 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The synthesis of specifically isotope labelled fluorotryptophan and its use in mammalian cell-based protein expression for 19F-NMR applications†
Giorgia
Toscano‡
 abc,
Martina
Rosati‡
abc,
Martina
Rosati‡
 d,
Letizia
Barbieri
d,
Letizia
Barbieri
 de,
Katharina
Maier
a,
Lucia
Banci
de,
Katharina
Maier
a,
Lucia
Banci
 def,
Enrico
Luchinat
def,
Enrico
Luchinat
 def,
Robert
Konrat
def,
Robert
Konrat
 bgh and
Roman J.
Lichtenecker
bgh and
Roman J.
Lichtenecker
 *abh
*abh
aInstitute of Organic Chemistry, University of Vienna, Währinger Str. 38, 1090-Vienna, Austria. E-mail: roman.lichtenecker@univie.ac.at
bChristian Doppler Laboratory for High-Content Structural Biology and Biotechnology, Max Perutz Laboratories, University of Vienna, Campus Vienna Biocenter 5, 1030 Vienna, Austria
cVienna Doctoral School in Chemistry (DoSChem), University of Vienna, Währingerstraße 42, 1090 Vienna, Austria
dCERM Magnetic Resonance Center, Università degli Studi di Firenze, Sesto Fiorentino, Italy
eConsorzio Interuniversitario Risonanze Magnetiche di Metallo Proteine CIRMMP, Sesto Fiorentino, Italy
fDipartimento di Chimica, Università degli Studi di Firenze, Sesto Fiorentino, Italy
gDepartment of Structural and Computational Biology, Max Perutz Laboratories, University of Vienna, Dr-Bohr-Gasse 9, 1030-Vienna, Austria
hMag-Lab, Karl-Farkas-Gasse 22, 1030 Vienna, Austria
First published on 4th November 2024
Abstract
19F nuclei serve as versatile sensors for detecting protein interactions and dynamics in biomolecular NMR spectroscopy. Although various methods have been developed to incorporate fluorine-containing aromatic residues into proteins using E. coli or cell-free expression techniques, similar approaches for protein production in mammalian cell lines remain limited. Here, we present a cost-effective synthetic route to obtain selectively deuterated, carbon-13 labeled fluorotryptophan and demonstrate its use in introducing 19F–13C spin pairs into carbonic anhydrase 2 and superoxide dismutase, following an expression protocol utilizing HEK cells.
NMR spectroscopy is a powerful tool for studying the conformational properties and interactions of biomolecules under conditions that closely mimic the natural cellular environment.1 The challenges associated with NMR spectroscopy, particularly in the case of large molecular weight samples, such as limited resolution and sensitivity, have been mitigated by advancements in NMR hardware (higher magnetic fields, cryogenic probes),2 improvements in experimental techniques (multidimensional NMR sequences, transverse relaxation optimized spectroscopy TROSY),3 and various selective isotope labeling strategies. In the context of these strategies, NMR-active nuclei such as carbon-13 or nitrogen-15 are only introduced in certain residue types, while protons are selectively replaced by deuteration.4 Proteins with such defined isotope patterns exhibit clear and unambiguous signals in simplified NMR spectra, which allow to follow more easily the changes in the chemical environment during interactions with binding partners. The resulting chemical shift changes can be analyzed to probe binding pockets and explore large interaction surfaces.5 NMR studies of largely unstructured, very large or even multicomponent biomolecular complexes may require the introduction of bioorthogonal NMR active nuclei, such as fluorine. 19F offers excellent NMR properties, like a high sensitivity (similar to 1H), and a wide chemical shift range (∼400 ppm compared to ∼15 ppm for 1H). Since fluorine is largely absent in biological systems, 19F-NMR serves as a powerful tool to analyze proteins in complex biological matrices, such as in-cell- or in-lysate NMR spectroscopy.6 However, introducing non-proteinogenic fluorine-containing amino acids might significantly alter a protein's conformation and function. Changing hydrogen for fluorine does not result in a drastic steric change, but being the most electronegative element in the periodic table fluorine polarizes bonds, changes pKa values and affects the sensitive networks of hydrogen bond interaction.7
Aryl-fluorinated aromatic residues have been shown to have minimal impact on the function of the target proteins in various applications.7 Additionally, fluorophenylalanines, fluorotyrosines, and fluorotryptophans can be easily incorporated by substituting the natural amino acids with their fluorinated counterparts in E. coli-based expression media.8 This can be achieved by either using auxotrophic host organisms or adding small molecules as inhibitors of the relevant biosynthetic pathways. A notable example of enhancing the uptake efficiency of non-natural amino acids through inhibition is the addition of N-(phosphonomethyl)glycine (glyphosate®) to block the shikimate pathway.9 In the case of fluorotryptophans (fTrp), even metabolic precursors like fluoroindoles are accepted by overexpressing bacterial microorganisms. Alternatively, in-vivo incorporation can be achieved by site-selective fluorination using F-amino acid/tRNA/aminoacyl tRNA synthetase combinations orthogonal to the translation machinery of the host organism.10 This extended genetic encoding strategy reduces the number of fluorine atoms within the target protein sequence, helping to preserve its natural properties. This approach was recently implemented for the site-selective incorporation of various fTrp isomers.10b
For many large proteins, especially for those modified by post-translational modifications, protein production in prokaryotic organisms is not an option. Consequently, protocols for mammalian cell expression have been developed and implemented.11 Recently, Costantino et al. reported on a robust method to incorporate differently fluorinated aromatic amino acids by substituting the target residue with its corresponding fluorinated derivative in HEK cell cultures, transiently transfected with the corresponding gene of interest.12 Building on these findings, we aimed to synthesize a fluorinated tryptophan with a selective 13C/2H isotope pattern (compound 1 in Fig. 1) to be used in mammalian cell overexpression systems.
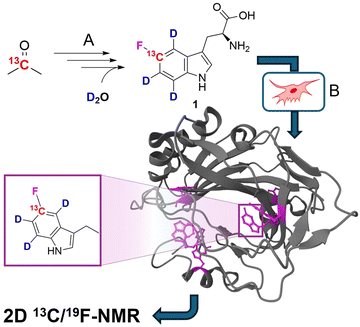 | ||
| Fig. 1 General strategy of selective 19F/13C/2H tryptophan labelling using mammalian cell lines. (A) Synthesis of labelled fTrp 1 by multistep organic synthesis from simple, commercially available isotope sources. (B) Expression of the target protein in mammalian cells by replacing the natural amino acid Trp with compound 1 in the corresponding media. The protein structure shows fTrp-containing carbonic anhydrase 2 (CA2; PDB structure: 8P6U) with fTrp shown in purple. | ||
5-Fluorotryptophan 1 (hereafter referred to as fTrp) contains a 13C–19F spin pair, which enables the additional resolution of overlapping fluorine resonances in the carbon-13 dimension, particularly in large proteins with multiple tryptophan residues. Moreover, Boeszoermenyi et al. reported the observation of 13C signals of remarkable narrow linewidths by exploiting the 19F–13C TROSY effect in large proteins.13 Corresponding synthetic procedures to incorporate such spin pairs have been reported for fluorinated tyrosine, -phenylalanine and -tryptophan.14 The authors of the latter used 13C/19F/2H indoles as precursors in E. coli overexpression – a compound that is not applicable in mammalian-based systems. Therefore, we wanted to close this methodological gap by introducing the corresponding fluorinated amino acid 1. FTrp is especially interesting in this regard, because Trp is generally less abundant compared to Phe and Tyr meaning a fluorinated Trp therefore alters the protein's properties to a lesser extent than fluorinated versions of the other aromatic residues. Additionally, Trp is frequently found at interaction sites, where the large, apolar indole side chain plays a crucial part in intermolecular interaction, contributing to π–π, cation–π or CH–π forces.15 We started our synthetic route with commercially available [2-13C] acetone 2, which we used to assemble the aryl ring by condensation with sodium nitromalonaldehyde as reported in earlier work (Fig. 2).16 The resulting [13C] nitrophenol 3 was selectively deuterated ortho to the OH-moiety under acidic conditions at high temperature (→4). In the next step, dehydroxyfluorination (4 → 5) using phenofluoromix® 11, as described by the group of Ritter, was performed.17 This reaction works best with electron deficient aryl rings, which makes nitrophenol an ideal substrate for this conversion. The electron density of the aromatic ring is drastically changed within the scope of the next reaction, which is the reduction of the nitro-group to the amine 6. This conversion activates the protons ortho to the NH2-group for the exchange with deuterium at elevated temperatures. While reviewing the literature for effective methods to construct the indole system from aniline derivatives and to introduce the asymmetric carbon atom of the target compound in a straightforward and efficient manner, we discovered a method developed by James Cook's group involving alkyne 12.18 This compound can be easily synthesized from the classic chiral Val/Gly Schöllkopf auxiliary. To implement this synthetic approach, we successfully prepared the isotope-labeled fluoro-ortho-iodoaniline 8 using iodine, achieving a satisfactory yield. With compounds 8 and 12 in hand we could apply the palladium catalyzed heteroannulation reaction to prepare the indole-bislactim ether 9. This compound was hydrolyzed to the amino acid ester with concomitant cleavage of the silyl group.18 Final saponification of the ester gave the target compound 1 with high deuteration grades in the aromatic positions (95% for ε-, 96% for ζ- and 93% for η-position, respectively, according to NMR analysis – see spectral characterization in the ESI†). In total, the target compound was prepared in nine consecutive steps with an overall yield of 19%. Notably, the costs for chemicals and reagents are low, at less than €3 per mg of the final compound, with potential for scale-up to gram quantities. The compound was then applied to the expression of two different model proteins using HEK293T cells, namely the 29.3 kDa carbonic anhydrase II (CA2), containing 7 tryptophans and superoxide dismutase (SOD1), with a molecular weight of 32 kDa, containing a single tryptophan. Both proteins were expressed following methods outlined in previous studies, assuming approximately 70% incorporation rates based on these findings; detailed experimental procedures are provided in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 In short, cells were transiently transfected with previously employed mammalian expression vectors19 using polyethylenimine and incubated for 24 h in commercial Dulbecco's modified Eagle's medium (DMEM), and for further 24 h in a modified DMEM prepared in-house containing compound 1 (80 μM) instead of L-tryptophan. After harvesting, the cells were lysed in PBS buffer by freeze–thaw cycles and the soluble fraction was analyzed by 13C–19F NMR spectroscopy. The carbonic anhydrase enzyme family catalyzes the zinc-mediated reversible hydration of carbon dioxide, playing a crucial role in physiological processes such as pH regulation, calcification, signal transduction, and gluconeogenesis.20 Due to their significance in metabolic regulation, there has been growing interest in targeting these enzymes for treating various diseases. Recently, extensive studies have explored the potential of carbonic anhydrase inhibitors as anti-cancer and anti-infective agents.21 Previous work demonstrated that the incorporation of fTrp using the same expression system could reach up to ∼70%, as confirmed by mass spectrometry and NMR, with X-ray analysis revealing negligible conformational changes.12 SOD1, an antioxidant enzyme responsible for catalyzing superoxide degradation, is associated with several central nervous system diseases, such as amyotrophic lateral sclerosis (ALS), Parkinson's, and Alzheimer's diseases, due to its misfolding and aggregation.22 In both cases, CA2 and SOD1, proper function and development of pathological states are strongly affected by Trp residues. In CA2, a histidine proton shuttle is stabilized by a tryptophan side chain, whereas tryptophan mediates aggregation in the case of SOD1.23 Our new labeling pattern enables the observation of 2D signals in both the 19F and 13C dimensions. Fig. 2 shows the 13C–19F TROSY spectra signals of CA2 and SOD1, recorded directly in cell lysates without prior purification. The spectrum in the 19F dimension of Fig. 2A closely resembles that reported in previous studies.12 While four of the signals were previously clustered within a narrow range of −122 to −123 ppm in the 1D fluorine NMR spectrum, they are now clearly resolved into distinct signals in 2D spectra. In the lysate, signals from six tryptophans are detected, while additional signals, which show splitting most probably due to changes in the local protein geometry caused by partial fluorine incorporation, could be detected in a sample of purified CA2 (Fig. S1, ESI†). This observation is in accordance with previously recorded X-ray crystallography data of fTrp-containing CA2 (PDB entry: 8P6U), which indicates that a fluorine atom on the Trp16 side-chain is sterically hindered, although the consequences for the overall structure remain minor.
12 In short, cells were transiently transfected with previously employed mammalian expression vectors19 using polyethylenimine and incubated for 24 h in commercial Dulbecco's modified Eagle's medium (DMEM), and for further 24 h in a modified DMEM prepared in-house containing compound 1 (80 μM) instead of L-tryptophan. After harvesting, the cells were lysed in PBS buffer by freeze–thaw cycles and the soluble fraction was analyzed by 13C–19F NMR spectroscopy. The carbonic anhydrase enzyme family catalyzes the zinc-mediated reversible hydration of carbon dioxide, playing a crucial role in physiological processes such as pH regulation, calcification, signal transduction, and gluconeogenesis.20 Due to their significance in metabolic regulation, there has been growing interest in targeting these enzymes for treating various diseases. Recently, extensive studies have explored the potential of carbonic anhydrase inhibitors as anti-cancer and anti-infective agents.21 Previous work demonstrated that the incorporation of fTrp using the same expression system could reach up to ∼70%, as confirmed by mass spectrometry and NMR, with X-ray analysis revealing negligible conformational changes.12 SOD1, an antioxidant enzyme responsible for catalyzing superoxide degradation, is associated with several central nervous system diseases, such as amyotrophic lateral sclerosis (ALS), Parkinson's, and Alzheimer's diseases, due to its misfolding and aggregation.22 In both cases, CA2 and SOD1, proper function and development of pathological states are strongly affected by Trp residues. In CA2, a histidine proton shuttle is stabilized by a tryptophan side chain, whereas tryptophan mediates aggregation in the case of SOD1.23 Our new labeling pattern enables the observation of 2D signals in both the 19F and 13C dimensions. Fig. 2 shows the 13C–19F TROSY spectra signals of CA2 and SOD1, recorded directly in cell lysates without prior purification. The spectrum in the 19F dimension of Fig. 2A closely resembles that reported in previous studies.12 While four of the signals were previously clustered within a narrow range of −122 to −123 ppm in the 1D fluorine NMR spectrum, they are now clearly resolved into distinct signals in 2D spectra. In the lysate, signals from six tryptophans are detected, while additional signals, which show splitting most probably due to changes in the local protein geometry caused by partial fluorine incorporation, could be detected in a sample of purified CA2 (Fig. S1, ESI†). This observation is in accordance with previously recorded X-ray crystallography data of fTrp-containing CA2 (PDB entry: 8P6U), which indicates that a fluorine atom on the Trp16 side-chain is sterically hindered, although the consequences for the overall structure remain minor.
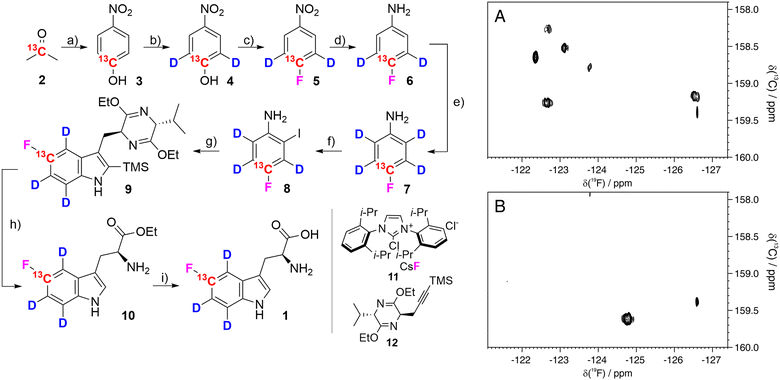 | ||
Fig. 2 Left: Synthesis of [5-13C,4,6,7-2H3]-5-fluoro-L-tryptophan 1: (a) sodium nitromalonoaldehyde, NaOHaq, 0 °C to 4 °C, 6 days; then HCl 6M, 63%; (b) DCl, MW 170 °C, 1.5 h, 98%; (c) phenofluor mix (11), toluene, 110 °C, 24 h, 76%; (d) 10% Pd/C, H2, methanol, RT, overnight, 95%; (e) DCl, MW 180 °C, 1.5 h, 95%; (f) I2, NaHCO3, H2O/dichloromethane 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, RT, 30 h, 87%; (g) compound 12, Pd(OAc)2, LiCl, Na2CO3, DMF, 100 °C, 30 h, 70%; (h) 6 N HCl, THF, 0 °C, 4h; then ice, NH4OH, 79%; (i) NaOH 1N, ethanol, 50 °C, 2 h; then HCl 2N, 94%. Right: 13C–19F TROSY spectra of lysates from HEK293T cells expressing CA2 (A) and SOD1 (B). Experimental details are given in the ESI.† 2, RT, 30 h, 87%; (g) compound 12, Pd(OAc)2, LiCl, Na2CO3, DMF, 100 °C, 30 h, 70%; (h) 6 N HCl, THF, 0 °C, 4h; then ice, NH4OH, 79%; (i) NaOH 1N, ethanol, 50 °C, 2 h; then HCl 2N, 94%. Right: 13C–19F TROSY spectra of lysates from HEK293T cells expressing CA2 (A) and SOD1 (B). Experimental details are given in the ESI.† | ||
As expected, the spectrum of SOD1 (Fig. 2B) contains only the signal from Trp32. An additional sharp signal is present in both spectra at −126.6 ppm and likely arises from free fTrp which could interact with the protein and is then slowly released in the lysate, as shown by the increase of its signal intensity over time. In summary, we developed an efficient synthetic route to produce [13C/2H3] fTrp 1 and successfully used it to express two model proteins (CA2 and SOD1) in HEK cells. The corresponding 2D-TROSY spectra of the cell lysates display well-resolved signals, demonstrating the clear advantage of adding a 13C label adjacent to the fluorine atom. These spectra can be utilized for interaction studies, analysis of conformational properties, and investigation of protein dynamics. The specific effect of deuteration on 19F−13C TROSY linewidths is still under investigation but is expected to improve transverse relaxation in the 13C evolution period and remove 2JCH couplings. Moreover, compound 1 is not limited to mammalian overexpression systems but can also be used in E. coli, yeast, cell-free and even genetic encoding methods. Additional applications, particularly involving in-cell NMR, are currently part of our ongoing research.
This work was supported by the Christian Doppler Laboratory for High-Content Structural Biology and Biotechnology (to G. T.), by Instruct-ERIC and specifically the CERM/CIRMMP Italian Instruct Centre, by Fragment-Screen, No. 101094131, funded by the Horizon Europe program of the EC (to L. B.), and by ITACA. SB “Potentiating the Italian Capacity for Structural Biology Services in Instruct-ERIC” (IR0000009) within the call MUR 3264/2021 PNRR M4/C2/L3.1.1, funded by the European Union–Next Generation EU.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
RJL and RK are shareholders of the company Mag-Lab, which is mentioned in the affiliations. There are no other conflicts to declare.Notes and references
- (a) I. R. Kleckner and M. P. Foster, Biochim. Biophys. Acta, 2011, 1814(8), 942 CrossRef CAS PubMed; (b) A. Bax and G. M. Clore, J. Magn. Reson., 2019, 306, 187 CrossRef CAS PubMed; (c) J. A. Purslow, B. Khatiwada, M. J. Bayro and V. Venditti, Front. Mol. Biosci., 2020, 7, 9 CrossRef CAS PubMed; (d) Y. Hu, K. Cheng, L. He, X. Zhang, B. Jiang, L. Jiang, C. Li, G. Wang, Y. Yang and M. Liu, Anal. Chem., 2021, 93, 1866 CrossRef CAS PubMed; (e) E. Luchinat, M. Cremonini and L. Banci, Chem. Rev., 2022, 122(10), 9267 CrossRef CAS PubMed.
- (a) E. Moser, E. Laistler, F. Schmitt and G. Kontaxis, Front. Phys., 2017, 5, 33, DOI:10.3389/fphy.2017.00033; (b) H. Kovacs, D. Moskau and M. Spraul, Prog. Nucl. Magn. Reson. Spectrosc., 2005, 2–3, 131 CrossRef.
- (a) M. Salzmann, K. Pervushin, G. Wider, H. Senn and K. Wüthrich, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13585 CrossRef CAS PubMed; (b) M. Sattler, J. Schleucher and C. Griesinger, Prog. Nucl. Magn. Reson. Spectrosc., 1999, 34(2), 93 CrossRef CAS; (c) C. Fernández and G. Wider, Curr. Opin. Struct. Biol., 2003, 13(5), 570 CrossRef PubMed.
- (a) M. Kainosho, T. Torizawa, Y. Iwashita, T. Terauchi, A. M. Ono and P. Güntert, Nature, 2006, 440, 52 CrossRef CAS PubMed; (b) S.-Y. Ohki and M. Kainosho, Prog. Nucl. Magn. Reson. Spectrosc., 2008, 53(4), 208 CrossRef CAS; (c) D. Lacabanne, B. H. Meier and A. Böckmann, J. Biomol. NMR, 2018, 71, 141 CrossRef CAS PubMed; (d) J. Schörghuber, L. Geist, G. Platzer, M. Feichtinger, M. Bisaccia, L. Scheibelberger, F. Weber, R. Konrat and R. J. Lichtenecker, J. Biomol. NMR, 2018, 71(3), 129 CrossRef PubMed; (e) B. Rowlinson, E. Crublet, R. Kerfah and M. J. Plevin, Biochem. Soc. Trans., 2022, 50(6), 1555 CrossRef CAS PubMed.
- (a) M. P. Williamson, Prog. Nucl. Magn. Reson. Spectrosc., 2013, 73, 1 CrossRef CAS PubMed; (b) W. Becker, K. C. Bhattiprolu, N. Gubensäk and K. Zangger, Chem. Phys. Chem., 2018, 19(8), 895 CrossRef CAS PubMed.
- (a) J. L. Kitevski-LeBlanc and R. S. Prosser, Prog. Nucl. Magn. Reason. Spectrosc., 2012, 62, 1 CrossRef CAS PubMed; (b) H. Chen, S. Viel, F. Ziarelli and L. Peng, Chem. Soc. Rev., 2013, 42(20), 7971 RSC; (c) K. E. Arntson and W. C. K. Pomerantz, J. Med. Chem., 2016, 59(11), 5158 CrossRef CAS PubMed; (d) C. T. Gee, K. E. Arntson, A. K. Urick, N. K. Mishra, L. M. L. Hawk, A. J. Wisniewski and W. C. K. Pomerantz, Nat. Protoc., 2016, 11(8), 1414 CrossRef PubMed; (e) A. Divakaran, S. E. Kirberger and W. C. K. Pomerantz, Acc. Chem. Res., 2019, 52(12), 3407 CrossRef CAS PubMed; (f) D. Gimenez, A. Phelan, C. D. Murphy and S. L. Cobb, Beilstein J. Org. Chem., 2021, 17(1), 293 CrossRef CAS PubMed; (g) A. M. Gronenborn, Structure, 2022, 30(1), 6 CrossRef CAS PubMed; (h) W. Zhu, A. J. Guseman, F. Bhinderwala, M. Lu, X.-C. Su and A. M. Grunenborn, Angew. Chem., Int. Ed., 2022, 61(23), e202201097 CrossRef CAS PubMed.
- (a) M. Salwiczek, E. K. Nyakatura, U. I. M. Gerling, S. Ye and B. Koksch, Chem. Soc. Rev., 2012, 41, 2135 RSC; (b) H. Welte, T. Zhou, X. Mihajlenko, O. Mayans and M. Kovermann, Sci. Rep., 2020, 10, 2640 CrossRef CAS PubMed; (c) D. T. Yang, A. M. Gronenborn and L. T. Chong, J. Phys. Chem. A, 2022, 126, 2286 CrossRef CAS PubMed.
- (a) C. Li, G.-F. Wang, Y. Wang, R. Creager-Allen, E. A. Lutz, H. Scronce, K. M. Slade, R. A. S. Ruf, R. A. Mehl and G. J. Pielak, J. Am. Chem. Soc., 2010, 132(1), 321 CrossRef CAS PubMed; (b) P. B. Crowley, C. Kyne and W. B. Monteith, Chem. Commun., 2012, 48(86), 10681 RSC; (c) M. G. Lete, A. Franconetti, S. Bertuzzi, S. Delgado, M. Azkargorta, F. Elortza, O. Millet, G. Jiménez-Osés, A. Arda and J. Jiménez-Barbero, Chem. – Eur. J., 2023, 29(5), e202202208 CrossRef CAS PubMed.
- H.-W. Kim, J. A. Perez, S. J. Ferguson and I. D. Campbell, FEBS Lett., 1990, 1(2), 34 CrossRef PubMed.
- (a) J. C. Jackson, J. T. Hammill and R. A. Mehl, J. Am. Chem. Soc., 2007, 129(5), 1160 CrossRef CAS PubMed; (b) H. Qianzhu, E. H. Abdelkader, G. Otting and T. Huber, J. Am. Chem. Soc., 2024, 146(19), 13641 CrossRef CAS PubMed.
- (a) L. Barbieri, E. Luchinat and L. Banci, Nat. Protoc., 2016, 11(6), 1101 CrossRef CAS PubMed; (b) A. R. Aricescu, W. Lu and E. Y. Jones, Acta. Cryst. D, 2006, 62(10), 1243 CrossRef PubMed.
- (a) L. B. T. Pham, A. Costantino, L. Barbieri, V. Calderone, E. Luchinat and L. Banci, J. Am. Chem. Soc., 2023, 145(2), 1389 CrossRef CAS PubMed; (b) A. Costantino, L. B. T. Pham, L. Barberi, V. Calderone, G. Ben-Nissan, M. Sharon, L. Banci and E. Luchinat, Protein Sci., 2024, 33(3), e4910 CrossRef CAS PubMed.
- (a) A. Boeszoermenyi, S. Chhabra, A. Dubey, D. L. Radeva, N. T. Burdzhiev, C. D. Chanev, O. I. Petrov, V. M. Gelev, M. Zhang, C. Anklin, H. Kovacs, G. Wagner, I. Kuprov, K. Takeuchi and H. Arthanari, Nat. Methods, 2019, 16, 333 CrossRef CAS PubMed; (b) A. Boeszoermenyi, B. Ogórek, A. Jain, H. Arthanari and G. Wagner, J. Biomol. NMR, 2020, 74, 365 CrossRef CAS PubMed.
- (a) A. Maleckis, I. D. Herath and G. Otting, Org. Biomol. Chem., 2021, 19, 5133 RSC; (b) G. Toscano, J. Holzinger, B. Nagl, G. Kontaxis, H. Kählig, R. Konrat and R. J. Lichtenecker, J. Biomol. NMR, 2024, 78, 139 CrossRef CAS PubMed.
- J. Shao, B. P. Kuiper, A.-M. W. H. Thunnissen, R. H. Cool, L. Zhou, C. Huang, B. W. Dijkstra and J. Broos, J. Am. Chem. Soc., 2022, 144(30), 13815 CrossRef CAS PubMed.
- R. J. Lichtenecker, Org. Biomol. Chem., 2014, 38(12), 7551 RSC.
- T. Fujimoto and T. Ritter, Org. Lett., 2015, 17(3), 544 CrossRef CAS PubMed.
- (a) C. Ma, S. Yu, X. He, X. Liu and J. M. Cook, Tetrahedron Lett., 2000, 41(16), 2781 CrossRef CAS; (b) C. Ma, X. Liu, X. Li, J. Flippen-Anderson, S. Yu and J. M. Cook, J. Org. Chem., 2001, 66(13), 4525 CrossRef CAS PubMed.
- E. Luchinat, L. Barbieri, M. Cremonini, A. Nocentini, C. T. Supuran and L. Banci, Angew. Chem., Int. Ed., 2020, 59(16), 6535 CrossRef CAS PubMed.
- S. Lindskog, Pharmacol. Ther., 1997, 74(1), 1 CrossRef CAS PubMed.
- S. Kumar, S. Rulhania, S. Jaswal and V. Monga, Eur. J. Med. Chem., 2021, 209, 112923 CrossRef CAS PubMed.
- M. Berdyński, P. Miszta, K. Safranow, P. M. Andersen, M. Morita, S. Filipek, C. Źekanowski and M. Kuźma-Kozakiewicz, Sci. Rep., 2022, 12, 103 CrossRef PubMed.
- (a) R. Mikulski, J. F. Domsic, G. Ling, C. Tu, A. H. Robbins, D. N. Silverman and R. McKenna, Arch. Biochem. Biophys., 2011, 516(2), 97 CrossRef CAS PubMed; (b) E. Pokrishevsky, L. McAlary, N. E. Farrawell, B. Zhao, M. Sher, J. J. Yerbury and N. R. Cashman, Sci. Rep., 2018, 8, 15590 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details concerning organic synthesis, protein expression and NMR spectroscopy. See DOI: https://doi.org/10.1039/d4cc04789c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |
