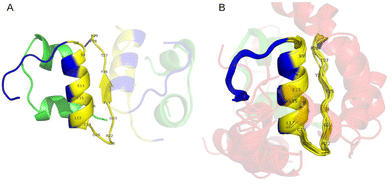Open Access Article
Open Access ArticleCytotoxic Staphylococcus aureus PSMα3 inhibits the aggregation of human insulin in vitro†
Aleksandra
Kalitnik
 *a,
Monika
Szefczyk
*a,
Monika
Szefczyk
 b,
Alicja W.
Wojciechowska
b,
Alicja W.
Wojciechowska
 a,
Jakub W.
Wojciechowski
a,
Marlena
Gąsior-Głogowska
a,
Joanna
Olesiak-Bańska
c and
Małgorzata
Kotulska
a,
Jakub W.
Wojciechowski
a,
Marlena
Gąsior-Głogowska
a,
Joanna
Olesiak-Bańska
c and
Małgorzata
Kotulska
 *a
*a
aDepartment of Biomedical Engineering, Faculty of Fundamental Problems of Technology, Wroclaw University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wroclaw, Poland. E-mail: aleksandra.kalitnik@pwr.edu.pl; malgorzata.kotulska@pwr.edu.pl
bDepartment of Bioorganic Chemistry, Faculty of Chemistry, Wroclaw University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wroclaw, Poland
cInstitute of Advanced Materials, Wroclaw University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wroclaw, Poland
First published on 17th May 2024
Abstract
Phenol-soluble modulins (PSMs) are extracellular short amphipathic peptides secreted by the bacteria Staphylococcus aureus (S. aureus). They play an essential role in the bacterial lifecycle, biofilm formation, and stabilisation. From the PSM family, PSMα3 has been of special interest recently due to its cytotoxicity and highly stable α-helical conformation, which also remains in its amyloid fibrils. In particular, PSMα3 fibrils were shown to be composed of self-associating “sheets” of α-helices oriented perpendicular to the fibril axis, mimicking the architecture of canonical cross-β fibrils. Therefore, they were called cross-α-fibrils. PSMα3 was synthesised and verified for identity with wild-type sequences (S. aureus). Then, using several experimental techniques, we evaluated its propensity for in vitro aggregation. According to our findings, synthetic PSMα3 (which lacks the N-terminal formyl groups found in bacteria) does not form amyloid fibrils and maintains α-helical conformation in a soluble monomeric form for several days of incubation. We also evaluated the influence of PSMα3 on human insulin fibrillation in vitro, using a variety of experimental approaches in combination with computational molecular studies. First, it was shown that PSMα3 drastically inhibits the fibrillation of human insulin. The anti-fibrillation effect of PSMα3 was concentration-dependent and required a concentration ratio of PSMα3: insulin equal to or above 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. Molecular modelling revealed that PSMα3 most likely inhibits the production of insulin primary nuclei by competing for residues involved in its dimerization.
100. Molecular modelling revealed that PSMα3 most likely inhibits the production of insulin primary nuclei by competing for residues involved in its dimerization.
Introduction
Phenol-soluble modulins (PSMs) are amyloid functional peptides produced by the Gram-positive bacteria Staphylococcus aureus (S. aureus). The amyloidogenic properties of the PSMs play an essential role in bacterial biofilm formation and stabilisation.1 The multifunctional PSM family is represented by several small amphipathic peptides characterised by differences in their sequences, fibrillation predispositions, structural plasticity, and the architecture of self-produced amyloid-like structures.2 Whereas other PSM peptides undergo conformational changes during aggregation, PSMα3 has been previously characterised by extremely high stability in maintaining its α-helical conformation even up to several days of incubation3 in the cases of soluble monomeric forms,3–5 oligomers,6 or mature fibrils.1,7,8 Several findings, based on the results obtained using different experimental methods, clearly demonstrated that amyloid fibrils of PSMα3 are composed of unique cross-α fibrils, where helices compile together into firm layers, imitating the cross-β structures.6–8 It is known that many external and internal factors influence peptide aggregation behaviour; for example, a slight modification of the peptide's primary structure can considerably affect its fibrillation behaviour.1,9–11 It has been previously shown that the synthesised PSMα3 peptide does not form amyloid fibrils in vitro during incubation for up to several days.9 The recombinant N-terminally formylated PSMα3 peptide, on the other hand, was demonstrated to have the highest in vitro aggregation propensity of all PSMs over a wide concentration range.1,10 Furthermore, because PSMs often retain their N-terminal formyl groups throughout secretion in bacteria, N-terminus formylation is likely responsible for PSMα3 contribution to in vivo PSM biofilm development, as well as playing a major role in high PSMα3 toxicity.12,13 It has also been discussed previously that optimisation of experimental procedures, such as monomerization and solubilization with different solvents, agitation, and changes in pH and temperature, allowed regulation of the assembly states of amyloid β during in vitro incubation and allowed an oligomeric or fibrillar fraction to be obtained.14 Furthermore, pH changes have been shown to have a considerable impact on the aggregation kinetics of particular PSM peptides in vitro, with the effects varying depending on the peptide.10 Particularly for N-formylated PSMα3, the fastest aggregation was shown at a slightly basic pH, and despite extending the incubation period at an acidic pH, no aggregation took place.10 Moreover, many other factors were shown to affect the aggregation behaviour of PSM peptides, such as extracellular DNA,15,16 as well as the presence of glucose or a high content of sodium chloride in the incubation medium.17 It is also well known that initial steps in experimental procedures, such as pre-treatment and solubilization of peptides prior to aggregation assays, are critical to control the process of peptide self-assembly.10,15 However, in some studies, the details of the experimental protocol for preparation of the studied peptide before the aggregation assay are not clearly indicated, which significantly hinders the correct estimation of the influence of external factors on the aggregation properties.6,18PSM peptides are also well recognised for their predisposition to cross-interact with other amyloidogenic proteins, modulating fibrillation of each other, which can result in both aggregation acceleration and inhibition.19–21 Furthermore, extensive polymorphism of PSM-composed amyloid-like structures was also shown to be determined by their cross-seeded interactions during the biofilm formation process.9,19 There is quite ambiguous data concerning the effects of the cross-interaction of PSMs with some human amyloids.20,21 For example, N-terminally formylated PSMα peptides were described previously to accelerate fibrillation of α-synuclein (α-syn) in human cells as well as in vitro under conditions that mimic physiological.20 On the other hand, the opposite effect of PSMα3 on α-syn fibril formation was described previously.21 Moreover, the α-helical structure of PSMα3-like peptides was revealed to be a crucial factor in inhibiting α-syn aggregation, and mutations in the α-helix scaffold resulted in full loss of the anti-fibrillation effect.21 Several short α-helical synthetic peptides were shown to effectively suppress the in vitro fibrillation of human insulin.22 Insulin is a vital polypeptide that serves a range of biological functions, the most essential of which is the treatment of diabetes patients. This pathological condition leads to the formation of a hard subcutaneous mass present in insulin amyloid plaques at the injection site that can trigger a local or systemic inflammatory response, among other side effects.23 Localised insulin fibril formation causes serious problems in insulin therapy, such as insufficient glycemic control due to impaired insulin absorption and catheter blockage during sustained subcutaneous insulin infusion.24 Since amyloids share a common secondary structure, one common therapeutic strategy for any type of amyloidosis is focused on developing protein aggregation inhibitors that reduce the aggregation propensity of precursor proteins, prevent or remodel toxic amyloids, or alleviate their multi-toxic effects.25
In this work, we used several experimental approaches, including the thioflavin T (ThT) fluorescence assay, circular dichroism (CD), attenuated total reflectance Fourier-transform infrared (ATR-FTIR) spectroscopy, and atomic force microscopy (AFM), to assess the aggregation propensity of PSMα3 during in vitro incubation, using slight changes in experimental conditions and pre-treatment procedures. We also evaluated the modulation of the fibrillation process of human insulin upon PSMα3 addition. The aggregation kinetics of insulin in vitro after the addition of the peptide at various concentrations were studied, along with the aggregation mechanism, as well as the secondary structure distribution and morphological features of the produced insulin aggregates. Furthermore, we used molecular modelling to examine the mechanism of PSMα3 interaction with human insulin. We predicted the structure of the insulin dimer using AlphaFold, and also performed molecular docking of PSM to insulin, which revealed a possible mechanism of cross-interactions.
Results
Studied peptides
The full-length, 22-residue PSMα3 peptide was synthesised using solid-phase peptide synthesis (SPPS) on a fully automated synthesiser with microwave heating by applying Fmoc/tBu chemistry and then purified using a preparative reverse phase high-performance liquid chromatography (RP-HPLC) system with a C18 column according to a previously described protocol.26 Quality analysis of the synthesised and purified peptide was conducted to prove the structural correlation with the wild-type PSMα3. The analytical data for the obtained PSMα3 peptide, as well as the mass spectrum and the analytical RP-HPLC chromatogram, can be found in the ESI† (Table S1 and Fig. S1, Fig. S2, ESI†). The insulin sample, used in the current study, was purchased from Sigma Aldrich, and all the characteristics were provided by the producer.PSMα3 do not form amyloid-like aggregates under the applied experimental conditions
Initially, UV circular dichroism (CD) spectroscopy was applied to evaluate the primarily spatial conformation of the synthesised PSMα3 peptide, as well as possible incubation-induced alterations of its secondary structure. The CD spectra of PSMα3, directly after dissolving and after 72 h of incubation at 37 °C, are presented in Fig. 1. As shown in Fig. 1, the CD spectrum of PSMα3 freshly dissolved in water shows significant minima at 208 and 222 nm, which are typically assigned to α-helical conformation.27 The calculated ratio of mean residue ellipticity within the characteristic wavelength ranges of θ222/θ208 was equal to 0.78, which is attributed to single-stranded non-interacting α-helices and indicates the absence of self-interacting monomers.28,29Fig. 1 demonstrates that according to the obtained data, PSMα3 maintained α-helical conformation after incubation at 37 °C up to 72 h. The CD spectrum of PSMα3 incubated for 72 h exhibits the same minima at 208 and 222 nm, which are characteristics of α-helical structures (Fig. 1). Moreover, the calculated ratio of mean residue ellipticity between the typical wavelength ranges of θ222/θ208 was equal to 0.85 and higher compared to freshly dissolved peptide (0.78), indicating stabilisation of helical conformation during incubation.6,30 | ||
| Fig. 1 CD spectra of PSMα3 before the aggregation assay (red) and after incubation for 72 h at 37 °C (blue). The CD intensity is given as the mean residue ellipticity (θ [deg × cm2 × dmol−1]). | ||
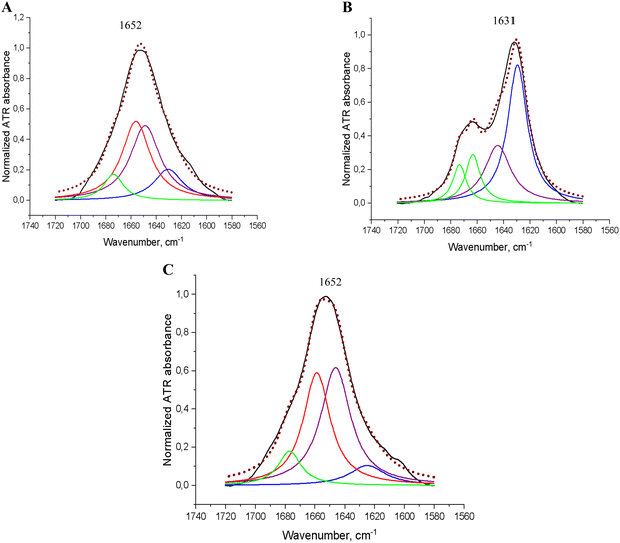
The secondary structure evaluation of the obtained PSMα3 was also conducted by attenuated total reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy. It was found that the results of the ATR-FTIR analysis were consistent with the CD spectroscopy data. The main absorption band at 1655 cm−1 in the amide I region of the FTIR spectra of freshly dissolved PSMα3 clearly proved its primarily α-helical structure (Fig. 2A), in accordance with the standard interpretation methodology.31 ATR-FTIR spectroscopy was applied for further studies of the possible secondary structure changes during the incubation process. The differences in fractional changes of PSMα3 during incubation were monitored by analysing the characteristic absorption bands in the amide I region, which is especially sensitive to protein secondary structural features.32 The ATR-FTIR spectrum of PSMα3 after 72 h of incubation at 37 °C is presented in Fig. 2B. Similarly, as in the case of freshly dissolved peptides, the ATR-FTIR spectrum of incubated PSMα3 showed the main absorption band in the amide I region at 1655 cm−1, which is assigned to the α-helical conformation (Fig. 2). Raw FTIR spectra of freshly dissolved PSMα3 and after 72 h of incubation at 37 °C are presented in the ESI† (Fig. S3).
 | ||
| Fig. 2 ATR-FTIR spectra of monomerized PSMα3 (A) before the aggregation assay, and (B) after incubation for 72 h at 37 °C. R-Square (COD): 0.99974 (A), and 0.99769 (B). | ||
The secondary structure fractional distribution for PSMα3 freshly dissolved, as well as after the incubation (72 h, 37 °C), was estimated using deconvolution of the FTIR spectra in the amide I region (1600–1700 cm−1), and the results are listed in Table 1.
| Peptide sample | Average peak position (cm−1) | Secondary structure assignment | Average composition (%) |
|---|---|---|---|
| PSMα3 monomerized, freshly dissolved in water | 1631 | β-Sheets (1–4) | 19, 5 |
| 1656 | α-Helix (1, 3) | 42 | |
| 1674 | β-Turn (1, 3, 5) | 26 | |
| 1694 | β-Sheet (1)/β-turn (3, 5) | 12, 5 | |
| PSMα3 incubated for 72 h at 37 °C | 1629 | β-Sheets (1–4) | 15, 3 |
| 1656 | α-Helix (1, 3) | 52, 7 | |
| 1679 | β-Turn (1, 3, 5) | 24 | |
| 1693 | β-Sheet (1)/β-turn (3, 5) | 8 | |
The primary peaks contributing to the ATR-FTIR overlapping signal in this region were identified using the secondary derivatives of each curve, which were calculated and visualised. According to the data of spectra deconvolution, followed by classical assignment of the peaks,31,33–36 the dominant conformational state of PSMα3, freshly dissolved and after 72 h of incubation, is α-helix (42% and 53%, respectively). These findings were confirmed by the presence of the main peaks at 1656 cm−1 (assigned to α-helix) with less contribution of β-sheets (up to 32%) due to the presence of absorption bands at 1629–1628 cm−1 and 1692–1691 cm−1, as well as β-turn components at 1679–1674 cm−1 (Table 1). Our data are consistent with a previously described conformation of monomeric PSMα3 in solution.1,6,18,19
The fibrillation propensity of PSMα3 was examined using the thioflavin T (ThT) fluorescence assay. Prior to the aggregation kinetics assay, the peptide was treated with a mixture of (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) TFA and HFIP for monomerization. The monomerized peptide was dissolved in water directly before analysis. The ThT kinetics assay is a quantitative method that indicates the progress of amyloid assembly over time since ThT has the capability of binding to β-sheets or other amyloid-like structures, leading to a significant increase in its fluorescence intensity, which correlates with the degree of fibril formation.37–39
1) TFA and HFIP for monomerization. The monomerized peptide was dissolved in water directly before analysis. The ThT kinetics assay is a quantitative method that indicates the progress of amyloid assembly over time since ThT has the capability of binding to β-sheets or other amyloid-like structures, leading to a significant increase in its fluorescence intensity, which correlates with the degree of fibril formation.37–39
We used a wide range of concentrations of PSMα3 (0.0625–5 mg mL−1). The kinetics of PSMα3 aggregation was monitored over a period of 48 h, recording ThT fluorescence at 30-minute intervals using a microplate reader. The data of the ThT kinetics assay for all used concentrations of PSMα3 are presented in Fig. 3. As shown in Fig. 3, no increase in ThT fluorescence intensity was registered for all concentrations of PSMα3 after incubation up to 48 h at 37 °C.
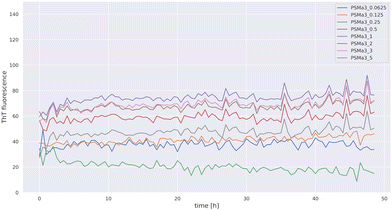 | ||
| Fig. 3 Time-dependent ThT fluorescence of PSMα3 incubated for 48 h at 37 °C in the concentration range from 5 to 0.0625 mg mL−1. | ||
We also studied PSMα3 aggregation behaviour with AFM and found that the results of AFM analysis clearly proved the data of the kinetics assay (Fig. 4). In particular, in the case where the same experimental procedure was applied prior to incubation (the same plates, pre-treatment procedure, and parameters of incubation), no aggregates were observed up to 72 h of incubation at 37 °C (Fig. 4A), and only a few oligomeric species were detected after 7 days of incubation (Fig. 4B).
We also evaluated if the monomerization procedure essentially influences PSMα3 aggregation potential under the studied experimental conditions. The AFM images of PSMα3, prepared without treatment with monomerizing agents before and after incubation, are presented in Fig. S5 in the ESI.† We found that without prior monomerization (Fig. S5A, ESI†), after incubation for 72 h under identical conditions, only a few small oligomeric structures were formed (Fig. S5B, ESI†). According to our results, under certain experimental conditions, PSMα3 does not form any amyloid aggregates and maintains a soluble α-helical conformation up to 72 h. Additionally, we evaluated how changes in the experimental procedure could modify the PSMα3 self-assembly process. We conducted incubation of monomerized PSMα3 simultaneously in small tubes and in plates. In the first case, we prepared the peptide by dissolving it in DMSO to a concentration of 10 mg mL−1 and then diluting the stock solution with water; in another case, the monomerized peptide was directly dissolved in water. The parameters of incubation (temperature, shaking) were the same for all the cases. After incubation for 72 h, we checked the assembly states of the studied samples using AFM analysis. The AFM images of PSMα3 incubated under different experimental conditions are presented in Fig. S6 in the ESI.† After 72 h of incubation in tubes, a few heterogeneous oligomers were detected. Moreover, solubilization of PSMα3 by DMSO resulted in the formation of bigger oligomer aggregates when compared to dissolving PSMα3 directly in water (Fig. S6A and B, ESI†). At the same time, in the case of dissolving the peptide first in DMSO, fewer oligomers were formed after 72 h of incubation in 96-well plates, and no aggregates could be detected in the case of using only water as the solvent (Fig. S6C and D, ESI†). However, it should be noted that no fibrils were ever found up to 72 hours after incubating PSMα3 under the same experimental conditions.
PSMα3 inhibits the aggregation of insulin
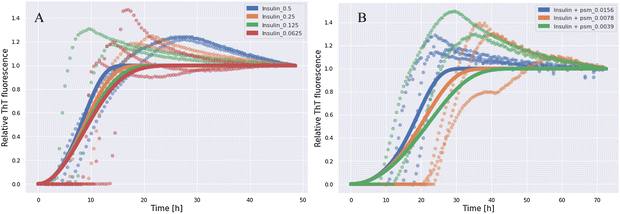
We studied the effect of co-incubation of PSMα3 and human insulin on the aggregation propensity of each other under certain experimental conditions in different concentration ranges. To exclude any influence of preformed aggregates, which can sometimes exist as impurities together with the insulin monomeric protein, both peptides were monomerized with the HFIP/TFA mixture prior to aggregation, according to the previously described protocol.1,14 The monomeric state of the peptides before aggregation was proven using atomic force microscopy (AFM). The AFM images of monomeric PSMα3 and insulin are presented in Fig. S4 in the ESI.† The possible influence of the peptides on the aggregation of each other was examined using the aggregation kinetics assay with ThT. The results of the ThT kinetics assay for insulin incubated alone at different concentrations are presented in Fig. 5A. The ThT kinetics data for insulin at 0.5 mg mL−1 co-incubated with PSMα3 in different concentrations are illustrated in Fig. 5B.As shown in Fig. 5A, a significant ThT signal increase was registered for insulin incubated at concentrations of 0.5–0.0625 mg mL−1, which gradually rose with time. According to the results of the ThT fluorescence assay, the formation of insulin aggregates was completely inhibited in the case of insulin/PSMα3 concentration ratios ranging from 0.5/0.5 mg mL−1 to 0.5/0.0325 mg mL−1. Partial inhibition occurs at the insulin/PSMα3 ratios of 0.5/0.0156 mg mL−1 and 0.5/0.0078 mg mL−1, whereas no inhibition was shown at the insulin/PSMα3 ratio of 0.5/0.0039 mg mL−1 (Fig. 5B). The results of the ThT fluorescence kinetics assay for insulin co-incubated at 0.5 mg mL−1 with PSMα3 at concentrations ranging from 0.0625 mg mL−1 to 0.0039 mg mL−1 are presented in Fig. S7 in the ESI.† The results suggest the existence of a PSMα3 concentration threshold for inhibition of the insulin aggregation. Interestingly, there is low dependence of half-time on the concentration in log(concentration)–log(half-time) plots presented in Fig. S8 (ESI†) for both insulin and insulin with the addition of PSMα3, suggesting that once the aggregation is induced it seems to follow the same scheme without saturation mechanisms. These observations suggest the critical role of secondary processes during the aggregation in both cases. Indeed, insulin kinetics best fits a “fragmentation dominated” model, the mean residual error, MRE = 0.0609, according to Amylofit software. The estimated model parameters were the following: k + kn = 0.109 (elongation rate constant × primary nucleation constant), nc = 0.166 (nucleus size), k + k− = 34.5 (elongation rate constant × fragmentation rate constant). Our data are consistent with other studies, where this model suggested the formation of secondary nuclei and fibril fragmentation and branching, as described earlier.22,40 The addition of PSMα3 does not change the type of the model and still the best fit is obtained for the “fragmentation dominated” model (MRE = 0.0569; the values of the model parameters were k + kn = 0.000292, nc = 0.00000658, and k + k− = 2.2 × 105). The decrease in parameter values associated with primary nucleation and increase in fragmentation coefficient were observed, in comparison to aggregation processes without PSMα3. This suggests that PSMα3 may be capable of inhibiting the formation of primary nuclei.
The data from the AFM analysis are strongly consistent with the findings of the kinetics aggregation assay. AFM images of insulin incubated alone at 0.5 mg mL−1 and co-incubated with PSMα3 in different concentrations, in which full inhibition, partial inhibition, or no inhibition was observed, are illustrated in Fig. 6. With regard to the co-incubation of insulin and PSMα3, we specifically observed almost complete inhibition of the aggregation process in the ratios of insulin/PSMα3 from 0.5/0.5 mg mL−1 to 0.5/0.031 mg mL−1, partial inhibition in the ratios between 0.5/0.01625 mg mL−1 and 0.5/0.0078 mg mL−1, and no inhibition in the ratio between 0.5/0.0039 (Fig. 6). AFM images of insulin co-incubated with PSMα3 in all used concentrations can be found in the ESI† (Fig. S9).
 | ||
| Fig. 6 AFM images of insulin incubated for 72 h at 37 °C alone at 0.5 mg mL−1 (A), and with PSMα3 at 0.031 mg mL−1 (B), at 0.0156 mg mL−1 (C), at 0.0078 mg mL−1 (D), and 0.0039 mg mL−1 (E). | ||
We analysed the FTIR spectra of insulin (0.5 mg mL−1) before and after incubation for 72 h at 37 °C, alone or with PSMα3 at different concentrations, where the highest (near full suppression of insulin aggregation) inhibitory effect was observed using the kinetics assay and microscopic analysis (0.031 mg mL−1). The obtained FTIR spectra are presented in Fig. 7.
Raw ATR-FTIR spectra of insulin before and after incubation, alone and with PSMα3, are presented in the ESI† (Fig. S10). We also performed deconvolution of the obtained FTIR spectra in the amide I region (1600–1700 cm−1) to analyse the secondary structure fractional distribution for the studied proteins in the case of insulin incubation alone or co-incubated with PSMα3. The results of the ATR-FTIR spectra deconvolution analysis are listed in Table 2.
| Peptide sample | Average peak position (cm−1) | Secondary structure assignment | Average composition (%) |
|---|---|---|---|
| Insulin (0.5 mg mL−1) freshly dissolved | 1630 | β-Sheets | 15 |
| 1648 | Unordered/random coil | 38 | |
| 1656 | α-Helix | 38 | |
| 1674 | β-Turn | 9 | |
| Insulin (0.5 mg mL−1) after incubation | 1629 | β-Sheets | 50 |
| 1644 | Unordered/random coil | 27 | |
| 1663 | β-Turn | 13 | |
| 1673 | β-Sheet/β-turn | 10 | |
| Insulin (0.5 mg mL−1) after incubation with PSMα3 at 0.031 mg mL−1 | 1624 | β-Sheets | 8 |
| 1646 | Unordered/random coil | 44 | |
| 1658 | α-Helix | 38 | |
| 1677 | β-Sheet/β-turn | 10 | |
As shown in Fig. 7A, in the FTIR spectrum of freshly dissolved insulin, the main absorption band in the amide I region is displayed at 1652 cm−1. This band, after deconvolution analysis (Table 2), was shown to consist of overlapped peaks at 1656 cm−1 and at 1648 cm−1 due to the co-existence of α-helices and disordered/random coil conformations.31,41 Our data are consistent with the previously described conformational characteristics of a native monomeric insulin in aqueous solutions with a pH close to neutral.42–45 Then, the structural transformation of insulin during incubation for 72 h at 37 °C to β-sheet-enriched structures is easily noticeable from the spectrum presented in Fig. 7B due to an intensive absorption band at 1631 cm−1. Deconvolution analysis demonstrated that the secondary structure of insulin after incubation for 72 h at 37 °C is mostly (>50%) presented in aggregated species (Table 2). This is confirmed by the main peak in the amide I region at 1629 cm−1, assigned to β-sheets, with simultaneous full loss of the α-helical fraction and a considerable decrease in the content of the random coil conformation when compared to the native α-helical/random conformation (Fig. 7A and Table 2). As shown in Fig. 7A, in the FTIR spectrum of insulin co-incubated with PSMα3, the main absorption band in the amide I region was observed at 1652 cm−1 assigned to overlapping peaks of α-helix and unordered structures.41,44 Deconvolution analysis (Table 2) demonstrated that the primary conformation state for insulin co-incubated with PSMα3 was represented in a random coil suggested by a major peak at 1646 cm−1 with less contribution of α-helix due to the obtained peak at 1658 cm−1, indicating a pronounced inhibitory effect of PSMα3 on insulin transformation into β-enriched aggregates.
Modelling
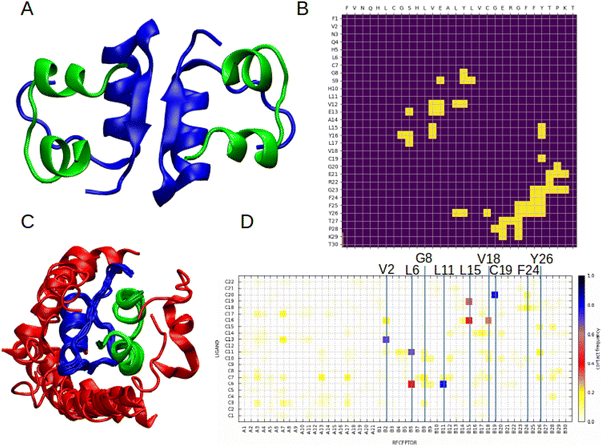
To better understand the mechanism of interactions between PSMα3 and insulin, we performed molecular modelling of the complex. Based on the results of ThT assays and spectroscopic analysis, it is most likely that PSM interferes with the early stages of insulin aggregation. Therefore, we first modelled the structure of the human insulin dimer using AlphaFold246 (Fig. 8A). The obtained model is consistent with the experimental structure obtained for engineered insulin mutants, as well as with a detailed molecular study indicating chain B as more involved in stabilising weak inter-chain interactions.86,87 Based on the model dimer structure we calculated contact maps for a pair of interacting insulin B chains (Fig. 8B) in order to identify which amino acids are likely to take part in the interaction. In the next step, we performed molecular docking to an experimental structure of the insulin monomer using the CABSdock server.47 A majority of the best-scoring conformations regarding binding of PSMα3 to insulin occurred on chain B (Fig. 8C). We also analysed the frequency of residue–residue contacts between insulin and PSMα3 in the top 1000 obtained models (Fig. 8D). The most numerous interactions can be observed for V2, L6, G8, L11, L15, V18, C19, F24, and Y26 in chain B of insulin. The majority of them are located in the region between positions 8–29, involved in the formation of insulin dimers. This includes the fragment F24-Y26 previously identified as significantly contributing to the insulin dimerisation.87 Also, some of these residues were recently identified as important for stabilising interactions of both insulin chains in the flat conformation observed in the cryo-EM structure of the full length amyloid fibril.88 This includes cysteine 19 in chain B, which forms a disulphide bridge and interacts with alanine 18 in chain A in the filament structure, and also has one of the highest contact frequencies with PSMα3. Another important residue is leucine 15 in chain B, which is located in a hydrophobic patch, stabilising both chains in the flat conformation. The same study highlights the importance of phenylalanine 24 and tyrosine B26, which becomes more exposed in fibrillar confirmation and likely promotes lateral inter-protofilament interactions.88 Visualisation of the contacts is presented in Fig. 9 and Fig. S11 (ESI†).These results suggest that PSMα3 could compete for binding sites with insulin monomers, therefore inhibiting the formation of insulin oligomers, as well as hampering conformational changes leading to fibril formation. The result is in good agreement with the kinetic data, suggesting inhibition of primary nucleus formation. This mechanism can also explain the decrease in β-sheets revealed by ATR-FTIR analysis, and as such structures appear upon the formation of oligomers and aggregates.
Discussion
In the presented work, we studied the aggregation characteristics of the PSMα3 peptide and its possible structural transformation during incubation under selected experimental conditions using different experimental methods widely used to study the process of amyloid aggregation.48 According to the data obtained simultaneously using several experimental techniques, such as the fluorescence-based kinetics assay, microscopic analysis (AFM), CD spectroscopy, and ATR-FTIR, PSMα3 does not form any amyloid-like aggregates in aqueous solutions under the used experimental conditions and after incubation for up to several days. The results of CD and ATR-FTIR analysis conclusively demonstrated that the secondary structure of the investigated PSMα3 did not significantly change during incubation, and the peptide maintained a primarily α-helical conformation with only minor changes in fraction distribution. This further indicates that the helical structure was stabilised after incubation, which is consistent with the previously described findings,1,6 demonstrating that PSMα3 maintains an α-helical conformation for up to several days of incubation.1,6 Moreover, equilibrium between α-helical/β-sheets transitions in aqueous solutions with a prevalence of α-helical conformation is typical of PSMα3 in vitro conditions.1,6,18,19 Deconvolution of ATR-FTIR spectra revealed a significant β-turn contribution that is retained for the peptide after incubation. This correlated with previous findings indicating substantial roles of β-turns in stabilising the native conformation of proteins with a high predisposition to the formation of a stable secondary structure49,50 and proved that PSMα3 has a tendency to stabilise its α-helical conformation following the incubation. The high β-turn contribution is determined by the presence of typical turn formers in the PSMα3 sequence, such as Asp and Asn amino acids,51–53 which arise for physical reasons (e.g., hydrogen bonding). Moreover, FTIR spectra analysis indicates that β-sheet components marginally decrease after incubation with a simultaneous slight increase in α-helical fraction and the maintenance of a high β-turn content that is consistent with inhibition of protein aggregation and stabilisation of helical conformation, according to previously described analysis of the roles of β-turns in protein folding.49,50According to the ThT kinetics assay, for PSMα3 in a wide range of concentrations, no changes in ThT fluorescence intensity were manifested following incubation for up to 25 days. The kinetics aggregation data were clearly proven by AFM analysis. Our findings are consistent with another study where PSMα3 was studied with a combination of methods of fluorescence staining and high-resolution imaging techniques and demonstrated to not form any amyloid aggregates after several days of incubation.9 At the same time, a few other studies demonstrated that the PSMα3 peptide produces amyloid fibrils in vitro after several hours of incubation.1,10 Crucially, nevertheless, all these experiments were conducted on recombinant N-terminally modified (N-formylated) PSMα3.1,10 N-Terminal formylation was found to have a significant impact on peptide aggregation properties.11 In particular, the N-terminally formylated δ-toxin, which is the most structurally similar to PSMα3, was shown to self-assemble into amyloid fibrils, while the deformylated peptide forms functional oligomeric complexes.11 Moreover, N-terminus formylation is likely responsible for PSMα3 contribution to in vivo PSM biofilm formation as well as playing an important role in high PSMα3 toxicity since PSMs usually retain their N-terminal formyl groups during secretion in bacteria,12,13 in contrast to synthetic peptides, where formylation can be conducted only as an additional stage if required for the objectives of the experiment.54 It is well known that initial steps in experimental procedures, such as pre-treatment and solubilization of peptides prior to aggregation assays, are critical to control the process of peptide self-assembly.10 Therefore, we applied different experimental conditions for the aggregation analysis of the studied PSMα3. In particular, we incubated the peptides in various ways (on plates or in Eppendorf tubes), did not apply monomerization before the aggregation assay, and also solubilized the peptide using DMSO with a subsequent dilution in water or just by dissolving it in water. We discovered that using an experimental setup that forgoes monomerization steps in addition to previously solubilized PSMα3 in DMSO slightly modifies its aggregation behaviour and results in faster formation of small oligomers detected by AFM (Fig. S6, ESI†). Our results are in agreement with other experimental investigations, which showed that faster fibrillation happens in the absence of monomerization because there may be some pre-formed assemblies present that might seed the aggregation and speed up the fibril growth.10,14,55 Additionally, in the case of using DMSO as a prior solvent, the chemical reaction of disulfide bond formation, which plays a significant role in peptide self-assembly and can accelerate the fibrillation process, has been previously discussed in several papers.56–59 Furthermore, DMSO was used as the first solvent in the majority of studies on protein aggregation in vitro, including the study on the fibrillation propensity of PSMα3.57,60,61 However, despite the fact that the aggregation kinetics of PSMα3 depends on experimental procedure manipulation, in the current study, PSMα3 was not found to possess the ability to self-assemble into amyloid fibrils after incubation for up to 7 days, and only a few spherical oligomers were formed after incubation for 72 h without pre-treatment for monomerization and solubilization with DMSO. Since the peptide aggregation process in vitro has been frequently demonstrated to require a significant amount of time (up to several weeks) for monomeric peptides to self-assemble into mature amyloid structures, it is likely that in our case, the incubation time was insufficient for fibril formation under the experimental conditions used.1,9,62 PSM peptides are known to cross-interact with other amyloidogenic proteins, resulting in regulation of each other's fibrillation, which can appear as both aggregation acceleration and inhibition.20,21,63 Despite this fact, the exact molecular mechanisms of such interactions remain elusive. In this work we assessed the effect of PSMα3 on human insulin fibril formation under co-incubation in a wide concentration range and using a variety of experimental techniques in combination with computational analysis. In our study, we found that PSMα3 possessed a pronounced concentration-dependent inhibitory effect on insulin aggregation during co-incubation under the experimental conditions used here. The obtained results are clearly illustrated using the data from the ThT kinetics assay and confirmed by the AFM analysis. Insulin with and without the addition of PSMα3 follows a fragmentation model, though the mix has a smaller size of predicted primary nuclei as well as a higher fragmentation rate, according to results obtained with Amylofit. The effect on primary nucleation might be caused by competition for binding sites between PSMα3 and insulin, as revealed by our computational molecular study. PSMα3 tends to bind to the same residues that participate in the formation of insulin dimers and, therefore, can inhibit aggregation by hampering the formation of primary nuclei. These findings might also explain the observed inhibition of β-sheet formation, as the discussed region forms β-sheets upon dimerization.77 The ATR-FTIR analysis data is consistent with the results obtained by other experimental methods as well as computational molecular studies. The obtained ATR-FTIR spectra clearly demonstrated insulin transformation over time from an α-helical/random coil structure to a predominantly aggregated species. In contrast, in the case of co-incubation of insulin with PSMα3, the dominant secondary structure is presented in α-helix/random coil conformation with a considerably lower content of aggregated species (Fig. 7 and Table 2), demonstrating the prevention of insulin transition into amyloid under PSMα3 influence. Our findings strongly correlated with the study of Santos et al.,21 which demonstrated a considerable inhibitory effect of PSMα3-like peptide on the fibrillation of α-syn in nanomolar concentration ratios. Moreover, in this work, the crucial role of the generic hydrophobic character of a highly stable PSMαs-like α-helical face scaffold in anti-aggregation effects was demonstrated.21 In our case, we also demonstrated that PSMα3 maintains α-helical conformation up to several days of incubation at 37 °C, wherein α-helical-designed peptides were shown before to suppress amyloid aggregation with high efficiency, including both bacterial and mammalian peptides and proteins.64–67 This is probably determined by the fact that the formation of intermediate cross-α-sheet-like structures is a universally adopted amyloidogenic motif during the amyloid self-assembly process, which has been shown to be associated with high toxicity and demonstrated before for the different pathogenic amyloids, such as Aβ, α-syn, transferrin, amylin, and others.64,68–71 In particular, these kinds of transient oligomer species, composed primarily of α-sheets, were also described before for human insulin.72 It was also recently demonstrated that the mechanism of inhibition of insulin fibrillation by short α-helical peptides is based on hydrogen bond formation with amino acid residues in both insulin chains that interfere with the insulin conformational transition from α-helix to β-sheet.22,73 Several findings demonstrated that α-helical or cross-α-sheets scaffolds, due to their morphological similarity to conventional β-sheets but at the same time higher plasticity through the alignment of main chain carbonyl groups on one side of the α-strand and NH-groups on the other, preferentially tend to bind intermediate oligomer species, which are usually enriched in α-sheets, interfering with further transformation into mature cross-β-fibrils.67,74–76
Conclusions
In the current study, we have demonstrated that under the experimental conditions used in the presented work and in a wide range of concentrations, synthetic PSMα3 does not form amyloid fibrils and maintains stable α-helical conformation in soluble form up to several days of incubation. Nevertheless, manipulations of the test procedure, such as monomerization, contact material, pre-treatment, and dissolving process, noticeably affect the aggregation properties of PSMα3. We have also shown that co-incubation of PSMα3 with human insulin leads to significant inhibition of insulin fibrillation. The anti-fibrillation effect of PSMα3 is concentration-dependent and starts with the concentration ratio of PSMα3: insulin equal to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. PSMα3 apparently blocks the formation of primary nuclei by insulin by competing for residues involved in dimerization. Overall, the results of our study demonstrated the great potential of PSMα3 from S. aureus as an inhibitor of human insulin fibrillation. This expands the potential applications of bacterial α-helical peptides and might help with the creation of novel therapeutic strategies for insulin-derived amyloidosis.
100. PSMα3 apparently blocks the formation of primary nuclei by insulin by competing for residues involved in dimerization. Overall, the results of our study demonstrated the great potential of PSMα3 from S. aureus as an inhibitor of human insulin fibrillation. This expands the potential applications of bacterial α-helical peptides and might help with the creation of novel therapeutic strategies for insulin-derived amyloidosis.
Materials and methods
Peptide synthesis and purification
All commercially available reagents and solvents were purchased from Sigma-Aldrich and Merck, and used without further purification. The studied peptide PSMα3 was obtained using an automated solid-phase peptide synthesiser (Liberty Blue, CEM) with microwave heating by applying Fmoc/tBu chemistry and using H-Rink amide ChemMatrix resin with 35–100 mesh particle size and 0.59 mmol g−1 loading. Fmoc deprotection was performed using 20% piperidine in DMF for 1 min at 90 °C. A single-coupling was achieved with a 0.5 M solution of N,N′-diisopropylcarbodiimide (DIC) and a 0.5 M solution of OXYMA Pure Novabiochem® (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in DMF for 4 min at 90 °C. The cleavage of the peptide from the resin was carried out with a mixture of TFA, TIS, and H2O (95
1) in DMF for 4 min at 90 °C. The cleavage of the peptide from the resin was carried out with a mixture of TFA, TIS, and H2O (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) after 3 h of shaking. The crude peptide was precipitated with ice-cold Et2O and centrifuged (8000 rpm, 10 min, 4 °C). The peptide was purified using preparative RP-HPLC (Knauer AZURA ASM 2.1 L) with a C18 column (Thermo Scientific, Hypersil Gold 12 μm, 250 mm × 20 mm) with a water/acetonitrile (0.05% TFA) eluent system. The purified peptide fractions were lyophilized, aliquoted to 1–5 mg, and stored at −20 °C prior to use.
2.5) after 3 h of shaking. The crude peptide was precipitated with ice-cold Et2O and centrifuged (8000 rpm, 10 min, 4 °C). The peptide was purified using preparative RP-HPLC (Knauer AZURA ASM 2.1 L) with a C18 column (Thermo Scientific, Hypersil Gold 12 μm, 250 mm × 20 mm) with a water/acetonitrile (0.05% TFA) eluent system. The purified peptide fractions were lyophilized, aliquoted to 1–5 mg, and stored at −20 °C prior to use.
Insulin
Insulin human (recombinant, yeast) was purchased from Sigma Aldrich (CAS Number 11061-68-0, MW 5807.57 g mol−1) with a purity >98%. All the analytical data were provided by the producer.Peptide pre-treatment
Lyophilized peptides were dissolved to a final peptide concentration of 1 mM in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP),1,14 sonicated for 10 min, and left overnight to evaporate at room temperature. Dried peptide stocks were stored at −20 °C prior to use. Before assays, peptide aliquots were re-suspended in pure Milli-Q water (resistivity of 18.2 MΩ cm at 25 °C).
1 mixture of trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP),1,14 sonicated for 10 min, and left overnight to evaporate at room temperature. Dried peptide stocks were stored at −20 °C prior to use. Before assays, peptide aliquots were re-suspended in pure Milli-Q water (resistivity of 18.2 MΩ cm at 25 °C).
Analytical high-performance liquid chromatography (HPLC)
Analytical RP-HPLC of PSMα3 was performed using the Shimadzu System and CHROMSERVIS® CromShell® C18-XB, 2.6 μm, 75 × 4.6 mm column. Programme (eluent A: 0.05% TFA in H2O, eluent B: 0.05% TFA in acetonitrile, flow 0.9 mL min−1): A: t = 0 min, 90% A; t = 30 min, 10% A.Mass spectrometry (MS)
The PSMα3 peptide was studied using the WATERS LCT Premier XE System, consisting of a high-resolution mass spectrometer with time of flight (TOF) using electrospray ionisation (ESI).Circular dichroism (CD)
CD spectra were recorded on the JASCO J-815 at 20 °C between 260 and 190 nm in pure Milli-Q water with the following parameters: 0.2 nm resolution, 1.0 nm bandwidth, 20 mdeg sensitivity, 0.25 s responses, and 50 nm min−1 scanning speed, 5 scans, and 0.02 cm cuvette path length. The CD spectra of the solvents alone were recorded and subtracted from the raw data. Typically, the samples were prepared by dilution of the peptide stock solution in Milli-Q water to obtain a peptide concentration of around 0.5 mg mL−1. The CD intensity is given as mean residue ellipticity (θ [deg cm2 dmol−1]) calculated using the equation: θ = MθMRE/10cln, where θ = mean residue ellipticity; θMRE = ellipticity; c = concentration; l = path length; n = number of residues.Fourier-transform infrared (FTIR) spectroscopy
All spectra were collected using a Nicolet 6700 FTIR spectrometer (Thermo Scientific, USA) with an attenuated total reflectance (ATR) accessory and heated diamond top plate (PIKE Technologies), continuously purged with dry air. Each sample of 10 μL of peptide aqueous solution was dropped directly on the diamond surface and allowed to dry out. Spectra were obtained in the range of 3600–400 cm−1. For each spectrum, 128 interferograms were co-added with 4 cm−1 resolution at a constant temperature of 22 °C. Directly before sampling, the background spectrum of diamond/air was recorded as a reference (512 scans, 4 cm−1). ATR-FTIR spectra were initially pre-processed with OMNIC 8.3.103 using automated atmospheric correction. The spectra were analysed using OriginPro 9.0 (OriginLab Corporation, USA). The analysis included baseline correction and smoothing using the Savitzky–Golay filter (polynomial order 2, points of window 20),78 normalisation of spectra to the absorbance of the amide I band, and deconvolution into subcomponents using the Gaussian or Lorentz function based on the minima of second derivative spectra with R-square (COD) > 0.988.In vitro aggregation kinetics studies with ThT
Peptide solutions were prepared in low-binding Eppendorf tubes (DNA LoBind® Tubes, Eppendorf). Aliquots of purified peptides were dissolved directly in MilliQ water containing 0.04 mM ThT or in DMSO to a concentration of 10 mg mL−1 and then diluted in water containing ThT to the desired concentrations. Aggregation kinetics for insulin was estimated in a concentration range of 0.5–0.0625 and for PSMα3 in a concentration range of 5–0.0625 mg mL−1. The effect of co-incubation of insulin with PSMα3 was evaluated by mixing the peptide solutions to the final insulin concentration of 0.5 mg mL−1 and different concentrations of PSMα3 from 0.5 mg mL−1 to 0.0039 mg mL−1. The PSMα3 concentration range was obtained by twofold serial dilution. A 96-well black plate (Brand plates, REF781608, polystyrene, medium binding, non-sterile) was used to read the ThT fluorescence emitted from the samples. Each well was loaded with 100 μL, and the plate was sealed with a polyester film (Excel Scientific, Seal plate film, 100-SEAL-PLT, non-sterile) to prevent evaporation. The ThT fluorescence was measured with three repeats for each sample. The plate was placed in a multi-mode microplate reader (CLARIO Star Plus plate reader, BMG Labtech) and incubated at 37 °C with constant shaking at 400 rpm. The fluorescence intensity signal was read from the top every 30 min up to 72 h with the excitation and emission wavelengths set to 440 and 480 nm, respectively. The half-time (t0.5) was estimated by taking the values half-way between the start and the end baseline. The background fluorescence intensity was automatically subtracted from those containing peptides. The data were representative of three independent experiments. Fitting of kinetics models was performed according to the protocol described in Meisl et al.79 4 different concentrations were considered for insulin (0.5, 0.25, 0.125, 0.0625) and 3 different concentrations for insulin with the addition of PSMα3 (0.0156, 0.0078, 0.0039) due to a lack of observed aggregation for higher concentrations of PSMα3 (0.0625 and 0.031).Kinetic analysis
In the first step, the relationship between half time and the monomer concentration was inspected according to the log(concentration)–log(half time) plot. Then, the slope of the relationship, gamma, was estimated using a linear regression fitter scipy.stats.linregress implemented in Python 3.9. In the next step, the kinetics data was uploaded to Amylofit software. The MRE (mean residual error) of all unseeded models without saturation was calculated and compared (basin hops = 3). The model with the lowest MRE was chosen as the final one. The fitting for the best model was repeated with basin hops = 7 and the results for this value are reported.Atomic force microscopy (AFM)
AFM images were collected using the Dimension V Veeco AFM instrument (Classone Equipment Inc, Decatur, GA, United States). The topography was measured in the tapping mode with the SS probe mounted. AFM imaging was performed according to a previously published protocol.80,81 In detail, AFM-grade mica in 10 mm by 10 mm pieces (Agar Scientific) and magnetised-stainless steel coin-like sample holders (14 mm diameter) were used. Approximately 25 μL of the sample was placed on a mica sheet, and after 2 minutes of adsorption, it was rinsed with Milli-Q water and dried. The resolution of the AFM scans was 512 × 512 pixels, with topographic image sizes of 10 × 10 μm. Images were collected several times across the mica surface. For each sample at least three images (4.2 μm × 4.2 μm) were analysed and processed using Gwyddion software (Czech Metrology Institute). The dimensions of the structures were calculated according to the three-dimensional profiles measured.Modelling
We started by modelling the structure of the insulin dimer using the Colabfold82 version of AlphaFold246 with the default parameters. For modelling the molecular docking of PSMα3 to the insulin structure (PDB: 5ENA), CABSdock server47 was used. The calculation of residue–residue contacts was performed using python scripts with NumPy83 and Matplotlib84 libraries. We defined residue–residue contacts as a pair of amino acids whose C-beta carbons (C-alpha in the case of glycine) were closer than 8 Angstroms in space, which is one of the classical definitions of “contacts” in bioinformatics. Calculations of contact frequencies were performed using the CABSdock server as the number of contacts between a given pair of residues divided by the total number of generated models. For visualisation of the molecular structures, VMD software was used.85Abbreviations
| AFM | Atomic force microscopy |
| ATR | Attenuated total reflectance |
| CD | Circular dichroism |
| DIC | N,N′-Diisopropylcarbodiimide |
| DMF | Dimethylformamide |
| DMSO | Dimethyl sulfoxide |
| FTIR | Fourier transform infrared spectroscopy |
| HFIP | Hexafluoroisopropanol |
| MS | Mass spectrometry |
| PSM | Phenol-soluble modulin |
| RP-HPLC | Reverse phase-high-performance liquid chromatography |
| TFA | Trifluoroacetic acid |
| ThT | Thioflavin T |
| TIS | Triisopropylsilane |
Data availability
Any additional information required to reanalyse the data reported in this paper is available upon request.Author contributions
Conceptualization, A. K. and M. K; methodology, A. K., M. S., M. G. G., J. W. and A. N.; investigation, A. K., M. S., M. G. G., A. N., and J. W.; writing, A. K., M. S., A. N., J. W; editing, A. K., M. S., M. G. G., A. N., J. O. B and M. K.; visualisation, A. K., A. N., and J. W.; software, A. N. and J. W.; supervision, M. K.; funding acquisition, M. K.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The work was financially supported by the National Science Centre, Poland, Grant No. 2019/35/B/NZ2/03997.References
- M. Zaman and M. Andreasen, eLife, 2020, 9, e59776 CrossRef CAS PubMed.
- H. Lade, et al. , Biomed Res. Int., 2022, 8221622 CAS.
- G. Y. Cheung, et al. , FASEB J, 2014, 28(1), 153 CrossRef CAS PubMed.
- M. Laabei, W. D. Jamieson, Y. Yang, J. Van Den Elsen and A. T. A. Jenkins, Biochim. Biophys. Acta, Biomembr., 2014, 1838(12), 3153–3161 CrossRef CAS PubMed.
- K. M. Towle, C. T. Lohans, M. Miskolzie, J. Z. Acedo, M. J. van Belkum and J. C. Vedera, Biochem., 2016, 55(34), 4798–4806 CrossRef CAS PubMed.
- E. Tayeb-Fligelman, N. Salinas, O. Tabachnikov and M. Landau, Struct., 2020, 28(3), 301–313 CrossRef CAS PubMed.
- E. Tayeb-Fligelman, et al. , Sci., 2017, 355(6327), 831–833 CrossRef PubMed.
- R. Malishev, E. Tayeb-Fligelman, S. David, M. Meijler, M. Landau and R. Jelinek, J. Mol. Bio., 2018, 430(10), 1431–1441 CrossRef CAS PubMed.
- P. Marinelli, I. Pallares, S. Navarro and S. Ventura, Sci. Rep., 2016, 6(1), 34552 CrossRef CAS PubMed.
- M. Zaman and M. Andreasen, Microorganisms, 2021, 9(1), 117 CrossRef CAS PubMed.
- X. Zhou, et al. , Virulence, 2021, 12(1), 1418–1437 CrossRef CAS PubMed.
- R. Wang, et al. , Nat. Med., 2007, 13(12), 1510–1514 CrossRef CAS PubMed.
- D. Mader, M. Liebeke, V. Winstel, K. Methling, M. Leibig, F. Götz and A. Peschel, BMC Microbiol., 2013, 13, 1–9 CrossRef PubMed.
- W. B. Stine, L. Jungbauer, C. Yu and M. J. LaDu, Methods Mol. Biol., 2013, 13–32 Search PubMed.
- K. Schwartz, M. Ganesan, D. E. Payne, M. J. Solomon and B. R. Boles, Mol. Microbiol., 2016, 99(1), 123–134 CrossRef CAS PubMed.
- A. Taglialegna, I. Lasa and J. Valle, J. Bacteriol., 2016, 198(19), 2579–2588 CrossRef CAS PubMed.
- H. Lade, J. H. Park, S. H. Chung, I. H. Kim, J. M. Kim, H. S. Joo and J. S. Kim, J. Clin. Med., 2019, 8(11), 1853 CrossRef CAS PubMed.
- O. M. Cracchiolo, D. N. Edun, V. M. Betti, J. M. Goldberg and A. L. Serrano, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(5), e2114923119 CrossRef CAS PubMed.
- N. Salinas, J. P. Colletier, A. Moshe and M. Landau, Nat. Commun., 2018, 9(1), 1–9 CrossRef CAS PubMed.
- C. Haikal, et al. , Int. J. Mol. Sci., 2021, 22(21), 11594 CrossRef CAS PubMed.
- J. Santos, P. Gracia, S. Navarro, S. Peña-Díaz, J. Pujols, N. Cremades, I. Pallarès and S. Ventura, Nat. Commun., 2021, 12(1), 3752 CrossRef CAS PubMed.
- B. Rosetti and S. Marchesan, Int. J. Mol. Sci., 2023, 24(2), 1306 CrossRef CAS PubMed.
- C. Samlaska, S. Reber and T. Murry, JAADCR, 2020, 6(4), 351–353 Search PubMed.
- J. He, E. Renard, P. Lord, D. Cohen, E. Cem-Duranty, J. Place and D. J. Burgess, JCR, 2021, 336, 1–15 CAS.
- M. A. Gertz and A. Dispenzieri, JAMA, 2020, 324(1), 79–89 CrossRef CAS PubMed.
- M. Szefczyk, N. Szulc, M. Gąsior-Głogowska, A. Modrak-Wójcik, A. Bzowska, W. Majstrzyk and Ł. Berlicki, Nanoscale, 2021, 13(7), 4000–4015 RSC.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89(5), 392–400 CrossRef CAS PubMed.
- N. E. Zhou, C. M. Kay and R. S. Hodges, JBC, 1992, 267(4), 2664–2670 CrossRef CAS.
- M. Szefczyk, E. Węglarz-Tomczak, P. Fortuna, A. Krzysztoń, E. Rudzińska-Szostak and Ł. Berlicki, Angew. Chem., Int. Ed., 2017, 56(8), 2087–2091 CrossRef CAS PubMed.
- K. M. Towle, C. T. Lohans, M. Miskolzie, J. Z. Acedo, M. J. van Belkum and J. C. Vederas, Biochem., 2016, 55(34), 4798–4806 CrossRef CAS PubMed.
- J. Kong and S. Yu, ABBS, 2007, 39(8), 549–559 CrossRef CAS PubMed.
- H. Li, R. Lantz and D. Du, Molecules, 2019, 24(1), 186 CrossRef PubMed.
- B. R. Singh, Infrared analysis of peptides and proteins, ACS, 2000 Search PubMed.
- A. Adochitei and G. Drochioiu, Rev. Roum. Chim., 2011, 56(8), 783–791 CAS.
- V. Adochitei, W. Liu and S. Franzen, Biophys. J., 2007, 93(7), 2429–2435 CrossRef PubMed.
- E. Vass, M. Hollósi, F. Besson and R. Buchet, Chem. Rev., 2003, 103(5), 1917–1954 CrossRef CAS PubMed.
- R. Khurana, C. Coleman, C. Ionescu-Zanetti, S. A. Carter, V. Krishna, R. K. Grover and S. Singh, J. Struct. Biol., 2005, 151(3), 229–238 CrossRef CAS PubMed.
- M. Groenning, M. Norrman, J. M. Flink, M. van de Weert, J. T. Bukrinsky, G. Schluckebier and S. Frokjaer, J. Struct. Biol., 2007, 159(3), 483–497 CrossRef CAS PubMed.
- K. Gade Malmos, L. M. Blancas-Mejia, B. Weber, J. Buchner, M. Ramirez-Alvarado, H. Naiki and D. Otzen, Amyloid, 2017, 24(1), 1–16 CrossRef CAS PubMed.
- M. H. Fagihi and G. Bhattacharjee, ACS Pharmacol. Transl. Sci., 2022, 5(11), 1050–1061 CrossRef CAS PubMed.
- M. Grechko and M. T. Zanni, J. Chem. Phys., 2012, 137(18), 184202 CrossRef PubMed.
- M. Bardhan, S. Dolui, S. Chaudhuri, U. Paul, G. Bhattacharjee, M. Ghosal and D. Senapati, RSC advances, 2021, 11(6), 3354–3362 RSC.
- F. Mastrotto, F. Bellato, V. Andretto, A. Malfanti, M. Garofalo, S. Salmaso and P. Caliceti, J. Pharm. Sci., 2020, 109(1), 900–910 CrossRef CAS PubMed.
- S. Delbeck and H. M. Heise, JDST, 2021, 15(4), 865–873 CAS.
- S. Delbeck and H. M. Heise, Biomedical Vibrational Spectroscopy 2020: Advances in Research and Industry, SPIE, 2020, 11236, pp. 53–61 Search PubMed.
- J. Jumper, et al. , Nature, 2021, 596(7873), 583–589 CrossRef CAS PubMed.
- M. Kurcinski, M. Jamroz, M. Blaszczyk, A. Kolinski and S. Kmiecik, Nucleic Acids Res., 2015, 43(W1), W419–W424 CrossRef CAS PubMed.
- M. R. Nilsson, Methods, 2004, 34(1), 151–160 CrossRef CAS PubMed.
- S. Deike, S. Rothemund, B. Voigt, S. Samantray, B. Strodel and W. H. Binder, Bioorg. Chem., 2020, 101, 104012 CrossRef CAS PubMed.
- A. M. C. Marcelino and L. M. Gierasch, Biopolymers, 2008, 89(5), 380–391 CrossRef CAS PubMed.
- G. D. Rose, L. M. Glerasch and J. A. Smith, Adv. Protein Chem., 1985, 37, 1–109 CrossRef CAS PubMed.
- P. Y. Chou and G. D. Fasman, Biophys. J., 1979, 26(3), 367–383 CrossRef CAS PubMed.
- J. A. Smith, L. G. Pease and K. D. Kopple, Crit. Rev. Biochem., 1980, 8(4), 315–399 CrossRef CAS PubMed.
- A. L. Tornesello, M. Sanseverino and F. M. Buonaguro, Molecules, 2016, 21(6), 736 CrossRef PubMed.
- H. Chung, E. J. Crooks, M. Ziliox and S. O. Smith, Methods Mol. Biol., 2018, 321–330 CrossRef CAS PubMed.
- J. P. Tam, C. R. Wu, W. Liu and J. W. Zhang, JACS, 1991, 113(17), 6657–6662 CrossRef CAS.
- A. Tjernberg, N. Markova, W. J. Griffiths and D. Hallén, SLAS Discovery, 2006, 11(2), 131–137 CrossRef CAS PubMed.
- A. Giugliarelli, L. Urbanelli, M. Ricci, M. Paolantoni, C. Emiliani, R. Saccardi and P. Sassi, J. Phys. Chem. A, 2016, 120(27), 5065–5070 CrossRef CAS PubMed.
- S. Nishinami, A. Hirano, T. Arakawa and K. Shiraki, Int. J. Biol. Macromol., 2018, 119, 180–185 CrossRef CAS PubMed.
- C. L. Shen and R. M. Murphy, Biophys. J., 1995, 69(2), 640–651 CrossRef CAS PubMed.
- G. Houen, et al. , Acta Chem. Scand., 1996, 50(1), 68–70 CrossRef CAS.
- G. A. Siddiqui and A. Naeem, Int. J. Biol. Macromol., 2018, 108, 360–366 CrossRef CAS PubMed.
- N. Nair, R. Biswas, F. Götz and L. Biswas, Infect. Immun., 2014, 82(6), 2162–2169 CrossRef PubMed.
- T. Prosswimmer and V. Daggett, Open Biol., 2022, 12(11), 220261 CrossRef CAS PubMed.
- A. Bleem, T. Prosswimmer, R. Chen, T. F. Hady, J. Li, J. D. Bryers and V. Daggett, Sci. Rep., 2023, 13(1), 9272 CrossRef CAS PubMed.
- N. Paranjapye and V. Daggett, J. Mol. Biol., 2018, 430(20), 3764–3773 CrossRef CAS PubMed.
- A. Bleem, R. Francisco, J. D. Bryers and V. Daggett, npj Biofilms Microbiomes, 2017, 3(1), 16 CrossRef PubMed.
- D. Shea, C. C. Hsu, T. M. Bi, N. Paranjapye, M. C. Childers, J. Cochran and V. Daggett, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(18), 8895–8900 CrossRef CAS PubMed.
- R. S. Armen, M. L. DeMarco, D. O. Alonso and V. Daggett, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(32), 11622–11627 CrossRef CAS PubMed.
- R. S. Armen, D. O. Alonso and V. Daggett, Structure, 2004, 12(10), 1847–1863 CrossRef CAS PubMed.
- V. Daggett, Acc. Chem. Res., 2006, 39(9), 594–602 CrossRef CAS PubMed.
- R. Kayed, E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman and C. G. Glabe, Science, 2003, 300(5618), 486–489 CrossRef CAS PubMed.
- M. K. Siddiqi, et al. , Int. J. Biol. Macromol., 2020, 143, 102–111 CrossRef CAS PubMed.
- N. L. Maris, D. Shea, A. Bleem, J. D. Bryers and V. Daggett, Biochemistry, 2018, 57(5), 507–510 CrossRef CAS PubMed.
- T. M. Bi and V. Daggett, YJBM, 2018, 91(3), 247 CAS.
- G. Hopping, J. Kellock, R. P. Barnwal, P. Law, J. Bryers, G. Varani and V. Daggett, eLife, 2014, 3, e01681 CrossRef PubMed.
- T. C. Michaels, D. Qian, A. Šarić, M. Vendruscolo, S. Linse and T. P. Knowles, Nat. Rev. Phys., 2023, 5(7), 379–397 CrossRef CAS.
- A. Savitzky and M. J. Golay, J. Anal. Chem., 1964, 36(8), 1627–1639 CAS.
- G. Meisl, J. B. Kirkegaard, P. Arosio, T. C. Michaels, M. Vendruscolo, C. M. Dobson and T. P. Knowles, Nat. Protoc., 2016, 11(2), 252–272 CrossRef CAS PubMed.
- M. Grelich-Mucha, M. Lipok, M. Różycka, M. Samoć and J. Olesiak-Bańska, J. Phys. Chem. Lett., 2022, 13(21), 4673–4681 CrossRef CAS PubMed.
- P. Obstarczyk, M. Lipok, M. Grelich-Mucha, M. Samoć and J. Olesiak-Bańska, J. Phys. Chem. Lett., 2021, 12(5), 1432–1437 CrossRef CAS PubMed.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, Nat. Methods, 2022, 19(6), 679–682 CrossRef CAS PubMed.
- C. R. Harris, et al. , Nature, 2020, 585(7825), 357–362 CrossRef CAS PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9(03), 90–95 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14(1), 33–38 CrossRef CAS PubMed.
- M. Falconi, M. T. Cambria, A. Cambria and A. Desideri, J. Biomol. Struct. Dyn., 2001, 18(5), 761–772 CrossRef CAS PubMed.
- B. Gorai and H. Vashisth, Front. Endocrinol., 2022, 13, 908724 CrossRef PubMed.
- L. Wang, et al. , Sci. Adv., 2023, 9(37), 1057 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp00669k |
| This journal is © the Owner Societies 2024 |