitoring of the progression acute lung injury through the imaging of the dynamic redox state (Fig. 20).
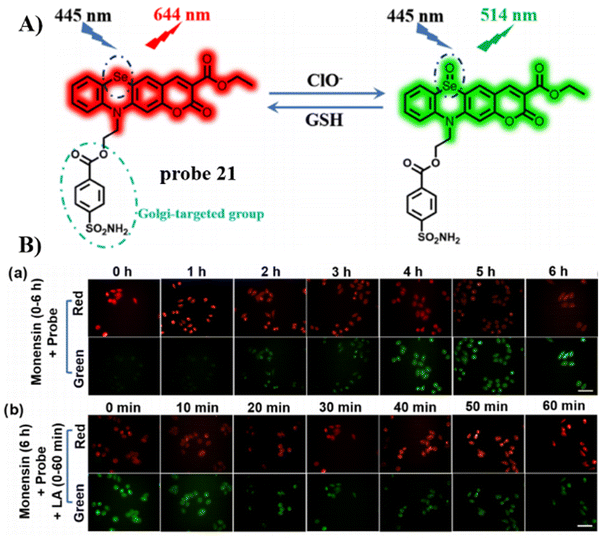 |
| | Fig. 20 (A) Possible reaction mechanism of 21 for sensing ClO−/GSH redox pair. (B) Fluorescence images of HeLa cells undergoing different treatments. Reproduced with permission from ref. 75. Copyright 2023, American Chemical Society. | |
The Ma group incorporated a 4-sulfamoylphenylamide group as a Golgi-targeting moiety with 6-carboxyrhodamine B containing an o-diaminobenzene NO recognition moiety to generate the Golgi-targeting fluorescent probe 22 for imaging and monitoring NO in the Golgi apparatus.76 Probe 22 exhibits accurate Golgi-targeting and high selectivity towards NO, and thus was used to monitor increases of NO in the Golgi apparatus of an AD model, providing an appropriate means for studying the function of NO in the Golgi apparatus during disease progression (Fig. 21).
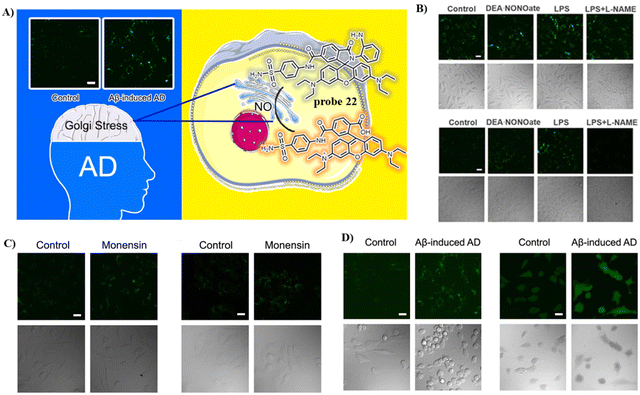 |
| | Fig. 21 (A) Structure and possible reaction mechanism of 22 for monitoring NO. (B) Confocal fluorescence images exogenous NO of SH-SY5Y (upper) cells and HepG2 (down) cells. (C) Confocal fluorescence images of SH-SY5Y (left) cells and HepG2 (right) cells during Golgi stress. (D) Confocal fluorescence images of SH-SH5Y cells with Golgi-NO (left) and DAF-DA (right) in the cellular AD model. Reproduced with permission from ref. 76. Copyright 2022, American Chemical Society. | |
Phenylsulfonamide-modified and bovine serum albumin (BSA) encapsulated probes have been shown to exhibit excellent Golgi-targeting ability. Recently, the Tang group developed a Golgi-targeting NIR fluorescent probe GSO for the in situ detection of O2˙− in the Golgi which was constructed using phenylsulfonamide as the Golgi-targeting group, a hemi-cyanine as the NIR fluorophore, and a trifluoromethanesulfonate group as the recognition site for O2˙−. The fluorescent probe 23 (GolRos) was then prepared by encapsulating GSO and Np-Golgi (probe 8) with BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2).77 The probe was successfully used for the simultaneous multicolored detection of both Golgi H2O2 and O2˙− in cells and in hypertensive mice, for the evaluation of various antihypertensive drugs, and for the identification of post-transcriptional modification sites on dihydrofolate reductase (DHFR) induced by the ROS. Moreover, the system is suitable for exploring the eNOS-associated oxidative stress signaling pathway in inflammation-induced hypertension (Fig. 22).
2).77 The probe was successfully used for the simultaneous multicolored detection of both Golgi H2O2 and O2˙− in cells and in hypertensive mice, for the evaluation of various antihypertensive drugs, and for the identification of post-transcriptional modification sites on dihydrofolate reductase (DHFR) induced by the ROS. Moreover, the system is suitable for exploring the eNOS-associated oxidative stress signaling pathway in inflammation-induced hypertension (Fig. 22).
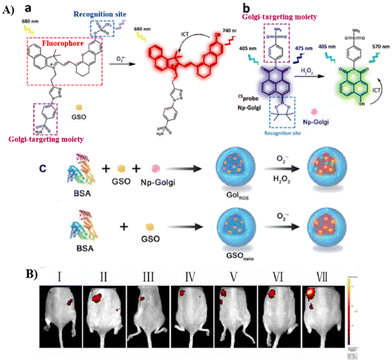 |
| | Fig. 22 (A) (a) Hemicyanine probe GSO and response mechanism towards O2˙−. (b) The structure of Np-Golgi (probe 8) and the response towards H2O2. (c) 23 and GSOnano preparation and response mechanism. (B) Fluorescence imaging of control mice (I), therapeutic group mice (II: hydrochlorothiazide, III: hydrochlorothiazide + tiron, IV: tiron, V: reserpine + tiron, VI: reserpine) and hypertension mice (VII). Reproduced with permission from ref. 77. Copyright 2022, Elsevier. | |
3.3 Probes with 2-trifluoromethyl-7-aminoquinoline group
Quinoline derivatives have been extensively used for the assembly of fluorescent probes owing to the excellent fluorescent properties. However, applications in probing organelles have been rarely explored. Quinoline primarily fluoresces in the near ultraviolet region, and its absorption and emission signals can be migrated to longer wavelengths by structural adjustment. Moreover, for quinoline derivatives the pyridyl moiety perfectly matches the slightly acidic (6.0–6.7) environment in the Golgi apparatus, making it excellent for Golgi-targeting.90 In 2019, the Chen group developed a dual-functional 2-trifluoromethyl-7-aminoquinoline fluorophore, which exhibits strong intramolecular charge-transfer fluorescence with large Stokes shifts due to the presence of amino group and trifluoromethyl group. The 2-trifluoromethyl-7-aminoquinoline group is not only Golgi-targeting but also contains an amine group that can be used to construct a wide range of fluorescent probes 24/25 (Fig. 23).91 Both Scheme 4 and Table 3 highlight Golgi-targeting probes using 2-trifluoromethyl-7-aminoquinoline as a functional motif and their applications.
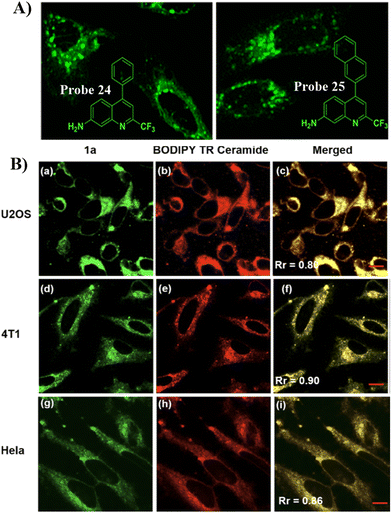 |
| | Fig. 23 (A) Schematic illustration of GA targeting quinoline fluorophore. (B) Co-localization cell imaging of 24 and BODIPY TR ceramide. Reproduced with permission from ref. 91. Copyright 2019, American Chemical Society. | |
 |
| | Scheme 4 Golgi-targeting probes with 2-trifluoromethyl-7-aminoquinoline. Red indicates the Golgi-targeting unit. | |
Table 3 Golgi-targeting fluorescent probes with 2-trifluoromethyl-7-aminoquinoline as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 24, 25 |
— |
389 nm/507 nm |
— |
— |
0.90 |
Golgi apparatus localization |
91 |
| 392 nm/513 nm |
| 26 |
Cys |
390 nm/515 nm |
0–10 μM |
105 nM |
0.92 |
Imaging in live cells and zebrafish |
92 |
| 27 |
Cys |
390 nm/518 nm |
0–40 μM |
51 nM |
0.91 |
Imaging in live cells and zebrafish |
93 |
| 28 |
H2S |
375 nm/515 nm |
0–30 μM |
0.11 μM |
0.94 |
Imaging in live cells and zebrafish |
94 |
| 29 |
Furin |
350, 470 nm/420, 640 nm |
0–500 U mL−1 |
0.265 U mL−1 |
0.84, 0.80 |
Imaging in live cells |
95 |
| 30 |
GSH |
320 nm/425, 510 nm |
0–75 μM |
0.388 U mL−1 |
0.92, 0.85, 0.92 |
Imaging in live cells |
96 |
| 31 |
CO |
360 nm/425, 520 nm |
0–10 μM |
0.49 μM |
0.81 |
Imaging in live cells and zebrafish |
97 |
| 32 |
Cys |
405 nm/520 nm |
0–40 μM |
41 nM |
0.913, 0.906, 0.924 |
Imaging in live cells and zebrafish |
98 |
| 33 |
ONOO− |
405 nm/490 nm |
0–30 μM |
115 nM |
0.81 |
Imaging in live cells and mice |
99 |
| 34 |
— |
405 nm/450 nm |
— |
57 nM |
0.99 |
Monitoring and assessing treatment response of sepsis-induced ALI |
100 |
| 35 |
H2O2 |
760 nm/820 nm |
0–40 μM |
54.8 ng mL−1 |
0.98 |
Imaging in live cells and zebrafish |
101 |
In 2020, the Zhu group developed a Golgi-targeting and Cys-specific probe 26 where a dithioester was selected as the receptor for Cys, and a quinoline derivative was selected as the fluorophore as well as the targeting group for the Golgi apparatus (Fig. 24).92 In addition to exhibiting sensitivity to native Cys in zebrafish and cells, the monitoring of Cys levels during Golgi stress was achieved.
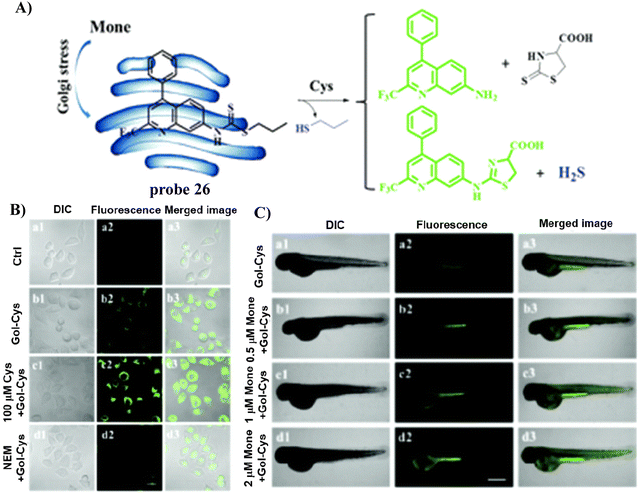 |
| | Fig. 24 (A) Probe 26 response mechanism for Cys. (B) Fluorescence images of HeLa cells. (C) Confocal microscopic images of Cys changes during Golgi stress in zebrafish. Reproduced with permission from ref. 92. Copyright 2020, The Royal Society of Chemistry. | |
By substituting the dithioester with thionobenzoate, the Zhu group explored a Golgi targeting Cys-specific probe 27 for detecting the generation of Cys in response to Golgi stress. Thionobenzoate was used as the recognition group, with the 4-CF3-7-aminoquinoline dye chosen as the fluorophore and Golgi targeting unit (Fig. 25).93 Probe 27 exhibited good sensitivity to both exogenous and endogenous Cys during biological imaging.
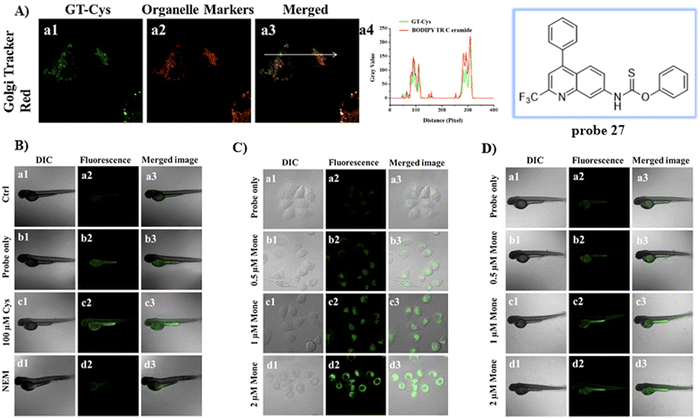 |
| | Fig. 25 (A) Co-localization cell fluorescence imaging of 27 and Golgi Tracker Red. (B) Confocal fluorescence images of zebrafish. (C) Confocal microscopic images for detection of Cys production during Golgi stress in HeLa cells. (D) Confocal microscopic images for detection of Cys production during Golgi stress in zebrafish. Reproduced with permission from ref. 93. Copyright 2020, Elsevier. | |
Similarly, by using the quinoline moiety as the Golgi-targeting group/fluorophore and, and azido unit as a H2S recognition site, the group of Zhu developed a Golgi-targeting H2S probe 28, which responds sensitively to H2S in the Golgi apparatus of both zebrafish and cells. In situ visualization of H2S under Golgi stress was achieved using a specific-Golgi stressor (Fig. 26).94
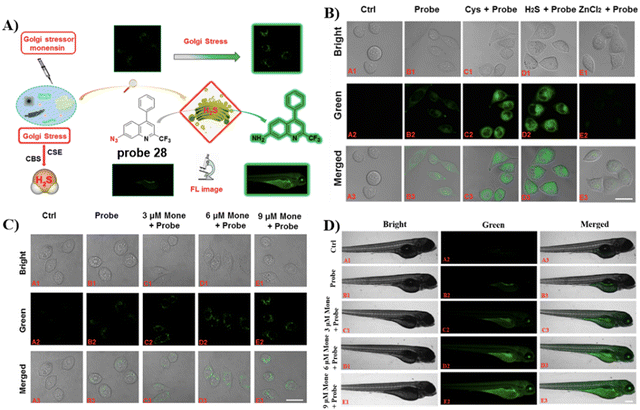 |
| | Fig. 26 (A) Response of probe 28 for H2S. (B) Cell fluorescence imaging by probe 28. (C) Golgi stress response results in cells. (D) Zebrafish imaging of probe 28. (E) Golgi stress response results in zebrafish. Reproduced with permission from ref. 94. Copyright 2020, American Chemical Society. | |
In 2021, the Liang group integrated the principles of intramolecular charge transfer (ICT) and fluorescence resonance energy transfer (FRET). A quinoline dye was chosen as a Golgi-targeting fluorophore and donor, while a dicyanomethylene-4H-pyran (DCM) was chosen as the acceptor fluorophore. The quinoline and DCM were linked using a Arg-Val-Arg-Arg (RVRR) peptide, to generate a FRET system. Which resulted in a Golgi-targeting and dual-emission “Turn-On” probe 29 for the imaging of intracellular furin. As a result of FRET between quinoline and DCM and inhibition of the intramolecular charge transfer (ICT), probe 29 was initially nonfluorescent but was activated upon amide bond cleavage, generating Q-RVRR and DCM-NH2, which resulted in fluorescence “Turn-On” at 420 and 640 nm (Fig. 27).95
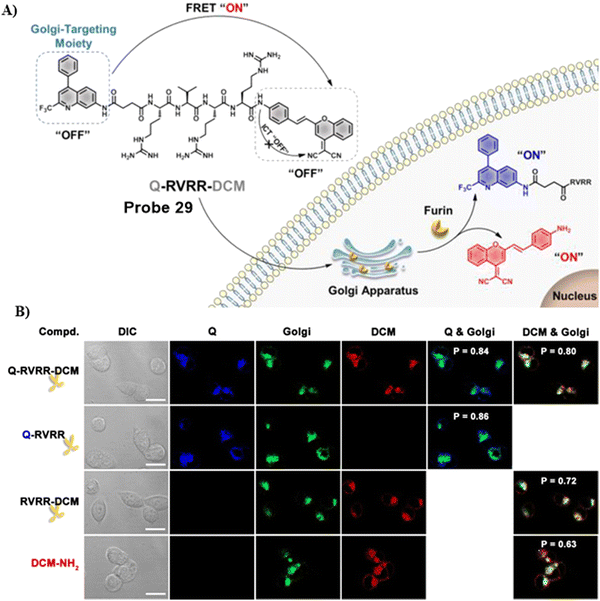 |
| | Fig. 27 (A) Response of probe 29 for Furin. (B) Co-localization cell imaging. Reproduced with permission from ref. 95. Copyright 2021, American Chemical Society. | |
In 2021, the Zhu group developed a ratiometric fluorescent Golgi-targeting probe 30 for GSH constructed using 4-CF3-7-aminoquinoline as a Golgi-targeting fluorophore containing an anticancer drug and GSH responsive receptor. Probe 30 reacted with GSH, releasing the drug in the Golgi to kill cancerous cells. Ratiometric probe 30 was able to selectively and sensitively trace changes of GSH concentration during Golgi stress in living cells. This GSH-responsive fluorescent probe exhibiting Golgi-targeting and therapeutic effects provides an approach for monitoring changes of GSH concentration in the Golgi apparatus and for precise disease treatments (Fig. 28).96
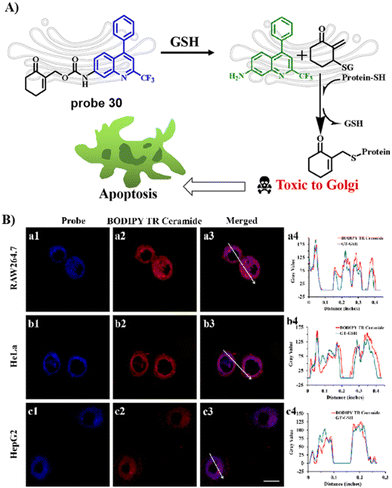 |
| | Fig. 28 (A) Response and cancer therapeutic effect of probe 30 towards GSH. (B) Fluorescence imaging of HeLa cells. Reproduced with permission from ref. 96. Copyright 2021, American Chemical Society. | |
In 2021, the group of Feng developed a Golgi-targeting fluorescent probe 31 using a aminoquinoline as both the Golgi-targeting group and fluorophore for the selective and sensitive imaging of CO based on the Tsuji–Trost reaction. The probe exhibited excellent Golgi-targeting ability and was used to ratiometricaly image exogenous and endogenous CO in zebrafish and cells, Moreover, probe 31 could be used for visually monitoring increases in CO of living cells under oxidative stress (Fig. 29).97
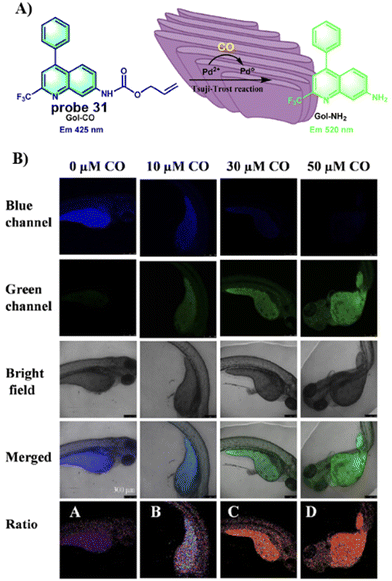 |
| | Fig. 29 (A) Response of probe 31 for CO. (B) CO fluorescence imaging in zebrafish with 31. Reproduced with permission from ref. 97. Copyright 2021, Elsevier. | |
By integrating an isothiocyanate (cysteine recognition site) with an aminoquinoline (fluorophore and Glogi-targeting group), the Zhu group developed a specific and sensitive fluorescent cysteine (Cys) probe 32 in 2022, which exhibited low cytotoxicity for the imaging of exogenous and endogenous cysteine in zebrafish and cells. In addition, it was used to confirm that cystathionine γ-lyase enzyme plays a critical role in the generation of Cysteine (Fig. 30).98
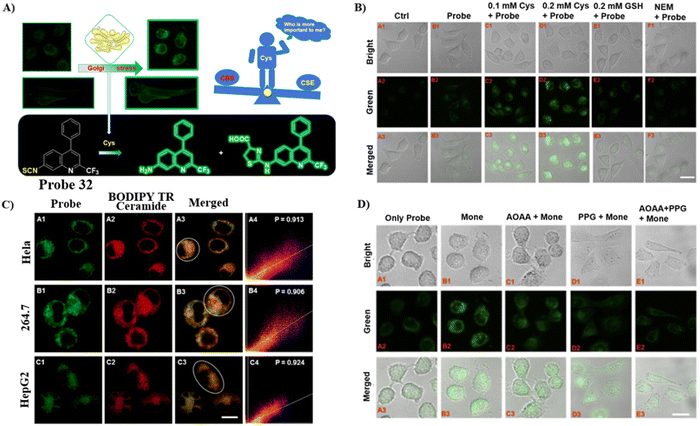 |
| | Fig. 30 (A) Response of probe 32 to Cys. (B) Cys imaging in cells using 32. (C) Co-localization cell imaging. (D) Fluorescence imaging for the inhibition of endogenous Cys with 32. Cells treated with probe 32 only (A). Cells treated with monensin and probe 32 in turn (B). Cells treated by AOAA, monensin and probe 32 in turn (C). Cells treated by PPG, monensin and probe 32 in turn (D). Cells treated by AOAA, PPG then incubated by monensin, probe 32. Reproduced with permission from ref. 98. Copyright 2022, Elsevier. | |
By combining the Golgi-targeting aminoquinoline fluorophore with α-keto acid ONOO− reaction site, the Jiang group developed a Golgi-targeting fluorescent probe 33 for monitoring ONOO− production during Golgi stress in a sepsis-induced acute lung injury (ALI) model.99 The probe exhibited good selectivity and high sensitivity towards ONOO− and as such was used for monitoring increases of ONOO− in the Golgi apparatus of live cells generated by different stimuli. Moreover, significant increases in ONOO− levels in the Golgi apparatus were observed in mice during sepsis-induced ALI (Fig. 31).
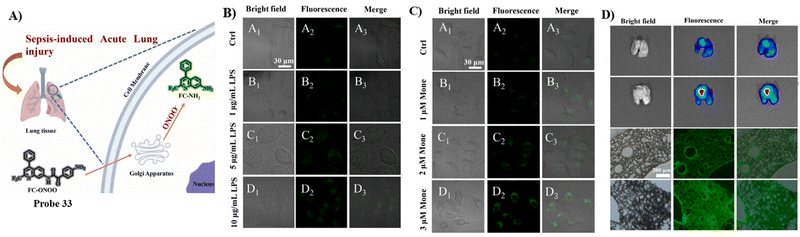 |
| | Fig. 31 (A) Probe 33 response for ONOO−. (B) Imaging of exogenous and endogenous ONOO− in A549 cells. (C) Imaging of ONOO− under Golgi stress in A549 cells. (D) Fluorescence images of 33 stained lung organs and slices of control group mice and LPS group mice. Reproduced with permission from ref. 99. Copyright 2022, Elsevier. | |
The Jiang group developed another Golgi-targeting fluorescent probe 34 by anchoring a NTR-sensitive p-nitrobenzyl moiety to a Golgi-targeting aminoquinoline fluorophore, for the accurate monitoring and assessment of sepsis-induced ALI. The probe exhibited good selectivity and high sensitivity with a LOD of 54.8 ng mL−1.100 As such, the probe could be used for tracking the enhancement NTR in the Golgi of zebrafish and cells under the action various stimuli. Golgi NTR production in sepsis-induced ALI and the prolyl hydroxylase (PHD) inhibitor (dimethyloxalylglycine, DMOG) for combating sepsis-induced ALI could be monitored using Gol-NTR (Fig. 32).
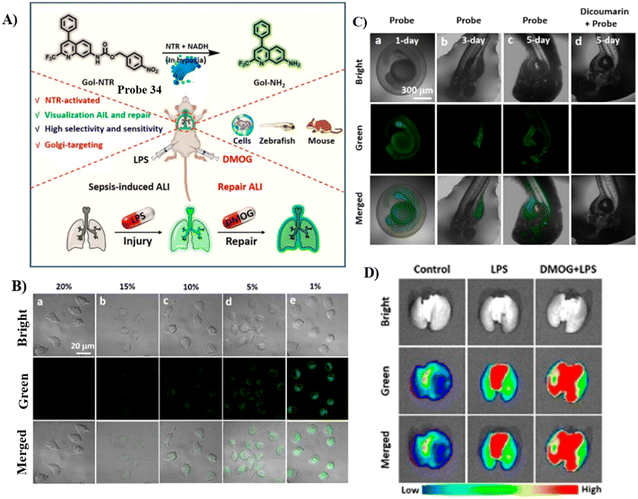 |
| | Fig. 32 (A) Probe 34 response for NTR. (B) Fluorescence images of NTR under normoxic (20% O2) and various hypoxic (15%, 10%, 5%, and 1% O2) conditions for 8 h, respectively. (C) Fluorescence images of NTR in living zebrafish cultured for different days (1–5 days). (D) Fluorescence imaging of lungs from the control group, LPS group, and DMOG + LPS group. Reproduced with permission from ref. 100. Copyright 2022, American Chemical Society. | |
Recently, the Wu group developed a novel Golgi-targeting and H2O2-specific ratiometric fluorescent probe 35 where a borate ester was chosen as the H2O2 recognition site, and an aminoquinoline derivative was selected as the fluorophore as well as the targeting group for the Golgi apparatus.101 the probe could be employed to monitor endogenous/exogenous H2O2 in zebrafish and cells using one-photon or two photon confocal imaging, and exhibited high selectivity, low toxicity and exceptional photostability. As such probe 35 represents an useful tool for monitoring H2O2 levels in biological environments (Fig. 33).
 |
| | Fig. 33 (A) Probe 35 response mechanism. (B) Fluorescence images of probe 35 in HeLa cells. (C) Two-photon fluorescence images of probe 35 in HeLa cells. (D) Two-photon fluorescence images of probe 35 in zebrafish. Reproduced with permission from ref. 101. Copyright 2023, Elsevier. | |
3.4 Probes with L-Cys group
Cys is the main biothiol found in living organisms, involved in many physiological and pathological processes, and plays an important role in maintaining the redox stability of biological systems. Some physiological and pathological processes (such as aging, neurological decline) are associated with changes in oxidation/antioxidant balance and Cys content. It has been reported that both galactosyltransferase and protein kinase D are able to anchor to the Golgi apparatus using Cys residues or Cys-rich domains,102,103 since the Cys thiol group is able to bind with a thiol groups of the Golgi through the generation of disulphide bonds in the oxidizing environment of the Golgi lumen,22,104 thus inspiring researchers to use a cysteine targeting strategy to develop optical imaging probes for the early disease diagnosis and Golgi-related disease therapeutics. Scheme 5 and Table 4 highlight Golgi-targeting probes using Cys as the functional motif and illustrates their applications.
 |
| | Scheme 5 Golgi-targeting probes with L-Cys as functional motif. Red indicates the Golgi-targeting unit. | |
Table 4 Golgi-targeting fluorescent probes with L-Cys as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 36 |
O2˙− |
OP 370 nm/495 nm |
— |
— |
0.93 |
Imaging in live cells and mice |
105 |
| TP 800 nm/495 nm |
| 37 |
Polarity |
700 nm/800–825 nm |
0–5 μM |
18 nM |
0.93 |
Imaging in live cells |
106 |
| 38 |
Zn2+ |
346 nm/414 nm |
|
|
0.92 |
Imaging in live cells |
107 |
Using L-Cys as the Golgi-targeting group, and a caffeic acid group as the O2˙− response site, the Tang group in 2019 developed a fluorescent Golgi-targeting probe 36 for the dynamic and reversible detection of O2˙−. Enhanced levels of O2˙− in addition to an increase of caspase-2 activity and apoptosis were identified in the Golgi apparatus of an ischaemia-reperfcfusion (IR) mouse liver, indicating that the tumor necrosis factor (TNF-α) acts as a positive mediator for O2˙− generation. These results illustrate how the Golgi could potentially mediate O2˙− variations in IR mice and illuminated the related molecular mechanisms (Fig. 34).105
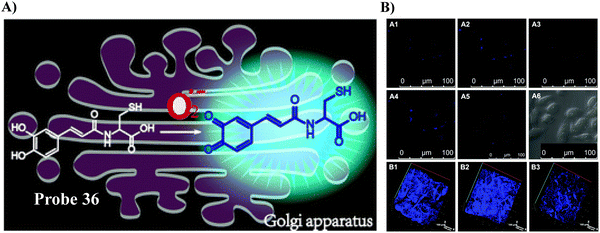 |
| | Fig. 34 (A) Response of probe 36 towards O2˙−. (B) Validation of 36 for the imaging of living cells and mice. Reproduced with permission from ref. 105. Copyright 2019, The Royal Society of Chemistry. | |
In 2019, the Tang group developed a NIR fluorescent Golgi-targeting probe 37 for visualizing polarity, in which 37 can aggregate in the Golgi apparatus with the help of cysteine, and the benzoyl difluoroboronate and merocyanine units are able to detect changes of polarity. Probe 37 exhibited not only excellent sensitivity and selectivity towards changes in polarity, but also good biocompatibility. Probe 37 was used for monitoring polarity differences between normal and cancerous cell lines and tracking changes of polarity on glutamate-stimulation of PC12 cells, in order to discover the inter-relationship between the polarity of the Golgi and the depression phenotype. Furthermore, the results indicated that an elevation of polarity may be caused by the reduced synthesis of brain-derived neurotrophic factor (BDNF) (Fig. 35).106
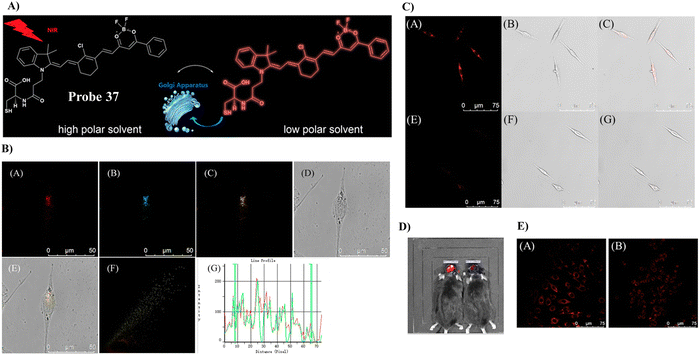 |
| | Fig. 35 (A) Response of probe 37 towards polarity. (B) Co-localization cell imaging of 37 and Golgi Tracker Red. (C) Imaging of polarity fluctuation in PC12 cells induced by excess glutamate. (D) Fluorescence image of the brains of mice with depression-like behavior. (E) Fluorescence images of PC12 cells using small interfering RNA (siRNA) to silence BDNF genes. Reproduced with permission from ref. 106. Copyright 2019, American Chemical Society. | |
Using a trityl protected cysteine as the Golgi-targeting group, which can be cleaved to release a latent Golgi-targeting L-Cys group, the Watkinson group developed a Golgi-targeting probe 38 to image mobile Zn2+, which contains an S-trityl protected cysteine moiety introduced using ‘click’ chemistry, for the increased hydrophobicity and membrane permeability of probe 38. The S-trityl protecting group was removed within 24 hours to release free Cysteine, and the probe was then anchored to the Golgi apparatus. The probe exhibited good Golgi-targeting ability, selectivity, and low cellular toxicity, good photophysical properties, and was used to monitor changes of mobile Zn2+ levels in the the Golgi apparatus during H2O2 induced oxidative stress (Fig. 36).107
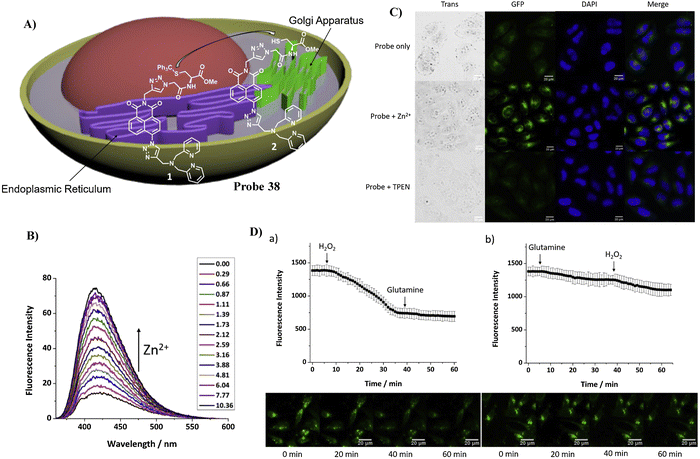 |
| | Fig. 36 (A) Response of probe 38 for Zn2+. (B) Fluorescence spectra of 38 at varying concentrations of Zn2+. (C) Fluorescence microscopic images of HeLa cells treated with probe 38, probe 38 with Zn2+, and probe 38 with TPEN. (D) Zn2+ level fluctuations during oxidative stress. Reproduced with permission from ref. 107. Copyright 2021, Elsevier. | |
3.5 Probes with targeting peptides
Peptides are privileged biomolecules in nature and play essential roles in living systems. Because of their unique molecular structure, peptides have been widely used in drug discovery, disease diagnosis, fluorescence detection and materials science. Peptides with organelle-specific import or retention sequences, are able to enable probes to target the subcellular apparatus such as nucleus, the trans-Golgi network (TGN) or endoplasmic reticulum (ER), when anchored to the fluorophore of the probes. In addition, furin is a Golgi-resident protein that is overexpressed in cancer cells and is a good target for lighting up the Golgi of cancer cells. Furin can cleave the C-terminal of the amino acid motif Arg-X-Lys/Arg-Arg (X any amino acid), and therefore, fluorescence imaging of the Golgi apparatus can be performed using a furin cleavage strategy.108–113 Both Scheme 6 and Table 5 highlight Golgi-targeting probes using peptides as functional motifs and their applications.
 |
| | Scheme 6 Golgi-targeting probes with peptides as functional unit. Red indicates Golgi-targeting unit. | |
Table 5 Golgi-targeting fluorescent probes with peptide as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 39 |
Cu+ |
400 nm/510 nm |
0–60 nM |
8 nM |
0.9 |
Imaging in live cells |
114 |
| 40 |
Furin |
365 nm/500 nm |
— |
— |
0.91 |
Imaging in live cells and tumor tissue slices of mice |
115 |
| 41 |
Furin |
680 nm/708 nm |
— |
— |
0.930 |
Imaging in live cells and tumor bearing mice |
116 |
| 42 |
Furin |
470 nm/549 nm |
2.0–10.0 ng μL−1 |
0.11 ng μL−1 |
— |
Imaging in live cells |
117 |
| 29 |
Furin |
350, 470 nm/420, 640 nm |
0–500 U mL−1 |
0.265 U mL−1 |
|
Imaging in live cells |
95 |
| 0.388 U mL−1 |
| 43 |
— |
— |
— |
— |
— |
Localization cell imaging |
118 |
| 44, 45 |
— |
— |
— |
— |
— |
Localization cell imaging |
119 |
In 2016, the Lee group developed a Golgi-targeting probe 39 to detect Cu+ by conjugating the N-terminal of a tripeptide (Leu-Leu-Cys) with a dansyl fluorophore, probe 39 formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with Cu+ resulting in fluorescence “turn-on” response for Cu+. Evaluation of the binding mode indicated the critical role played by the thiol group of the peptide receptor in the strong binding with Cu+, with additional support from the sulfonamide and amide groups. Meanwhile, probe 39 was able to penetrate living RKO cells and detect Cu+ in the Golgi apparatus (Fig. 37).114
1 complex with Cu+ resulting in fluorescence “turn-on” response for Cu+. Evaluation of the binding mode indicated the critical role played by the thiol group of the peptide receptor in the strong binding with Cu+, with additional support from the sulfonamide and amide groups. Meanwhile, probe 39 was able to penetrate living RKO cells and detect Cu+ in the Golgi apparatus (Fig. 37).114
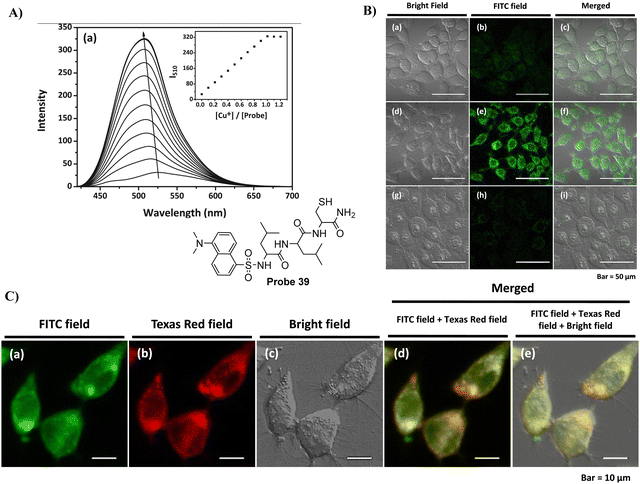 |
| | Fig. 37 (A) Fluorescence spectra of 39 at different concentrations of Cu+. (B) Cu+ response of probe 39 in cells. (C) Co-localization cell imaging of 39 and BODIPY TR ceramide. Reproduced with permission from ref. 114. Copyright 2016, Elsevier. | |
In 2018, the Zhang group developed a Golgi-targeting probe 40 for the long-term in situ detection of endogenous furin activity. A furin-specific peptide (RVRR) was integrated with an insoluble solid-state fluorophore 6-chloro-2-(2-hydroxyphenyl)quinazolin-4(3H)-one through a self-immolative linker. Probe 40 exhibited high sensitivity and selectivity towards furin and released the free fluorophore, which precipitated in proximity to the enzyme active site and emitted bright solid-state fluorescence enabling in situ imaging. Probe 40 was able to realize the long-term bioimaging of intracellular furin, and was used to monitor the changes of furin under the stabilization of hypoxia-inducible factor (HIF) regulated by cobalt chloride (CoCl2) in addition to facilitating the imaging of furin in MDA-MB-468 cells and tumor tissues (Fig. 38).115
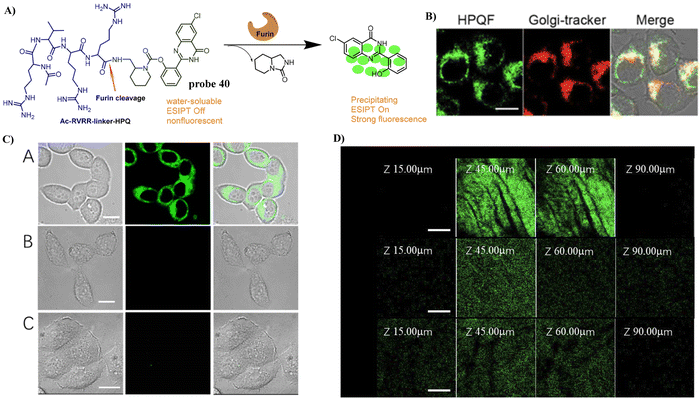 |
| | Fig. 38 (A) Response mechanism of 40 towards furin. (B) Co-localization cell fluorescence imaging of 40 and Golgi-tracker Red. (C) Fluoresence imaging of endogenous furin enzyme in Live Cells. (D) Fluorescence imaging of endogenous furin enzyme in cancer tissues. Reproduced with permission from ref. 115. Copyright 2018, American Chemical Society. | |
In 2019, the Zhang group reported on a Golgi-targeting furin-activatable NIR fluorescent probe 41 by substituting the fluorophore 6-chloro-2-(2-hydroxyphenyl) quinazolin-4(3H)-one linked through a self-eliminating linker to a furin specific peptide substrate as Golgi-targeting group, for the specific, sensitive, real-time imaging of furin in several tumor tissues, cells, and tumor-bearing mouse models. Moreover, probe 41 was used to unveil the relationship between furin content and CoCl2 stabilization of HIF-1 (Fig. 39).116
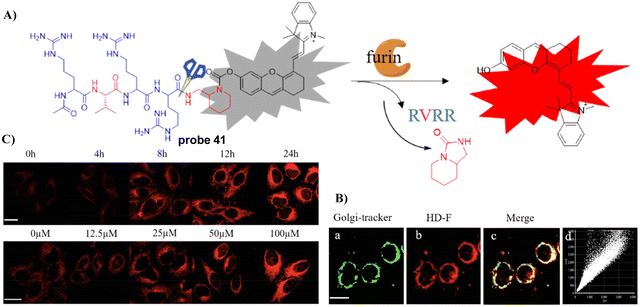 |
| | Fig. 39 (A) Response mechanism of 41 towards furin. (B) Co-localization cell imaging of 41 and Golgi-tracker Red. (C) Visualization of the furin fluctuations induced by HIF-1. Reproduced with permission from ref. 116. Copyright 2019, American Chemical Society. | |
In 2019, the Yi group developed a biocompatible amphiphilic peptide probe 42 with a Golgi-targeting RVRRFFF sequence and a nitrobenzoxadiazole (NBD) fluorophore that self-assemble into micelles for the detection of furin, where the as-formed β-sheet nanostructured probe 42 could respond to furin rapidly due to the RVRR moiety, and was able to distinguish high furin-expressing cancerous cells from normal cells. (Fig. 40).117
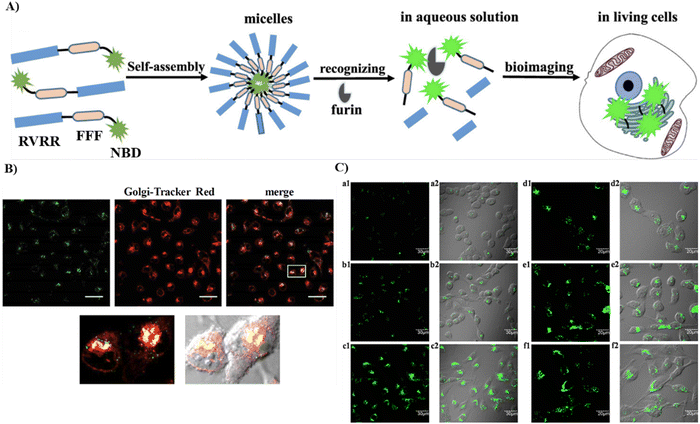 |
| | Fig. 40 (A) Schematic of the self-assembly process and furin enzyme response of 42. (B) Co-localization cell imaging of 42 and Golgi Tracker Red. (C) Fluorescence and long-term imaging by 42 of furin in living cells. Reproduced with permission from ref. 117. Copyright 2019, American Chemical Society. | |
By changing an oxygen for a sulphur atom in the phosphoester bond of phosphopeptides, the as-generated thiophosphopeptide undergoes rapid alkaline phosphatase (ALP)-catalyzed dephosphorylation to form a self-assembled thiopeptide. This assembly can target the GA and selectively kill cancer cells. Using fast enzymatic kinetics and redox reactions in the oxidative environment of the Golgi, the Xu group have developed a Golgi-targeting fluorescent probe 43, consisting of three units: (i) a 4-nitro-2,1,3-benzoxadiazole (NBD) group, which in a hydrophobic environment emits green fluorescence; (ii) a hydrophobic D-diphenylalanine (ff) that facilitates self-assembly and withstands proteolysis; (iii) a thiophosphate as enzymatic self-assembly trigger group. The probe enters the cells through caveolin-mediated endocytosis and macropinocytosis, and accumulates in the GA assisted by dephosphorylation and generation of disulfide bonds to selectively kill cancer cells (Fig. 41).118
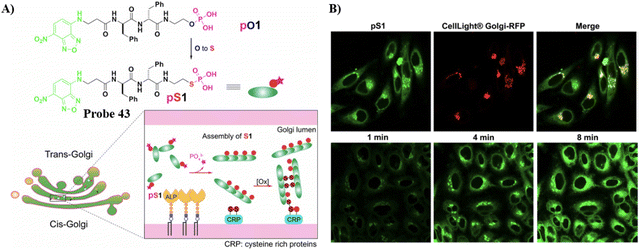 |
| | Fig. 41 (A) Thiophosphopeptide 43 targeting the Golgi apparatus. (B) CLSM images of HeLa cells (top) stained with Golgi-tracker Golgi-RFP after treating with 43 for 8 minutes and (bottom) treated with 43 for 1, 4, and 8 min. Reproduced with permission from ref. 118. Copyright 2021, Wiley. | |
Replacing the thiophosphate group with different sulfur-containing groups such as peptide thioesters, the Xu group have developed Golgi-targeting fluorescent probes 44 and 45. The probes contain nitrobenzoxadiazole (NBD) as the fluorophore and an enzyme-responsive aminoethyl thioester locking group attached to the self-assembling D-diphenylalanine (ff), for targeting the GA in a wide range of cells. Peptide thioesters enter cells via caveolin-mediated endocytosis or macropinocytosis, the thiopeptides generated then aggregate in the GA after being hydrolyzed by GA-associated thioesterases, resulting in cell death (Fig. 42).119
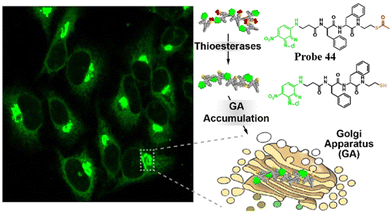 |
| | Fig. 42 Thioesterase-responsive fluorescent probe 44 in Golgi apparatus. Reproduced with permission from ref. 119. Copyright 2022, American Chemical Society. | |
3.6 Probes containing lipids
Ceramide is a precursor of complex sphingolipids which are synthesized in the ER and transported to the GA through the ceramide transporter protein (CERT) in mammalian cells for further processing into complex sphingolipids.120 As such, small molecular fluorophores containing lipid-based tagging groups such as NBD-ceramide and Bodipy-ceramide have been used for Golgi apparatus labeling.19,121 Scheme 7 and Table 6 highlight current Golgi-targeting probes using lipids as a functional motif and their applications.
 |
| | Scheme 7 Golgi-targeting probes with lipids as the functional motif. Red indicates the Golgi-targeting unit. | |
Table 6 Golgi-targeting fluorescent probes with lipids as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 46 |
Fe2+ |
630 nm/665 nm |
0–10 μM |
50 nM |
0.64 |
Imaging in live cells |
122 |
| 47 |
pH |
570 nm/600 nm |
pH 3.80–5.40 |
pKa 4.32 |
0.9 |
Imaging in live cells and mice |
123 |
| 48 |
Zn2+ |
440 nm/507 nm |
— |
— |
0.85 |
Imaging in live cells |
124 |
| 49 |
O2˙− |
405 nm/450 nm |
0–100 μM |
0.39 μM |
0.99 |
Imaging in live cells and zebrafish |
125 |
| 50 |
Fe2+ |
370 nm/460 nm |
0–140 μM |
52 nM |
0.99 |
Imaging in live cells |
126 |
In 2019, the Hirayama group developed fluorescent probe 46 for the detection of Fe(II) ions in the Golgi by combining their N-oxide-based Fe(II)-specific chemical switch, with a Golgi targeting unit, and polarity-sensitive fluorogenic unit. Probe 46 confirmed that the intracellular Fe(II) ion distribution is greater in the Golgi apparatus than lysosomes, using a synchronous imaging study with a previously reported lysosome-specific fluorescent probe LysoRhoNox, whereas the elevation of Fe(II) in lysosomes was observed in cells with the induced dysfunction of vacuolar protein sorting-associated protein 35 (VPS35). It is found that the treatment of dysfunctional VPS35 by a molecular chaperone (R55) restored the Golgi-dominant distribution of Fe(II) ions, indicating that the impairment of the divalent metal transporter 1 (DMT1) traffic machinery influences subcellular iron homeostasis and promotes Fe(II) escape at the Golgi and accumulation of Fe(II) at the lysosome (Fig. 43).122
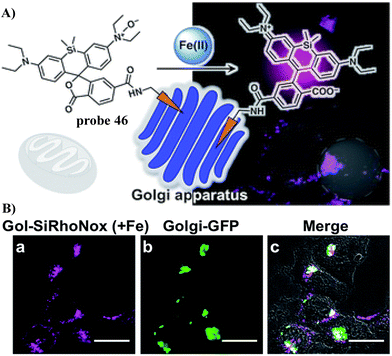 |
| | Fig. 43 (A) The response mechanism of 46 towards Fe2+. (B) Co-localization experiment of HepG2 cells with Golgi-specific GFP and 46. Reproduced with permission from ref. 122. Copyright 2019, The Royal Society of Chemistry. | |
In 2019, the Dong group developed a Golgi-targeting fluorescent pH-responsive probe 47 based on the rhodamine B skeleton with sphingosine as the Golgi-targeting group, generating an off–on fluorescence response to the reduced pH in the Golgi resulting in spirocyclic ring opening with high specificity and sensitivity. The probe was used to monitor pH changes of the Golgi caused by LPS-mediated inflammation and drug treatment in vivo (Fig. 44).123
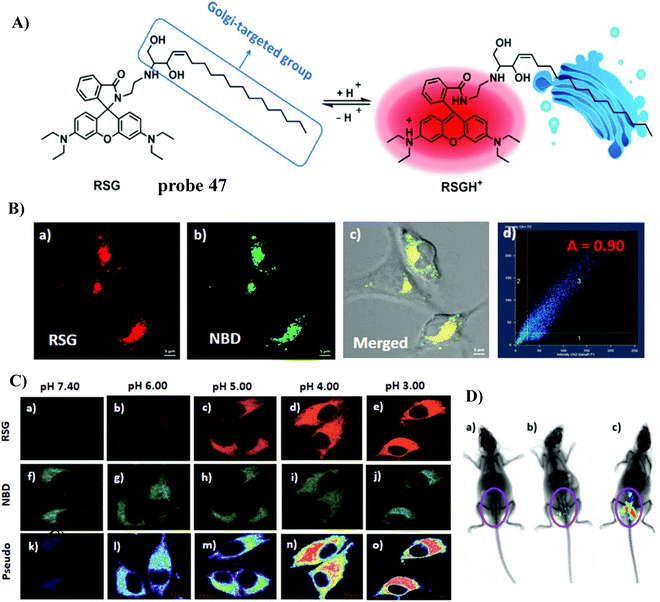 |
| | Fig. 44 (A) Response mechanism of 47 towards pH. (B) Co-staining experiment of SMMC-7721 cells with NBD and 47. (C) pH fluorescence imaging using 47 in living cells. (D) In vivo fluorescence images of 47 in mice stimulated using LPS. Reproduced with permission from ref. 123. Copyright 2019, The Royal Society of Chemistry. | |
In 2020, the Mizukami group developed a Golgi-targeting Zn2+-selective probe 48 that exhibits high Zn2+ selectivity and low sensitivity to pH. The HaloTag labeling technology ensured that the probe targeted organelles such as the mitochondria, nucleus, Golgi apparatus and endoplasmic reticulum. In response to labile Zn2+ the probe exhibited reversible fluorescence response and as such was used for monitoring the concentration of Zn2+ in the Golgi apparatus (Fig. 45).124 Subsequently, a protocol for the use of probe 48 in live cells for quantifying labile Zn2+ in the Golgi apparatus was reported.127
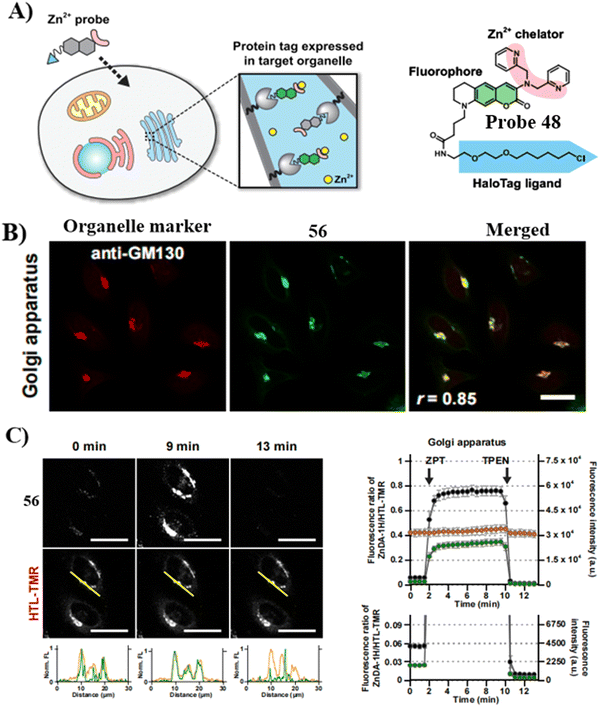 |
| | Fig. 45 (A) Illustration of organelle-targeting by small-molecule fluorescent probes using a HaloTag. (B) Fluorescent images of HeLa cells using 48 and anti-GM130 antibody. (C) Detection of intracellular [Zn2+] changes in live HeLa cells in the Golgi apparatus. Reproduced with permission from ref. 124. Copyright 2020, Elsevier. | |
Probes containing a tetradecyl chain exhibit Golgi targeting ability in cell imaging. Recently, the Chen group developed two Golgi-targeting fluorescent probes 49 and 50 using myristoyl as the Golgi targeting group and coumarin as the fluorophore. Probe 49 was able to detect endogenous and exogenous O2˙− in zebrafish and cells. The probe was then used to monitor the O2˙− levels up-regulated during ischemia-reperfusion (I/R) processes in cardiomyocytes. As such, probe 49 provides a diagnostic tool for myocardial oxidative stress injury and could be used to provide insight for role of GOLPH3 (a potential Golgi stress marker) in myocardial I/R injury (Fig. 46).125 While, probe 50 is a turn-on Golgi-targeting fluorescent probe for the sensitive and selective detection of Fe2+. Probe 50 could be used to detect exogenous and endogenous Fe2+ in HUVEC and HepG2 cells (Fig. 47).126
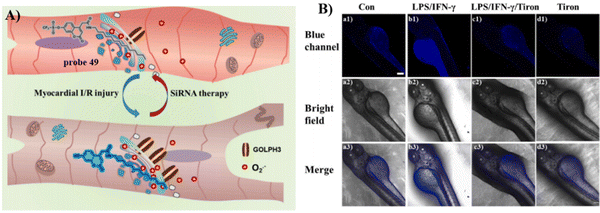 |
| | Fig. 46 (A) Probe 49 for detecting O2˙− levels in myocardial ischemia-reperfusion injury and SiRNA therapy and elucidating the function of GOLPH3. (B) Fluorescence imaging (blue scale) of probe 49 responding to O2˙− in zebrafish. Reproduced with permission from ref. 125. Copyright 2023, Elsevier. | |
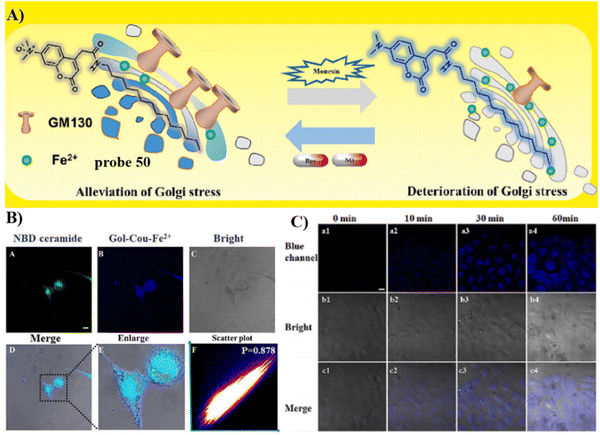 |
| | Fig. 47 (A) Response mechanism of probe 50. (B) Confocal fluorescence co-stained experiment in HUVEC co-incubated with 50 (10 μM) pre-treated with Fe2+ (50 μM) and NBD-ceramide (5 μM). (C) Confocal fluorescence image of HUVEC cell at various times under hypoxic conditions. Reproduced with permission from ref. 126. Copyright 2023, Elsevier. | |
3.7 Probes encapsulated by BSA
Bovine serum albumin (BSA) is a complex serum globular protein with a single chain of 582 amino acids cross-linked with 17 cystine residues, that plays an important role in the transportation of exogenous and endogenous ligands (such as drugs) to specific targets.128 Probes encapsulated with BSA can form large nanoparticulate structures through hydrophobic interactions and the system can preferentially aggregate in the Golgi apparatus.35,77,129 In our opinion, the Golgi-targeting ability of BSA can be attributed to the abundant cysteine residues in BSA, which can form disulphide bonds through binding with thiol groups of the Golgi. Scheme 8 and Table 7 highlight Golgi-targeting probes using BSA as a functional motif and their applications.
 |
| | Scheme 8 Golgi-targeting probes using BSA as a functional motif. | |
Table 7 Golgi-targeting fluorescent probes with BSA as functional motif
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 51 |
pH |
540 nm/808 nm |
pH 5.39–7.97 |
pKa 6.92 |
— |
Killing of cancer cells, photothermal therapy in mice |
130 |
| 52 |
HNO |
480 nm/520 nm |
0–30 μM |
130 nM |
0.84 |
Imaging in live cells and DILI mice |
131 |
| 23 |
O2˙−, H2O2 |
633 nm/740 nm |
0–35 μM |
62 nM |
0.827, 0.839 |
Imaging in live cells and mice |
77 |
In 2017, the Yi group developed a pH-responsive photothermal ablation agent (pH-PTT) for the Golgi apparatus using asymmetric cyanine dyes for photothermal therapy (PTT), which was highly pH sensitive from 6.2–7.5. pH-PTT efficiently generates thermal energy due to 808 nm light irradiation and formation of extended conjugated structure at low pH conditions. Nanoparticles (51) assembled using BSA with pH-PTT preferentially accumulate in the Golgi apparatus of cancerous cells. Thus, the system could be exclusively activated by the acidic microenvironment of the Golgi apparatus of cancer cells resulting in efficient PTT (Fig. 48).130
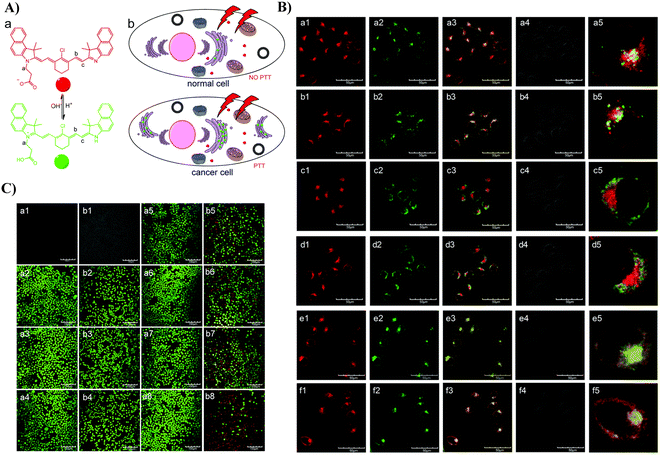 |
| | Fig. 48 (A) Illustration of pH-PTT as an intelligent drug for specific PTT activation in the Golgi apparatus. (B) CLSM images of HepG2 (a), (c) and (e) and HL-7702 (b), (d) and (f) cells co-stained with 51/Lyso tracker (a) and (b), Mito tracker (c) and (d) and 51/Golgi green. (C) CLSM images of Calcein AM/PI stained (a) HL-7702 and (b) HepG2 cells with or without 51 treated after exposure to an 808 nm laser (1.45 W cm−2) for different times. Reproduced with permission from ref. 130. Copyright 2017, The Royal Society of Chemistry. | |
In 2021, the Tang group developed a fluorescent probe Golgi-HNO (52), using 2-(diphenylphosphino)-benzoate as the recognition group and fluorescein as the fluorophore, for the imaging and detection of nitroxyl (HNO) in the Golgi apparatus. Probe 52 was encapsulated using Golgi targeting BSA. Biological results indicated that an increase in HNO levels was closely associated with drug-induced liver injury, and catalase was associated HNO synthesis, indicating that HNO is suitable biomarker for DILI diagnosis, and is suitable for providing guidance on the design and development of therapeutic drugs. (Fig. 49).131
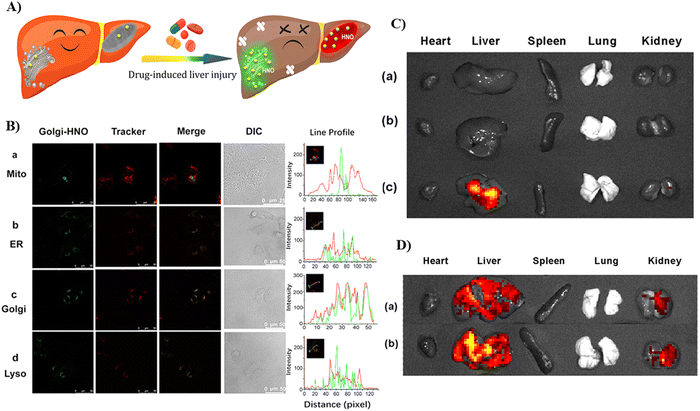 |
| | Fig. 49 (A) Green-colored fluorescence imaging of Golgi-HNO 52 in Mice with DILI. (B) Co-localization imaging of 52 with different commercial localization dyes. (C) Fluorescence imaging of APAP-induced mice organs, where the mice were injected with probe 52. Control group mice (a); 75 mg mL−1 low-dose (LD) APAP-induced mice (b); 150 mg mL−1 high-dose (HD) APAP-induced mice (c). (D) Fluorescence imaging of organs from bleomycin-induced mice, where the mice were injected with probe 52. Control group mice (a) and bleomycin-induced mice (b). Reproduced with permission from ref. 131. Copyright 2021, American Chemical Society. | |
3.8 Probes with other targeting groups
In addition to the Golgi imaging strategies described above, there are several other types of fluorescent probes used for Golgi imaging. Unlike the usual Golgi-targeting groups mentioned above, the precise mode of targeting is unknown. Therefore, such fluorescent probes do not fully elucidate what part of the probe is Golgi-targeting or the localization mechanism. As such we provide appropriate suggestions on how the probes specifically target the Golgi apparatus following our current understanding of the GA, in order to better understand how to design Golgi-targeting fluorescent probes. (Scheme 9 and Table 8).
 |
| | Scheme 9 Golgi-targeting probes using other groups. | |
Table 8 Golgi-targeting probes with other groups
| Probe |
Analyte |
λex/λem (nm) |
Detection range |
LOD |
Pearson's correlation coefficient |
Application |
Ref. |
| 53 |
— |
375 nm/450 nm |
— |
— |
0.86 |
Imaging in live cells |
132 |
| 54 |
Zn2+ |
507 nm/— nm |
— |
— |
— |
Imaging in live cells |
133 |
| 55 |
Zn2+ |
469 nm/550 nm |
— |
— |
— |
Imaging in live cells and zebrafish |
134 |
| 56 |
Zn2+ |
492 nm/515 nm |
— |
— |
— |
Imaging in live cells |
135 |
| 57 |
Zn2+ |
364 nm/497 nm |
— |
4.2 × 10−14 M |
— |
Localization cell imaging |
136 |
| 58 |
Fe2+ |
540 nm/575 nm |
— |
0.2 μM |
— |
Localization cell imaging |
137 |
| 59 |
Zn2+ |
388 nm/518 nm |
— |
— |
0.92 |
3D distributions in live cells and tissues |
138 |
| 60 |
HOCl |
550 nm/586 nm |
— |
— |
0.98 |
SIM and 3D-SIM imaging in living cells |
139 |
| 61 |
Mn2+ |
— |
— |
— |
— |
Confocal microscopy and nanosynchrotron X-ray fluorescence imaging |
140 |
In 2021, using adenosine (Ade) as a Golgi-targeting group and tetraphenylethylene (TPE) with AIE characteristics as fluorophore, the Zhao group developed fluorescent probe 53, that exhibited excellent Golgi apparatus co-localization efficacy and photostability (Fig. 50).132
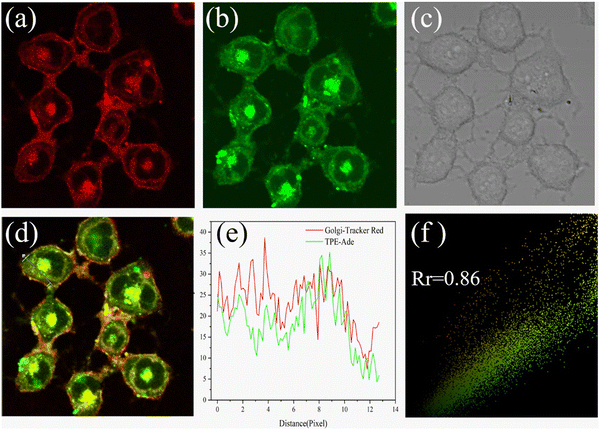 |
| | Fig. 50 Co-localization fluorescence images of 53 with commercial Golgi-tracker Red in HL-7402 cells. Reproduced with permission from ref. 132. Copyright 2021, Elsevier. | |
By using a pyridyl moiety to match with the weakly acidic microenvironment in the Golgi, some reported fluorescent probes have been developed over the past several decades to target the Golgi. In 2000, the Tsien group developed a fluorescent Zn2+ probe (54) based on fluorescein derivatives with a Zn2+ chelating group bis(2-pyridylmethyl)amine (DPA), to preferentially stain Golgi-associated vesicles and the Golgi.133 In 2009, the Guo group created a Zn2+ visible light excitable fluorescent probe (55) using a standard ICT fluorophore, 4-amino-7-nitro-2,1,3-benzoxadiazole (ANBD), which was integrated with a N,N,N′-tri(pyridin-2-ylmethyl)ethane-1,2-diamine (TPEA) group and a bis(pyridin-2-ylmethyl)amine (BPA) group.134 Probe 55 exhibits a large Stokes shift, visible ICT absorption band, and good biocompatibility. Preferential accumulation in the lysosomes and Golgi was observed, enabling Zn2+ imaging of zebrafish and cells (Fig. 51).
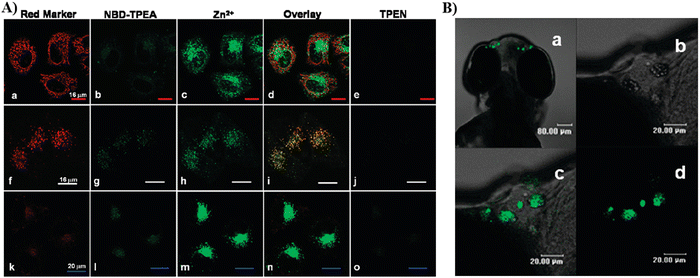 |
| | Fig. 51 (A) Co-localization imaging of 55 and MitoTracker Red CMXRos (top row), LysoTracker Red DND-99 (middle row), BODIPY TR ceramide (bottom row) in HeLa cells. (B) Confocal fluorescence images of the head of a 4-day-old zebrafish larva fed Zn2+ solution (5 μM) at 28.5 °C for 12 h. Reproduced with permission from ref. 134. Copyright 2009, American Chemical Society. | |
In 2010, the Lippard group developed a fluorescent cell-permeable probe 56 based on similar fluorescein derivatives with dramatic fluorescence enhancements upon addition of Zn2+. Probe 56 localizes in the Golgi apparatus of living cells, due to the generation of a negative charge after intracellular hydrolysis of the ester groups (Fig. 52).135
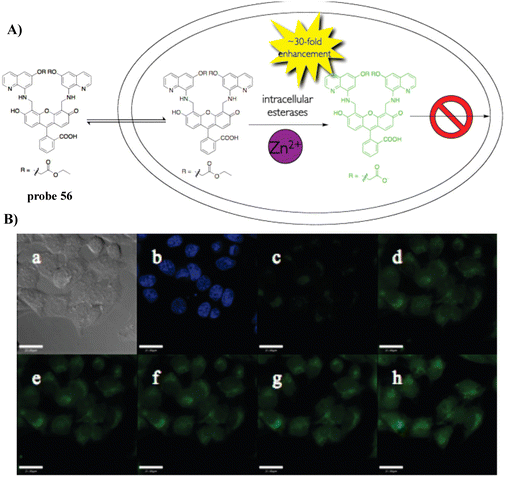 |
| | Fig. 52 (A) The structure and possible reaction mechanism of 56 for identifying Zn2+. (B) Cell imaging results confirming the retention of 56 in HeLa cells coincubated with 56 and Hoechst 33258 for 18 h. Reproduced with permission from ref. 135. Copyright 2010, American Chemical Society. | |
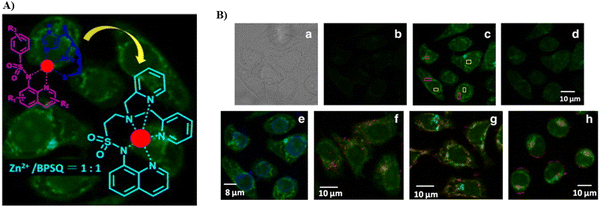
Also utilizing the pyridyl group to closely match the Golgi environment, in 2011, taking advantage of the synergistic coordination of Zn2+ by bis(pyridin-2-ylmethyl)amine (BPA) and 8-sulfonamidoquinoline (SQ) motifs, and by integrating BPA with SQ, the Guo group developed fluorescent probe (57), which exhibits Golgi-targeting and physiological pH-independent Zn2+ response, that was used for live cell imaging (Fig. 53).136
 |
| | Fig. 53 (A) Structure and possible mechanism of 57 for monitoring Zn2+. (B) Confocal microscopic images of HeLa cells stained by 57 with Zn2+ (a)–(d). Co-stained images of cells incubated with Zn2+ followed by colocalization with 57 and organelle dye. (e) Hochest 33342, (f) LysoTracker Red DND-99, (g) MitoTracker Red CMXRos, (h) Golgi marker BODIPY TR ceramide. Reproduced with permission from ref. 136. Copyright 2011, Elsevier. | |
In 2013, the Nagasawa group reported on a fluorescent reaction-based turn-on probe (RhoNox-1, 58) using a rhodamine as the fluorophore and N-oxide for the selective detection of Fe2+. The initial fluorescence was quenched due to non-radiative deactivation by twisted internal charge transfer (TICT) of the excited state of the tertiary amine N-oxide substituted rhodamine and photo-induced electron transfer (PeT). Probe 58 exhibited significant fluorescence enhancements in Fe2+-loaded cells due to N-oxide deoxygenation mediated by Fe2+, and was successfully used for monitoring basal and endogenous labile Fe2+ in live cells (Fig. 54).137
 |
| | Fig. 54 (A) Probe 58 reaction mechanism for Fe2+. (B) Confocal fluorescence microscopic images of Fe2+ detection in HepG2 cells using 58. (C) Co-stained fluorescence images of 58 treated HepG2 cells with BODIPY-FL-ceramide. Reproduced with permission from ref. 137. Copyright 2013, The Royal Society of Chemistry. | |
In 2015, the Kim group developed a fluorescent Golgi-targeting turn-on probe (SZnC, 59) for the detection of Zn2+, that was insensitive to pH changes, exhibited easy cell loading, low cytotoxicity, good Golgi-localization and high photostability. Using two-photon microscopic imaging the probe facilitated the real-time monitoring of Golgi Zn2+ changes in addition to the 3D distribution in tissues and live cells (Fig. 55).138 More importantly, it is worth noting that both the lipophilicity and the weakly basic pyridyl moiety of probe 59 are probably responsible for the specific Golgi apparatus localization.
 |
| | Fig. 55 (A) Mechanism of 59 for identifying Zn2+. (B) TPM images of a rat hippocampal slice stained with 20 μM SZnC for 1 h. (a) 150 TPM images along the z-direction at the depth of approximately 100–200 μM, accumulated at 10× magnification. (b) Bright-field image. (c)–(f) 20× magnification in the dentate gyrus (DG) regions white box in (a) at a depth of 120 μM. (c) Bright-field image, (d)–(f) TPM images before (d) and after (e) addition of 200 mM Zn2+/pyrithione to the imaging solution. (f) After addition of 1 mM TPEN to (e). (g) Relative TPEF intensity of (d)–(f) as a function of time. Reproduced with permission from ref. 138. Copyright 2015, The Royal Society of Chemistry. | |
In 2018, the Das group reported a fluorescent BODIPY-based HOCl turn-on probe (SF-1, 60), that was used to image HOCl that was generated endogenously in RAW 264.7 macrophages using structured-illumination microscopy (SIM). Due to a low charge/intermediate amphiphilicity, the probe localized in the Golgi and lysosomes which was confirmed by widefield and super resolution structured illumination microscopy which enabled the monitoring of HOCl generated in these organelles. Moreover, 3D-SIM images of individual cells were also constructed (Fig. 56).139
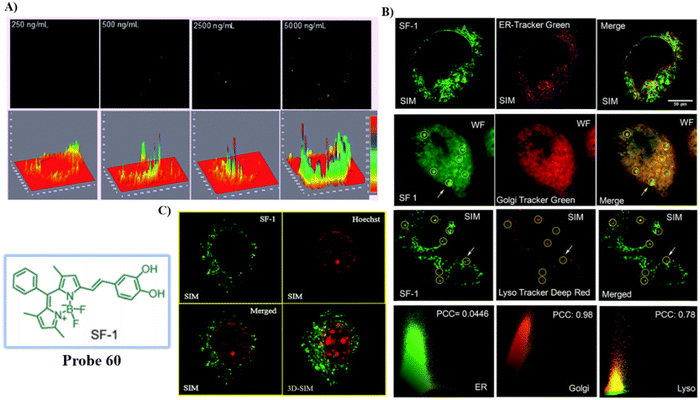 |
| | Fig. 56 (A) Response of probe 60 for endogenous HOCl: SIM images and emission intensity profiles of 60 in the presence of different concentration of LPS incubated for 12 h in fixed RAW 264.7 cells. (B) Co-stained cell imaging of 60 and ER-Green, Golgi Tracker Green and Lyso Tracker Deep Red. (C) Dual colour SIM and 3D-SIM of 60 in the presence of LPS and Hoechst 33442. Reproduced with permission from ref. 139. Copyright 2018, The Royal Society of Chemistry. | |
In 2019, the Datta group reported a sensitive “turn-on” fluorescent manganese sensor (61) to monitor the storage locations of manganese ions in the Golgi apparatus under exposure to micromolar manganese using X-ray fluorescence imaging and confocal microscopy. The experimental results, suggest the important role of the Golgi apparatus for manganese trafficking and storage at sub-cytotoxic levels (Fig. 57).140
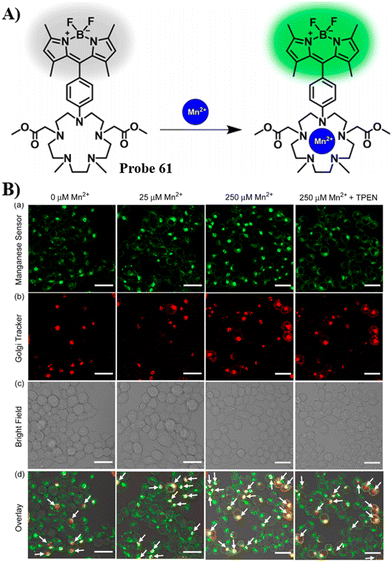 |
| | Fig. 57 (A) Mechanism of 61 for identifying Mn2+. (B) Left to right: Confocal fluorescence images of live HEK293T cells with different concentrations of MnCl2, followed by a cell-permeable divalent metal-ion chelator, TPEN. Cells were tranduced with CellLight Golgi-RFP and incubated with 61. Probe 61 emission in the green channel (λex = 488 nm) (a). Golgi-RFP emission in the red channel (λex = 543 nm) (b). Bright-field images (c). Bright-field images of cells overlaid with confocal images (d). Reproduced with permission from ref. 140. Copyright 2019, American Chemical Society. | |
4. Conclusion and outlook
The Golgi apparatus is widely distributed in eukaryotic cells and is dynamically regulated during cell cycles, and as such plays an important role in a variety of metabolic pathways and cell signaling mechanisms, including collecting, processing, sorting, transporting and secreting some important proteins/enzymes from the endoplasmic reticulum to intra- and extra-cellular destinations. Organelle-targeting fluorescent probes have made significant advances recently, due to their numerous advantages in monitoring biological systems. Amongst these probes, fluorescent Golgi-targeting probes have provided an opportunity to monitor the physiological and pathological changes and to further diagnose and treat relevant diseases associated with the Golgi apparatus. Here, we have extensively covered Golgi-targeting fluorescent probes and highlighted their application. Specifically, appropriate design strategies and Golgi-targeting mechanisms have been discussed to provide guidance for developing improved systems. Subsequently, examples of probes suitable for sensing different targets and with diverse characteristics have been elucidated, including probes containing cyclooxygenase-2 (COX-2) inhibitors, probes containing a phenylsulfonamide group, probes with a 2-trifluoromethyl-7-aminoquinoline group, probes with a L-Cys group, probes with targeting peptides, probes containing lipids, probes encapsulated by BSA, and probes with other targeting groups.
While many selective systems have been developed significant improvements are required, since current Golgi-targeting strategies need to overcome some remaining challenges including a better understanding of the biological mechanisms, provide a single highly Golgi selective response, as well as providing extended targeting duration, etc., while advances have been made in the development of Golgi-targeting probes. Going forward, we propose that the following areas should be particularly targeted: (1) explore Golgi targeting functional groups with improved optical properties such as multi-photon excitation and high quantum yield, that exhibit enhanced precision of localization in the Golgi apparatus. (2) Elucidate the exact targeting mechanism of current targeting motifs to enhance the function of Golgi-targeting fluorescent probes. (3) Develop multiplexing probes with different signal modes to enable simultaneous dynamic monitoring of multiple physiological changes in the same cell. This will help determine the intricate dynamics and physiological parameters of the complex cellular environment. (4) Develop novel multifunctional fluorescent probes with both Golgi targeted imaging and disease therapy by integrating Golgi-targeted imaging with therapeutic systems. The combination of targeted imaging and disease treatment systems will help enhance our understanding of the Golgi apparatus in the pathological state, whilst simultaneously achieving the desired therapeutic effect. (5) Develop Golgi-targeting probes for other important biomarkers such as protein glycosylation and phosphorylation, which will help understand the relationship between these biomarkers and disease development. (6) Improve the performance of the probes, by enhancing biocompatibility, incorporating reversibility, reducing toxicity, and improving optical performance. We believe that the design and development of high-precision and multi-functional fluorescent probes will be an important research direction in the field of Golgi targeting fluorescent probes in the future, and this review will provide guidance for researchers working on probe design and construction.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
We acknowledge the Joint Program for Fostering Talents of National Natural Science Foundation of China and Henan Province (U1304202), State Scholarship Fund (CSC 201908410366), Natural Science Foundation of Henan Province (232300420402), Henan Province Science and Technology Project (222300420523) (212102210646) and The Scientific Research Innovation Team of Xuchang University (2022CXTD001) for financial support. T. D. J. wishes to thank the University of Bath and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University (2020ZD01) for support.
References
- C. P. Satori, M. M. Henderson, E. A. Krautkramer, V. Kostal, M. D. Distefano and E. A. Arriaga, Chem. Rev., 2013, 113, 2733–2811 CrossRef CAS PubMed.
- R. Heald and O. Cohen-Fix, Curr. Opin. Cell Biol., 2014, 26, 79–86 CrossRef CAS PubMed.
- J. Li, E. Ahat and Y. Wang, Results Probl. Cell Differ., 2019, 67, 441–485 CAS.
- J. H. Lin, P. Walter and T. S. B. Yen, Annu. Rev. Pathol.: Mech. Dis., 2008, 3, 399–425 CrossRef CAS PubMed.
- F. Zappa, M. Failli and M. A. De Matteis, Curr. Opin. Cell Biol., 2018, 50, 102–116 CrossRef CAS PubMed.
- A. Eisenberg-Lerner, R. Benyair, N. Hizkiahou, N. Nudel, R. Maor, M. P. Kramer, M. D. Shmueli, I. Zigdon, M. C. Lev, A. Ulman, J. Y. Sagiv, M. Dayan, B. Dassa, M. Rosenwald, I. Shachar, J. Li, Y. Wang, N. Dezorella, S. Khan, Z. Porat, E. Shimoni, O. Avinoam and Y. Merbl, Nat. Commun., 2020, 11, 409 CrossRef CAS PubMed.
- J. Gao, A. Gao, H. Zhou and L. Chen, Cell Biol. Int., 2022, 46, 1309–1319 CrossRef CAS PubMed.
- Y. Zhang, Y. Wang, E. Read, M. Fu, Y. Pei, L. Wu, R. Wang and G. Yang, Antioxid. Redox Signaling, 2019, 32, 583–601 CrossRef PubMed.
- J. Li and Y. Wang, Cells, 2022, 11, 289 CrossRef CAS PubMed.
- S. Kellokumpu, Front. Cell Dev. Biol., 2019, 7, 93 CrossRef PubMed.
- C. Preisinger, B. Short, V. De Corte, E. Bruyneel, A. Haas, R. Kopajtich, J. Gettemans and F. A. Barr, J. Cell Biol., 2004, 164, 1009–1020 CrossRef CAS PubMed.
- J. I. Sbodio, B. D. Paul, C. E. Machamer and S. H. Snyder, Cell Rep., 2013, 4, 890–897 CrossRef CAS PubMed.
- L. Lu, Q. Zhou, Z. Chen and L. Chen, J. Cell. Physiol., 2018, 233, 2911–2919 CrossRef CAS PubMed.
- J. I. Sbodio, S. H. Snyder and B. D. Paul, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 780–785 CrossRef CAS PubMed.
- C. Kienzle and J. von Blume, Trends Cell Biol., 2014, 24, 584–593 CrossRef CAS PubMed.
- H. Singh, D. Sareen, J. M. George, V. Bhardwaj, S. Rha, S. J. Lee, S. Sharma, A. Sharma and J. S. Kim, Coord. Chem. Rev., 2022, 452, 214283 CrossRef CAS.
- M. Tian, J. Zhan and W. Lin, Coord. Chem. Rev., 2022, 451, 214266 CrossRef CAS.
- Y. Wei, Y. Liu, Y. He and Y. Wang, J. Mater. Chem. B, 2021, 9, 908–920 RSC.
- J. Lin, K. Yang and E. J. New, Org. Biomol. Chem., 2021, 19, 9339–9357 RSC.
- S. K. Pramanik and Amitva Das, Chem. Commun., 2021, 57, 12058–12073 RSC.
- J. Yin, L. Huang, L. Wu, J. Li, T. D. James and W. Lin, Chem. Soc. Rev., 2021, 50, 12098–12150 RSC.
- H. Yu, Y. Guo, W. Zhu, K. Havener and X. Zheng, Coord. Chem. Rev., 2021, 444, 214019 CrossRef CAS.
- D. Singh, D. Rajput and S. Kanvah, Chem. Commun., 2022, 58, 2413–2429 RSC.
- C. Ma, F. Xia and S. O. Kelley, Bioconjugate Chem., 2020, 31, 2650–2667 CrossRef PubMed.
- K. Yu, J. Pan, E. Husamelden, H. Zhang, Q. He, Y. Wei and M. Tian, Chem. – Asian J., 2020, 15, 3942–3960 CrossRef CAS PubMed.
- L. Fang and Michael Watkinson, Chem. Sci., 2020, 11, 11366–11379 RSC.
- S. Samanta, Y. He, A. Sharma, J. Kim, W. Pan, Z. Yang, J. Li, W. Yan, L. Liu, J. Qu and J. S. Kim, Chem, 2019, 5, 1697–1726 CAS.
- Y. Wen, F. Huo and C. Yin, Chin. Chem. Lett., 2019, 30, 1834–1842 CrossRef CAS.
- X. Zhang, Q. Sun, Z. Huang, L. Huang and Y. Xiao, J. Mater. Chem. B, 2019, 7, 2749–2758 RSC.
- P. Gao, W. Pan, N. Li and B. Tang, Chem. Sci., 2019, 10, 6035–6071 RSC.
- J. Zhu, Z. Xu, Y. Yang and L. Xu, Chem. Commun., 2019, 55, 6629–6671 RSC.
- H. Zhu, J. Fan, J. Du and X. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS PubMed.
- W. Xu, Z. Zeng, J. Jiang, Y. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658–13699 CrossRef CAS PubMed.
- G. H. Luo, T. Z. Xu, X. Li, W. Jiang, Y. H. Duo and B. Z. Tang, Coord. Chem. Rev., 2022, 462, 214508 CrossRef.
- C. Liu, H. Zhu, Y. Zhang, M. Su, M. Liu, X. Zhang, X. Wang, X. Rong, K. Wang, X. Li and B. Zhu, Coord. Chem. Rev., 2022, 462, 214504 CrossRef CAS.
- M. G. Farquhar and G. E. Palade, Trends Cell Biol., 1998, 8, 2–10 CrossRef CAS PubMed.
- N. Lipsky and R. Pagano, Science, 1985, 228, 745–747 CrossRef CAS PubMed.
- N. B. Cole, C. L. Smith, N. Sciaky, M. Terasaki, M. Edidin and J. Lippincott-Schwartz, Science, 1996, 273, 794–797 CrossRef PubMed.
- A. Dragulescu-Andrasi, G. Liang and J. Rao, Bioconjugate Chem., 2009, 20, 1660–1666 CrossRef CAS PubMed.
- H. Wang, X. T. Zhang, T. C. Xiu, H. T. Wang, P. Li and B. Tang, Coord. Chem. Rev., 2024, 502, 215618 CrossRef CAS.
- C. Denkert, K. J. Winzer, B. M. Muller, W. Weichert, S. Pest, M. Kobel, G. Kristiansen, A. Reles, A. Siegert, H. Guski and S. Hauptmann, Cancer, 2003, 97, 2978–2987 CrossRef CAS PubMed.
- M. J. Uddin, B. C. Crews, K. Ghebreselasie and L. J. Marnett, Bioconjugate Chem., 2013, 24, 712–723 CrossRef CAS PubMed.
- L. Fourriere, S. Divoux, M. Roceri, F. Perez and G. Boncompain, J. Cell Sci., 2016, 129, 3238–3250 CrossRef CAS PubMed.
- J. Kim, H. R. Cho, H. Jeon, D. Kim, C. Song, N. Lee, S. H. Choi and T. Hyeon, J. Am. Chem. Soc., 2017, 139, 10992–10995 CrossRef CAS PubMed.
- J. L. Masferrer, K. M. Leahy, A. T. Koki, B. S. Zweifel, S. L. Settle, B. M. Woerner, D. A. Edwards, A. G. Flickinger, R. J. Moore and K. Seibert, Cancer Res., 2000, 60, 1306–1311 CAS.
- E. Richardsen, R. D. Uglehus, J. Due, C. Busch and L. T. Busund, Cancer Epidemiol., 2010, 34, 316–322 CrossRef CAS PubMed.
- M. J. Uddin, B. C. Crews, A. L. Blobaum, P. J. Kingsley, D. L. Gorden, J. O. McIntyre, L. M. Matrisian, K. Subbaramaiah, A. J. Dannenberg, D. W. Piston and L. J. Marnett, Cancer Res., 2010, 70, 3618–3627 CrossRef CAS PubMed.
- M. T. Rizzo, Clin. Chim. Acta, 2011, 412, 671–687 CrossRef CAS PubMed.
- C. Denkert, M. Kobel, S. Berger, A. Siegert, A. Leclere, U. Trefzer and S. Hauptmann, Cancer Res., 2001, 61, 303–308 CAS.
- H. M. Kandil, G. Tanner, W. Smalley, S. Halter, A. Radhika and R. N. Dubois, Dig. Dis. Sci., 2001, 46, 785–789 CrossRef CAS PubMed.
- C. E. Eberhart, R. J. Coffey, A. Radhika, F. M. Giardiello, S. Ferrenbach and R. N. DuBois, Gastroenterology, 1994, 107, 1183–1188 CrossRef CAS PubMed.
- A. Bhardwaj, J. Kaur, F. Wuest and E. E. Knaus, ChemMedChem, 2014, 9, 109–116 CrossRef CAS PubMed.
- A. Bhardwaj, J. Kaur, S. K. Sharma, Z. Huang, F. Wuest and E. E. Knaus, Bioorg. Med. Chem. Lett., 2013, 23, 163–168 CrossRef CAS PubMed.
- W. F. Hood, J. K. Gierse, P. C. Isakson, J. R. Kiefer, R. G. Kurumbail, K. Seibert and J. B. Monahan, Mol. Pharmacol., 2003, 63, 870–877 CrossRef CAS PubMed.
- H. Zhang, J. Fan, J. Wang, S. Zhang, B. Dou and X. Peng, J. Am. Chem. Soc., 2013, 135, 11663–11669 CrossRef CAS PubMed.
- B. Wang, J. Fan, X. Wang, H. Zhu, J. Wang, H. Mu and X. Peng, Chem. Commun., 2015, 51, 792–795 RSC.
- B. Gurram, S. Zhang, M. Li, H. Li, Y. Xie, H. Cui, J. Du, J. Fan, J. Wang and X. Peng, Anal. Chem., 2018, 90, 5187–5193 CrossRef CAS PubMed.
- B. Gurram, M. Li, J. Fan, J. Wang and X. Peng, Front. Chem. Sci. Eng., 2020, 14(1), 41–52 CrossRef CAS.
- Y. Luo, S. Zhang, H. Wang, Q. Luo, Z. Xie, B. Xu and W. Tian, CCS Chem., 2022, 4, 456–463 CrossRef CAS.
- K. Huang, H. Zhang, M. Yan, J. Xue and J. Chen, Dyes Pigm., 2022, 198, 109997 CrossRef CAS.
- C. Liu, L. Zhou, Y. Zheng, H. Man, Z. Ye, X. Zhang, L. Xie and Y. Xiao, Chem. Commun., 2022, 58, 10052–10055 RSC.
- R. G. Kurumbail, A. M. Stevens, J. K. Gierse, J. J. McDonald, R. A. Stegeman, J. Y. Pak, D. Gildehaus, J. M. Miyashiro, T. D. Penning, K. Seibert, P. C. Isakson and W. C. Stallings, Nature, 1996, 384, 644–648 CrossRef CAS PubMed.
- H. Wang, Z. He, Y. Yang, J. Zhang, W. Zhang, W. Zhang, P. Li and B. Tang, Chem. Sci., 2019, 10, 10876–10880 RSC.
- H. Zhu, C. Liu, C. Liang, B. Tian, H. Zhang, X. Zhang, W. Sheng, Y. Yu, S. Huang and B. Zhu, Chem. Commun., 2020, 56, 4086–4089 RSC.
- H. Wang, Y. Yang, F. Huang, Z. He, P. Li, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2020, 92, 3103–3110 CrossRef CAS PubMed.
- H. Wang, M. Dong, H. Wang, F. Huang, P. Li, W. Zhang, W. Zhang and B. Tang, Chem. Commun., 2021, 57, 5838–5841 RSC.
- P. Xiao, K. Ma, M. Kang, L. Huang, Q. Wu, N. Song, J. Ge, D. Li, J. Dong, L. Wang, D. Wang and B. Z. Tang, Chem. Sci., 2021, 12, 13949–13957 RSC.
- M. M. Fortibui, W. Lim, S. Lee, S. Park and J. Kim, Molecules, 2021, 26, 4980 CrossRef CAS PubMed.
- S. Li, K. Yang, J. Zeng, Y. Ding, D. Cheng and L. He, ACS Omega, 2022, 7, 9929–9935 CrossRef CAS PubMed.
- S. Feng, Z. Zheng, S. Gong and G. Feng, Sens. Actuators, B, 2022, 361, 131751 CrossRef CAS.
- J. Li, J. Tang, X. Yang, P. Xie, J. Liu, D. Zhang and Y. Ye, Sens. Actuators, B, 2022, 358, 131513 CrossRef CAS.
- X. Zhang, L. Chen, Y. Y. Wei, J. L. Du, S. P. Yu, X. G. Liu, W. Liu, Y.-J. Liu, Y.-Z. Yang and Q. Li, Dyes Pigm., 2022, 201, 110213 CrossRef CAS.
- Z. Feng, J. Wu, M. Jiang, J. Sha, W. Liu, H. Ren, W. Zhang, C.-S. Lee and P. Wang, Sens. Actuators, B, 2022, 366, 131963 CrossRef CAS.
- Q. Zhang, Y. Tang, S. Song, J. Peng and W. Y. Lin, New J. Chem., 2022, 46, 14333–14337 RSC.
- S. Han, H. Wang, H. Li, K. Li, J. Wang and X. Song, Anal. Chem., 2023, 95, 8002–8010 CrossRef CAS PubMed.
- Z. He, D. Liu, Y. Liu, X. Li, W. Shi and H. Ma, Anal. Chem., 2022, 94, 10256–10262 CrossRef CAS PubMed.
- H. Wang, C. Liu, X. Zhang, T. Xiu, P. Li, W. Zhang, W. Zhang, X. Wang, Z. Liu and B. Tang, Biosens. Bioelectron., 2022, 213, 114480 CrossRef CAS PubMed.
- G. Fei, S. Ma, C. Wang, T. Chen, Y. Li, Y. Liu, B. Tang, T. D. James and G. Chen, Coord. Chem. Rev., 2021, 447, 214134 CrossRef CAS.
- S. M. Usama, F. Inagaki, H. Kobayashi and M. J. Schnermann, J. Am. Chem. Soc., 2021, 143, 5674–5679 CrossRef CAS PubMed.
- R. M. Exner, F. Cortezon-Tamarit and S. I. Pascu, Angew. Chem., Int. Ed., 2021, 60, 6230–6241 CrossRef CAS PubMed.
- Z. Xu, H. Wang, X. Hou, W. Xu, T. Xiang and C. Wu, Sens. Actuators, B, 2014, 201, 469–474 CrossRef CAS.
- L. Feng, W. Chen, X. Ma, S. H. Liu and J. Yin, Org. Biomol. Chem., 2020, 18, 9385–9397 RSC.
- N. Zhou, F. Huo, Y. Yue and C. Yin, J. Am. Chem. Soc., 2020, 142, 17751–17755 CrossRef CAS PubMed.
- H. Mu, K. Miki, H. Harada, K. Tanaka, K. Nogita and K. Ohe, ACS Sens., 2021, 6, 123–129 CrossRef CAS PubMed.
- C. Zhang, Z. Qiu, L. Zhang, Q. Pang, Z. Yang, J.-K. Qin, H. Liang and S. Zhao, Chem. Sci., 2021, 12, 4883–4888 RSC.
- L. Chen, J. Chen, Y. Fang, F. Zeng and S. Wu, Chem. Commun., 2021, 57, 7842–7845 RSC.
- H. Fang, Y. Chen, Y. Wang, S. Geng, S. Yao, D. Song, W. He and Z. Guo, Sci. China: Chem., 2020, 63, 699–706 CrossRef CAS.
- J. Xiang, C. Liu, L. Zhou, X. Yang, Y. Li, Y. Jiang, T. Mahmood, P. Zhang, P. Gong and L. Cai, Anal. Chem., 2020, 92, 4721–4725 CrossRef CAS PubMed.
- Y. Zhou, P. Li, X. Wang, C. Wu, N. Fan, X. Liu, L. Wu, W. Zhang, W. Zhang, Z. Liu and B. Tang, Chem. Sci., 2020, 11, 12149–12156 RSC.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- J. Chen, H. Liu, L. Yang, J. Jiang, G. Bi, G. Zhang, G. Li and X. Chen, ACS Med. Chem. Lett., 2019, 10, 954–959 CrossRef CAS PubMed.
- X. Zhang, C. Liu, Y. Chen, X. Cai, W. Sheng, H. Zhu, P. Jia, Z. Li, S. Huang and B. Zhu, Chem. Commun., 2020, 56, 1807–1810 RSC.
- X. Zhang, C. Liu, X. Cai, B. Tian, H. Zhu, Y. Chen, W. Sheng, P. Jia, Z. Li, Y. Yu, S. Huang and B. Zhu, Sens. Actuators, B, 2020, 310, 127820 CrossRef CAS.
- H. Zhu, C. Liang, X. Cai, H. Zhang, C. Liu, P. Jia, Z. Li, Y. Yu, X. Zhang, W. Sheng and B. Zhu, Anal. Chem., 2020, 92, 1883–1889 CrossRef CAS PubMed.
- X. Hu, Z. Hai, C. Wu, W. Zhan and G. Liang, Anal. Chem., 2021, 93, 1636–1642 CrossRef CAS PubMed.
- X. Rong, C. Liu, M. Li, H. Zhu, Y. Zhang, M. Su, X. Wang, X. Li, K. Wang, M. Yu, W. Sheng and B. Zhu, Anal. Chem., 2021, 93, 16105–16112 CrossRef CAS PubMed.
- Z. Zheng, S. Feng, S. Gong and G. Feng, Sens. Actuators, B, 2021, 347, 130631 CrossRef CAS.
- H. Zhu, C. Liu, X. Rong, Y. Zhang, M. Su, X. Wang, M. Liu, X. Zhang, W. Sheng and B. Zhu, Bioorg. Chem., 2022, 122, 105741 CrossRef CAS PubMed.
- Z. Yan, Z. Tang, X. Wang, Z. Zheng, Z. Tian, X. Geng, Y. Li and H. Jiang, Sens. Actuators, B, 2022, 369, 132352 CrossRef CAS.
- Z. Tang, Z. Yan, L. Gong, L. Zhang, X. Yin, J. Sun, K. Wu, W. Yang, G. Fan, Y. Li and H. Jiang, Anal. Chem., 2022, 94, 14778–14784 CrossRef CAS PubMed.
- Z. Feng, J. Wu, W. Liu, H. Ren, X. Zheng, W. Zhang, C.-S. Lee and P. Wang, Sens. Actuators, B, 2023, 384, 133668 CrossRef CAS.
- D. Aoki, N. Lee, N. Yamaguchi, C. Dubois and M. N. Fukuda, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 4319–4323 CrossRef CAS PubMed.
- Y. Maeda, G. V. Beznoussenko, J. Van Lint, A. A. Mironov and V. Malhotra, EMBO J., 2001, 20, 5982–5990 CrossRef CAS PubMed.
- K. Maeda, P. Hägglund, C. Finnie, B. Svensson and A. Henriksen, Structure, 2006, 14, 1701–1710 CrossRef CAS PubMed.
- W. Zhang, J. Zhang, P. Li, J. Liu, D. Su and B. Tang, Chem. Sci., 2019, 10, 879–883 RSC.
- P. Li, X. Guo, X. Bai, X. Wang, Q. Ding, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2019, 91, 3382–3388 CrossRef CAS PubMed.
- L. Fang, R. Crespo-Otero, C. R. Jones and M. Watkinson, Sens. Actuators, B, 2021, 338, 129850 CrossRef CAS.
- N. Yamaguchi and M. N. Fukuda, J. Biol. Chem., 1995, 270, 12170–12176 CrossRef CAS PubMed.
- M. Kneen, J. Farinas, Y. Li and A. S. Verkman, Biophys.
J., 1998, 74, 1591–1599 CrossRef CAS PubMed.
- S. H. Wong and W. Hong, J. Biol. Chem., 1993, 268, 22853–22862 CrossRef CAS PubMed.
- K. Bos, C. Wraight and K. K. Stanley, EMBO J., 1993, 12, 2219–2228 CrossRef CAS PubMed.
- J. S. Humphrey, P. J. Peters, L. C. Yuan and J. S. Bonifacino, J. Cell Biol., 1993, 120, 1123–1135 CrossRef CAS PubMed.
- E. H. W. Pap, T. B. Dansen, R. van Summeren and K. W. A. Wirtz, Exp. Cell Res., 2001, 265, 288–293 CrossRef CAS PubMed.
- K. H. Jung, E. T. Oh, H. J. Park and K. H. Lee, Biosens. Bioelectron., 2016, 85, 437–444 CrossRef CAS PubMed.
- K. Li, X. X. Hu, H. W. Liu, S. Xu, S. Y. Huan, J. B. Li, T. G. Deng and X.-B. Zhang, Anal. Chem., 2018, 90, 11680–11687 CrossRef CAS PubMed.
- L. Zhu, H. W. Liu, Y. Yang, X. X. Hu, K. Li, S. Xu, J. B. Li, G. Ke and X. B. Zhang, Anal. Chem., 2019, 91, 9682–9689 CrossRef CAS PubMed.
- X. Li, C. Cao, P. Wei, M. Xu, Z. Liu, L. Liu, Y. Zhong, R. Li, Y. Zhou and T. Yi, ACS Appl. Mater. Interfaces, 2019, 11, 12327–12334 CrossRef CAS PubMed.
- W. Tan, Q. Zhang, J. Wang, M. Yi, H. He and B. Xu, Angew. Chem., Int. Ed., 2021, 60, 12796–12801 CrossRef CAS PubMed.
- W. Tan, Q. Zhang, M. C. Quiñones-Frías, A. Y. Hsu, Y. Zhang, A. Rodal, P. Hong, H. R. Luo and B. Xu, J. Am. Chem. Soc., 2022, 144, 6709–6713 CrossRef CAS PubMed.
- S. Limar, C. Körner, F. Martínez-Montañés, V. G. Stancheva, V. N. Wolf, S. Walter, E. A. Miller, C. S. Ejsing, V. V. Galassi and F. Fröhlich, J. Cell Biol., 2023, 222, e202109162 CAS.
- R. P. Rao, C. Yuan, J. C. Allegood, S. S. Rawat, M. B. Edwards, X. Wang, A. H. Merrill, U. Acharya and J. K. Acharya, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11364–11369 CrossRef CAS PubMed.
- T. Hirayama, M. Inden, H. Tsuboi, M. Niwa, Y. Uchida, Y. Naka, I. Hozumib and H. Nagasawa, Chem. Sci., 2019, 10, 1514–1521 RSC.
- L. Fan, X. Wang, J. Ge, F. Li, C. Zhang, B. Lin, S. Shuang and C. Dong, Chem. Commun., 2019, 55, 6685–6688 RSC.
- T. Kowada, T. Watanabe, Y. Amagai, R. Liu, M. Yamada, H. Takahashi, T. Matsui, K. Inaba and S. Mizukami, Cell Chem. Biol., 2020, 27, 1521–1531 CrossRef CAS PubMed.
- A. Li, Y. Liu, Labapuchi, Z. Chen, S. Li, R. Zhong, D. Cheng, L. Chen and L. He, Anal. Chim. Acta, 2023, 1255, 341100 CrossRef CAS PubMed.
- A. Li, Y. Liu, Labapuchi, Z. Chen, S. Li, R. Zhong, D. Cheng, L. Chen and L. He, Spectrochim. Acta, Part A, 2023, 294, 122560 CrossRef CAS PubMed.
- T. Kowada, T. Watanabe, R. Liu and S. Mizukami, STAR Protoc., 2021, 2, 100395 CrossRef CAS PubMed.
- X. Zeng, X. Zhang, B. Zhu, H. Jia, Y. Li and J. Xue, Analyst, 2011, 136, 4008–4012 RSC.
- R. W. Horobin and F. R. Doubell, Biotech. Histochem., 2013, 88, 461–476 CrossRef CAS PubMed.
- F. Xue, Y. Wen, P. Wei, Y. Gao, Z. Zhou, S. Xiao and T. Yi, Chem. Commun., 2017, 53, 6424–6427 RSC.
- H. Wang, C. Liu, Z. He, P. Li, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2021, 93, 6551–6558 CrossRef CAS PubMed.
- X. Xing, Y. Jia, J. Zhang, Z. Wu, M. Qin, P. Li, X. Feng, Y. Sun and G. Zhao, Sens. Actuators, B, 2021, 329, 129245 CrossRef CAS.
- G. K. Walkup, S. C. Burdette, S. J. Lippard and R. Y. Tsien, J. Am. Chem. Soc., 2000, 122, 5644–5645 CrossRef CAS.
- F. Qian, C. Zhang, Y. Zhang, W. He, X. Gao, P. Hu and Z. Guo, J. Am. Chem. Soc., 2009, 131, 1460–1468 CrossRef CAS PubMed.
- L. E. McQuade and S. J. Lippard, Inorg. Chem., 2010, 49, 9535–9545 CrossRef CAS PubMed.
- C. Zhang, Y. Zhang, Y. Chen, Z. Xie, Z. Liu, X. Dong, W. He, C. Shen and Z. Guo, Inorg. Chem. Commun., 2011, 14, 304–307 CrossRef CAS.
- T. Hirayama, K. Okuda and H. Nagasawa, Chem. Sci., 2013, 4, 1250–1256 RSC.
- H. Singh, H. W. Lee, C. H. Heo, J. W. Byun, A. R. Sarkar and H. M. Kim, Chem. Commun., 2015, 51, 12099–12102 RSC.
- F. Ali, S. Aute, S. Sreedharan, H. A. Anila, H. K. Saeed, C. G. Smythe, J. A. Thomas and A. Das, Chem. Commun., 2018, 54, 1849–1852 RSC.
- S. Das, A. Carmona, K. Khatua, F. Porcaro, A. Somogyi, R. Ortega and A. Datta, Inorg. Chem., 2019, 58, 13724–13732 CrossRef CAS PubMed.
Footnote |
| † These authors are equally contributed to this work. |
|
| This journal is © The Royal Society of Chemistry 2024 |
 Open Access Article
Open Access Article c,
Zhi-Hong Xu
c,
Zhi-Hong Xu *ab,
Yuan Wang
*ab,
Yuan Wang *d and
Tony D. James
*d and
Tony D. James
 *ce
*ce























![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2).77 The probe was successfully used for the simultaneous multicolored detection of both Golgi H2O2 and O2˙− in cells and in hypertensive mice, for the evaluation of various antihypertensive drugs, and for the identification of post-transcriptional modification sites on dihydrofolate reductase (DHFR) induced by the ROS. Moreover, the system is suitable for exploring the eNOS-associated oxidative stress signaling pathway in inflammation-induced hypertension (Fig. 22).
2).77 The probe was successfully used for the simultaneous multicolored detection of both Golgi H2O2 and O2˙− in cells and in hypertensive mice, for the evaluation of various antihypertensive drugs, and for the identification of post-transcriptional modification sites on dihydrofolate reductase (DHFR) induced by the ROS. Moreover, the system is suitable for exploring the eNOS-associated oxidative stress signaling pathway in inflammation-induced hypertension (Fig. 22).

















![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with Cu+ resulting in fluorescence “turn-on” response for Cu+. Evaluation of the binding mode indicated the critical role played by the thiol group of the peptide receptor in the strong binding with Cu+, with additional support from the sulfonamide and amide groups. Meanwhile, probe 39 was able to penetrate living RKO cells and detect Cu+ in the Golgi apparatus (Fig. 37).114
1 complex with Cu+ resulting in fluorescence “turn-on” response for Cu+. Evaluation of the binding mode indicated the critical role played by the thiol group of the peptide receptor in the strong binding with Cu+, with additional support from the sulfonamide and amide groups. Meanwhile, probe 39 was able to penetrate living RKO cells and detect Cu+ in the Golgi apparatus (Fig. 37).114



























