Open Access Article
Open Access ArticleUnveiling structural and energetic characterization of the emissive RNA alphabet anchored in the methylthieno[3,4-d]pyrimidine heterocycle core
Mohit
Chawla
 *a,
Albert
Poater
*a,
Albert
Poater
 b,
Romina
Oliva
c and
Luigi
Cavallo
b,
Romina
Oliva
c and
Luigi
Cavallo
 *a
*a
aKing Abdullah University of Science and Technology (KAUST), Physical Sciences and Engineering Division, KAUST Catalysis Center, Thuwal 23955-6900, Saudi Arabia. E-mail: mohit.chawla@kaust.edu.sa; luigi.cavallo@kaust.edu.sa
bInstitut de Química Computacional i Catàlisi and Departament de Química, Universitat de Girona, c/Ma Aurèlia Capmany 69, Girona 17003, Catalonia, Spain
cDepartment of Sciences and Technologies, University Parthenope of Naples, Centro Direzionale Isola C4, Naples, I-80143, Italy
First published on 20th May 2024
Abstract
This study presents a comprehensive theoretical exploration of the fluorescent non-natural emissive nucleobases- mthA, mthG, mthC, and mthU derived from the methylthieno[3,4-d]pyrimidine heterocycle. Our calculations, aligning with experimental findings, reveal that these non-natural bases exert minimal influence on the geometry of classical Watson–Crick base pairs within an RNA duplex, maintaining H-bonding akin to natural bases. In terms of energy, the impact of the modified bases, but for mthG, is also found to be little significant. We delved into an in-depth analysis of the photophysical properties of these non-natural bases. This investigation unveiled a correlation between their absorption/emission peaks and the substantial impact of the modification on the energy levels of the highest unoccupied molecular orbitals (HOMO) and the lowest unoccupied molecular orbital (LUMO). Notably, this alteration in energy levels resulted in a significant reduction of the HOMO–LUMO gap, from approximately 5.4–5.5 eV in the natural bases, to roughly 3.9–4.7 eV in the modified bases. This shift led to a consequential change in absorption and emission spectra towards longer wavelengths, elucidating their bathochromic shift.
Introduction
In the realm of synthetic biology, a prominent objective involves engineering biological systems to cater to diverse applications.1–3 One such endeavor revolves around the manipulation of natural nucleobases, traditionally devoid of fluorescence, into fluorescent bases. These modified bases serve as invaluable labeling tags, facilitating the investigation of nucleic acid interactions with various biomolecules.4 Such approach has already witnessed successful application across a spectrum of biotechnological and biomedical domains.5–15 Fluorescent nucleobase analogues are utilized for exploring the photophysical properties within duplex and G-quadruplex structures,16 developing Fluorescent Molecular Rotors (FMRs), which are crucial for investigating nucleic acids structure and function, and designing nucleobase analogs as multiphoton fluorescent probes.17 Additionally, they play a role in creating antisense oligonucleotide-incorporated fluorescent reporters for live cell imaging.18The ideal engineered fluorescent nucleobases aim for isomorphism with their parent nucleobases while retaining selective base-pairing, thereby ensuring minimal disruption to the structure of nucleic acids. These developments mark significant strides in enabling advanced research and applications within this domain.13,19,20 Interestingly, the scientific community has documented over a hundred distinct fluorescent nucleobase analogs in literature.21
One of the extensively utilized synthetically altered fluorescent nucleobases is 2-aminopurine, akin to natural adenine. This compound exhibits fluorescence both in its singular form and when integrated into duplex DNA in a solution.22 However, its presence tends to compromise the stability of the DNA duplex structure and prompts mispairing interactions with thymine within the DNA framework.23 An alternative approach revolves around modifications at the C8 or N7 positions of adenine, involving the incorporation of a triazole functionality. This strategy showcases fluorescent properties while inducing only a moderate destabilization of duplex DNA structures.24 This avenue of exploration has led to the investigation of various natural base analogs, some commercially available, which display moderate fluorescence upon integration into duplex DNA structures.20,25–39
In this context, Tor and colleagues devised two comprehensive sets of fluorescent analogs for RNA nucleosides. These analogs, derived from the thieno[3,4-d]-pyrimidine and isothiazolo[4,3-d]pyrimidine heterocycle core, form a complete alphabet and are both isomorphic and isofunctional to the natural nucleosides.20 The first set of synthesized nucleosides based on thieno[3,4-d]-pyrimidine core lack the N7 atom of the purine skeleton, whose basicity and coordinating ability is required for many biomolecular interactions.20 Therefore, these emissive nucleosides were not expected to be fully compatible with the RNA function.
To tackle this challenge and strive toward the objective of creating an emissive alphabet that closely mimics the natural nucleosides in both structure and function, the same authors developed a new generation of nucleosides. This updated set is based on the isothiazolo[4,3-d]-pyrimidine heterocyclic core, resulting in tzA, tzG, tzC, and tzU nucleosides. Notably, this series maintains the N7 atom within the purine skeleton, ensuring structural fidelity while enhancing their emissive properties.19 Thus, these new emissive nucleosides are isomorphic and possess structural similarity to natural purines, and have been shown to be also ‘isofunctional’ to natural nucleosides, since the adenosine deaminase is able to deaminate the adenosine analogue mthA as effectively as the natural adenosine.40 However, this alphabet is associated with a clear disadvantage deriving from hypochromic shifts and reduced emission quantum yields.
Hence, quite recently, third series of emissive ribonucleoside analogs was developed.41 This new set is based on the methylthieno[3,4-d]-pyrimidine core and was designed to explore the photophysical characteristics of this novel alphabet, see Fig. 1. Intriguingly, these synthesized nucleosides exhibit improved photophysical properties. Despite inducing structural changes at the Hoogsteen edge through the introduction of a methyl group, this updated emissive alphabet holds promise for advancing the biophysical analysis of specific RNA-related processes.41
To complement this experimental information, here we performed electronic structure calculations to evaluate the impact of the modification on the structure and energy of the base pairs containing the methylthieno[3,4-d]-pyrimidine analogues mthA, mthG, mthC, and mthU. Following an approach that we have used to evaluate the impact of natural and non-natural modifications on H-bonding base pairing,42–48 we initially focused on the effect of the modification on the geometry and energetics of the H-bonded base pairs they participate in. Initially, our focus was directed towards the classical cis Watson–Crick pairing (cWW), given that the Watson–Crick edge remains unaffected across all four modified nucleobases. Additionally, we conducted a comparative assessment of the aromatic characteristics between natural and non-natural nucleobases. This investigation aimed to explore potential effects on their stacking properties, thereby influencing the stability of the encompassing nucleic acid structure. Furthermore, we used time-dependent density functional theory (TD-DFT), already successfully employed in literature for similar applications,16,49–52 to analyze the impact of these modifications on the UV-Vis properties of natural nucleosides and to calculate the optical absorption properties of both modified nucleobases and nucleosides. These findings were then rationalized in terms of alterations in the molecular orbitals caused by the modifications.
Models and computational details
To investigate the geometry and stability of the non-natural modified base pairs under examination, we conducted modeling of the H-bonded modified base pairs. This involved employing density functional theory (DFT) and post Hartree–Fock methods to calculate their binding energy. To allow straightforward comparison with results previously published, we performed the calculations at exactly the same level of theory.Modeling the non-natural base pairs
We first study the impact on H-bonding of the non-natural mthA, mthG, mthU, and mthC bases forming classical Watson–Crick base pairs in combination with the classical A, G, C, and U bases, by taking into account the mthA:U, A:mthU, mthG:C, and G:mthC base pairs. Further, we also modeled the H-bonding interactions where both the bases were modified, that is the mthA:mthU and mthG:mthC base pairs. In addition, we also modeled the canonical A:U and G:C base pair geometries as well for the sake of comparative analysis. Thus, in total, we modeled eight different combinations of the non-natural modified base pair systems along with their natural base pair counterparts. For all the model systems described above, the base pairs are truncated at the C1′ atom of the ribose. This is the standard approach used in literature.43,44,47,53–56QM calculations of the modeled base pairs
Geometry optimizations were performed within a DFT approach, based on the hybrid B3LYP functional as implemented in the Gaussian 09 package.57–59 The correlation-consistent polarized valence triple-ζ cc-pvTZ basis set60 was used for all the geometry optimizations in gas phase as well as in water, modeled with the C-PCM continuum solvation model.61 As dispersion interactions could impact the stability of the examined base pairs in varying degrees, we included the Grimme D3 correction term with the Becke–Johnson damping scheme to account for these contributions to the electronic energy.62 Interaction energies were calculated on the B3LYP-D3/cc-pvTZ optimized geometries at the second order Møller–Plesset (MP2)63 level of theory using the augmented aug-cc-pvTZ basis set. For these calculations we took advantage of the faster RIMP264 method as implemented in Turbomole 6.1 package, with water modeled with the continuum solvation model COSMO.61 All the interaction energies were corrected for the basis set super position error (BSSE),65 using the counterpoise procedure.65 Thus, the binding energy EBind is calculated as in eqn (1):| EBind = EComplex – (EM1 + EM2) + BSSE; | (1) |
To have an immediate and intuitive understanding of the impact of a specific modification, we introduced the modification energy, EMod, defined as the energy difference between the binding energy of the modified and of the corresponding natural base pair (in this specific case A:U and G:C base pair),42,43,70,71 as shown in eqn (2).
| EMod = EBind (modified base pair) – EBind (natural base pair). | (2) |
Absorption and emission spectra were calculated using time dependent density functional theory (TD-DFT),72–74 with the B3LYP functional as implemented in the Gaussian09 package. A similar approach has already been performed on the non-natural emissive nucleobases derived from thieno[3,4-d]-pyrimidine heterocycle and isothiazolo[4,3-d]-pyrimidine heterocycle core.75,76 Geometry optimization of the singlet ground state S0 (for adsorption) and of the first singlet excited state S1 (for emission) were performed without symmetry constraints. For the absorption peaks, we always assumed a transition between the single ground state and the first excited singlet state. To facilitate a comparison between the predicted absorption and emission wavelengths and experimental findings, we incorporated water as an implicit solvent in our calculations.
Electron density analysis
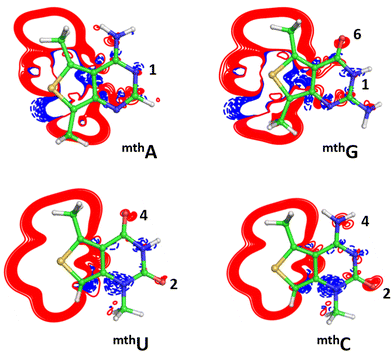
Comparative analysis of the electron density of natural (A/U/G/C) and modified (mthA, mthU, mthG, mthC) bases was performed as follows. For example, first, the geometry of the mthA base was optimized at the B3LYP/cc-pvTZ level of theory. For the sake of easier analysis, CS symmetry with the symmetry plane coincident with the purine plane was imposed, and the electron density analysis was performed in the symmetry plane. After optimization, we compared the RI-MP2/aug-cc-pvTZ electron densities of the modified mthA base, ρmthA, and that of the unmodified A base with the geometry it has in the mthA base, ρA/mthA. In other words, we took the optimized geometry of the mthA base, and we replaced the C9 carbon of mthA with nitrogen, plus we replaced the S8 atom of mthA with a carbon, further, we replaced the C7 atom in mthA with N atom and replaced the methyl group with a hydrogen atom. The geometry of this natural and non-modified A base was optimized by fixing the atoms of the 6-membered ring that is common among the modified mthA and the natural A base. With this approach, the heavy atom skeleton of the mthA and A bases is identical and can be perfectly superimposed. This is fundamental to avoid noise in the analysis of the electron density difference, ρmthA−A/mthA = ρmthA − ρA/mthA. A similar procedure was followed for the other bases.Aromaticity analysis
Aromaticity of the nucleobases was quantified using the nucleus-independent chemical shifts (NICS) values proposed by Schleyer and co-workers,77 computed using the gauge-including atomic orbital method78 (GIAO) and including the solvent contribution by C-PCM. The NICS is defined as the negative of the absolute shielding computed at specific points around the molecular system, in particular at ring centres, NICS(0) values, or d Å above such ring centres, NICS(d) values.79 In the current work, NICS(1) is defined as the average value of the shielding calculated 1 Å above and below the plane of the overall planar bases.Results and discussion
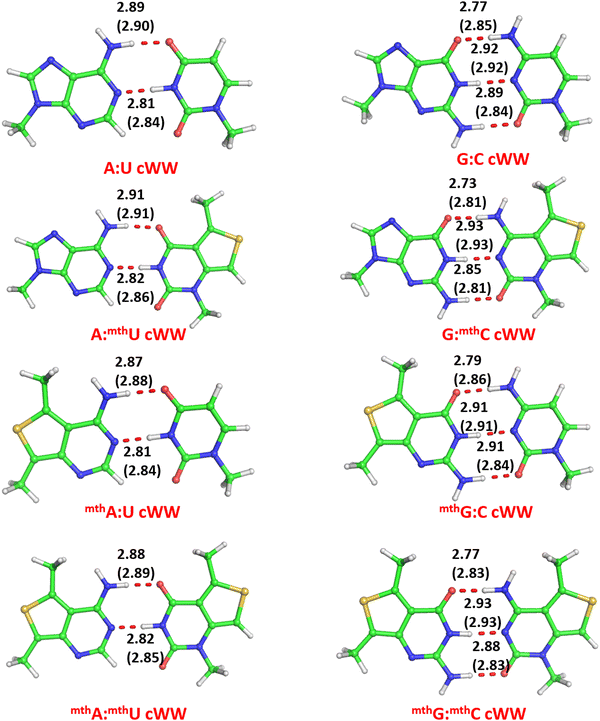
To investigate the influence of the fluorescent modified nucleobases (mthA, mthU, mthG, and mthC) on the structure and stability of nucleic acid base pairs, we examined eight distinct base pairs characterized by a classical antiparallel Watson–Crick (cWW) geometry typical of duplex structures. The optimized geometry and binding energies were calculated for these base pairs in both the gas phase and aqueous environments. Similarly to our previous studies,75,76 in this work we have calculated the modification energy, EMod, defined as the difference between the binding energy of the modified and of the corresponding unmodified base pair, for instance cWW mthA:U or cWW mthA:mthU vs. cWW A:U (see Methods), which will give an immediate effect of the impact of specific modification on the base pair. Negative and positive EMod values delineate whether the modified base pair exhibits greater or lesser stability than its unmodified counterpart. Table 1 compiles the calculated interaction energies and EMod values for the analyzed base pairs. Additionally, Fig. 2 illustrates the optimal geometries of the base pairs forming ‘Watson–Crick’ arrangements, showcasing H-bonding distances in both gas phase and in water.| System | Geometry | E Int | E Def | E Bind (gas) | E mod (gas) | E Bind (water) | E mod (water) |
|---|---|---|---|---|---|---|---|
| A:U | cWW | −16.51 | 1.50 | −15.01 | — | −7.93 | — |
| A:mthU | cWW | −15.73 | 1.38 | −14.34 | 0.67 | −7.69 | 0.25 |
| mthA:U | cWW | −17.37 | 1.77 | −15.60 | −0.59 | −8.41 | −0.47 |
| mthA:mthU | cWW | −16.42 | 1.64 | −14.78 | 0.23 | −8.12 | −0.19 |
| G:C | cWW | −30.74 | 2.75 | −27.99 | — | −12.49 | — |
| G:mthC | cWW | −32.10 | 3.34 | −28.76 | −0.77 | −13.00 | −0.51 |
| mthG:C | cWW | −28.04 | 3.32 | −24.72 | 3.26 | −11.89 | 0.61 |
| mthG:mthC | cWW | −28.87 | 3.23 | −25.64 | 2.35 | −12.21 | 0.29 |
Structure and energy impact of the modified nucleobases on H-bonded base pairs
Eight cWW base pairs were considered, four related to the A:U and four to the G:C pair, namely the unmodified A:U and G:C cWW and the fully modified mthA:mthU and mthG:mthC pairs, along with the base pairs hosting a single modified base, i.e.mthA:U/A:mthU and mthG:C/G:mthC (see Fig. 2). The optimized structure of the modified mthA:U, A:mthU and mthA:mthU cWW base pairs is extremely similar to that of the classical A:U cWW pair, with differences in H-bond lengths within 0.02 Å. Similarly, the C1′–C1′ distances (which gives an indication of the isostericity of H-bonded base pairs)80,81 are almost identical in the modified and unmodified A:U pairs (10.52 Å for A:U versus 10.51/10.49/10.48 Å for A:mthU/mthA:U/mthA:mthU). An analogous trend is observed for base pairs involving the mthG and mthC modifications. The geometry of mthG:C, G:mthC, and mthG:mthC base pairs closely mirrors that of the natural G:C cWW base pair, with differences in H-bond distances within 0.04 Å. Likewise, the C1′–C1′ distances are very similar (10.71 Å for G:C versus 10.78/10.78/10.79 Å for G:mthC/mthG:C/mthG:mthC).The binding energy of the modified pairs involving mthA and mthU bases have a slight destabilizing effect with an EMod of 0.67/025 kcal mol−1 for cWW A:mthU base pair in gas phase/water. This can be attributed to the decreased electron density around the N3 atom of mthU, making it a less effective donor compared to the canonical U base (Fig. 3). Conversely, a slight stabilization is observed for mthA:U, with an EMod of −0.59/−0.47 kcal mol−1 in gas phase/water, respectively. This stabilization is attributed to a slight increase in electron density around N6 of mthA, making it a better H-bond donor, and N1 of mthA, a superior H-bond acceptor, explaining a stabilization effect of mthA:U compared to canonical A:U base pair.
In contrast, the modified base pairs involving mthG exhibit a significant destabilizing effect, with EMod values ranging from +3.26/+0.61 to +2.35/+0.29 kcal mol−1 for mthG:C and mthG:mthC in gas phase/water, respectively. Electron density difference analysis reveals that the O6 of mthG has increased electron density, while N1 of mthG becomes a poor H-bond donor, leading to an overall less stable base pair compared to the canonical G:C pair (Fig. 3). For G:mthC pairs, a slight stabilization effect of −0.77/−0.51 kcal mol−1 is observed, attributed to the enhanced electron density around O2 and N4 of mthC, making them better H-bond acceptors and donors, respectively, compared to the canonical G:C base pair.
In conclusion, our analysis confirms the absence of deformation or strain in RNA duplexes due to modified bases. They maintain indeed an H-bonding geometry comparable to that of the natural bases.
Nevertheless, we observe a specific destabilization trend in base pairs containing the mthG modification, particularly pronounced in gas phase calculations. This warrants further investigation, particularly within the context of complex RNA structures.
Aromaticity analysis
To assess the impact of aromaticity on the 6 and 5-membered rings in the non-natural methylthieno bases in comparison to their natural counterparts, we utilized the nucleus-independent chemical shift (NICS) as a metric.77 The NICS is defined as the negative of the absolute shielding calculated at specific points within the molecular system, specifically at the ring centers, known as NICS(0) values, or at positions 1 Å above the ring centers, referred to as NICS(1) values.79 Larger NICS values, i.e., more negative values, indicate a higher level of ring aromaticity. In these computations, the NICS values of benzene served as the reference point. The data presented in Table 2 highlights a consistent trend: the NICS index for the 6-membered ring consistently shows lower values compared to benzene. This suggests a reduced level of aromaticity within the 6-membered ring, observed in both natural and non-natural bases.| 6 membered ring | 5 membered ring | ||||
|---|---|---|---|---|---|
| NICS(0) | NICS(1) | HOMA | NICS(0) | NICS(1) | |
| C6H6 | −8.227 | −10.370 | 0.995 | — | — |
| A | −6.032 | −8.006 | 0.978 | −11.201 | −9.709 |
| mthA | −2.246 | −5.380 | −0.693 | −13.475 | −11.059 |
| C | −1.347 | −3.173 | 0.755 | — | — |
| mthC | 0.320 | −2.041 | −0.563 | −12.577 | −9.276 |
| G | −2.909 | −3.426 | 0.803 | −10.611 | −8.821 |
| mthG | −0.046 | −1.864 | −0.523 | −11.848 | −9.240 |
| U | −1.089 | −1.786 | 0.640 | — | — |
| mthU | 0.150 | −0.891 | −0.533 | −12.332 | −9.050 |
Comparing the NICS(1) indices between natural and non-natural bases, we observe a further decrease in aromaticity of the 6-membered ring in purine and pyrimidine bases for the non-natural variants. For instance, the NICS(1) value for the 6-membered ring of mthA is only −6.06, while it is −8.01 for A. This sharp decrease in aromaticity is notable in the non-natural base. This trend is consistent across the other bases. Conversely, the aromaticity of the 5-membered ring of non-natural purines mthA/mthG, is higher, where the C8–H group in the natural base is replaced by a S8, and N7 is replaced with C–CH3. Specifically, the NICS(1) of the 5-membered ring of mthA is −11.63, compared to −9.71 for A. A similar trend is observed for mthG and G. In summary, this analysis indicates that in the 6-membered ring of non-natural bases, the delocalization of electrons in the π-conjugated aromatic system is less extensive compared to natural nucleobases. Conversely, electron delocalization is more pronounced in the 5-membered ring of non-natural purines compared to natural purines.
To corroborate the findings from the NICS comparison, the harmonic oscillator model of aromaticity (HOMA), which is defined as the normalized sum of squared deviations of bond lengths from the optimal value in benzene, was also calculated, as shown in Table 2. This alternative analysis further supports the decrease in aromaticity of the 6-membered ring in the non-natural mthA, mthU, mthG, and mthC bases, when compared to the natural A, U, G and C bases. Interestingly, the substitution in the unnatural bases in the additional 7 position, with respect to the results of our previous studies imposes an additional destabilization of both rings of different size75 reducing the NICS values by an amount significant but never higher than 0.1 units in the case of the 6-membered ring, but already more pronounced up to 1.5 units in the 5-membered ring. Therefore, the 5-membered ring has greater aromaticity if there is a N atom in the ring, rather than a C–CH3 group. Furthermore, the reduction in aromaticity is more pronounced for the mthU and mthG bases. This is due to the repulsion between two pairs of hydrogen atoms that are placed at 2.223, 2.169 Å for the bases mthA and mthC between the C–CH3 group and one hydrogen of the NH2 group since the whole group is placed in the same plane of the 6-membered ring. While there are attractive H-bonds of those two hydrogens of the methyl fragment C–CH3 and the oxygen of the keto group of the 6-membered ring of mthG and mthU, but at a relatively long distance of 2.856 and 2.818 Å. In addition, these interactions are relevant between the hydrogen atoms of the C–CH3 group of the 5-membered ring with the NH2 or the O moieties of the 6-membered ring because the decrease in aromaticity is more pronounced for in NICS(1) instead of NICS(0) due to the hydrogen atoms of the C–CH3 group lying out of the plane of the rings.
To assess the potential changes in the aromaticity of bases upon the formation of base pairs, we computed the NICS (1) for individual bases when incorporated into base pairs. Analysis of Table 3 indicates a clear trend where the aromaticity values closely resemble those of isolated bases. However, a subtle elevation in aromaticity is observed, averaging approximately 0.5 ppm, and reaching up to 1 ppm in agreement with our prior studies.75 It should be noted that it is the mthU base that sees its aromaticity most altered, and even more the mthC base in G:mthC, but only up to a loss of aromaticity of 1.111 ppm.
| Base left | Base right | |||
|---|---|---|---|---|
| 6 membered ring | 5 membered ring | 6 membered ring | 5 membered ring | |
| A:U | −7.417 (−0.589) | −9.618 (−0.091) | −1.750 (−0.036) | — |
| A:mthU | −7.380 (−0.626) | −9.550 (−0.159) | −0.942 (−0.922) | −8.962 (−0.088) |
| mthA:mthU | −4.717 (−0.663) | −10.736 (−0.323) | −0.962 (−0.902) | −9.010 (−0.040) |
| mthA:U | −4.689 (−0.691) | −10.809 (−0.250) | −1.803 (0.017) | — |
| G:C | −3.519 (0.093) | −8.704 (−0.117) | −2.677 (−0.496) | — |
| G:mthC | −3.492 (0.066) | −8.559 (−0.262) | −1.770 (−0.271) | −10.387 (1.111) |
| mthG:mthC | −2.037 (0.173) | −9.109 (−0.131) | −1.783 (−0.258) | −9.344 (0.068) |
| mthG:C | −2.057 (0.193) | −9.070 (−0.170) | −2.755 (−0.418) | — |
Photophysical properties of methylthieno[3,4-d]-pyrimidine nucleobase analogues
Time-dependent density functional theory (TD-DFT) has emerged as a highly reliable technique for predicting properties such as absorption, emission, quantum yield, and transition dipole moment, in previous studies where various fluorescent base analogues were characterized as monomers, as well as inside RNA and/or DNA strands. It has also proven instrumental in the rational design of fluorescent nucleobase analogues. For example, TD-DFT calculations conducted on nine quadracyclic adenine derivatives successfully described their relative fluorescence quantum yield and brightness, indicating the potential for employing a TD-DFT-based rational design strategy to develop bright fluorophores from a common scaffold, thereby streamlining the otherwise costly and time-consuming screening process.52 In a subsequent study by the same authors, TD-DFT calculations were utilized to predict the transition dipole moment orientation for qAN4 and qAnitro bases, revealing an associated phase angle of 25 degrees.51 Additionally, the fluorescent base analogue tCO was successfully incorporated into RNA, maintaining the A-form duplex structure, enhancing stability, and exhibiting high fluorescence quantum yield, positioning it as a potentially valuable fluorescent RNA base analogue with superior brightness compared to existing internal RNA analogues.50 Additionally, Manderville, Wetmore, and their team investigated fluorescent donor–acceptor (D–A) 8-aryl-dG probes incorporated within the thrombin binding aptamer (TBA), impacting the stability of G-quadruplex (GQ) structures. These probes, which feature diverse moieties, demonstrate potential in distinguishing between duplex and GQ configurations, thus facilitating fluorescence-based applications such as molecular sensing and diagnostics. Their ability to be selectively excited and their sensitivity to environmental conditions position them as promising tools for monitoring molecular interactions and structural changes.16In this study, we also used the TD-DFT studies to calculate the photophysical properties of natural and non-natural fluorescent nucleobases, see Table 4, since our previous studies showed that TD-DFT reproduces the absorption/emission peaks better than the RI-CC2 approach.75,76 Calculating the energy difference between the ground (S0) and the first excited singlet state (S1), we find the energy absorption peaks at 354, 333, 333 and 302 nm for mthA, mthG, mthC and mthU, respectively, which are in good agreement with the experimentally reported absorption peaks of 353, 327, 323 and 316 nm.41 We report all the peaks using TD-DFT in the present work. Although S0 and S1 involve contributions from neighbor orbitals, peaks are dominated by the π to π* HOMO to LUMO transition. For comparison, we also calculated the same absorption peaks for the natural nucleobases, 251, 248, 256 and 246 nm for A, G, C and U bases, which compare with experimental values of 259, 252, 271, and 262 nm, respectively.82 This indicates that the methylthieno[3,4-d]-pyrimidine modification results in a bathochromic shift of roughly 102/90 nm for mthG/mthA, and of roughly 80/58 nm for mthC/mthU, as compared to their natural counterparts. Slightly larger bathochromic shifts were calculated in the emission spectra,83 with bathochromic displacements of 160, 158, 143 and 109 nm for mthA, mthG, mthC and mthU with respect to natural bases. We find the energy emission peaks at 463, 452, 465 and 393 nm for mthA, mthG, mthC and mthU, respectively, which are in good agreement with the experimentally reported peaks of 467, 456, 455 and 427 nm. For the sake of comparison, we calculated here the absorption/emission peaks also for first generation non-natural alphabet proposed by Tor and co-workers,75,76thA, thG, thU, thC. As compared to thA, thG, thU, thC, the third generation mthA, mthG, mthC and mthU, analysed here, also exhibited a batochromic shift, by 10, 3, 3, 2 nm, respectively, for absorption, and by 28, 18, 26, 19 nm, respectively, for emission.41
| Base | Absorption | Emission | H | L | ΔEHL | ||
|---|---|---|---|---|---|---|---|
| λ abs | os. str. | λ em | os. str. | ||||
| mthA | 353.9 (353) | 0.168 | 462.6 (467) | 0.195 | −5.76 | −1.82 | −3.94 |
| mthG | 332.7 (327) | 0.109 | 451.7 (456) | 0.126 | −5.96 | −1.64 | −4.32 |
| mthC | 332.7 (323) | 0.093 | 465.4 (455) | 0.103 | −6.10 | −1.88 | −4.22 |
| mthU | 301.6 (306) | 0.071 | 393.4 (427) | 0.094 | −6.54 | −1.81 | −4.73 |
| thA | 343.7 (341) | 0.177 | 434.2 (420) | 0.219 | −6.05 | −1.97 | −4.08 |
| thG | 329.9 (321) | 0.111 | 433.2 (453) | 0.134 | −6.02 | −1.72 | −4.30 |
| thC | 329.7 (320) | 0.095 | 439.6 (429) | 0.119 | −6.19 | −1.87 | −4.32 |
| thU | 299.3 (304) | 0.073 | 374.4 (409) | 0.106 | −6.57 | −1.85 | −4.72 |
| A | 251.5 | 0.268 | 302.86 | 0.467 | −6.29 | −0.91 | −5.38 |
| G | 252.8 | 0.128 | 293.50 | 0.103 | −6.25 | −0.79 | −5.46 |
| C | 252.9 | 0.197 | 321.97 | 0.202 | −6.68 | −1.23 | −5.45 |
| U | 243.6 | 0.185 | 284.26 | 0.271 | −7.14 | −1.61 | −5.53 |
As expected, and in light of the above results, the HOMO and LUMO play a major role in the absorption/emission properties of the bases featuring the methylthieno[3,4-d]-pyrimidine modification, therefore we decided to further analyse how the above modification impacts these orbitals. Previously, we had shown a clear correlation between the bathochromic shifts of the tz- and ts-bases and the HOMO to LUMO energy gap.75,76
Data reported in Table 4 shows that the energy of both the HOMO and LUMO is substantially altered by the modification, with differences between a non-natural base and its natural analogue within roughly 0.29–0.58 eV for HOMO and 0.37–0.91 for LUMO. This reduces the HOMO–LUMO gap from roughly 5.4–5.5 eV in the natural bases to roughly 3.9–4.7 eV in the modified methylthieno[3,4-d]-pyrimidine bases, with the consequent bathochromic shift in the absorption and emission spectra. Considering that both the HOMO and LUMO energies are substantially changed from the natural bases, we report their shapes in Fig. 4. As illustrative examples, the LUMOs of A, G, C and U are compared to those of the homologous modified mthA, mthG, mthC and mthU in Fig. 4. Inspection of the shape of the HOMO of the modified bases shows higher participation of the 5-membered ring for the mthG base, with a strong participation of the S8 atom with its low energy π orbitals, which destabilize the HOMO. Since the LUMO is also stabilized, less quantitatively than the destabilization of the HOMO, the HOMO–LUMO gap reduces significantly, and thus, the stability of the modified bases decreases. Finally, the reduced bathochromic shift of modified pyrimidines can also be rationalized in terms of reduced aromaticity of modified and natural pyrimidines relative to purines, see Table 2, which lowers the impact of the S8 atom on the absorption/emission properties of the modified bases. In addition, we calculated the non-covalent interaction (NCI) plots,84,85 to determine whether the greater or lesser stability of the base pairs was due to base modification. As can be seen in Fig. 5, these analyses allow us to conclude that the non-covalent interaction between the natural and non-natural pairs do not differ qualitatively. Even though the interaction between the base pairs seems to be identical according to the 3D NCI plots in Fig. 5, this intermolecular interaction means a weakening of the intramolecular H-bonds as can be seen in the 2D NCI plots Fig. 5, particularly for A:mthU and also for the mthG;mthC and mthG:C pairs. Oppositely, the favourable interactions are especially important for the mthA:U, and even more for and G:mthC, in perfect agreement with results in Table 1.
 | ||
| Fig. 4 Graphic representation of the frontier molecular orbitals of the natural bases (top) and the modified ones (bottom). | ||
Conclusions
Our investigation delved into the electronic properties and the influence on base-pair stability of a novel emissive RNA alphabet founded on methylthieno[3,4-d]-pyrimidine (mthA, mthU, mthG, and mthC), proposed as highly isomorphic and isofunctional counterparts to the natural nucleobases. Our study demonstrated that these emissive nucleobases exert minimal effects on the geometry of classical Watson–Crick base pairs, suggesting that RNA duplexes can be capable to accommodate them, especially the purine derivatives, without significant alterations in H-bonding. However, the introduction of mthG in particular may impact their stability. The potential steric implications of the additional methylthieno ring in pyrimidine derivatives also warrant further investigation.As the stacking interactions play a significant role in the structural stability of nucleic acids, we evaluated the aromaticity of both non-natural emissive and natural nucleobases. Our analysis revealed a subtle reduction in the aromaticity of the 6-membered ring of emissive purines and pyrimidines as compared to their natural counterparts. In contrast, non-natural purines (mthA/mthG) exhibited a significant increase in aromaticity in the 5-membered ring, attributed to the presence of the S8 substitution. The influence of this modified aromaticity on relative stacking interactions would need to be further inquired. Moreover, our assessment of molecular orbitals facilitated a rationalization of the absorption and emission properties of the modified nucleobases, accounting for their fluorescence.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). For computer time, this research used the resources of the Supercomputing Laboratory at King Abdullah University of Science & Technology (KAUST) in Thuwal, Saudi Arabia. A. P. is grateful for financial support from the Ministerio de Ciencia e Innovación (PID2021-127423NB-I00); Generalitat de Catalunya for project 2021SGR623 and ICREA Academia prize 2019. A. P. is a Serra Húnter fellow. R. O. acknowledges financial support under the National Recovery and Resilience Plan (NRRP), Mission 4, Component 2, Investment 1.1, Call for tender No. 104 published on 2.2.2022 by the Italian Ministry of University and Research (MUR), funded by the European Union – NextGenerationEU– Project Title Unraveling The moonLIghtinG AcTivity of the PFKFB bIfunctiOnal eNzyme with phospholipase C-gamma (LIGATION) – CUP D53D23010710006- Grant Assignment Decree No. 1017 adopted on 07/07/2023 by the Italian Ministry of Ministry of University and Research (MUR).References
- S. A. Benner and A. M. Sismour, Nat. Rev. Genet., 2005, 6, 533–543 CrossRef CAS PubMed
.
- S. A. Benner, Chimia, 1987, 41, 142–148 CAS
.
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS PubMed
.
- W. Liu, D. Shin, Y. Tor and B. S. Cooperman, ACS Chem. Biol., 2013, 8, 2017–2023 CrossRef CAS PubMed
.
- S. G. Srivatsan, N. J. Greco and Y. Tor, Angew. Chem., Int. Ed., 2008, 47, 6661–6665 CrossRef CAS PubMed
.
- S. G. Srivatsan and Y. Tor, Tetrahedron, 2007, 63, 3601–3607 CrossRef CAS PubMed
.
- S. G. Srivatsan and Y. Tor, Nat. Protoc., 2007, 2, 1547–1555 CrossRef CAS PubMed
.
- S. G. Srivatsan and Y. Tor, J. Am. Chem. Soc., 2007, 129, 2044–2053 CrossRef CAS PubMed
.
- S. G. Srivatsan, H. Weizman and Y. Tor, Org. Biomol. Chem., 2008, 6, 1334–1338 RSC
.
- N. J. Greco and Y. Tor, J. Am. Chem. Soc., 2005, 127, 10784–10785 CrossRef CAS PubMed
.
- N. J. Greco and Y. Tor, Nat. Protoc., 2007, 2, 305–316 CrossRef CAS PubMed
.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579–2619 CrossRef CAS PubMed
.
- R. W. Sinkeldam, P. A. Hopkins and Y. Tor, Chem. Phys. Chem., 2012, 13, 3350–3356 CrossRef CAS PubMed
.
- R. W. Sinkeldam, L. S. McCoy, D. Shin and Y. Tor, Angew. Chem., Int. Ed., 2013, 52, 14026–14030 CrossRef CAS PubMed
.
- L. M. Wilhelmsson, Q. Rev. Biophys., 2010, 43, 159–183 CrossRef CAS PubMed
.
- D. J. M. Blanchard, K. L. Fadock, M. Sproviero, P. S. Deore, T. Z. Cservenyi, R. A. Manderville, P. Sharma and S. D. Wetmore, J. Mater. Chem. C, 2016, 4, 2915–2924 RSC
.
- R. E. Johnson, M. T. Murray, L. J. Bycraft, S. D. Wetmore and R. A. Manderville, Chem. Sci., 2023, 14, 4832–4844 RSC
.
- J. R. Nilsson, C. Benitez-Martin, H. G. Sansom, P. Pfeiffer, T. Baladi, H. N. Le, A. Dahlén, S. W. Magennis and L. M. Wilhelmsson, PCCP, 2023, 25, 20218–20224 RSC
.
- A. R. Rovira, A. Fin and Y. Tor, J. Am. Chem. Soc., 2015, 137, 14602–14605 CrossRef CAS PubMed
.
- D. Shin, R. W. Sinkeldam and Y. Tor, J. Am. Chem. Soc., 2011, 133, 14912–14915 CrossRef CAS PubMed
.
- W. Xu, K. M. Chan and E. T. Kool, Nat. Chem., 2017, 9, 1043–1055 CrossRef CAS PubMed
.
- D. C. Ward, E. Reich and L. Stryer, J. Biol. Chem., 1969, 244, 1228–1237 CrossRef CAS PubMed
.
- D. Xu, K. O. Evans and T. M. Nordlund, Biochemistry, 1994, 33, 9592–9599 CrossRef CAS PubMed
.
- C. Dyrager, K. Borjesson, P. Diner, A. Elf, B. Albinsson, L. M. Wilhelmsson and M. Grotli, Eur. J. Org. Chem., 2009, 1515–1521, DOI:10.1002/ejoc.200900018
.
- M. E. Hawkins, Cell Biochem. Biophys., 2001, 34, 257–281 CrossRef CAS PubMed
.
- M. E. Hawkins, W. Pfleiderer, F. M. Balis, D. Porter and J. R. Knutson, Anal. Biochem., 1997, 244, 86–95 CrossRef CAS PubMed
.
- A. R. Hernandez and E. T. Kool, Org. Lett., 2011, 13, 676–679 CrossRef CAS PubMed
.
- A. R. Hernandez, L. W. Peterson and E. T. Kool, ACS Chem. Biol., 2012, 7, 1454–1461 CrossRef CAS PubMed
.
- A. T. Krueger and E. T. Kool, J. Am. Chem. Soc., 2008, 130, 3989–3999 CrossRef CAS PubMed
.
- A. H. Lee and E. T. Kool, J. Am. Chem. Soc., 2006, 128, 9219–9230 CrossRef CAS PubMed
.
- C. Liu and C. T. Martin, J. Mol. Biol., 2001, 308, 465–475 CrossRef CAS PubMed
.
- C. Liu and C. T. Martin, J. Biol. Chem., 2002, 277, 2725–2731 CrossRef CAS PubMed
.
- H. Liu, J. Gao, S. R. Lynch, Y. D. Saito, L. Maynard and E. T. Kool, Science, 2003, 302, 868–871 CrossRef CAS PubMed
.
- H. Lu, K. He and E. T. Kool, Angew. Chem., Int. Ed., 2004, 43, 5834–5836 CrossRef CAS PubMed
.
- E. L. Rachofsky, E. Seibert, J. T. Stivers, R. Osman and J. B. Ross, Biochemistry, 2001, 40, 957–967 CrossRef CAS PubMed
.
- P. K. Samanta, A. K. Manna and S. K. Pati, Chem. – Asian J., 2012, 7, 2718–2728 CrossRef CAS PubMed
.
- P. K. Samanta, A. K. Manna and S. K. Pati, J. Phys. Chem. B, 2012, 116, 7618–7626 CrossRef CAS PubMed
.
- D. I. Scopes, J. R. Barrio and N. J. Leonard, Science, 1977, 195, 296–298 CrossRef CAS PubMed
.
- J. T. Stivers, Nucleic Acids Res., 1998, 26, 3837–3844 CrossRef CAS PubMed
.
- R. A. Mizrahi, D. Shin, R. W. Sinkeldam, K. J. Phelps, A. Fin, D. J. Tantillo, Y. Tor and P. A. Beal, Angew. Chem., Int. Ed., 2015, 54, 8713–8716 CrossRef CAS PubMed
.
- P. T. Ludford, 3rd, S. Yang, M. S. Bucardo and Y. Tor, Chemistry, 2022, 28, e202104472 CrossRef PubMed
.
- M. Chawla, R. Credendino, R. Oliva and L. Cavallo, J. Phys. Chem. B, 2015, 119, 12982–12989 CrossRef CAS PubMed
.
- M. Chawla, R. Oliva, J. M. Bujnicki and L. Cavallo, Nucleic Acids Res., 2015, 43, 9573 CrossRef CAS PubMed
.
- R. Oliva, L. Cavallo and A. Tramontano, Nucleic Acids Res., 2006, 34, 865–879 CrossRef CAS PubMed
.
- R. Oliva, A. Tramontano and L. Cavallo, RNA, 2007, 13, 1427–1436 CrossRef CAS PubMed
.
- M. Chawla, R. Credendino, E. Chermak, R. Oliva and L. Cavallo, J. Phys. Chem. B, 2016, 120, 2216–2224 CrossRef CAS PubMed
.
- M. Chawla, S. Abdel-Azeim, R. Oliva and L. Cavallo, Nucleic Acids Res., 2014, 42, 714–726 CrossRef CAS PubMed
.
- M. Chawla, R. Credendino, A. Poater, R. Oliva and L. Cavallo, J. Am. Chem. Soc., 2015, 137, 299–306 CrossRef CAS PubMed
.
- H. N. Le, J. Kuchlyan, T. Baladi, B. Albinsson, A. Dahlen and L. M. Wilhelmsson, Chemistry, 2024, e202303539, DOI:10.1002/chem.202303539
.
- A. F. Fuchtbauer, S. Preus, K. Borjesson, S. A. McPhee, D. M. J. Lilley and L. M. Wilhelmsson, Sci. Rep., 2017, 7, 2393 CrossRef PubMed
.
- A. F. Fuchtbauer, M. S. Wranne, S. Sarangamath, M. Bood, A. H. El-Sagheer, T. Brown, H. Graden, M. Grotli and L. M. Wilhelmsson, ChemPlusChem, 2020, 85, 319–326 CrossRef PubMed
.
- A. Foller Larsen, B. Dumat, M. S. Wranne, C. P. Lawson, S. Preus, M. Bood, H. Graden, L. M. Wilhelmsson and M. Grotli, Sci. Rep., 2015, 5, 12653 CrossRef CAS PubMed
.
- J. E. Sponer, K. Reblova, A. Mokdad, V. Sychrovsky, J. Leszczynski and J. Sponer, J. Phys. Chem. B, 2007, 111, 9153–9164 CrossRef CAS PubMed
.
- J. E. Sponer, N. Spackova, P. Kulhanek, J. Leszczynski and J. Sponer, J. Phys. Chem. A, 2005, 109, 2292–2301 CrossRef CAS PubMed
.
- J. E. Sponer, N. Spackova, J. Leszczynski and J. Sponer, J. Phys. Chem. B, 2005, 109, 11399–11410 CrossRef CAS PubMed
.
- P. Sharma, L. A. Lait and S. D. Wetmore, PCCP, 2013, 15, 2435–2448 RSC
.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS
.
- A. D. Becke, Abstr. Pap., Jt. Conf. – Chem. Inst. can. Am. Chem. Soc., 1996, 212, 112 Search PubMed
.
- C. T. Lee, W. T. Yang and R. G. Parr, Phys Rev B: Solid State, 1988, 37, 785–789 CrossRef CAS PubMed
.
- T. H. Dunning, J. Chem. Phys., 1989, 90, 1007–1023 CrossRef CAS
.
- A. Klamt and G. Schuurmann, J. Chem. Soc., Perkin Trans. 2, 1993, 799–805 RSC
.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed
.
- C. Moller and M. S. Plesset, Phys. Rev., 1934, 46, 0618–0622 CrossRef CAS
.
- F. Weigend and M. Haser, Theor. Chem. Acc., 1997, 97, 331–340 Search PubMed
.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553 CrossRef CAS
.
- P. Sharma, S. Sharma, M. Chawla and A. Mitra, J. Mol. Model., 2009, 15, 633–649 CrossRef CAS PubMed
.
- M. Chawla, P. Sharma, S. Halder, D. Bhattacharyya and A. Mitra, J. Phys. Chem. B, 2011, 115, 1469–1484 CrossRef CAS PubMed
.
- C. L. Zirbel, J. E. Sponer, J. Sponer, J. Stombaugh and N. B. Leontis, Nucleic Acids Res., 2009, 37, 4898–4918 CrossRef CAS PubMed
.
- A. Mladek, P. Sharma, A. Mitra, D. Bhattacharyya, J. Sponer and J. E. Sponer, J. Phys. Chem. B, 2009, 113, 1743–1755 CrossRef CAS PubMed
.
- M. Chawla, Y. Minenkov, K. B. Vu, R. Oliva and L. Cavallo, ChemBioChem, 2019, 20, 2262–2270 CrossRef CAS PubMed
.
- M. Chawla, I. Autiero, R. Oliva and L. Cavallo, Phys. Chem. Chem. Phys., 2018, 20, 3699–3709 RSC
.
- R. Bauernschmitt and R. Ahlrichs, Chem. Phys. Lett., 1996, 256, 454–464 CrossRef CAS
.
- M. E. Casida, C. Jamorski, K. C. Casida and D. R. Salahub, J. Chem. Phys., 1998, 108, 4439–4449 CrossRef CAS
.
- R. E. Stratmann, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 1998, 109, 8218–8224 CrossRef CAS
.
- M. Chawla, A. Poater, P. Besalu-Sala, K. Kalra, R. Oliva and L. Cavallo, Phys. Chem. Chem. Phys., 2018, 20, 7676–7685 RSC
.
- M. Chawla, A. Poater, R. Oliva and L. Cavallo, Phys. Chem. Chem. Phys., 2016, 18, 18045–18053 RSC
.
- P. V. Schleyer, C. Maerker, A. Dransfeld, H. J. Jiao and N. J. R. V. Hommes, J. Am. Chem. Soc., 1996, 118, 6317–6318 CrossRef CAS PubMed
.
- K. Wolinski, J. F. Hinton and P. Pulay, J. Am. Chem. Soc., 1990, 112, 8251–8260 CrossRef CAS
.
- J. Poater, M. Duran, M. Sola and B. Silvi, Chem. Rev., 2005, 105, 3911–3947 CrossRef CAS PubMed
.
- N. B. Leontis, J. Stombaugh and E. Westhof, Nucleic Acids Res., 2002, 30, 3497–3531 CrossRef CAS PubMed
.
- N. B. Leontis and E. Westhof, RNA, 2001, 7, 499–512 CrossRef CAS PubMed
.
- M. J. Cavaluzzi and P. N. Borer, Nucleic Acids Res., 2004, 32, e13 CrossRef PubMed
.
- D. Onidas, D. Markovitsi, S. Marguet, A. Sharonov and T. Gustavsson, J. Phys. Chem. B, 2002, 106, 11367–11374 CrossRef CAS
.
- E. R. Johnson, S. Keinan, P. Mori-Sanchez, J. Contreras-Garcia, A. J. Cohen and W. Yang, J. Am. Chem. Soc., 2010, 132, 6498–6506 CrossRef CAS PubMed
.
- R. A. Boto, F. Peccati, R. Laplaza, C. Quan, A. Carbone, J. P. Piquemal, Y. Maday and A. J. Contreras-Garci, J. Chem. Theory Comput., 2020, 16, 4150–4158 CrossRef CAS PubMed
.
| This journal is © the Owner Societies 2024 |