Open Access Article
Open Access ArticleImproving electrochemical hybridization assays with restriction enzymes†
Xingcheng
Zhou‡
 a,
Marjon
Zamani‡
a,
Marjon
Zamani‡
 ab,
Katherine
Austin
a,
Marieke
De Bock
a,
Joshua
Chaj Ullola‡
a,
Smah
Riki
a and
Ariel L.
Furst
ab,
Katherine
Austin
a,
Marieke
De Bock
a,
Joshua
Chaj Ullola‡
a,
Smah
Riki
a and
Ariel L.
Furst
 *ab
*ab
aDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. E-mail: afurst@mit.edu
bCenter for Environmental Health Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
First published on 29th January 2024
Abstract
Nucleic acids in blood are early indicators of disease that could be detected by point-of-care biosensors if sufficiently sensitive and facile sensors existed. Electrochemical hybridization assays are sensitive and specific but are limited to very short nucleic acids. We have developed a restriction enzyme-assisted electrochemical hybridization (REH) assay for improved nucleic acid detection. By incorporating target-specific restriction enzymes, we detect long nucleic acids, with performance dependent on the location of the cut site relative to the electrode surface. Thus, we have further established guidelines for REH design to serve as a generalizable platform for robust electrochemical detection of long nucleic acids.
Liquid biopsies are minimally-invasive diagnostics that can detect circulating cell-free nucleic acids (ccfNAs) in blood and have been applied to diagnose cancer.1 ccfNAs are pieces of DNA and RNA released into the bloodstream that can indicate disease2,3 but must be detected with quantitative polymerase chain reaction (qPCR),4,5 which has burdensome equipment and reagent requirements.6 Many technologies including qPCR rely on fluorescent or colorimetric readout, requiring costly instrumentation.7 ccfNA detection at the point of care (POC) would enable broad implementation of these tests, accelerating diagnosis and treatment to improve patient outcomes.
Electrochemical nucleic acid (NA) detection is both sensitive and portable, while maintaining a low cost for instrumentation.8,9 Yet, most electrochemical NA detection focuses on short sequences (less than 20 bases),10,11 making them unsuitable for ccfNA detection, as ccfNAs are generally between 40 and 500 bases.12,13 Therefore, it is imperative to improve the NA recognition mechanism to improve the range of detectable targets.
Hybridization assays are often used to detect NAs, as their sequence-specificity affords improved sensitivity.14,15 These assays generally consist of an electrode-immobilized, redox-labeled oligonucleotide probe complementary to the target sequence.16,17 Probes can either be linear or stem-loop, both of which cause a signal decrease upon target binding.10,18 Target binding to a linear probe rigidifies the oligonucleotides, while stem-loop binding unwinds the probe. In both cases, the redox-active molecule ends up farther from the electrode,19 with linear probes generally providing larger signal differential.20,21 Linear probes are sufficiently sensitive to distinguish single-nucleotide mismatches in the target.22 However, these assays are limited to a narrow range of operating and interrogation conditions. Thus, stem-loop probes are favourable for more robust NA detection than linear probes.
Despite the robustness of stem-loop probes, they show comparatively limited signal suppression in most cases.22 A study by Lubin et al. investigated the influence of oligo length, geometry, and redox tag placement on sensor performance.22 For all possible configurations, longer probes were found to exhibit decreased specificity and mismatch discrimination as compared to shorter probes.23 Thus, a critical roadblock in electrochemical detection of ccfNAs is the limited signal change conventionally observed for hybridization-based assays.
Here, we report a novel restriction enzyme-assisted electrochemical hybridization (REH) assay through the incorporation of enzymatic cleavage into a conventional stem-loop assay (Fig. 1a). Upon target DNA binding to the stem-loop, hybridized probe-target is specifically cleaved by a restriction enzyme to both enhance signal suppression and improve specificity. We show that REH assay performance is dependent on the location of the restriction site, enabling guidance for optimal probe design. Taken together, we demonstrate significant improvement to hybridization-based assays through the incorporation of a simple enzymatic treatment. We use our learning to develop guidelines for improved electrochemical ccfNA detection.
Electrode platform: before integrating biological and electrochemical systems, we must confirm that our probe biomolecules are in relevant, accessible conformations on our electrode surfaces. Here, we employed our previously-reported gold leaf electrodes that demonstrate improved biosensing as compared to commercial screen-printed gold electrodes.24 Our electrodes were modified with either linear or stem-loop DNA containing a terminal thiol to enable oligonucleotide self-assembly. Stem-loop structures were formed prior to immobilization through 95 °C heating followed by snap-cooling. These DNA-modified surfaces were then used to assay hybridization and enzymatic cleavage.
Hybridization assay efficiency: hybridization assays based on DNA hairpin or stem-loop unwinding are well-established for electrochemical nucleic acid detection. Generally, a sample containing the target is added to an electrode modified with the stem-loop probe DNA. Upon hybridization in the loop region, target nucleic acids unwind the stem and form a fully complementary duplex (Fig. 1a). However, these sensors often suffer from small signal changes upon hybridization, which we observed here. The performance of the long stem-loop probe was monitored in a hybridization assay through addition of either complementary (34 bases) or non-complementary target (22 bases) nucleic acids (Fig. 1b and c). Two sets of square wave voltammetry (SWV) measurements were made: one following surface modification and one after target addition and hybridization. The ratio of the two voltammogram peaks was used to determine the assay efficiency.
The methylene blue (MB) redox signal ratio was ∼1 for both samples with no nucleic acid added (buffer-only control) and the non-complementary target, as expected. Incubation with the complementary target resulted in a slight signal decrease (signal ratio of ∼0.7), indicating a change at the electrode surface that causes the signal suppression. However, the decrease is not statistically significant (Fig. 1), which is consistent with literature reports for this sequence (reported decrease of ∼35%).2 From these results, we conclude that either the complementary target does not hybridize to the immobilized probe or the conformational change upon hybridization is insufficient to impact electron transfer between the MB and the electrode. In either case, these results confirm that this detection scheme is not amenable to long nucleic acid detection.
Incorporating enzymatic cleavage: as the conventional hybridization assay did not yield statistically significant results, we investigated whether inefficient hybridization or insufficient conformational change upon hybridization is responsible for the small decrease. We treated electrodes exposed to target oligonucleotide with a restriction enzyme that sequence-specifically cleaves duplexed DNA. We selected two restriction enzymes (Hpy188I and AlwI) that cut sites present in duplexed, target-bound DNA but not in the stem-loop probe structure (Fig. 2a and b). The Hpy188I cleavage site is farther from the electrode and closer to the MB tag, while Alwl cleaves DNA adjacent to the electrode surface. We verified that the enzyme cleavage was successful through gel electrophoresis (Fig S1, ESI†).
Treatment of electrodes with low concentrations of each of the restriction enzymes (either 100 U mL−1 or 200 U mL−1) resulted in significant MB signal decreases (Fig. 2c). Both Hpy188I concentrations exhibited significant MB signal decreases as compared to the buffer only control (p < 0.01 and 0.0001, respectively, according to a t-test). In contrast, only the lower concentration of AlwI (100 U mL−1) yielded a statistically-significant decrease in the MB signal as compared to the buffer-only control (p < 0.001 according to a t-test). The higher AlwI concentration did not cause a decrease in the MB signal as compared to the control.
Further, when additional hybridization controls were evaluated, no significant signal decreases were observed. Following either buffer-only or non-complementary DNA treatment of the electrode (Fig. 3a), electrodes were exposed to either Hpy188I or AlwI. No statistically-significant signal decreases were observed for any control conditions. The MB signal decrease from restriction enzyme treatment establishes that proper B-form duplexed DNA in a biologically-relevant and accessible conformation is generated on the surface. These results further confirm that we obtain hybridized probe-target DNA on the surface when the target is complementary to the probe (Fig. 3). As demonstrated by the controls, if the DNA remains in the stem-loop form, no signal decrease is observed.
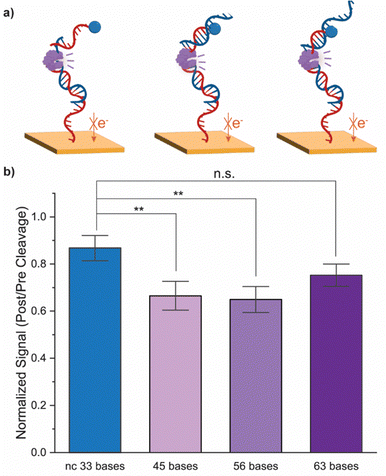
We further assessed the impact of target sequence length on cleavage efficacy. We increased the target nucleic acid from 34 bases to complementary sequences of 45 bases, 56 bases, and 63 bases, and a non-complementary sequence of 33 bases. Following hybridization, the enzyme with the highest cleavage performance as determined earlier, 100 U mL−1 of Hpy188I, was applied to the electrode. As seen in Fig. 4, for complementary sequences up to 56 bases, the MB redox signal significantly decreases as compared to the non-complementary control (p < 0.01 for both). No statistical significance was observed for the 63 base complementary sequence. This is likely due to increased steric hindrance and electrostatic repulsion as the target length increases.25 As ccfNAs generally have a minimum length of 40 bases, this assay is compatible for liquid biopsy applications.
The restriction enzyme cleavage results further confirm the importance of sequence design for DNA detection assays. The length and conformation of stem-loop probes have been shown to impact the resultant signal change from hybridization (Fig. 1). Additionally, we found that the location of the restriction site within the probe-target duplex impacts the efficacy of enzymatic cleavage. Hpy188l was found to cleave DNA at both low and high protein concentrations, while Alwl only cleaves at low concentrations, indicating that protein-DNA sterics must be considered in sensor design. This observation is consistent with our previous work demonstrating that binding site accessibility for electrode-immobilized DNA determines the relative activity of the protein.24
As the importance of ccfNAs increases, new methods for their point-of-use monitoring are needed. Stem-loop hybridization assays are popular electrochemical platforms to detect these biomarkers but are limited by the length of the target sequence and necessary operating conditions. By incorporating a post-hybridization restriction enzyme treatment step, we obtain statistically-significant signal decreases from this assay. In fact, by introducing restriction enzyme cleavage, we detect a previously-undetectable hybridization event. We further increased the assay specificity through the application of sequence-specific restriction enzymes that only cleave the target sequence in properly-hybridized probe-target duplex. Furthermore, existing electrochemical detection of ccfNAs of longer sequences generally involve the amplification of target sequences or application of expensive monoclonal antibodies, our assay demonstrates a simplified workflow towards analysing liquid biopsy samples.26,27
These assay improvements highlight critical design considerations for hybridization-based nucleic acid detection assays. Considerations include the length of the stem-loop probe, the location of the restriction site within the resultant duplex, and the concentration of restriction enzyme used. Taken together, our results demonstrate the power of our novel REH assay to expand the scope of targets detectable with stem-loop hybridization assays, while increasing assay specificity by leveraging the precision of restriction enzymes. This assay represents a significant step towards point-of-care diagnostics based on ccfNA detection.
We would like to acknowledge the MIT Abdul Latif Jameel Water and Food Systems Seed Funding program for supporting this work. Support for this research was also provided by a core center grant P30-ES002109 from the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH) and an agreement between MIT and a USGA. M. Z. was supported by an NIEHS Training Grant, Grant # T32-ES007020. X. Z. was supported by a fellowship through the MathWorks Corporation.
Conflicts of interest
There are no conflicts to declare.References
- S. N. Lone, S. Nisar, T. Masoodi, M. Singh, A. Rizwan, S. Hashem, W. El-Rifai, D. Bedognetti, S. K. Batra, M. Haris, A. A. Bhat and M. A. Macha, Mol. Cancer, 2022, 21, 1–22 CrossRef PubMed.
- H. Schwarzenbach, D. S. B. Hoon and K. Pantel, Nat. Rev. Cancer, 2011, 11, 426–437 CrossRef PubMed.
- O. Pös, O. Biró, T. Szemes and B. Nagy, Eur. J. Hum. Genet., 2018, 26, 937–945 Search PubMed.
- S. Holdenrieder, P. Stieber, L. Y. S. Chan, S. Geiger, A. Kremer, D. Nagel and Y. M. D. Lo, Clin. Chem., 2005, 51, 1544–1546 CrossRef CAS.
- C. E. Jin, B. Koo, T. Y. Lee, K. Han, S. B. Lim, I. J. Park and Y. Shin, Adv. Sci., 2018, 5, 1800614 CrossRef PubMed.
- H. Lee, W. Na, C. Park, K. H. Park and S. Shin, Sci. Rep., 2018, 8, 5467 CrossRef PubMed.
- G. C. O’Connell, P. D. Chantler and T. L. Barr, Lab. Med, 2017, 48, 332–338 CrossRef PubMed.
- M. Zamani, J. M. Robson, A. Fan, M. S. Bono, A. L. Furst and C. M. Klapperich, ACS Cent. Sci., 2021, 7, 963–972 CrossRef PubMed.
- X. Zhou, D. A. Schuh, L. M. Castle and A. L. Furst, Front. Chem., 2022, 0, 614 Search PubMed.
- M. Mastali, J. T. Babbitt, Y. Li, E. M. Landaw, V. Gau, B. M. Churchill and D. A. Haake, J. Clin. Microbiol., 2008, 46, 2707–2716 CrossRef PubMed.
- A. Karbelkar, R. Ahlmark, X. Zhou, K. Austin, G. Fan, V. Y. Yang and A. Furst, Bioconjug. Chem., 2023, 34, 358–365 CrossRef CAS PubMed.
- F. Li, F. Wei, W. L. Huang, C. C. Lin, L. Li, M. M. Shen, Q. Yan, W. Liao, D. Chia, M. Tu, J. H. Tang, Z. Feng, Y. Kim, W. C. Su and D. T. W. Wong, Cancers, 2020, 12, 1–15 Search PubMed.
- H. R. Underhill, Mol. Diagn. Ther., 2021, 25, 389–408 CrossRef.
- M. Bartosik, R. Hrstka, E. Palecek and B. Vojtesek, Anal. Chim. Acta, 2014, 813, 35–40 CrossRef.
- J. I. A. Rashid and N. A. Yusof, Sens. Bio-Sensing Res., 2017, 16, 19–31 CrossRef.
- E. E. Ferapontova, Curr. Opin. Electrochem., 2017, 5, 218–225 CrossRef.
- J. Wang, Anal. Chim. Acta, 2002, 469, 63–71 CrossRef.
- W. Yang and R. Y. Lai, Langmuir, 2011, 27, 14669–14677 CrossRef CAS PubMed.
- J. C. Cunningham, N. J. Brenes and R. M. Crooks, Anal. Chem., 2014, 86, 6166–6170 CrossRef CAS PubMed.
- R. Y. Lai, D. S. Seferos, A. J. Heeger, G. C. Bazan and K. W. Plaxco, Langmuir, 2006, 22, 10796–10800 CrossRef PubMed.
- V. Ruiz-Valdepenas Montiel, E. Povedano, E. Vargas, R. M. Torrente-Rodríguez, M. Pedrero, A. J. Reviejo, S. Campuzano and J. M. Pingarrón, ACS Sens., 2018, 3, 211–221 CrossRef.
- A. A. Lubin, B. V. S. Hunt, R. J. White and K. W. Plaxco, Anal. Chem., 2009, 81, 2150–2158 CrossRef.
- D. Kang, X. Zuo, R. Yang, F. Xia, K. W. Plaxco and R. J. White, Anal. Chem., 2009, 81, 9109–9113 CrossRef PubMed.
- M. Zamani, V. Yang, L. Maziashvili, G. Fan, C. M. Klapperich and A. L. Furst, ACS Meas. Sci. Au, 2022, 2, 91–95 CrossRef PubMed.
- H. Ravan, S. Kashanian, N. Sanadgol, A. Badoei-Dalfard and Z. Karami, Anal. Biochem., 2014, 444, 41–46 CrossRef PubMed.
- H.-F. Wang, R.-N. Ma, F. Sun, L.-P. Jia, W. Zhang, L. Shang, Q.-W. Xue, W.-L. Jia and H.-S. Wang, Biosens. Bioelectron., 2018, 122, 224–230 CrossRef CAS PubMed.
- C. Cai, Z. Guo, Y. Cao, W. Zhang and Y. Chen, Nanotheranostics, 2018, 2, 12–20 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc06192b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |