Open Access Article
Open Access ArticleUnexpectedly stable homopurine parallel triplex of SNA:RNA*SNA and L-aTNA:RNA*L-aTNA†
Yukiko
Kamiya
 *abc,
Siyuan
Lao
a,
Jumpei
Ariyoshi
b,
Fuminori
Sato
a and
Hiroyuki
Asanuma
*abc,
Siyuan
Lao
a,
Jumpei
Ariyoshi
b,
Fuminori
Sato
a and
Hiroyuki
Asanuma
 *a
*a
aDepartment of Biomolecular Engineering, Graduate School of Engineering, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8603, Japan. E-mail: asanuma@chembio.nagoya-u.ac.jp
bLaboratory of Bioanalytical Chemistry, Kobe Pharmaceutical University, Higashinada-Ku, Kobe, 658-8558, Japan. E-mail: y-kamiya@kobepharma-u.ac.jp
cInstitute for Molecular Science, National Institutes of Natural Sciences, Okazaki, Aichi 444-8585, Japan
First published on 21st December 2023
Abstract
Homopurine strands are known to form antiparallel triplexes stabilized by G*G and A*A Hoogsteen pairs, which have two hydrogen bonds. But there has been no report on the parallel triplex formation of homopurine involving both adenosine and guanosine to the duplex. In this paper, we first report parallel triplex formation between a homopurine serinol nucleic acid (SNA) strand and an RNA/SNA duplex. Melting profiles revealed that the parallel SNA:RNA*SNA triplex was remarkably stable, even though the A*A pair has a single hydrogen bond. An L-acyclic threoninol nucleic acid (L-aTNA) homopurine strand also formed a stable parallel triplex with an L-aTNA/RNA duplex.
Non-canonical base-pairing enables formation of multistranded nucleic acid structures such as triplexes,1–6 G-quadruplexes,7–10 and i-motifs.11,12 Parallel triplexes form under slightly acidic conditions when a homopyrimidine strand recognizes a Watson–Crick duplex through Hoogsteen pairing.4–6 Antiparallel triplexes with the third strand composed of only thymidine or homopurine have also been characterised.13,14 Non-natural nucleobases and xeno nucleic acids (XNAs) with non-ribose scaffolds also form multi-stranded structures. Homo-adenine is reported to form triplex mediated by cyanuric acid.15 Two peptide nucleic acids (PNAs) with homopyrimidines bind single-stranded homopurine DNA to form a parallel triplex.16,17 Tight binding of PNAs enables strand-invasive recognition of DNA duplex. Our group has synthesized and characterized the duplex formed by serinol nucleic acid (SNA)18 and acyclic threoninol nucleic acid (aTNA) with either D- or L-threoninol as a scaffold.19,20L-aTNA with a homopyrimidine sequence is reported to form parallel triplex with DNA similar to PNA.21
Thymidine- and cytidine-containing oligonucleotides form parallel triplexes but homothymidine triplex-forming oligonucleotide (TFO) may also form parallel or antiparallel triplexes (Scheme 1a).4–6,13,14 However, homopurine strands involving both guanosine and adenosine have only been reported to form antiparallel triplexes (Scheme 1b).13,14 This may be because if the TFO involves adenine taking anti conformation, only a single hydrogen bond is possible with the duplex (Scheme 1c).14 Furthermore, as the sequences of the duplex-forming homopurine strand and the TFO are the same, analysis of the triplex is difficult.
 | ||
| Scheme 1 Hydrogen bonds formed by triplex structures with (a) a parallel homopyrimidine TFO, (b) an antiparallel homopurine TFO, and (c) a parallel homopurine TFO. | ||
Recently, our group has successfully resolved the crystal structures of L-aTNA:RNA and SNA:RNA heteroduplexes.22 We found that both heteroduplexes were unwound relative to the RNA helical structure and that base pairs aligned perpendicularly to the helical axis, different from canonical B- or A-type duplexes. Interestingly and unexpectedly, in the crystals, we observed triplex structures at the termini of both heteroduplexes (Fig. S1, ESI†). These triplex structures involved C:G*G Watson–Crick and Hoogsteen pairs with RNA in a parallel orientation. This result prompted us to study XNA:RNA*XNA triplexes with the homopurine XNA TFO in the parallel orientation.
To examine parallel triplex formation of SNA-TFO, we used an RNA–SNA oligonucleotide that forms a hairpin (S:R-HP) as a template to avoid formation of an SNA:SNA duplex (Scheme 2 and S2, ESI†), and a TFO designed to bind to S:R-HP in parallel fashion (P-SNA-TFO). The use of a hairpin was necessary because an SNA:SNA duplex is more stable than an SNA:RNA duplex,23,24 and unless a hairpin is used, strand invasion would lead to an SNA:SNA duplex rather than a triplex. The melting temperature (Tm) of the S:R-HP was 80.5 °C (Fig. S2, ESI†). An RNA–SNA oligonucleotide that cannot form a hairpin in which only the SNA region is complementary to P-SNA-TFO was used as a negative control (Non-S:R, Scheme 2). S:R-HP and Non-S:R were labelled at the 5′ termini with FAM (green); TAMRA (red) was conjugated to the (S) terminus of P-SNA-TFO. Note that the G-rich TFO might form anti-parallel G-quadruplexes (Fig. S3, ESI†).25
 | ||
| Scheme 2 Chemical structures of SNA and L-aTNA, and schematics and sequences of select oligonucleotides used in this study. For all oligonucleotide sequences, see Scheme S2 (ESI†). | ||
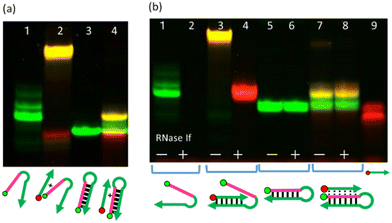
We analysed triplex formation by native polyacrylamide electrophoresis (PAGE). When P-SNA-TFO was mixed with Non-S:R, which should result in formation of a duplex, a yellow band with overlapping green and red bands was observed that migrated more slowly than Non-S:R (Fig. 1a, lanes 1 and 2). Mixing of S:R-HP and P-SNA-TFO also resulted in a yellow band that migrated more slowly than S:R-HP (Fig. 1a, lanes 3 and 4), but the position was different from that of the P-SNA-TFO:Non-S:R duplex. To rule out the possibility that strand invasion had resulted in a P-SNA-TFO:SNA duplex, we incubated samples with RNase If, which selectively digests single-stranded RNA.26
If a P-SNA-TFO:SNA duplex was formed, the single-stranded RNA should be digested by RNase If. Neither S:R-HP nor S:R-HP in the complex with P-SNA-TFO were digested by RNase If (Fig. 1b, lanes 5–8), whereas both Non-S:R and Non-S:R in the complex with P-SNA-TFO were completely digested (Fig. 1b, lanes 1–4), clearly demonstrating that a triplex was formed by P-SNA-TFO and the S:R-HP.
Next, we examined stoichiometries and Tms of complexes by replacing the TAMRA on the (S) terminus of the P-SNA-TFO with quencher Dabcyl (Scheme 2). When FAM-S:R-HP was mixed with Dabcyl-P-SNA-TFO, FAM fluorescence was considerably quenched (Fig. 2a). Fluorescent titration revealed that triplex formation proceeded via 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation of the S:R-HP and P-SNA-TFO (Fig. 2b). From the melting profile of FAM fluorescence, the Tm of the parallel triplex was 61.4 °C (Fig. 3, blue line), which was higher than the Tm of the antiparallel triplex (41.2 °C, Fig. 3, red line). It should be noted that A*A pair in parallel triplex has only single hydrogen bond (Scheme 1c), whereas it has two in the antiparallel triplex. Despite this, the Tm of the parallel SNA:RNA*SNA triplex was considerably higher than those of the antiparallel triplexes. Although homopurine TFO could form both parallel and antiparallel triplexes, they may have different base-stacking geometries. Additionally, we compared Tm of the homopurine triplex to that of the homopyrimidine triplex (Scheme S2, ESI†). As expected from the previous report,21 homopyrimidine TFO formed parallel triplex (Fig. 3b), but less stable than the homopurine parallel triplex (Fig. 3a). The homopyrimidine TFO did not form anti-parallel type triplex (Fig. 3b).
1 complexation of the S:R-HP and P-SNA-TFO (Fig. 2b). From the melting profile of FAM fluorescence, the Tm of the parallel triplex was 61.4 °C (Fig. 3, blue line), which was higher than the Tm of the antiparallel triplex (41.2 °C, Fig. 3, red line). It should be noted that A*A pair in parallel triplex has only single hydrogen bond (Scheme 1c), whereas it has two in the antiparallel triplex. Despite this, the Tm of the parallel SNA:RNA*SNA triplex was considerably higher than those of the antiparallel triplexes. Although homopurine TFO could form both parallel and antiparallel triplexes, they may have different base-stacking geometries. Additionally, we compared Tm of the homopurine triplex to that of the homopyrimidine triplex (Scheme S2, ESI†). As expected from the previous report,21 homopyrimidine TFO formed parallel triplex (Fig. 3b), but less stable than the homopurine parallel triplex (Fig. 3a). The homopyrimidine TFO did not form anti-parallel type triplex (Fig. 3b).
 | ||
| Fig. 2 (a) Fluorescence of FAM conjugated to S:R-HP in the presence and absence of Dabcyl-P-SNA-TFO. [S:R-HP] = 1.79 μM, [P-SNA-TFO] = 2.14 μM. 90 mM Tris-borate buffer, 10 mM MgCl2, pH 7.0, 4 °C. (b) Fluorescence of FAM-S:R-HP as a function of the ratio of [Dabcyl-P-SNA-TFO] to [S:R-HP]. Initial [S:R-HP] = 2.0 μM, 90 mM Tris-borate buffer, 10 mM MgCl2, pH 7.0, 4 °C. Excitation wavelength was 495 nm, and fluorescence intensity was monitored at 523 nm. See ESI† for the titration experiment. | ||
We next examined whether L-aTNA can form a parallel triplex with an RNA-L-aTNA hairpin (LT:R-HP, Scheme S2, ESI†). As observed in fluorescence experiments with the SNA-based system, fluorescence of FAM-LT:R-HP was considerably quenched in the presence of Dabcyl-P-L-aTNA-TFO, and the fluorescence titration showed that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex was formed (Fig. S4a, ESI†). The Tm of the triplex formed with the L-aTNA-TFO was 74.2 °C (Fig. S4b, ESI†), 13 °C higher than the Tm of the parallel SNA triplex. We also confirmed that homopyrimidine L-aTNA-TFO did not form antiparallel triplex but parallel triplex (Fig. S4c, ESI†). These results indicate that scaffold structures of SNA and L-aTNA result in stable parallel triplexes containing a single RNA strand.
1 complex was formed (Fig. S4a, ESI†). The Tm of the triplex formed with the L-aTNA-TFO was 74.2 °C (Fig. S4b, ESI†), 13 °C higher than the Tm of the parallel SNA triplex. We also confirmed that homopyrimidine L-aTNA-TFO did not form antiparallel triplex but parallel triplex (Fig. S4c, ESI†). These results indicate that scaffold structures of SNA and L-aTNA result in stable parallel triplexes containing a single RNA strand.
To examine the sequence specificity of parallel triplex formation, we measured Tms of mismatched SNA-TFOs. When a guanine near the centre of the TFO was replaced with a different nucleobase, the Tm remarkably decreased by 15–30 °C (Fig. 4a). When an adenine near the centre was replaced, the Tm decreased, but the decrease was around 10 °C (Fig. 4b), smaller in magnitude than observed when guanine was substituted. Similar sequence-specificity was also observed for the L-aTNA triplex (Fig. S5, ESI†). These results indicate that G*G and A*A pairs stabilize the triplex. The smaller decrease in Tm when adenine was replaced compared to guanine is likely because there is a single hydrogen bond in the A*A pair and two in the G*G pair.
Finally, we investigated triplex formation with DNA in the hairpin. Unlike RNA, quenching of FAM conjugated to S:D-HP by the P-SNA-TFO was inefficient (Fig. 5a). A titration showed an inflection point at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of S:D-HP to P-SNA-TFO (Fig. 5b), and a clear sigmoid curve of melting profile was observed (Fig. 5c). The Tm was 59.9 °C, which was comparable to that of RNA-containing parallel triplex (61.4 °C). PAGE analysis revealed the reason for inefficient quenching. In the sample containing FAM-S:D-HP and TAMRA-P-SNA-TFO two yellow bands were observed that were assigned to triplex and invaded duplex (Fig. 5d). Since the SNA:DNA duplex is less stable than the SNA:RNA duplex,20 some P-SNA-TFO invaded into the hairpin. When the DNA portion of S:D-HP is single stranded, the 5′ FAM is not quenched by the Dabcyl on the P-SNA-TFO. We also examined whether an SNA:SNA*SNA triplex was formed using the same strategy. Little quenching was detected (Fig. S6, ESI†). Thus, we concluded that a parallel triplex is formed only on DNA and RNA.
1 ratio of S:D-HP to P-SNA-TFO (Fig. 5b), and a clear sigmoid curve of melting profile was observed (Fig. 5c). The Tm was 59.9 °C, which was comparable to that of RNA-containing parallel triplex (61.4 °C). PAGE analysis revealed the reason for inefficient quenching. In the sample containing FAM-S:D-HP and TAMRA-P-SNA-TFO two yellow bands were observed that were assigned to triplex and invaded duplex (Fig. 5d). Since the SNA:DNA duplex is less stable than the SNA:RNA duplex,20 some P-SNA-TFO invaded into the hairpin. When the DNA portion of S:D-HP is single stranded, the 5′ FAM is not quenched by the Dabcyl on the P-SNA-TFO. We also examined whether an SNA:SNA*SNA triplex was formed using the same strategy. Little quenching was detected (Fig. S6, ESI†). Thus, we concluded that a parallel triplex is formed only on DNA and RNA.
SNA- and L-aTNA-TFOs formed stable parallel triplexes even though an A*A pair has a single hydrogen bond. Previous X-ray crystallographic analyses of SNA:RNA and L-aTNA:RNA duplexes revealed unique hydrogen bonds of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O with –CH2– and H8 of purine to fix the conformation of SNA and L-aTNA (Fig. 6a).22 Computer modelling with these hydrogen bonds suggest that these hydrogen bonds stabilize the parallel triplexes involving an RNA and SNA or L-aTNA duplex and an XNA TFO (Fig. 6b). In contrast, the antiparallel conformation does not allow these hydrogen bonds. Thus, the single hydrogen bond in A*A pair is compensated for by these additional hydrogen bonds, which stabilize the parallel triplex.
O with –CH2– and H8 of purine to fix the conformation of SNA and L-aTNA (Fig. 6a).22 Computer modelling with these hydrogen bonds suggest that these hydrogen bonds stabilize the parallel triplexes involving an RNA and SNA or L-aTNA duplex and an XNA TFO (Fig. 6b). In contrast, the antiparallel conformation does not allow these hydrogen bonds. Thus, the single hydrogen bond in A*A pair is compensated for by these additional hydrogen bonds, which stabilize the parallel triplex.
 | ||
Fig. 6 (a) Crystal structure of anti-parallel duplex of SNA and RNA. Hydrogen bonds between C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and both –CH2– and H8 of purine observed in the crystal structure of the RNA:SNA duplex (PDB: 7bpg). (b) Computer modelling of SNA:RNA*SNA triplex stabilized by hydrogen bonds between the carbonyl and adjacent hydrogens based on the crystal structure. Hydrogen atoms are omitted for clarity. O and both –CH2– and H8 of purine observed in the crystal structure of the RNA:SNA duplex (PDB: 7bpg). (b) Computer modelling of SNA:RNA*SNA triplex stabilized by hydrogen bonds between the carbonyl and adjacent hydrogens based on the crystal structure. Hydrogen atoms are omitted for clarity. | ||
In conclusion, we discovered that homopurine SNA and L-aTNA oligonucleotides form XNA:RNA*XNA and XNA:DNA*XNA parallel triplexes in a sequence-specific manner. Even though the A*A pair has a single hydrogen bonding, the parallel triplex was considerably more stable than the antiparallel triplex in which the A*A pair has two hydrogen bonds. The unexpectedly stable parallel triplex results from the interaction of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O with both –CH2– and H8 of purine. Note that interior linker that connects nucleobase and main chain is one atom longer for SNA and L-aTNA than DNA and PNA, which may give flexibility and allowed both parallel and antiparallel triplex with homopurine TFO. The parallel triplex is thus specific to SNA and L-aTNA and is a new supramolecular motif that should prove useful in XNA-based nanotechnology.27 This is the first example of parallel triplex formation with a homopurine sequence. Unlike conventional homopyrimidine TFO, homopurine TFO does not require acidic conditions. Therefore, parallel homopurine TFO will be able to capture purine-rich RNA in cells under physiological conditions. In order to target single-stranded RNA in cells, formation of SNA or L-aTNA homoduplexes must be suppressed. For this purpose, replacement of the A–T pair with a pseudo-complementary 2,6-diaminopurine-thiouracil pair will be effective.28,29 We have recently succeeded in the incorporation of diaminopurine and thiouracil into SNA and L-aTNA.23,24
O with both –CH2– and H8 of purine. Note that interior linker that connects nucleobase and main chain is one atom longer for SNA and L-aTNA than DNA and PNA, which may give flexibility and allowed both parallel and antiparallel triplex with homopurine TFO. The parallel triplex is thus specific to SNA and L-aTNA and is a new supramolecular motif that should prove useful in XNA-based nanotechnology.27 This is the first example of parallel triplex formation with a homopurine sequence. Unlike conventional homopyrimidine TFO, homopurine TFO does not require acidic conditions. Therefore, parallel homopurine TFO will be able to capture purine-rich RNA in cells under physiological conditions. In order to target single-stranded RNA in cells, formation of SNA or L-aTNA homoduplexes must be suppressed. For this purpose, replacement of the A–T pair with a pseudo-complementary 2,6-diaminopurine-thiouracil pair will be effective.28,29 We have recently succeeded in the incorporation of diaminopurine and thiouracil into SNA and L-aTNA.23,24
This research was supported by the Joint Research on ExCELLS (No. 23EXC202), by AMED (grant number 23am0401007), by Inamori Foundation (to Y. K.), and by JSPS KAKENHI (grants JP21H05025 to H. A. and 23H04067 to Y. K.). S. L. and F. S. thank the “Nagoya University Interdisciplinary Frontier Fellowship” supported by Nagoya University and JST, the establishment of university fellowships towards the creation of science technology innovation (grant number JPMJFS2120). We are grateful to Dr M. Yagi-Utsumi and Prof. K. Kato (ExCELLS) for NMR measurements on the early stage of this study.
H. A. and Y. K. planned and began this work. S.L. performed most of the experiments. J. A. and F. S. supported measurements of melting profile and organic synthesis. H. A. and Y. K. wrote the manuscript with contribution from all the authors.
Conflicts of interest
There are no conflicts to declare.References
- G. Felsenfeld, D. Davies and A. Rich, J. Am. Chem. Soc., 1957, 79, 2023 CrossRef CAS.
- K. Hoogsteen, Acta Crystallogr., 1959, 12, 822 CrossRef CAS.
- N. T. Thuong and C. Hélène, Angew. Chem., Int. Ed. Engl., 1993, 32, 666 CrossRef.
- L. Lavelle and J. Fresco, Nucleic Acids Res., 1995, 23, 2692 CrossRef CAS PubMed.
- F.-X. Barre, C. Glovannangeli, C. Hélène and A. Harel-Bellan, Nucleic Acids Res., 1999, 27, 743 CrossRef CAS PubMed.
- H. Okamura, Y. Taniguchi and S. Sasaki, Angew. Chem., Int. Ed., 2016, 55, 12445 CrossRef CAS PubMed.
- G. Gellert, M. N. Lipsett and D. R. Daveis, Proc. Natl. Acad. Sci. U. S. A., 1962, 48, 2013 CrossRef PubMed.
- R. K. Moyzis, J. M. Buckingham, L. S. Cram, M. Dani, L. L. Deaven, M. D. Jones, J. Meyne, R. L. Ratliff and J.-R. Wu, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 6622 CrossRef CAS PubMed.
- C. M. Azzalin, P. Reichenback, L. Khoriauli, E. Giulotto and J. Linger, Science, 2007, 318, 798 CrossRef CAS PubMed.
- Y. Wu, K. Kaminaga and M. Komiyama, J. Am. Chem. Soc., 2008, 130, 11179 CrossRef PubMed.
- K. Gehring, J.-L. Leroy and M. Guéron, Nature, 1993, 363, 561 CrossRef CAS PubMed.
- A. T. Phan and J.-L. Mergny, Nucleic Acids Res., 2002, 30, 4618 CrossRef CAS PubMed.
- P. A. Beal and P. B. Dervan, Science, 1991, 251, 1360 CrossRef CAS PubMed.
- P. A. Beal and P. B. Dervan, Nucleic Acids Res., 1992, 20, 2773 CrossRef CAS PubMed.
- N. Avakyan, A. A. Greschner, F. Aldaye, C. J. Serpell, V. Toader, A. Petitjean and H. F. Sleiman, Nat. Chem., 2016, 8, 368 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497 CrossRef CAS PubMed.
- P. E. Nielsen, Acc. Chem. Res., 1999, 32, 624 CrossRef CAS.
- H. Kashida, K. Murayama, T. Toda and H. Asanuma, Angew. Chem., Int. Ed., 2011, 50, 1285 CrossRef CAS PubMed.
- H. Asanuma, T. Toda, K. Murayama, X. G. Liang and H. Kashida, J. Am. Chem. Soc., 2010, 132, 14702 CrossRef CAS PubMed.
- K. Murayama, H. Kashida and H. Asanuma, Chem. Commun., 2015, 51, 6500 RSC.
- V. Kumar, V. Kesavan and K. V. Gothelf, Org. Biomol. Chem., 2015, 13, 2366 RSC.
- Y. Kamiya, T. Satoh, A. Kodama, T. Suzuki, K. Murayama, H. Kashida, S. Uchiyama, K. Kato and H. Asanuma, Commun. Chem., 2020, 3, 156 CrossRef CAS PubMed.
- Y. Kamiya, F. Sato, K. Murayama, A. Kodama, S. Uchiyama and H. Asanuma, Chem. – Asian J., 2020, 15, 1266 CrossRef CAS PubMed.
- F. Sato, Y. Kamiya and H. Asanuma, J. Org. Chem., 2023, 88, 796 CrossRef CAS PubMed.
- V. Kumar and K. V. Gothelf, Org. Biomol. Chem., 2016, 14, 1540 RSC.
- J. Meador, B. Cannnon, V. J. Cannistraro and D. Kennel, Eur. J. Biochem., 1990, 187, 549 CrossRef CAS PubMed.
- T. J. D. Nguyen, I. Manuguerra, V. Kumar and K. V. Gothelf, Chem. – Eur. J., 2019, 25, 12303 CrossRef CAS PubMed.
- H. B. Gamper, A. Gewirtz, J. Edwards and Y. M. Hou, Biochemistry, 2004, 43, 10224 CrossRef CAS PubMed.
- J. Lohse, O. Dahl and P. E. Nielsen, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11804 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc05555h |
| This journal is © The Royal Society of Chemistry 2024 |