 Open Access Article
Open Access ArticleRecent advances in nucleic acid signal amplification-based aptasensors for sensing mycotoxins
Dandan
Zhang
a,
Ting
Luo
a,
Xiangyue
Cai
a,
Ning-ning
Zhao
*b and
Chun-yang
Zhang
 *c
*c
aCollege of Materials and Chemistry & Chemical Engineering, Chengdu University of Technology, Chengdu 610059, Sichuan, China
bCollege of Chemistry, Chemical Engineering and Materials Science, Shandong Normal University, Jinan 250014, China. E-mail: zhaoningning1997@qq.com
cSchool of Chemistry and Chemical Engineering, Southeast University, Nanjing, 211189, China. E-mail: zhangcy@seu.edu.cn
First published on 9th April 2024
Abstract
Mycotoxin contamination in food products may cause serious health hazards and economic losses. The effective control and accurate detection of mycotoxins have become a global concern. Even though a variety of methods have been developed for mycotoxin detection, most conventional methods suffer from complicated operation procedures, low sensitivity, high cost, and long assay time. Therefore, the development of simple and sensitive methods for mycotoxin assay is highly needed. The introduction of nucleic acid signal amplification technology (NASAT) into aptasensors significantly improves the sensitivity and facilitates the detection of mycotoxins. Herein, we give a comprehensive review of the recent advances in NASAT-based aptasensors for assaying mycotoxins and summarize the principles, features, and applications of NASAT-based aptasensors. Moreover, we highlight the challenges and prospects in the field, including the simultaneous detection of multiple mycotoxins and the development of portable devices for field detection.
1. Introduction
Food safety has attracted global attention. Food contamination affects the multistep process of the food supply chain from farms to dining tables, and it may originate from agricultural practices, the storage and transportation steps of raw agricultural commodities, industrial food processing, and the storage of foods. The presence of mycotoxins in foodstuff is a most conspicuous and serious concern.1 Approximately 25% of the world's foods are tainted with mycotoxins every year, and their presence in food affects its quality and safety and may cause serious economic losses and safety risks.2,3Mycotoxins are secondary metabolites of fungal species (e.g., Aspergillus, Penicillium, Alternaria, Fusarium, Claviceps, and Stachybotrys species).4 Due to their teratogenic, mutagenic, carcinogenic, immunosuppressive, and endocrine-disrupting effects, low concentrations of mycotoxins can cause chronic toxicity to humans and other animals through the food chain.5–9 Most mycotoxins have unique chemical and thermal stability, making them difficult to remove during conventional food processing and disinfection.2,10 To date, about 400 different mycotoxins have been identified, but only few species are the concerns of food safety. Based on the carcinogenic risk of mycotoxins to human health, International Agency for Research on Cancer (IARC) classifies them into different categories, such as aflatoxins (AFs), zearalenone (ZEN), T2/HT2, deoxynivalenol (DON), patulin (PAT), and ochratoxin A (OTA).4,6,8,9,11–13 Among them, aflatoxin B1 (AFB1) is one of the most toxic and ubiquitous potential carcinogens, and it is classified into Group 1A. AFB1 may cause lung cancer, hepatocellular cancer, gallbladder cancer, and colon cancer in humans. Approximately 28.2% of liver cancers are induced by AFB1.14,15 OTA and fumonisin are possibly carcinogenic to humans and are classified into Group 2B.16–19Table 1 lists the chemical structures of major mycotoxins and the affected food commodities as well as their toxic effects on humans and animals. The European Union, the Food and Drug Administration of the USA, and the Chinese Pharmacopoeia Commission have established regulatory guidelines and maximum residue levels (MRLs) for the majority of toxic mycotoxins prevalent in certain commodities to protect human health (Table 1). To meet these strict requirements, the development of sensitive and reliable analytical methods to detect mycotoxins in diverse matrices at trace levels is extremely urgent.9
| Mycotoxins | Structure | Molar mass (g mol−1) | Commodities affected | Toxic effects | Country | Maximum permitted lever (μg kg−1) | Ref. |
|---|---|---|---|---|---|---|---|
| a Regulations (EC) No. 2002/32/EC, 1881/2006, 2021/1399. b 2013/165/EU: Commission. Recommendation. c The number in brackets refers to infant food and young children. | |||||||
| AFB1 |
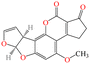
|
312.27 | Grains such as maize, rice, wheat, milk and milk products, etc. | Carcinogenic, teratogenic, mutagenic, immunosuppressive | China | 5–20(0.5)c | 55 |
| EU | 2–12(0.1)c | ||||||
| U.S. | 20 | ||||||
| OTA |
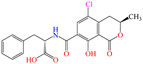
|
224.19 | Cereals, wine, coffee, cocoa, beans, dried fruits, nuts, spices, cheese, etc. | Nephrotoxic, hepatotoxic, neurotoxic, teratogenic, immunotoxic | China | 2–10 | 56 |
| EU | 2–80(0.5)c | ||||||
| U.S. | Not set | ||||||
| DON |
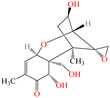
|
296.32 | Cereals, cereal products | Diarrhea, vomiting, anorexia, immune dysregulation | China | 1000 | 57 |
| EU | 500–1750, (200)c | ||||||
| U.S. | 1000 | ||||||
| ZEN |
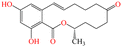
|
318.36 | Cereals, cereal products, maize, rice, beer, etc. | Hyperoestrogenic, hepatotoxic, haematotoxic, immunotoxic, genotoxic | China | 60 | 58 |
| EU | 50–400(20)c | ||||||
| U.S. | Not set | ||||||
| AFM1 |
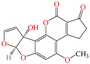
|
584.87 | Milk, dairy | Carcinogenic, cytotoxic, hepatotoxic | China | 0.5 | 55 |
| EU | 0.05(0.025)c | ||||||
| U.S. | 0.5 | ||||||
| T-2 |
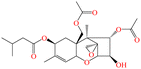
|
466.52 | Wheat, rye, maize, soybeans | Growth retardation, myelotoxic, hemotoxic, necrotic lesions on contact sites | China | Not set | 59 |
| EU | In preparationb | ||||||
| U.S. | 2000–4000 | ||||||
| PAT |
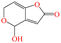
|
154.12 | Fruits and vegetables | Nausea, vomiting, and other gastrointestinal symptoms, kidney damage | China | 50 | 60 |
| EU | 25–50, (10)c | ||||||
| U.S. | 50 | ||||||
Chromatography is the earliest and most conventional method for mycotoxin assay,20 including high-performance liquid chromatography (HPLC), thin layer chromatography (TLC), gas chromatography (GC), liquid chromatography-tandem mass spectrometry (LC-MS), HPLC coupled with mass spectroscopy (HPLCMS).21–25 Despite the high sensitivity and accuracy of these methods, they often require expensive instruments, professional operators, and cumbersome pre-treatment steps.26 Antibody-based immunoassays (e.g., enzyme-linked immunosorbent assay (ELISA), lateral flow immunoassays (LFIA), and colloidal gold immune chromate graphic assay (GICA))27–29 can achieve the rapid and accurate quantification of mycotoxin.30,31 However, these methods suffer from semi-quantification, false positive results, poor repeatability, and long reaction times.32 In addition, the antibodies used in these methods suffer from complex preparation processes, high costs, poor stability, and low immunogenicity, which limits their wider application.
Aptamers are single-stranded oligonucleotides (DNA or RNA) that mimic monoclonal antibodies and are screened through a selection process, namely the systematic evolution of ligands by exponential enrichment (SELEX).33 Compared with antibodies, aptamers have distinct advantages of high binding affinity, good specificity, excellent stability, easy modification, low cost, little batch-to-batch differences, and a wide range of targets.33–36 As a new kind of highly selective biological recognition element, aptamers have been widely used in the construction of biosensors for mycotoxin assays.37–46 In terms of sensor components, aptasensors usually include an aptamer-based target recognizer and a signal transducer. Because mycotoxins are usually measured in ppb (parts per billion), the achievement of a detection limit in nanograms has become a great challenge.47 Therefore, the development of nucleic acid signal amplification-based aptasensors is crucial for the achievement of sensitive detection of mycotoxins in real samples.
2. Aptamer sequences used in mycotoxin detection
The capability of aptamers to specifically identify target molecules is critical to improving the selectivity and sensitivity of aptasensors, and the integration of SELEX with graphene oxide (GO) has been applied to screen mycotoxin aptamers with high affinity.48–53 The non-specific adsorption of the single-stranded DNA (ssDNA) random library on GO is the basis for separating unbound DNA in the GO-SELEX process.54 The individual steps involved in one GO-SELEX and GO-counter SELEX round are illustrated in Fig. 1. In the first step of affinity selection, a preincubated mixture of the random ssDNA library and the target is incubated with GO solution. Subsequently, the free unbound ssDNAs are adsorbed on GO and then removed by centrifugation. The supernatant with ssDNAs bound to the target is then recovered, amplified, and purified for a further GO-counter SELEX process to eliminate the false positive binding of aptamers. In the GO-counter SELEX process, affinity-based desorption is triggered by incubating GO with the target to restore the adsorbed ssDNA on GO. Eventually, the recovered ssDNAs are purified, cloned, and sequenced to obtain the final hit aptamers. A series of mycotoxin aptamers have been successfully screened (Table 2),61–68 including AFB1, OTA, Aflatoxin M1 (AFM1), and ZEN. Notably, the aptamers for other mycotoxins are rarely reported, except for the aptamers of the PAT and T-2 toxins. Due to the complexity of food substrates and the trace levels of mycotoxins in the substrates, the sensitive detection of mycotoxins remains a challenge.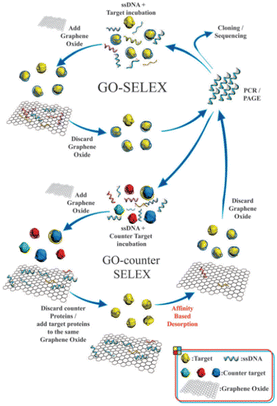 | ||
| Fig. 1 Schematic illustration of the steps involved in the GO-SELEX procedure of aptamer identification.54 | ||
| Target | Aptamer sequence (from 5′ to 3′) | Kd (nM) | Ref. |
|---|---|---|---|
| OTA | GATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACA | 0.36 | 52 |
| AGCCTCGTCTGTTCTCCCGGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTGGGGAAGACAAGCAGACGT | 290 ± 150 | 49 | |
| GGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTG | 406 ± 166 | 94 | |
| AFB1 | GTTGGGCACGTGTTGTCTCTCTGTGTCTCGTGCCCTTCGCTAGGCCCACA | 0.59 ± 0.33 | 95 |
| AGCAGCACAGAGGTCAGATGGTGCTATCATGCGCTCAATGGGAGACTTTAGCTGCCCCCACCTATGCGTGCTACCGTGAA | 11.29 ± 1.27 | 96 | |
| CTCGTCTCGTTCTCTCAGTCTGGTTAGCACGGGACCCAGGCTATTGGCAACTTCTAGTCCGACACGAAGAAGAAGGAGGA | 48.29 ± 9.45 | 97 | |
| AFM1 | GTTGGGCACGTGTTGTCTCTCTGTGTCTCGTGCCCTTCGCTAGGCCCACA | 10 | 98 |
| ATCCGTCACACCTGCTCTGACGCTGGGGTCGACCCGGAGAAATGCATTCCCCTGTGGTGTTGGCTCCCGTAT | 35.6 ± 2.9 | 99 | |
| ZEN | TCATCTATCTATGGTACATTACTATCTGTAATGTGATATG | 41 ± 5 | 100 |
| DON | GCATCACTACAGTCATTACGCATCGTAGGGGGGATCGTTAAGGAAGTGCCCGGAGGCGGTATCGTGTGAAGTGCTGTCCC | — | 101 |
| T-2 | CAGCTCAGAAGCTTGATCCTGTATATCAAGCATCGCGTGTTTACACATGCGAGAGGTGAAGACTCGAAGTCGTGCATCTG | 20.8 ± 3.1 | 48 |
| PAT | GGCCCGCCAACCCGCATCATCTACACTGATATTTTACCTT | 21.83 ± 5.022 | 102 |
3. Nucleic acid signal amplification techniques
To achieve the sensitive detection of trace mycotoxins in complex matrices, signal amplification strategies have been introduced into the construction of aptasensors.69,70 The nucleic acid signal amplification technique (NASAT) takes advantage of various enzymes, nucleases, and deoxyribozyme (DNAzyme) to amplify the signal,71,72 and the integration of NASAT with aptamers can greatly improve the sensitivity of the mycotoxin assay.66–80 NASAT is divided into enzyme-assisted nucleic acid amplification and enzyme-free amplification. Enzyme-assisted nucleic acid amplification includes polymerase chain reaction (PCR), rolling circle amplification (RCA), strand displacement amplification (SDA), nuclease-assisted amplification, and CRISPR/Cas system-related amplification. Enzyme-free amplification includes catalytic hairpin assembly (CHA), hybridization chain reaction (HCR), DNA walker, DNA tweezer, and DNAzyme.3.1. Enzymatic amplification-based aptasensors
In recent years, enzymatic amplification technology has been widely used for mycotoxin detection. The enzyme-assisted nucleic acid amplification techniques mainly focus on PCR, RCA, SDA, nuclease-assisted strategies, and the CRISPR/Cas system.Walter et al. developed a target-induced dissociation (TID)-based aptamer-assisted real-time PCR assay to detect OTA (Fig. 2A).75 The OTA aptamers are immobilized on the dT-modified magnetic beads (dT beads, where dT is d(T)25). When OTA is present, the aptamer is released from the dT beads as a result of TID, and the magnetic beads are separated with a magnetic stand. The released aptamers are amplified and quantified by qPCR. This assay can sensitively detect OTA with a limit of detection (LOD) of 0.009 ng mL−1. Moreover, the Apta-qPCR can be easily extended to detect other mycotoxins.
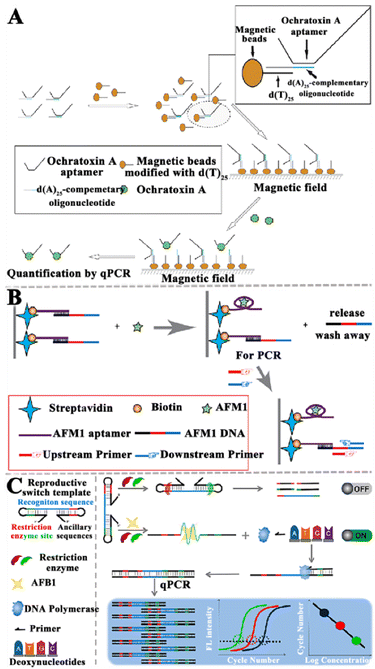 | ||
| Fig. 2 The construction of aptasensors based on PCR-assisted signal amplification for the mycotoxin assay. (A) The construction of an RT-qPCR-based aptasensor for the OTA assay.75 (B) The construction of an RT-qPCR-based aptasensor for the AFM1 assay.76 (C) Construction of a qPCR-based aptasensor for the AFB1 assay.77 | ||
Wang et al. constructed an aptamer-based biosensor to sensitively and selectively detect AFM1 (Fig. 2B).76 The aptamer is immobilized based on strong interactions between biotin and streptavidin. The presence of AFM1 induces the release of complementary ssDNA due to the formation of an aptamer/AFM1 complex, and then the released ssDNA acts as the template for RT-qPCR amplification. Finally, AFM1 can be quantified according to the linear relationship between the change in PCR amplification signal and the AFM1 level. This aptasensor exhibits high sensitivity with a LOD of 0.03 ng L−1, and it can be applied in the quantitative detection of AFM1 in infant rice cereal and infant milk powder samples. Notably, this approach is potentially useful for food safety analysis, and it can be extended to the detection of a large number of targets.
He et al. established a qPCR method-based aptasensor for the sensitive detection of AFB1 (Fig. 2C).77 When AFB1 is present, the template can be switched from a dumbbell-shaped structure to a linear structure. Due to the restriction, enzymes cannot act on the linear template, and the linear DNA templates are subjected to qPCR for the sensitive quantification of AFB1. This method achieves a LOD of 0.03 pg mL−1 and it can be applied for quantification of AFB1 in complex foods (e.g., soy sauce, milk, yellow wine, and peanut butter). This work extends the application of qPCR to non-nucleic acid detection, providing a new approach for the sensitive detection of small-molecule toxicants in foods. Although PCR is the most widely used technology for nucleic acid signal amplification, it still has some shortcomings. For instance, PCR requires thermal cycle amplification instruments and complex amplification procedures, which cannot be implemented at ambient temperature. In addition, some false-positive results and cross-contamination from amplicons often occur in PCR amplification.78,79
Various RCA-based aptasensors have been established for mycotoxin detection. Chen et al. developed an electrochemical aptasensor to detect OTA based on RCA-assisted signal amplification (Fig. 3A).103 In this aptasensor, the RCA primer is composed of a two-part sequence, including the aptamer sequence of OTA and a partially complementary sequence of the capture probe on the electrode surface. When OTA is present, it preferentially binds with the RCA primer, inducing the disassociation of primer from the padlock. Under these circumstances, RCA cannot be initiated, and the primer cannot be prolonged. As a result, methylene blue (MB) cannot bind to the guanine bases of DNA molecules, inducing a decrease in the electrochemical signal. This aptasensor shows good specificity and high sensitivity with a LOD of 0.065 pg mL−1, and it can accurately measure OTA in wine samples. Hu et al. constructed a colorimetric liquid crystal (LC)-based aptasensor for AFB1 detection (Fig. 3B). The microdroplets were formed by introducing the aqueous mixture solution of horseradish peroxidase (HRP) and sodium dodecyl sulfate (SDS) into the LCs, encapsulating HRP in the microdroplets and stabilizing with SDS. Upon the addition of cetyl trimethyl ammonium bromide (CTAB), HRP is ejected from the LCs into the overlaying aqueous solution due to the interfacial charge interaction, and it subsequently catalyses the colorless 3,3′,5,5′-tetramethylbenzidine (TMB) into yellow products. In the presence of AFB1, the binding of the aptamer with AFB1 induces the dissociation of the aptamer/primer duplex decorated on MBs, initiating the RCA reaction to generate long-chain ssDNAs on magnetic beads (MBs).104 Due to the complete capture of CTAB by the amplified ssDNA, the release of HRP into the aqueous solution is prevented and no color change is observed. This assay can sensitively detect AFB1 with a LOD of 0.014 pg mL−1, and it can measure AFB1 in rice and peanut oil. Wang and co-workers reported a competitive “signal-off” fluorescent aptasensor for AFM1 detection based on the combination of time-resolved fluorescent nanoparticles (TRFNPs) with RCA (Fig. 3C).105 In this assay, the AFM1 aptamer serves as an RCA primer for DNA amplification. In the absence of AFM1, the rolling circle template (RCT) is complementary to the aptamer and ligated by T4 DNA ligase to generate the RCA template. Upon the addition of phi29 DNA polymerase, the RCA reaction is initiated to generate long ssDNA roll-loop amplification products (RCAPs) containing multiple AFM1 aptamers. Since TRFNPs-cDNA complements the aptamer sequence to form an RCAP/TRFNPs-cDNA duplex that cannot be adsorbed on graphitic carbon nitride (g-C3N4) nanosheets, the fluorescence of TRFNPs is not quenched. When AFM1 is present, the aptamer recognizes the target AFM1 to form an aptamer-target complex, and there is neither the occurrence of RCA nor the formation of the duplex. The free signal probe TRFNPs-cDNA adsorbs on the g-C3N4, resulting in the quenching of fluorescence. The assay exhibited a LOD of 0.0194 pg mL−1 and a recovery ratio of 92–99.8%.
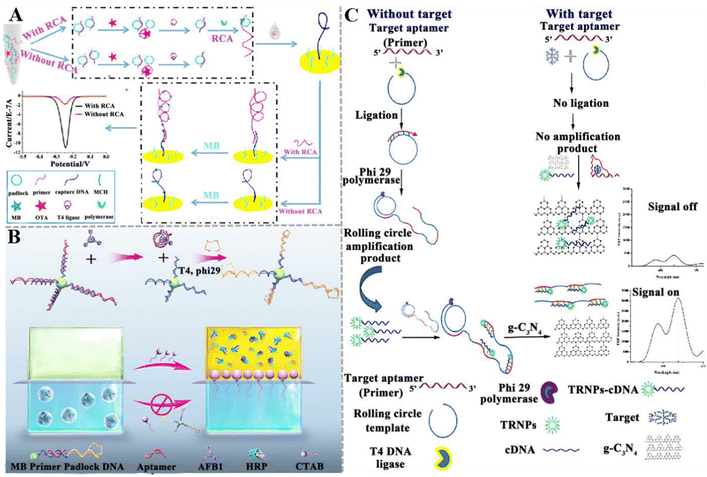 | ||
| Fig. 3 The construction of aptasensors based on RCA-assisted signal amplification for mycotoxin detection. (A) The construction of an electrochemical aptasensor for the OTA assay based on RCA-assisted signal amplification.103 (B) The construction of a colorimetric aptasensor for the AFB1 assay based on RCA-assisted signal amplification.104 (C) The construction of a fluorescent aptasensor for the AFM1 assay based on TRFNPs combined with RCA.105 | ||
Although a series of RCA-based aptasensors have been developed for the sensitive detection of mycotoxins, there are still some limitations. For example, the efficiency of RCA amplification is easily affected by the lock-probe hybridization and the efficiency of the probe-template connection and single-stranded template restriction. When the RCA template is linear DNA, the circularization of the RCA template requires additional enzymes and procedures, increasing the cost and operational complexity.
Xu et al. developed a target-induced SDA electrochemical aptasensor for the detection of ZEN (Fig. 4A).110 When ZEN is present, it binds with the aptamer section of multifunctional hairpin DNA anchored on the surface of polydopamine nanospheres (PDANSs, which act as a carrier), leading to the release of multifunctional hairpin DNAs into the solution. Then, the SDA reaction is initiated with the assistance of polymerase and endonuclease, producing abundant negatively charged dsDNA. Methylene blue (MB) molecules are embedded into the dsDNA grooves to obtain the complex with a negative charge. These complexes are electrostatically repelled on the ITO electrode with a negative charge, inducing a decrease in current intensity. This electrochemical aptasensor can detect ZEN in corn flour with a LOD of 0.18 pg mL−1. This electrochemical aptasensor can be used to monitor other mycotoxins by simply changing the corresponding aptamer. Gao et al. developed a fluorescent aptasensor based on a programmable DNA tweezer and SDA for the detection of AFB1 and ZEN (Fig. 4B).111 They synthesized the aptamer-functionalized magnetic nanoparticles (MNPs). When target AFB1 or ZEN is present, it binds with the aptamer, liberating the cDNA after magnetic separation. The hairpin-structure DNA is complementarily paired with the cDNA, inducing the opening of the initial hairpin structure to form another hairpin structure, initiating the SDA reaction with the aid of KF polymerase and Nb.BbvCI to achieve exponential amplification of cDNA. Subsequently, the amplification product cDNAs are added to the DNA tweezers system to generate a distinct signal output. This strategy shows high sensitivity with a LOD of 0.933 pg mL−1 for AFB1 and 1.07 pg mL−1 for ZEN. Moreover, this method can be easily adapted to detect other analytes by just substituting the aptamer and cDNA.
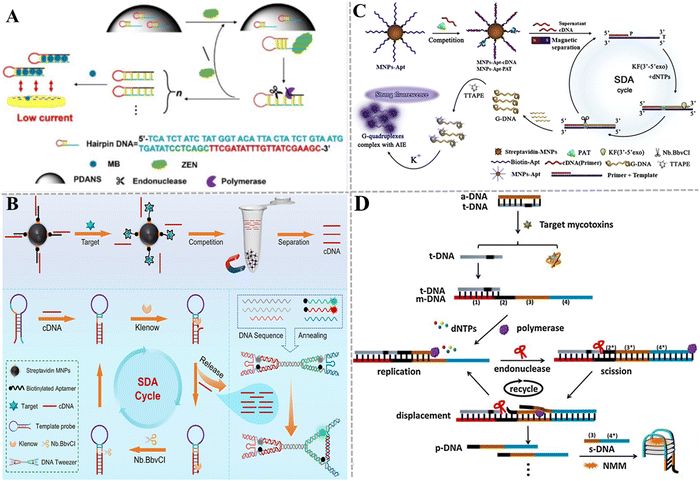 | ||
| Fig. 4 The construction of aptasensors based on SDA for mycotoxin detection. (A) The construction of an electrochemical aptasensor for the ZEN assay based on target-induced SDA.110 (B) The construction of a fluorescent aptasensor based on programmable DNA tweezers and SDA for the detection of AFB1 and ZEN.111 (C) The construction of an aptasensor based on SDA and DNA G-quadruplex for the PAT assay.112 (D) The construction of a label-free aptasensor for AFM1 and OTA detection.113 | ||
Liu et al. took advantage of SDA, DNA G-quadruplex, and aggregation-induced emission (AIE) to develop an aptasensor for the sensitive and rapid detection of PAT (Fig. 4C).112 The aptamer is modified on magnetic beads via biotin–streptavidin interaction. After the cDNA and PAT competitively bind with aptamers, the cDNA is separated by magnetic separation and serves as a primer of SDA to generate plenty of G-rich ssDNAs. The G-rich ssDNA can form the G-quadruplex that can be stained by TTAPE dye to produce a fluorescence signal in the presence of K+, and the aggregation of more TTAPEs generates a stronger fluorescence signal. This assay exhibits good specificity and high sensitivity with a LOD of 0.042 pg mL−1. Gao et al. presented a label-free method based on SDA and a G-quadruplex-specific fluorescence probe for AFM1 and OTA detection (Fig. 4D).113 In this assay, the aptamer (a-DNA) hybridizes partly with a DNA strand (t-DNA). The presence of target mycotoxin triggers SDA to generate plenty of DNA (p-DNA) products with the assistance of polymerase and nicking endonuclease. Subsequently, p-DNA and s-DNA form a G-quadruplex that can act as the signal-transducing element to enhance the fluorescence of N-methyl mesoporphyrin IX (NMM). This strategy can distinguish different target mycotoxins using the corresponding aptamers. The LOD is 17.79 ng kg−1 for AFM1 and 18.98 ng kg−1 for OTA.
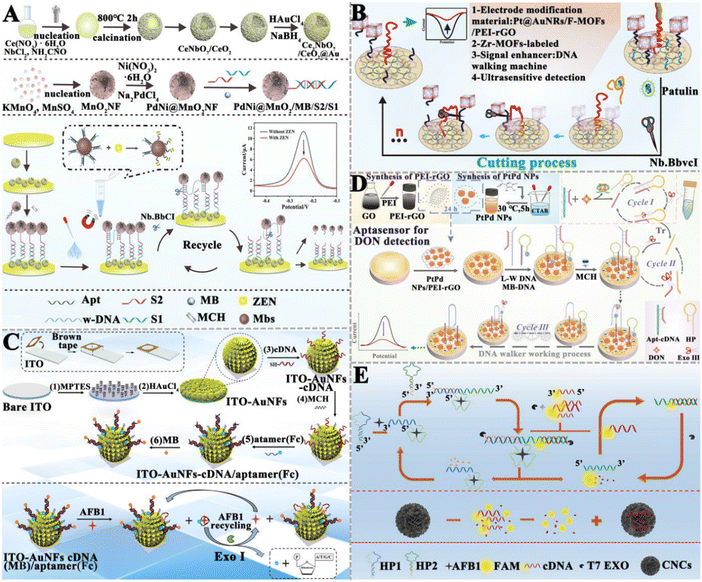 | ||
| Fig. 5 The construction of nuclease-based aptasensors for mycotoxin detection. (A) The construction of an electrochemical aptasensor based on the Nb.BbvCI-triggered and bipedal DNA walker amplification for the ZEN assay.115 (B) The construction of an electrochemical aptasensor based on the Nb.BbvCI-powered DNA walking for the PAT assay.116 (C) The construction of an electrochemical aptasensor based on EXO I-assisted target recycling amplification for the AFB1 assay.117 (D) The construction of an electrochemical aptasensor based on Exo III-powered WDNA for the DON assay.118 (E) The construction of a fluorescent aptasensor based on the T7 Exo double recycling amplification for the AFB1 assay.119 | ||
Exonucleases are a class of enzymes that can sequentially hydrolyze phosphodiester bonds from the ends of nucleic acid chains to generate single nucleotides. Compared with restriction endonucleases, exonucleases do not require the design of specific recognition sites. Wang et al. developed an electrochemical aptasensor based on exonuclease I (Exo I)-assisted target recycling amplification for AFB1 detection (Fig. 5C).117 Specifically, the ferrocene (Fc)-labeled aptamer forms a hybrid with the MB-labeled complementary DNA of the aptamer to form a duplex structure. Due to the rigid structure of the duplex, Fc molecules are close to the electrode surface and MB molecules are far away, generating a faded external response signal and an enhanced internal reference signal. When AFB1 is present in a sample, the aptamer specifically binds with the target, inducing the detachment of Fc from the electrode surface and a signal-off assay for Fc indicators. Meanwhile, more MB molecules are close to the electrode surface, inducing a signal-on assay for MB indicators. Consequently, the ratiometric electrochemical detection of AFB1 is achieved with the peak current intensity ratio (IMB/IFc) as the signal transducer. In this assay, Exo I is introduced to digest the detached aptamer, and the released AFB1 can subsequently initiate target recycling and signal amplification. This aptasensor exhibits a LOD of 0.032 pg mL−1 and can be used to detect AFB1 in actual peanut samples, and achieves results comparable to ELISA. Ren et al. designed an electrochemical aptasensor based on an Exo III-powered DNA walker (WDNA) for DON detection (Fig. 5D).118 In this electrochemical aptasensor, PtPd NPs/PEI-rGO as the substrate material can significantly increase the sensing area, improve the conductivity, and provide the binding sites for nucleic acid strands. The presence of DON can specifically bind to the aptamer, initiating Exo III-assisted cyclic amplification to generate triggers (Tr) (cycle I). Then, the hybridization of Tr with the locking probe (LP) activates the DNA walker (W-DNA) (cycle II). W-DNA runs continuously with the assistance of Exo III, inducing a decrease in the current signal (cycle III). This aptasensor has a LOD of 6.9 × 10−9 mg mL−1 and exhibits good stability in real sample analysis. Jin et al. constructed a carbon nanocage (CNC)-based fluorescent aptasensor based on T7 Exo-initiated double recycling amplification for AFB1 detection (Fig. 5E).119 The CNC is introduced as the fluorescence resonance energy transfer (FRET) receptors to quench the fluorophore-labeled DNA probe (FAM-cDNA). In the presence of target AFB1, T7 Exo triggers a double recycling amplification reaction, inducing the digestion of a large number of FAM-cDNAs and the release of FAM fluorophores. This aptasensor can sensitively detect AFB1 with a LOD of 1.4 pg mL−1, and it can be applied in real sample analysis with good recovery results. Notably, even though nucleases can catalyze the cyclic digestion of substrates to achieve the sensitive detection of target objects, there are some limitations. For example, restriction enzymes need the design of the corresponding cleavage sites, which increases the complexity of the nucleic acid sequence design. In addition, the exonuclease may contain a variety of other catalytic functions and the influence of non-specific catalytic activity should be carefully considered.
3.1.5.1. Integrated with the CRISPR/Cas system directly. The CRISPR/Cas system has shown great potential in nucleic acid biosensing.72,125 The combination of the CRISPR/Cas system with aptamers can be applied to detect trace mycotoxins. Upon the identification of mycotoxins by aptamers, the cleavage activity of the Cas protein is triggered to achieve signal amplification. Wu et al. fabricated a CRISPR/Cas12a-modulated luminescence resonance energy transfer (LRET) aptasensor for the detection of DON (Fig. 6A).121 The partial hybridization of the DON aptamer with crRNA activates the indiscriminate cleavage activity of Cas12a, resulting in the cleavage of adjacent ssDNA-UCNPs into small fragments. The cleaved ssDNA-UCNPs cannot adsorb on MXene-Au, and the upconversion luminescence (UCL) remains. In the presence of DON, the aptamer binds to DON, leading to the inhibition of the Cas12a activity. As a result, fewer ssDNA-UCNPs are cleaved and more ssDNA-UCNPs can easily adsorb to the MXene-Au surface, leading to the quenching of UCL by LRET. This aptasensor can sensitively detect DON with a LOD of 0.64 ng mL−1, and obtain satisfactory recoveries in corn flour and Tai Lake water. Wei et al. developed a fluorescent aptasensor based on CRISPR/Cas12a and MXenes for AFB1 detection (Fig. 6B).122 In this aptasensor, the ingeniously designed activator is locked by dual-AFB1 aptamers. As a result, the Cas12a and crRNA complexes are inactivated, and the fluorescence of FAM-labeled ssDNA (ssDNA-FAM) is quenched by MXenes. In the presence of AFB1, it binds to the aptamer, inducing the release of the activator. The released activator is complementary to crRNA, activating Cas12a and inducing the cleavage of ssDNA-FAM adsorbed on MXenes and the recovery of fluorescence. This fluorescent aptasensor exhibits a LOD of 0.92 pg mL−1 and good assay performance in real peanut samples. Zhu et al. constructed a dual-mode aptasensor based on target-regulated CRISPR/Cas12a for OTA detection (Fig. 6C).123 In the absence of OTA, the crRNA hybridizes with the aptamer to put the CRISPR/Cas12a system in an “active state”, inducing the cleavage of hairpin DNA on the electrode and the generation of a weak ECL signal and a high current response. When OTA is present, the aptamer preferentially binds to OTA, resulting in the inhibition of Cas12a activity. The hairpin DNA on the electrode remains intact, resulting in the recovery of the ECL signal and the attenuation of the current signal. This dual-mode aptasensor has high sensitivity and good anti-interference capability, with a wide range of application prospects.
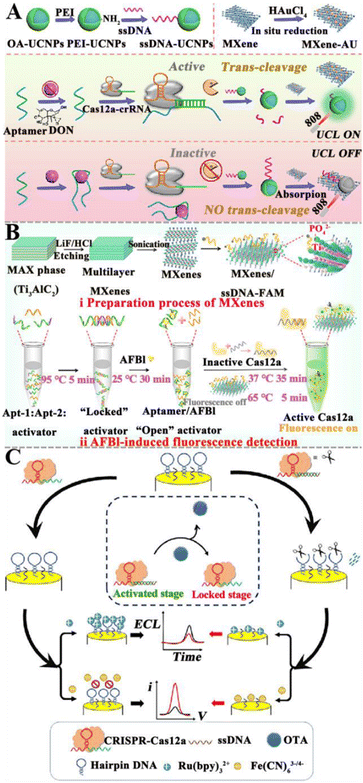 | ||
| Fig. 6 The construction of aptasensors based on the CRISPR/Cas system for mycotoxin detection. (A) The construction of a CRISPR/Cas12a-modulated LRET aptasensor for the DON assay.121 (B) The construction of an aptasensor based on the CRISPR/Cas system for the AFB1 assay.122 (C) The construction of a dual-mode aptasensor based on a target-regulated CRISPR/Cas12a for the OTA assay.123 | ||
3.1.5.2. NASAT-assisted CRISPR/Cas system. Recently, several NASAT-assisted CRISPR/Cas platforms have been developed for mycotoxin detection. Chen et al. constructed a CRISPR/Cas12a-assisted chemiluminescent aptasensor for AFB1 detection (Fig. 7A).126 This aptasensor takes advantage of the properties of polydopamine (PDA) to self-polymerize and self-assemble into a PDA film through a peroxidation process and subsequently, the biotin-DNA/padlock hybrids can be obtained through the biotin–streptavidin interaction, activating RCA to produce long linear RCA products with repeated sequences. Finally, a large number of complementary DNA-HRPs are assembled into long, linear RCA products to form the pipette-poly-HRP. In the presence of AFB1, the aptamer preferentially binds to AFB1, resulting in the dissociation of the pre-hybridized aptamer-activator duplex. The released activator hybridizes with the crRNA, initiating the collateral endonuclease activity of Cas12a to degrade the pipet-bound HRP-containing ssDNA linker. As a result, HRPs are released into the solution and quantified by chemiluminescence measurements. This chemiluminescent aptasensor exhibits high sensitivity with a LOD of 5.2 pg mL−1, and it can be applied to measure AFB1 in cereal samples.
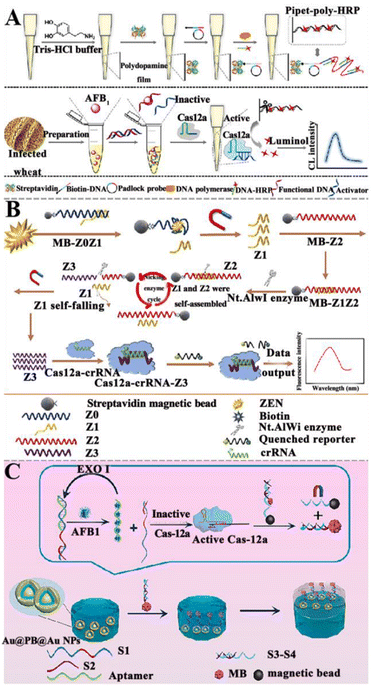 | ||
| Fig. 7 The construction of aptasensors based on CRISPR/Cas and NASAT for mycotoxin detection. (A) The construction of a CRISPR/Cas12a-assisted chemiluminescent aptasensor for the AFB1 assay.126 (B) The construction of an aptasensor based on CRISPR/Cas12a and the Nt.A1Wi enzyme for ZEN assay.127 (C) The construction of a SERS aptasensor based on the CRISPR/Cas system for the AFB1 assay.128 | ||
Alternatively, Wu et al. took advantage of the CRISPR/Cas12a and Nt.A1Wi enzyme to construct an aptasensor for ZEN detection (Fig. 7B).127 When ZEN is present, it competitively binds to the aptamer Z0, resulting in the release of Z1 into the solution. After magnetic separation, the MB-Z2 probe is added to the supernatant, and two ssDNAs (Z1 & Z2) form a dsDNA. Subsequently, the Nt.AlwI enzyme recognizes the dsDNA and cleaves only the Z2 ssDNA. After enzymatic hydrolysis, Z1 ssDNA and the obtained Z3 ssDNA product are separated from the magnetic beads. The separated Z1 ssDNA can continue to hybridize with Z2 ssDNA, inducing the cleavage of Z2 with the assistance of Nt.AlwI and producing large amounts of ssDNA Z3. The Z3 and CRISPR/Cas12a can form a CRISPR/Cas12a-Z3 complex, activating the trans-cleavage capability of Cas12a to cleave the quenched reporter and induce the recovery of fluorescence. This aptasensor can sensitively detect ZEN with a wide linear range of 1–1000 pg mL−1 and a LOD as low as 0.213 pg mL−1 and obtain good recovery rates in the corn-oil samples.
Yang et al. developed a SERS aptasensor based on CRISPR for the sensitive detection of AFB1 (Fig. 7C).128 DNA strands modified with –COOH (S4) are coated on the surface of Fe3O4 NPs, and the Fe3O4 NPs are modified with –NH2 through peptide bonds to form the magnetic core–shell structure nanoparticles. Prussian blue (PB) is embedded in the core–shell nanoparticles, and then the ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CN of the PB molecule and Au form an Au layer outside. When target AFB1 is present, it binds to the aptamer. Under the catalysis of the Exo I enzyme, the target AFB1 is recycled, and the released S1–S2 structure can activate cas12a, inducing the separation of double strands with beacon molecules from the magnetic beads. The double-strand DNA (dsDNA) with beacon molecules can conjugate with Au@PB@Au NPs through the Au–S bond, and then self-assemble at the water–oil interface. Cyclohexane is used as an oil-phase solvent to form a water–oil two-phase interface, facilitating the formation of nanoparticle film at the interface and the generation of a distinct SERS signal. This SERS aptasensor can sensitively measure AFB1 with a LOD of 3.55 pg mL−1, and lead to a good recovery rate in milk and soy sauce. Although the CRISPR/Cas-based aptasensor has the advantages of being fast, with low cost and high practicality, its accuracy and sensitivity still need to be systematically verified in real sample analyses. It is, therefore, necessary to use enzyme-linked immunosorbent or other mature methods to confirm the accuracy of results.
CN of the PB molecule and Au form an Au layer outside. When target AFB1 is present, it binds to the aptamer. Under the catalysis of the Exo I enzyme, the target AFB1 is recycled, and the released S1–S2 structure can activate cas12a, inducing the separation of double strands with beacon molecules from the magnetic beads. The double-strand DNA (dsDNA) with beacon molecules can conjugate with Au@PB@Au NPs through the Au–S bond, and then self-assemble at the water–oil interface. Cyclohexane is used as an oil-phase solvent to form a water–oil two-phase interface, facilitating the formation of nanoparticle film at the interface and the generation of a distinct SERS signal. This SERS aptasensor can sensitively measure AFB1 with a LOD of 3.55 pg mL−1, and lead to a good recovery rate in milk and soy sauce. Although the CRISPR/Cas-based aptasensor has the advantages of being fast, with low cost and high practicality, its accuracy and sensitivity still need to be systematically verified in real sample analyses. It is, therefore, necessary to use enzyme-linked immunosorbent or other mature methods to confirm the accuracy of results.
3.2. Enzyme-free nucleic acid amplification-based aptasensors
In comparison with enzyme-assisted nucleic acid amplification, enzyme-free amplification does not involve any protein enzymes (e.g., nucleases, ligases, and polymerases) to facilitate nucleic acid signal amplification,72 efficiently eliminating the issues of protein enzyme instability, high cost, and susceptibility to experimental conditions (e.g., ion concentration, temperature, and pH value).125 Enzyme-free amplification can be easily carried out in complex biological samples through programmable cascade hybridization reactions.72 The reported enzyme-free nucleic acid amplification for mycotoxin detection includes catalytic hairpin assembly (CHA), hybridization chain reaction (HCR), DNA tweezers, and DNAzymes.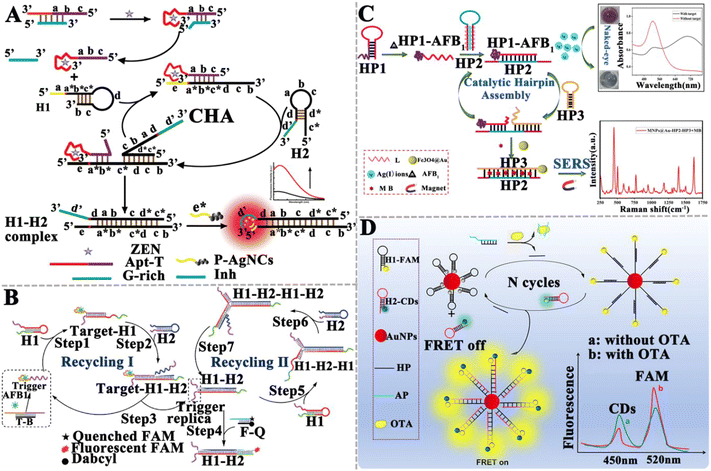 | ||
| Fig. 8 The construction of CHA-based aptasensors for mycotoxin detection. (A) The construction of a CHA-based fluorometric aptasensor for ZEN assay.129 (B) The construction of a CHA-based aptasensor for the AFB1 assay.130 (C) The construction of a dual-mode aptasensor for the AFB1 assay.131 (D) The construction of a CHA-based FRET aptasensor for the ratiometric detection of OTA.132 | ||
Li et al. developed a surface-enhanced Raman scattering (SERS) dual-mode aptasensor for the homogeneous detection of mycotoxin with the naked eye (Fig. 8C).131 The specific binding of the aptamer to AFB1 forms the HP1–AFB1 complex, and then the HP1–AFB1 complex hybridizes with Ag+-labeled hairpin DNA (HP2), inducing the release of Ag+ and initiating an enzyme-free CHA reaction to form the dsDNA (HP2–HP3), accompanied by the release of the HP1–AFB1 complex. The released HP1–AFB1 complexes can be subsequently recognized by HP2 and HP3, triggering the cascade recycling amplification and resulting in the accumulation of abundant free Ag+ and the HP2–HP3 duplex. As a result, MB is embedded into the HP2–HP3 duplex and acts as a Raman tag to generate a strong SERS signal with the aid of Fe3O4@Au. Meanwhile, the free Ag+ induces the aggregation of gold nanoparticles (AuNPs) and consequently, a visually distinguishable color change from red to black-blue. The SERS intensity and visualization signals are linearly correlated with the AFB1 concentration in the range of 0.0156–31.2 ng mL−1 and 0.61-39 ng mL−1, respectively. The LOD was determined to be 1.6 pg mL−1 for the SERS assay and 152 pg mL−1 for the colorimetric assay. The He group constructed a CHA-based Förster resonance energy transfer (FRET) aptasensor for the ratiometric detection of OTA (Fig. 8D).132 In the presence of OTA, the aptamer specifically interacts with OTA, inducing the release of helper DNA (HP). Subsequently, HP opens the hairpin structure of FAM-labeled hairpin DNA 1 (H1-FAM) modified on the AuNP surface. CHA between H1-FAM and hairpin H2 that is labeled with carbon quantum dots (H2-CDs) induces the release of HP, which initiates the next cycle reaction and the occurrence of FRET between CD and FAM. Taking advantage of the measurement of the FCDs/FFAM ratio, this aptasensor can sensitively detect OTA with a wide linear range from 5.0 pg mL−1 to 3.0 ng mL−1 and a LOD of 1.5 pg mL−1, and it has been successfully applied in the detection of OTA in rice.
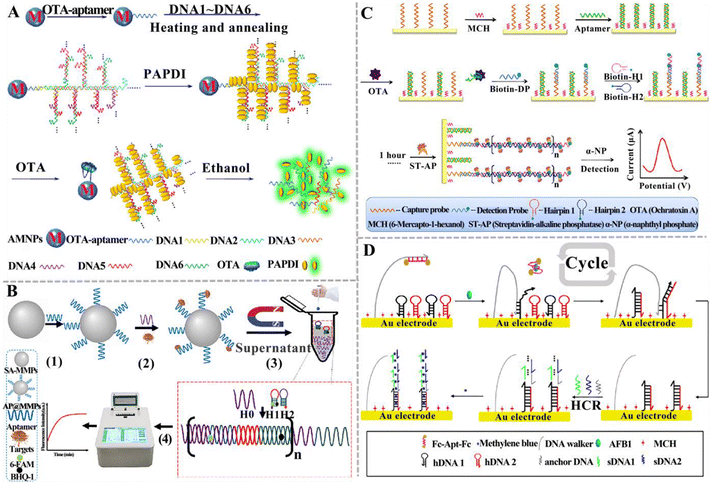 | ||
| Fig. 9 The construction of HCR-based aptasensors for mycotoxin detection. (A) The construction of an HCR-based aptasensor for the OTA assay.133 (B) The construction of an HCR-based fluorescent aptasensor for the simultaneous detection of the T-2 toxin and ZEN.134 (C) The construction of an HCR-based electrochemical aptasensor for the OTA assay.135 (D) The construction of an HCR-based electrochemical aptasensor for the ratiometric detection of AFB1.136 | ||
Qiu et al. constructed an electrochemical aptasensor based on HCR to detect OTA (Fig. 9C).135 In this assay, a capture probe is immobilized on the electrode and then hybridizes with the OTA aptamer (Apt) to form an Apt/DNA duplex. The presence of OTA induces the formation of an Apt-OTA complex and the dissociation of Apt from the electrode. Upon the addition of the detection probe and two hairpin-helper DNAs, HCR was activated to extend dsDNA polymers on the electrode surface. Finally, a streptavidin-alkaline phosphatase (ST-AP) conjugate was added to bind biotin-modified hairpin DNA. AP can hydrolyze a synthetic enzyme substrate (α-naphthyl phosphate) to produce a differential pulse voltammetry (DPV) current. The DPV current was enhanced with the increasing concentration of OTA. This electrochemical aptasensor can detect OTA with a LOD of 2 pg mL−1, and it can be applied to measure OTA in cereal samples. You and co-workers established an electrochemical aptasensor for the ratiometric detection of AFB1 (Fig. 9D).136 This assay involves a dual-amplification by coupling HCR with a DNA walker. The DNA walker strand, hairpin DNA 1 (hDNA1), and hairpin DNA 2 (hDNA2) were initially assembled on the surface of the gold electrode (AuE) via Au–S bonds. The DNA walker strand was blocked by the AFB1 aptamer that was modified with ferrocene (Fc). When AFB1 is present, it specifically binds to Fc-Apt-Fc, resulting in the release of the Fc-labelled aptamer (Fc-Apt-Fc) and the decrease in current (IFc). Subsequently, the DNA walker strand hybridizes with hDNA1 through a toehold-mediated strand exchange, and then hDNA2 hybridizes with hDNA1 to form the dsDNA, initiating a cyclic reaction with the assistance of the DNA walker strand. Meanwhile, the free hDNA1 hybridizes with hDNA2 to form the dsDNA, triggering the DNA walker-mediated amplification. The dsDNA subsequently hybridizes with the anchored DNA to activate HCR between sDNA1 and sDNA2, generating a longer dsDNA that can adsorb MB. AFB1 can be accurately quantified by measuring the current ratio of MB (IMB) and Fc (IFc). The LOD can reach 0.0008 pg mL−1. Consistent results were obtained for this aptasensor and gold-standard HPLC-MS/MS method for peanut sample analysis. Notably, even though HCR-based aptasensors have been widely used for mycotoxin detection, there are some limitations. Because HCR occurs through the random diffusion of DNA hairpins, the kinetics and efficiency are relatively low. The smooth opening of the hairpin structure heavily relies on the sequence design.
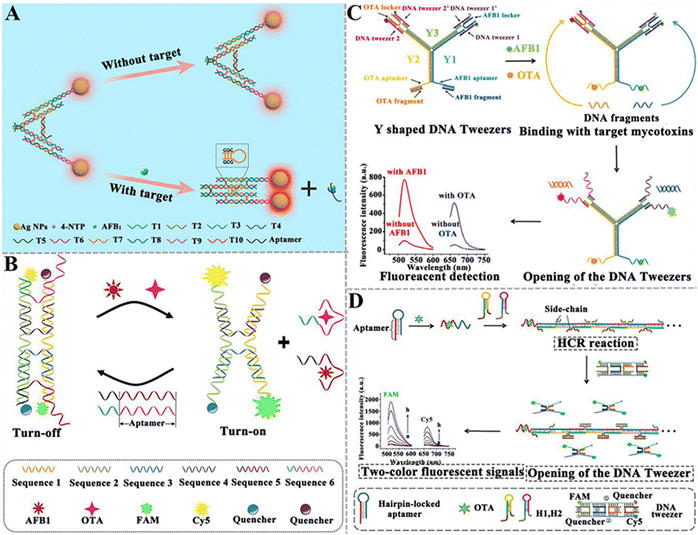 | ||
| Fig. 10 The construction of DNA tweezer-based aptasensors for mycotoxin detection. (A) The construction of a programmable DNA tweezer-actuated SERS aptasensor for the AFB1 assay.142 (B) The construction of a dual DNA tweezer-based aptasensor for the simultaneous detection of AFB1 and OTA.143 (C) The construction of a versatile Y-shaped DNA nanostructure-based aptasensor for the detection of AFB1 and OTA.144 (D) The construction of a two-color aptasensor based on HCR and DNA tweezers for the OTA assay.145 | ||
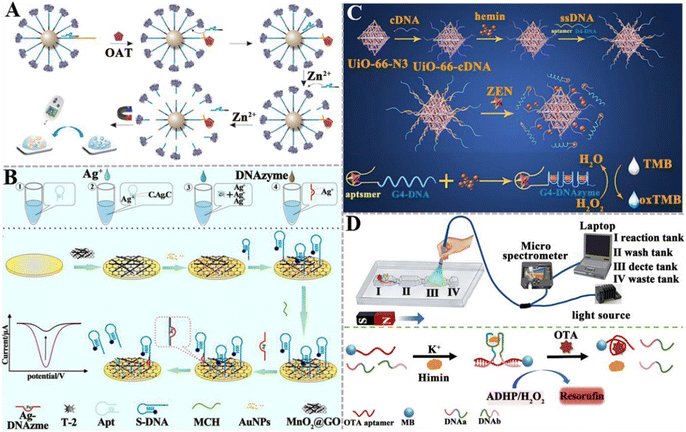 | ||
| Fig. 11 The construction of DNAzyme-based aptasensors for mycotoxin detection. (A) The construction of an aptasensor based on PGM and DNAzyme for OTA assay.151 (B) The construction of an electrochemical aptasensor based on Ag+-dependent DNAzyme-assisted signal amplification for the T-2 toxin assay.152 (C) The construction of a colorimetric aptasensor based on a stimuli-responsive MOF nano-container and DNAzyme for the ZEN assay.153 (D) The construction of a portable fluorescent/colorimetric aptasensor based on the G-quadruplex DNAzyme for the rapid and dual-mode detection of OTA.154 | ||
DNAzymes mimic peroxidase is an excellent alternative to natural peroxidase and has recently been explored for mycotoxin detection. Wang et al. developed a colorimetric aptasensor based on a stimuli-responsive metal–organic framework (MOF) nano-container and DNAzyme for ZEN detection (Fig. 11C).153 In this aptasensor, an azide-functionalized MOF (UiO-66-N3) functions as a nano-container to entrap hemin after being conjugated with DBCO-functionalized cDNA. The bio-gate composed of a DNA duplex acts as a gatekeeper to control the release. When ZEN is present, it competes for the binding sites of aptamers with cDNA, inducing the opening of hybrid duplexes and the unlocking of pores. The presence of ZEN induces the concentration-dependent release of hemin and ssDNA (containing trimeric CatG4) into the supernatant. Subsequently, hemin is embedded into the secondary structure of trimer CatG4, inducing its conversion to a trivalent G4-DNAzyme that possesses higher catalytical activity than hemin itself. Upon the addition of TMB, the liquid emits a strong colorimetric signal. This colorimetric aptasensor can detect ZEN with a LOD of 0.36 pg mL−1. Chen et al. constructed a portable fluorescent/colorimetric aptasensor based on G-quadruplex DNAzyme for the rapid and dual-mode detection of OTA (Fig. 11D).154 The OTA aptamers and MBs can self-assemble with two segments of DNA and hemin to form a G-quadruplex DNAzyme structure that can catalyse the oxidation of Amplex Red (ADHP) by H2O2, inducing the change in solution color to red and the generation of a strong fluorescence. When OTA is present, it specifically binds to aptamers, inducing the decomposition of the DNA enzyme structure and decreasing its catalytic activity. Under optimal conditions, this aptasensor can measure OTA with a LOD of 0.011 μg kg−1 for the fluorescence mode and 0.008 μg kg−1 for the colorimetric mode.
4. Conclusions and perspectives
The perennial hot topic of food safety and contamination has attracted more and more attention around the world due to its direct impact on human health and social harmony. The main assurance is to continuously develop rapid and sensitive detection methods for food pollutants in the entire food supply chain, from farm to fork/plate. However, conventional methods for mycotoxin assay are not suitable for rapid and sensitive assays. Alternatively, a variety of aptasensors have been constructed for mycotoxin detection in recent years. To overcome the limitation of low signals generated by mycotoxins, nucleic acid signal amplification technology (NASAT) has been introduced into the aptasensors to significantly improve the sensitivity and facilitate the detection of low-abundance mycotoxins. Herein, we have provided a comprehensive review of the recent advances in NASAT-based aptasensors for mycotoxin assay, and summarized the principles, features, and applications of NASAT-based aptasensors. The NASAT is divided into enzyme-assisted and enzyme-free nucleic acid amplification approaches. The working principles and practical applications of NASAT-based aptasensors for mycotoxin detection are summarized in Table 3.| Amplification strategy | Signal model | Target | Linear range (ng mL−1) | LOD (ng mL−1) | Real sample | Ref. |
|---|---|---|---|---|---|---|
| RT-qPCR | Fluorescence | AFM1 | 1.0 × 10−4–1.0 | 3 × 10−5 | Rice, milk | 76 |
| RT-qPCR | Fluorescence | AFB1 | 3.12 × 10−5–3.12 | 3 × 10−5 | Soy sauce, milk, yellow wine, and peanut butter | 77 |
| RT-qPCR | qPCR | OTA | 0.039–1000 | 0.009 | Beer | 75 |
| RCA | Electrochemical | OTA | 1.0 × 10−4–5.0 | 6.5 × 10−5 | Wine | 103 |
| RCA | Colorimetric | AFB1 | 1.0 × 10−5–1.2 × 10−4 | 1.4 × 10−5 | Rice, peanut oil | 104 |
| RCT | Fluorescence | OTA | 0.0001–0.5 | 1.94 × 10−5 | Milk | 105 |
| SDA | Electrochemical | ZEN | 1.0 × 10−3–0.15 | 1.8 × 10−4 | Corn | 110 |
| SDA | Fluorescence | AFB1, ZEN | 1.0 × 10−3–10, 1.0 × 10−2–100 | 9.33 × 10−4; 1.07 × 10−3 | Cornmeal | 111 |
| SDA | Fluorescence | PAT | 0.001–100 | 4.2 × 10−5 | Apple and grape juices | 112 |
| SDA | Fluorescence | AFM1, OTA | 1.968 × 10−2–0.1968 2.498 × 10−2–4.03 | 0.01779; 0.01898 | Milk | 113 |
| Nicking endonuclease-assisted amplification | Electrochemical | ZEN | 1.0 × 10−4–1.0 × 103 | 4.57 × 10−6 | Corn | 115 |
| Nicking endonuclease-assisted amplification | Electrochemical | PAT | 5.0 × 10−6–5.0 × 10−1 | 4.14 × 10−5 | Apple juice, apple wine | 116 |
| ExoI-assisted amplification | Electrochemical | AFB1 | 1.0 × 10−4–1.0 | 3.2 × 10−5 | Peanut | 117 |
| ExoIII-assisted amplification | Electrochemical | DON | 1.0 × 10−2–1.0 × 102 | 6.9 × 10−3 | Maize flour | 118 |
| T7 Exo-assisted amplification | Fluorescence | AFB1 | 0.01–500.0 | 1.4 × 10−3 | Instant grains, peanut, corn flour, Chinese herb (QC-522B-1) | 119 |
| CRISPR/Cas | Luminescence | DON | 1.0–500.0 | 0.64 | Corn flour, Tai Lake water | 121 |
| CRISPR/Cas | Fluorescence | AFB1 | 0.001–80.0 | 9.2 × 10−4 | Peanut | 122 |
| CRISPR/Cas | Electrochemiluminescent, Electrochemical | OTA | 1.0 × 10−3–5.0, 1.0 × 10−3–5.0 | 2.9 × 10−4; 3.7 × 10−4 | Morinda officinalis | 123 |
| CRISPR/Cas & Nt.AlwI | Fluorescence | ZEN | 1.0 × 10−3–1.0 | 2.13 × 10−4 | Corn oil | 127 |
| CRISPR-Cas12a & RCA | Chemiluminescence | AFB1 | 0.05–100.0 | 5.2 × 10−3 | Wheat | 126 |
| CRISPR/Cas & Exo I | SERS | AFB1 | 0.001–1.5 | 3.55 × 10−3 | Milk, soy sauce | 128 |
| CHA | Fluorescence | ZEN | 1.3 × 10−3–100 | 3.2 × 10−4 | Maize, beer | 129 |
| CHA | Fluorescence | AFB1 | 0.30–15.0 | 0.13 | Beer, infant rice cereal | 130 |
| CHA | SERS, colorimetric | AFB1 | 0.0156–31.2; 0.61–39.0 | 1.6 × 10−3; 0.152 | Wheat | 131 |
| CHA | Fluorescence | OTA | 5.0 × 10−3–3.0 | 1.5 × 10−3 | Rice | 132 |
| HCR | Fluorescence | OTA | 4.038 × 10−4–8.076 × 10−3 | 4.038 × 10−5 | Corn | 133 |
| HCR | Electrochemical | OTA | 0.005–100.0 | 2.0 × 10−3 | Corn, wheat | 135 |
| HCR | Electrochemical | AFB1 | 3.0 × 10−6–3 × 10−3 | 8.8 × 10−7 | Peanut | 136 |
| HCR | Fluorescence | T-2, ZEN | 0.001–10.0, 0.01–100.0 | 1.0 × 10−4; 1.2 × 10−3 | Corn, oat flour | 134 |
| DNA Tweezer | SERS | AFB1 | 1.0 × 10−5–1.0 | 5.07 × 10−6 | Maize | 142 |
| DNA Tweezer | Fluorescence | AFB1, OTA | 8.0 × 10−2–10.0, 0.5–50.0 | 3.5 × 10−2; 0.1 | Corn, peanut, coffee, olive oil, peanut oil | 143 |
| HCR & DNA Tweezer | Dual-fluorescence | OTA | 0.02–0.8 | 0.006 for FAM; 0.014 for Cy5 | Peanut, corn, sesame, coffee beans, peanut oil, Soybean oil | 145 |
| DNA Tweezer | Fluorescence | AFB1, OTA | 0.5–200.0, 4.0–300.0 | 0.1; 2.0 | Sesame, olive oil | 144 |
| DNAzyme | personal glucose meter | OTA | 1.0 × 10−3–300.0 | 8.8 × 10−4 | Wine | 151 |
| DNAzyme | Electrochemical | T-2 | 2.0 × 10−6–20.0 | 1.07 × 10−7 | Milk | 152 |
| DNAzyme | Colorimetric | ZEN | 0.01–100.0 | 3.6 × 10−4 | Maize, wheat | 153 |
| DNAzyme | Fluorescent, Colorimetric | OTA | 5.0–100.0, 0.05–10.0 | 0.011; 0.008 | Wheat | 154 |
Despite great progress made in the development of NASAT-based aptasensors for mycotoxin assay, several challenges remain to be solved. (1) Few mycotoxin aptamers have been reported so far. In addition, the aptamer matching degree is low in complex matrix environments, and some invalid sequences may affect the stability of aptamer binding and increase the synthesis cost.155 It is essential to improve the screening procedure by combining “positive SELEX” with “negative SELEX”, which can efficiently eliminate the influence of the matrix and obtain highly specific aptamer sequences.52 (2) At present, the detection of mycotoxins is challenged by the coexistence of multiple targets.156,157 The discovery of new aptamer molecules and an increase in target coverage is highly required, which facilitates the construction of efficient aptasensors for high-throughput and the simultaneous detection of multiple mycotoxins. (3) The reported aptasensors are limited to a single mode that suffers from false positives. The development of dual-mode aptasensors that combine different output modes (e.g., fluorometric, colorimetric, and electrochemical measurements) greatly improves the detection accuracy.136,154 (4) Due to the trace levels of mycotoxins in foods, NASATs are often involved in the construction of aptasensors for the ultrasensitive detection of mycotoxins.69 Notably, some amplification approaches have the limitations of non-specific amplification and high cost. The discovery of low-cost and low-background signal amplification strategies can significantly improve the detection sensitivity. (5) To overcome the inherent amplification bias and non-specific amplification in NASAT, the introduction of nanomaterials (e.g., metal nanoparticles, carbon nanotubes, GO, and UCNPs) into amplification reactions may minimize the amplification bias and eliminate the non-specific amplification.119,132,152,158,159 Compared with the nanosensors used directly for mycotoxin detection, the aptasensors based on nanomaterials and NASAT possess improved sensitivity, facilitating the ultra-sensitive monitoring of mycotoxins in complex food substrates from farm to table. (6) The NASAT-based aptasensors for mycotoxin detection reported above are largely limited to laboratory assays. Given the extensive and real-time food contamination occurrences, the rapid, accurate, modest, and reliable onsite detection system is imperative, particularly in the vast rural regions where resources are scarce. Portable instruments (e.g., side-flow analysis, microfluidic devices, and smartphone-based point-of-care testing (POCT)) have been applied in simple, rapid, and low-cost field inspections. However, the reported side-flow analysis, microfluidic devices, and smartphone-based POCT for mycotoxin detection usually rely on antibodies that are expensive to produce, and the use of animals for this purpose may raise ethical concerns.158,160–163 The introduction of NASAT-based aptasensors into the side-flow analysis and microfluidic system may significantly improve the sensitivity, overcoming the issue of low field detection sensitivity.164–168 In addition, the use of smartphones to analyze the output signal not only satisfies the portability of the field detection equipment but also ensures the reliability and stability of the result analysis as compared with the naked eye. Above all, the prospects of NASAT-based aptasensors include the following: (1) going from ultra-sensitive detection in the laboratory to rapid and sensitive detection in the field; (2) making the detection process more intelligent and automated; (3) high-throughput analysis of mycotoxins from single-factor analysis to multi-target detection. We believe that with the development of nanomaterials, microfluidics, and new technologies, NASAT-based aptasensors will play more and more important roles in food safety.
Author contributions
Dandan Zhang: writing the original draft, reviewing, and editing. Ting Luo: reviewing and editing. Xiangyue Cai: reviewing and editing. Ning-ning Zhao: reviewing and editing. Chun-yang Zhang: reviewing, editing, and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Teacher Development Research Start-up Fund of Chengdu University of Technology (10912-KYQD2020-08414).Notes and references
- W. Zhu, L. Li, Z. Zhou, X. Yang, N. Hao, Y. Guo and K. Wang, Food Chem., 2020, 319, 126544 CrossRef CAS PubMed.
- S. Marin, A. J. Ramos, G. Cano-Sancho and V. Sanchis, Food Chem. Toxicol., 2013, 60, 218–237 CrossRef CAS PubMed.
- Q. Jiang, J. Wu, K. Yao, Y. Yin, M. M. Gong, C. Yang and F. Lin, ACS Sens., 2019, 4, 3072–3079 CrossRef CAS PubMed.
- Y. Yang, G. Li, D. Wu, J. Liu, X. Li, P. Luo, N. Hu, H. Wang and Y. Wu, Trends Food Sci. Technol., 2020, 96, 233–252 CrossRef CAS.
- Q. Zhou and D. Tang, TrAC-Trend Anal. Chem., 2020, 124, 115814 CrossRef CAS.
- S. Brase, A. Encinas, J. Keck and C. F. Nising, Chem. Rev., 2009, 109, 3903–3990 CrossRef CAS PubMed.
- W. P. Liew and S. Mohd-Redzwan, Front. Cell. Infect. Microbiol., 2018, 8, 60 CrossRef PubMed.
- J. Wen, P. Mu and Y. Deng, Toxicol. Res., 2016, 5, 377–387 CrossRef CAS PubMed.
- M. Jia, X. Liao, L. Fang, B. Jia, M. Liu, D. Li, L. Zhou and W. Kong, TrAC-Trend Anal. Chem., 2021, 136, 116193 CrossRef CAS.
- Z. Gong, Y. Huang, X. Hu, J. Zhang, Q. Chen and H. Chen, Biosensors, 2023, 13, 140–173 CrossRef CAS PubMed.
- A. L. Capriotti, G. Caruso, C. Cavaliere, P. Foglia, R. Samperi and A. Lagana, Mass Spectrom. Rev., 2012, 31, 466–503 CrossRef CAS PubMed.
- S. Agriopoulou, E. Stamatelopoulou and T. Varzakas, Foods, 2020, 9, 518–540 CrossRef CAS PubMed.
- A. Mousavi Khaneghah, A. Farhadi, A. Nematollahi, Y. Vasseghian and Y. Fakhri, Trends Food Sci. Technol., 2020, 102, 193–202 CrossRef CAS.
- X. Tang, J. Zuo, C. Yang, J. Jiang, Q. Zhang, J. Ping and P. Li, TrAC-Trend Anal. Chem., 2023, 165, 117144 CrossRef CAS.
- Y. Liu and F. Wu, Environ. Health Perspect., 2010, 118, 818–824 CrossRef CAS PubMed.
- L. Xu, Z. Zhang, Q. Zhang and P. Li, Toxins, 2016, 8, 239–254 CrossRef PubMed.
- Z. Xue, Y. Zhang, W. Yu, J. Zhang, J. Wang, F. Wan, Y. Kim, Y. Liu and X. Kou, Anal. Chim. Acta, 2019, 1069, 1–27 CrossRef CAS PubMed.
- A. Gallo, G. Giuberti, J. C. Frisvad, T. Bertuzzi and K. F. Nielsen, Toxins, 2015, 7, 3057–3111 CrossRef CAS PubMed.
- R. Li, Y. Wen, F. Wang and P. He, J. Anim. Sci. Biotechnol., 2021, 12, 108 CrossRef CAS PubMed.
- L. Zhang, X. W. Dou, C. Zhang, A. F. Logrieco and M. H. Yang, Toxins, 2018, 10, 65–103 CrossRef PubMed.
- A. Alshannaq and J. H. Yu, Int. J. Environ. Res. Public Health, 2017, 14, 632–651 CrossRef PubMed.
- T. I. Ekwomadu, T. A. Dada, S. A. Akinola, N. Nleya and M. Mwanza, Separations, 2021, 8, 143–154 CrossRef CAS.
- J. He, B. Zhang, H. Zhang, L. L. Hao, T. Z. Ma, J. Wang and S. Y. Han, J. Food Sci., 2019, 84, 2688–2697 CrossRef CAS PubMed.
- M. Solfrizzo, L. Gambacorta, R. Bibi, M. Ciriaci, A. Paoloni and I. Pecorelli, J. AOAC Int., 2018, 101, 647–657 CrossRef CAS PubMed.
- Y. Fan, J. Li, K. Amin, H. Yu, H. Yang, Z. Guo and J. Liu, Food Res. Int., 2023, 170, 113022 CrossRef CAS PubMed.
- M. Majdinasab, S. Ben Aissa and J. L. Marty, Toxins, 2020, 13, 13–47 CrossRef PubMed.
- M. E. E. Alahi and S. C. Mukhopadhyay, Sensors, 2017, 17, 1885–1904 CrossRef PubMed.
- S.-C. Pei, W.-J. Lee, G.-P. Zhang, X.-F. Hu, S. A. Eremin and L.-J. Zhang, Food Control, 2013, 31, 65–70 CrossRef CAS.
- H. Peng, Y. Chang, R. C. Baker and G. Zhang, Food Addit. Contam.: Part A, 2020, 37, 496–506 CrossRef CAS PubMed.
- X. Liu, G. Ying, X. Liao, C. Sun, F. Wei, X. Xing, L. Shi, Y. Sun, W. Kong and L. Zhou, Anal. Chem., 2019, 91, 1194–1202 CrossRef CAS PubMed.
- C. Sun, X. Liao, P. Huang, G. Shan, X. Ma, L. Fu, L. Zhou and W. Kong, Food Chem., 2020, 315, 126289 CrossRef CAS PubMed.
- L. Qin, J. Y. Jiang, L. Zhang, X. W. Dou, Z. Ouyang, L. Wan and M. H. Yang, Mycology, 2020, 11, 126–146 CrossRef CAS PubMed.
- S. D. Jayasena, Clin. Chem., 1999, 45, 1628–1650 CrossRef CAS.
- N. Zhang, B. Liu, X. Cui, Y. Li, J. Tang, H. Wang, D. Zhang and Z. Li, Talanta, 2021, 223, 121729 CrossRef CAS PubMed.
- S. Ni, Z. Zhuo, Y. Pan, Y. Yu, F. Li, J. Liu, L. Wang, X. Wu, D. Li, Y. Wan, L. Zhang, Z. Yang, B. T. Zhang, A. Lu and G. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 9500–9519 CrossRef CAS PubMed.
- G. Zon, Anal. Biochem., 2022, 644, 113894 CrossRef CAS PubMed.
- J. Chen, X. Zhang, S. Cai, D. Wu, M. Chen, S. Wang and J. Zhang, Biosens. Bioelectron., 2014, 57, 226–231 CrossRef CAS PubMed.
- F. Costantini, C. Sberna, G. Petrucci, M. Reverberi, F. Domenici, C. Fanelli, C. Manetti, G. de Cesare, M. DeRosa, A. Nascetti and D. Caputo, Sens. Actuators, B, 2016, 230, 31–39 CrossRef CAS.
- S. Perrier, Z. Zhu, E. Fiore, C. Ravelet, V. Guieu and E. Peyrin, Anal. Chem., 2014, 86, 4233–4240 CrossRef CAS PubMed.
- A. Ruscito, M. Smith, D. N. Goudreau and M. C. DeRosa, J. AOAC Int., 2016, 99, 865–877 CrossRef CAS PubMed.
- C. Wang, J. Qian, K. Wang, K. Wang, Q. Liu, X. Dong, C. Wang and X. Huang, Biosens. Bioelectron., 2015, 68, 783–790 CrossRef CAS PubMed.
- Q. Zhao, Q. Lv and H. Wang, Anal. Chem., 2014, 86, 1238–1245 CrossRef CAS PubMed.
- S. Wu, N. Duan, X. Ma, Y. Xia, H. Wang, Z. Wang and Q. Zhang, Anal. Chem., 2012, 84, 6263–6270 CrossRef CAS PubMed.
- B. Wang, Y. Chen, Y. Wu, B. Weng, Y. Liu, Z. Lu, C. M. Li and C. Yu, Biosens. Bioelectron., 2016, 78, 23–30 CrossRef CAS PubMed.
- Y. Seok, J. Y. Byun, W. B. Shim and M. G. Kim, Anal. Chim. Acta, 2015, 886, 182–187 CrossRef CAS PubMed.
- W. B. Shim, M. J. Kim, H. Mun and M. G. Kim, Biosens. Bioelectron., 2014, 62, 288–294 CrossRef CAS PubMed.
- A. Shrivastava and R. K. Sharma, Toxin Rev., 2021, 41, 618–638 CrossRef.
- X. Chen, Y. Huang, N. Duan, S. Wu, Y. Xia, X. Ma, C. Zhu, Y. Jiang and Z. Wang, J. Agric. Food Chem., 2014, 62, 10368–10374 CrossRef CAS PubMed.
- M. McKeague, R. Velu, K. Hill, V. Bardoczy, T. Meszaros and M. C. DeRosa, Toxins, 2014, 6, 2435–2452 CrossRef PubMed.
- X. Wang, X. Gao, J. He, X. Hu, Y. Li, X. Li, L. Fan and H. Z. Yu, Analyst, 2019, 144, 3826–3835 RSC.
- X. Wei, P. Ma, K. Imran Mahmood, Y. Zhang and Z. Wang, J. Agric. Food Chem., 2023, 71, 7546–7556 CrossRef CAS PubMed.
- J. A. Cruz-Aguado and G. Penner, J. Agric. Food Chem., 2008, 56, 10456–10461 CrossRef CAS PubMed.
- M. McKeague, C. R. Bradley, A. De Girolamo, A. Visconti, J. D. Miller and M. C. Derosa, Int. J. Mol. Sci., 2010, 11, 4864–4881 CrossRef CAS PubMed.
- J. W. Park, R. Tatavarty, D. W. Kim, H. T. Jung and M. B. Gu, Chem. Commun., 2012, 48, 2071–2073 RSC.
- B. R. Rushing and M. I. Selim, Food Chem. Toxicol., 2019, 124, 81–100 CrossRef CAS PubMed.
- Y. Tao, S. Xie, F. Xu, A. Liu, Y. Wang, D. Chen, Y. Pan, L. Huang, D. Peng, X. Wang and Z. Yuan, Food Chem. Toxicol., 2018, 112, 320–331 CrossRef CAS PubMed.
- D. Liu, L. Ge, Q. Wang, J. Su, X. Chen, C. Wang and K. Huang, Environ. Int., 2020, 143, 105949 CrossRef CAS PubMed.
- T. Kuiper-Goodman, P. M. Scott and H. Watanabe, Regul. Toxicol. Pharmacol., 1987, 7, 253–306 CrossRef CAS PubMed.
- A. V. Nathanail, E. Varga, J. Meng-Reiterer, C. Bueschl, H. Michlmayr, A. Malachova, P. Fruhmann, M. Jestoi, K. Peltonen, G. Adam, M. Lemmens, R. Schuhmacher and F. Berthiller, J. Agric. Food Chem., 2015, 63, 7862–7872 CrossRef CAS PubMed.
- G. J. A. Speijers, M. A. M. Franken and F. X. R. V. Leeuwen, Food Chem. Toxicol., 1988, 26, 23–30 CrossRef CAS PubMed.
- D.-l Li, X. Zhang, Y. Ma, Y. Deng, R. Hu and Y. Yang, Anal. Methods, 2018, 10, 3273–3279 RSC.
- H. Guo, P. Ma, K. Li, S. Zhang, Y. Zhang, H. Guo and Z. Wang, Sens. Actuators, B, 2022, 358, 131484 CrossRef CAS.
- C. Zhu, D. Liu, Y. Li, S. Ma, M. Wang and T. You, Biosens. Bioelectron., 2021, 174, 112654 CrossRef CAS PubMed.
- D. Yang, Y. Hui, Y. Liu, W. Wang, C. He, A. Zhao, L. Wei and B. Wang, Food Chem., 2024, 433, 137362 CrossRef CAS PubMed.
- S. Xiang, J. Li, F. Wang, H. Yang, Y. Jiang, P. Zhang, R. Cai and W. Tan, Anal. Chem., 2023, 95, 15125–15132 CrossRef CAS PubMed.
- Z. Wu, E. Xu, M. F. J. Chughtai, Z. Jin and J. Irudayaraj, Food Chem., 2017, 230, 673–680 CrossRef CAS PubMed.
- W. Yu, X. Lin, N. Duan, Z. Wang and S. Wu, Anal. Chim. Acta, 2023, 1244, 340846 CrossRef CAS PubMed.
- X. Zhao, H. Shen, B. Huo, Y. Wang and Z. Gao, Biosens. Bioelectron., 2022, 211, 114383 CrossRef CAS PubMed.
- J. Zhou, T. Y. Wang, Z. Lan, H. J. Yang, X. J. Ye, R. Min, Z. H. Wang, Q. Huang, J. Cao, Y. E. Gao, W. L. Wang, X. L. Sun and Y. Zhang, Food Res. Int., 2023, 173, 113286 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- F. Ma, C. C. Li and C. Y. Zhang, Chem. Commun., 2021, 57, 13415–13428 RSC.
- W.-j Liu, X. Zhang, F. Ma and C.-y Zhang, TrAC-Trend Anal. Chem., 2023, 160, 116998 CrossRef CAS.
- M. Joshi and J. D. Deshpande., Int. J. Biomed. Res., 2011, 5, 81–97 Search PubMed.
- N. Arnheim and H. Erlich, Annu. Rev. Biochem., 1992, 61, 131–156 CrossRef CAS PubMed.
- H. Modh, T. Scheper and J. G. Walter, Eng. Life Sci., 2017, 17, 923–930 CrossRef CAS PubMed.
- X. Guo, F. Wen, N. Zheng, S. Li, M. L. Fauconnier and J. Wang, Anal. Bioanal. Chem., 2016, 408, 5577–5584 CrossRef CAS PubMed.
- J. Sun, X. Ning, L. Cui, W. Qin, W. Wang and S. He, Food Chem., 2024, 432, 137240 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- N. Zhang, J. Li, B. Liu, D. Zhang, C. Zhang, Y. Guo, X. Chu, W. Wang, H. Wang, X. Yan and Z. Li, Talanta, 2022, 236, 122866 CrossRef CAS PubMed.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D. K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- W. Zhao, C. H. Cui, S. Bose, D. Guo, C. Shen, W. P. Wong, K. Halvorsen, O. C. Farokhzad, G. S. Teo, J. A. Phillips, D. M. Dorfman, R. Karnik and J. M. Karp, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19626–19631 CrossRef CAS PubMed.
- Z. Zhang, M. M. Ali, M. A. Eckert, D. K. Kang, Y. Y. Chen, L. S. Sender, D. A. Fruman and W. Zhao, Biomaterials, 2013, 34, 9728–9735 CrossRef CAS PubMed.
- L. Tang, Y. Liu, M. M. Ali, D. K. Kang, W. Zhao and J. Li, Anal. Chem., 2012, 84, 4711–4717 CrossRef CAS PubMed.
- Z. Cheglakov, Y. Weizmann, B. Basnar and I. Willner, Org. Biomol. Chem., 2007, 5, 223–225 RSC.
- M. M. Ali and Y. Li, Angew. Chem., Int. Ed., 2009, 48, 3512–3515 CrossRef CAS PubMed.
- H. Dong, C. Wang, Y. Xiong, H. Lu, H. Ju and X. Zhang, Biosens. Bioelectron., 2013, 41, 348–353 CrossRef CAS PubMed.
- F. Dahl, J. Banér, M. Gullberg, M. Mendel-Hartvig, U. Landegren and M. Nilsson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4548–4553 CrossRef CAS PubMed.
- W. Zhao, Y. Gao, S. A. Kandadai, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2006, 45, 2409–2413 CrossRef CAS PubMed.
- S. Beyer, P. Nickels and F. C. Simmel, Nano Lett., 2005, 5, 719–722 CrossRef CAS PubMed.
- H. Su, R. Yuan, Y. Chai, L. Mao and Y. Zhuo, Biosens. Bioelectron., 2011, 26, 4601–4604 CrossRef CAS PubMed.
- L. Linck, E. Reiß, F. Bier and U. Resch-Genger, Anal. Methods, 2012, 4, 1215–1220 RSC.
- A. Berr and I. Schubert, Plant J., 2006, 45, 857–862 CrossRef CAS PubMed.
- M. McKeague, A. De Girolamo, S. Valenzano, M. Pascale, A. Ruscito, R. Velu, N. R. Frost, K. Hill, M. Smith, E. M. McConnell and M. C. DeRosa, Anal. Chem., 2015, 87, 8608–8612 CrossRef CAS PubMed.
- W.-B. Shim, H. Mun, H.-A. Joung, J. A. Ofori, D.-H. Chung and M.-G. Kim, Food Control, 2014, 36, 30–35 CrossRef CAS.
- X. Ma, W. Wang, X. Chen, Y. Xia, S. Wu, N. Duan and Z. Wang, Eur. Food Res. Technol., 2014, 238, 919–925 CrossRef CAS.
- K. Setlem, B. Mondal, S. Ramlal and J. Kingston, Front Microbiol., 2016, 7, 1909 Search PubMed.
- C. Hamula, J. Guthrie, H. Zhang, X. Li and X. Le, TrAC-Trend Anal. Chem., 2006, 25, 681–691 CrossRef CAS.
- S. Malhotra, A. K. Pandey, Y. S. Rajput and R. Sharma, J. Mol. Recognit., 2014, 27, 493–500 CrossRef CAS PubMed.
- X. Chen, Y. Huang, N. Duan, S. Wu, X. Ma, Y. Xia, C. Zhu, Y. Jiang and Z. Wang, Anal. Bioanal. Chem., 2013, 405, 6573–6581 CrossRef CAS PubMed.
- C. C. Ong, S. Siva Sangu, N. M. Illias, S. Chandra Bose Gopinath and M. S. M. Saheed, Biosens. Bioelectron., 2020, 154, 112088 CrossRef CAS PubMed.
- S. Wu, N. Duan, W. Zhang, S. Zhao and Z. Wang, Anal. Biochem., 2016, 508, 58–64 CrossRef CAS PubMed.
- L. Huang, J. Wu, L. Zheng, H. Qian, F. Xue, Y. Wu, D. Pan, S. B. Adeloju and W. Chen, Anal. Chem., 2013, 85, 10842–10849 CrossRef CAS PubMed.
- W. Wu, S. Xia, M. Zhao, J. Ping, J. M. Lin and Q. Hu, Anal. Chim. Acta, 2022, 1220, 340065 CrossRef CAS PubMed.
- S. Niazi, I. M. Khan, Y. Yu, I. Pasha, Y. Lv, A. Mohsin, B. S. Mushtaq and Z. Wang, Sens. Actuators, B, 2020, 315, 128049 CrossRef CAS.
- G. T. Walker, M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau and D. P. Malinowski, Nucleic Acids Res., 1992, 20, 1691–1696 CrossRef CAS PubMed.
- L. Zhang, H. Jiang, Z. Zhu, J. Liu and B. Li, Talanta, 2022, 243, 123388 CrossRef CAS PubMed.
- C. Yuan, J. Fang, M. L. de la Chapelle, Y. Zhang, X. Zeng, G. Huang, X. Yang and W. Fu, TrAC-Trend Anal. Chem., 2021, 143, 116401 CrossRef CAS.
- J. Liu, D. Wu, J. Chen, S. Jia, J. Chen, Y. Wu and G. Li, Crit. Rev. Food Sci. Nutr., 2022, 64, 2960–2985 CrossRef PubMed.
- X. Zhu, C. Yang, W. Quan, G. Yang, L. Guo and H. Xu, Anal. Methods, 2023, 15, 987–992 RSC.
- Y. Yang, Z. Zhou, Y. Guo, R. Chen, D. Tian, S. Ren, H. Zhou and Z. Gao, Anal. Chim. Acta, 2024, 1292, 342245 CrossRef CAS PubMed.
- M. Zhang, Y. Wang, X. Sun, J. Bai, Y. Peng, B. Ning, Z. Gao and B. Liu, Anal. Chim. Acta, 2020, 1106, 161–167 CrossRef CAS PubMed.
- J. Yin, Y. Liu, S. Wang, J. Deng, X. Lin and J. Gao, Sens. Actuators, B, 2018, 256, 573–579 CrossRef CAS.
- M. Yan, W. Bai, C. Zhu, Y. Huang, J. Yan and A. Chen, Biosens. Bioelectron., 2016, 77, 613–623 CrossRef CAS PubMed.
- H. Yan, B. He, R. Zhao, W. Ren, Z. Suo, Y. Xu, Y. Zhang, C. Bai, H. Yan and R. Liu, J. Hazard. Mater., 2022, 438, 129491 CrossRef CAS PubMed.
- B. He and X. Dong, Chem. Eng. J., 2021, 405, 126642 CrossRef CAS.
- H. Cui, K. An, C. Wang, Y. Chen, S. Jia, J. Qian, N. Hao, J. Wei and K. Wang, Sens. Actuators, B, 2022, 355, 131238 CrossRef CAS.
- K. Wang, B. He, L. Xie, L. Li, J. Yang, R. Liu, M. Wei, H. Jin and W. Ren, Sens. Actuators, B, 2021, 349, 130767 CrossRef CAS.
- X. Niu, J. Yang, Z. Suo, M. Wei, Y. Liu, B. He and H. Jin, Microchem. J., 2023, 187, 108418 CrossRef CAS.
- K. S. Makarova, D. H. Haft, R. Barrangou, S. J. Brouns, E. Charpentier, P. Horvath, S. Moineau, F. J. Mojica, Y. I. Wolf, A. F. Yakunin and J. Van Der Oost, Nat. Rev. Microbiol., 2011, 9, 467–477 CrossRef CAS PubMed.
- X. Lin, C. Li, X. Meng, W. Yu, N. Duan, Z. Wang and S. Wu, J. Hazard. Mater., 2022, 433, 128750 CrossRef CAS PubMed.
- Z. Wu, D. W. Sun, H. Pu and Q. Wei, Talanta, 2023, 252, 123773 CrossRef CAS PubMed.
- H. Xu, R. Pan, W. Huang and X. Zhu, Anal. Methods, 2023, 15, 4518–4523 RSC.
- K. S. Makarova, Y. I. Wolf, O. S. Alkhnbashi, F. Costa, S. A. Shah, S. J. Saunders, R. Barrangou, S. J. Brouns, E. Charpentier, D. H. Haft, P. Horvath, S. Moineau, F. J. Mojica, R. M. Terns, M. P. Terns, M. F. White, A. F. Yakunin, R. A. Garrett, J. van der Oost, R. Backofen and E. V. Koonin, Nat. Rev. Microbiol., 2015, 13, 722–736 CrossRef CAS PubMed.
- Z.-y Wang, P. Li, L. Cui, J.-G. Qiu, B. Jiang and C.-y Zhang, TrAC-Trend Anal. Chem., 2020, 129, 115959 CrossRef CAS.
- Z. Wang, L. Wei, S. Ruan and Y. Chen, J. Agric. Food Chem., 2023, 71, 4417–4425 CrossRef CAS PubMed.
- X. Yao, Q. Yang, Y. Wang, C. Bi, H. Du and W. Wu, Foods, 2022, 11, 487–499 CrossRef CAS PubMed.
- J. Zhang, L. Jiang, H. Li, R. Yuan and X. Yang, Food Chem., 2023, 415, 135768 CrossRef CAS PubMed.
- N. Yin, S. Yuan, M. Zhang, J. Wang, Y. Li, Y. Peng, J. Bai, B. Ning, J. Liang and Z. Gao, Microchim. Acta, 2019, 186, 765 CrossRef PubMed.
- L. Zhao, J. Mao, L. Hu, S. Zhang and X. Yang, Anal. Methods, 2021, 13, 222–226 RSC.
- Y. Yang, D. Wu, J. Liu, Z. Su, L. Li, Y. Wu and G. Li, Biosens. Bioelectron., 2022, 214, 114526 CrossRef CAS PubMed.
- H. Zhang, Y. Wang, Y. Lin, W. Chu, Z. Luo, M. Zhao, J. Hu, X. Miao and F. He, Anal. Bioanal. Chem., 2023, 415, 867–874 CrossRef CAS PubMed.
- B. Wang, Y. Wu, Y. Chen, B. Weng, L. Xu and C. Li, Biosens. Bioelectron., 2016, 81, 125–130 CrossRef CAS PubMed.
- W. Zhu, G. Ji, R. Chen, Y. Xiang, S. Ji, S. Zhang, Z. Gao, H. Liu, Y. Wang and T. Han, Talanta, 2023, 255, 124249 CrossRef CAS PubMed.
- Y. Qing, X. Li, S. Chen, X. Zhou, M. Luo, X. Xu, C. Li and J. Qiu, Microchim. Acta, 2017, 184, 863–870 CrossRef CAS.
- F. Jia, D. Liu, N. Dong, Y. Li, S. Meng and T. You, Biosens. Bioelectron., 2021, 182, 113169 CrossRef CAS PubMed.
- X. Wang, C. Lau, M. Kai and J. Lu, Analyst, 2013, 138, 2691–2697 RSC.
- Y. Lin, J. Wang, F. Luo, L. Guo, B. Qiu and Z. Lin, Electrochim. Acta, 2018, 283, 798–805 CrossRef CAS.
- J. Tang, Y. Cheng, J. Zheng, J. Li, Y. Sun, S. Peng and Z. Zhu, Anal. Methods, 2019, 11, 5638–5644 RSC.
- Y. Liu, H. Yan, J. Shangguan, X. Yang, M. Wang and W. Liu, Microchim. Acta, 2018, 185, 427 CrossRef PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills Jr, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- J. Li, W. Wang, H. Zhang, Z. Lu, W. Wu, M. Shu and H. Han, Anal. Chem., 2020, 92, 4900–4907 CrossRef CAS PubMed.
- Z. Xiong, Q. Wang, Y. Xie, N. Li, W. Yun and L. Yang, Food Chem., 2021, 338, 128122 CrossRef CAS PubMed.
- Q. Ma, D. Nie, X. Sun, Y. Xu, J. He, L. Yang and L. Yang, Spectrochim. Acta, Part A, 2022, 281, 121634 CrossRef CAS PubMed.
- G. Wu, Z. Xiong, S. H. Oh, Y. Ren, Q. Wang and L. Yang, Food Chem., 2021, 356, 129663 CrossRef CAS PubMed.
- D. Li, S. Song and C. Fan, Acc. Chem. Res., 2010, 43, 631–641 CrossRef CAS PubMed.
- D. Zhang, F. Ma, J. Leng and C. Y. Zhang, Chem. Commun., 2018, 54, 13678–13681 RSC.
- Q. Zhang, R. Zhao, C. C. Li, Y. Zhang, C. Tang, X. Luo, F. Ma and C. Y. Zhang, Anal. Chem., 2022, 94, 13978–13986 CrossRef CAS PubMed.
- N. N. Zhao, Y. Z. Liu, L. Zhang, W. Liu, X. Zou, Q. Xu and C. Y. Zhang, Anal. Chem., 2023, 95, 12974–12981 CrossRef CAS PubMed.
- L.-j Wang, Q. Liu, F. Ma and C.-y Zhang, TrAC-Trend Anal. Chem., 2023, 167, 117270 CrossRef CAS.
- S. Zhang, Y. Luan, M. Xiong, J. Zhang, R. Lake and Y. Lu, ACS Appl. Mater. Interfaces, 2021, 13, 9472–9481 CrossRef CAS PubMed.
- L. Wang, H. Jin, M. Wei, W. Ren, Y. Zhang, L. Jiang, T. Wei and B. He, Sens. Actuators, B, 2021, 328, 129063 CrossRef CAS.
- Y. Sun, Y. Lv, S. Qi, Y. Zhang and Z. Wang, Food Chem., 2022, 371, 131145 CrossRef CAS PubMed.
- X. Lin, Y. Fang, Q. Chen, Z. Guo, X. Chen and X. Chen, Talanta, 2024, 267, 125273 CrossRef CAS PubMed.
- E. Schax, M. Lönne, T. Scheper, S. Belkin and J.-G. Walter, Biotechnol. Bioprocess Eng., 2016, 20, 1016–1025 CrossRef.
- Z. Han, Z. Tang, K. Jiang, Q. Huang, J. Meng, D. Nie and Z. Zhao, Biosens. Bioelectron., 2020, 150, 111894 CrossRef CAS PubMed.
- G. Ge, T. Wang, Z. Liu, X. Liu, T. Li, Y. Chen, J. Fan, E. Bukye, X. Huang and L. Song, Talanta, 2023, 265, 124908 CrossRef CAS PubMed.
- K.-Y. Xing, S. Shan, D.-F. Liu and W.-H. Lai, TrAC-Trend Anal. Chem., 2020, 133, 116087 CrossRef CAS.
- A. Rhouati, G. Bulbul, U. Latif, A. Hayat, Z. H. Li and J. L. Marty, Toxins, 2017, 9, 349–371 CrossRef PubMed.
- X. Xiang, Q. Ye, Y. Shang, F. Li, B. Zhou, Y. Shao, C. Wang, J. Zhang, L. Xue, M. Chen, Y. Ding and Q. Wu, Biosens. Bioelectron., 2021, 190, 113394 CrossRef CAS PubMed.
- Z. Suo, X. Niu, M. Wei, H. Jin and B. He, Anal. Chim. Acta, 2023, 1246, 340888 CrossRef CAS PubMed.
- L. Shi, Y. Li, C. Jia, J. Shan, S. Wang, S. Liu, J. Sun, D. Zhang, Y. Ji and J. Wang, Trends Food Sci. Technol., 2023, 138, 100–115 CrossRef CAS.
- E. M. Khalaf, H. Sanaan Jabbar, R. Mireya Romero-Parra, G. Raheem Lateef Al-Awsi, H. Setia Budi, A. S. Altamimi, M. Abdulfadhil Gatea, K. T. Falih, K. Singh and K. A. Alkhuzai, Microchem. J., 2023, 190, 108692 CrossRef CAS.
- C. Wu, Y. Yue, B. Huang, H. Ji, L. Wu and H. Huang, Talanta, 2024, 269, 125414 CrossRef CAS PubMed.
- Y. Lan, B. He, C. S. Tan and D. Ming, Biosensors, 2022, 12, 477–488 CrossRef CAS PubMed.
- X. Xiang, M. Song, X. Xu, J. Lu, Y. Chen, S. Chen, Y. He and Y. Shang, Anal. Chem., 2023, 95, 7993–8001 CrossRef CAS PubMed.
- A. Jaisankar, S. Krishnan and L. Rangasamy, Anal. Biochem., 2022, 655, 114874 CrossRef CAS PubMed.
- W. K. Abdelbasset, S. V. Savina, D. Mavaluru, R. A. Shichiyakh, D. O. Bokov and Y. F. Mustafa, Talanta, 2023, 252, 123769 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |





