 Open Access Article
Open Access ArticleAffinity enhancement of polo-like kinase 1 polo box domain-binding ligands by a bivalent approach using a covalent kinase-binding component†
Kohei
Tsuji
 *ab,
Hirokazu
Tamamura
*ab,
Hirokazu
Tamamura
 b and
Terrence R.
Burke
Jr.
b and
Terrence R.
Burke
Jr.
 *a
*a
aChemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, MD 21702, USA. E-mail: burkete@mail.nih.gov
bDepartment of Medicinal Chemistry, Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University, Tokyo 101-0062, Japan. E-mail: ktsuji.mr@tmd.ac.jp
First published on 4th June 2024
Abstract
The polo-like kinase 1 (Plk1) is an important cell cycle regulator that is recognized as a target molecule for development of anti-cancer agents. Plk1 consists of a catalytic kinase domain (KD) and a polo-box domain (PBD), which engages in protein–protein interactions (PPIs) essential to proper Plk1 function. Recently, we developed extremely high-affinity PBD-binding inhibitors based on a bivalent approach using the Plk1 KD-binding inhibitor, BI2536, and a PBD-binding peptide. Certain of the resulting bivalent constructs exhibited more than 100-fold Plk1 affinity enhancement relative to the best monovalent PBD-binding ligands. Herein, we report an extensive investigation of bivalent ligands that utilize the non-selective kinase inhibitor Wortmannin as a Plk1 KD-binding component. We found that bivalent ligands incorporating Wortmannin demonstrated affinity enhancements that could be similar to what we had obtained with BI2536 and that they could tightly bind to the protein. This suggests that these tight binding ligands might be useful for structural analysis of full-length Plk1.
Introduction
In developing potent inhibitors against target proteins, bivalent-to-multivalent approaches may be particularly advantageous in that affinity, inhibitory potency, and target selectivity may be greater than those obtained with monovalent compounds.1 Protein kinases (PKs) are a family of enzymes that transfer a phosphate group from the γ-phosphate of adenosine triphosphate (ATP) to serine, threonine, or tyrosine residues in substrate proteins. Therefore, kinases have a substrate binding pocket and an ATP binding pocket, as well as other specific interaction sites.2,3 Productive phosphorylation requires simultaneous binding of ATP together with appropriate acceptor proteins.4 Accordingly, bivalent approaches that conjugate an ATP-binding cleft binder with a pseudosubstrate (bisubstrate ligands) are often employed to develop kinase inhibitors.5,6 When the ATP-binding components are linked with moieties that bind to ancillary regulatory domains, the resulting ligands may be described as being intramolecular bivalent ligands.5,7 This genre of bivalent kinase inhibitors have been classified as type V inhibitors.8The polo-like kinase 1 (Plk1) is a member of the serine/threonine family of kinases that functions as an important cell cycle regulator.9 This protein is overexpressed in numerous cancers, including breast cancers, colon cancers, non-small cell lung cancer, and prostate cancers.10–15 Therefore it is not surprising that Plk1 is an attractive target for anti-cancer therapy development.16–18 Functioning of Plk1 is spatiotemporally restricted by interdomain protein–protein interactions (PPIs) of an N-terminal catalytic kinase domain (KD) and a C-terminal polo-box domain (PBD) that recognizes phosphoserine (pS) and phosphothreonine (pT)-containing sequences.19 Multiple Plk1 KD-directed agents are currently in clinical trials for the treatment of cancers. However, cytotoxicity arising from off-target effects can be dose-limiting.16–18 This may be due in part to lack of selectivity against other kinases. Although ATP-binding sites in kinases are highly conserved among the approximately 500 members of the human kinome,20 PBDs are unique to the five members of the Plk family of kinases. Therefore, targeting PBDs may afford a more selective approach for down-regulating the Plk1 activity.21–23
Over the past decades we have been developing Plk1 PBD-binding inhibitors. We have started with the peptide, PLHSpT (1, Fig. 1), which arises from the polo-box domain interacting protein 1 (PBIP1), which contains the phosphorylated Thr78 (pT78) residue.24 We found that attaching alkylphenyl groups to the sidechain imidazole π-nitrogen of the His residue resulted in peptides such as PLH*SpT 2, H* indicates the presence of a –(CH2)8Ph group on the His N3(π)-nitrogen (i.e., H* = His-[N(π)-(CH2)8Ph], Fig. 1), which showed greater than 1000-fold higher affinity than the parent peptide 1. Based on the co-crystal structure of isolated Plk1 PBD and PLH*SpT (2, PDB: 3RQ7), we found that the alkylphenyl sidechain occupies a hydrophobic “cryptic binding pocket” formed by residues Y417, Y421, Y481, F482, Y485 and L478.25,26 We also found that this hydrophobic channel can be accessed by alkyl groups tethered from N-terminal regions of the peptide as well.27 Guided by the 3RQ7 crystal structure, we went on to develop several potent and highly selective inhibitor peptides by tethering aryl and heteroaryl moieties from the His N3(π) nitrogen,28–30 attachment of long-chain alkylphenyl groups to the side chains of other amino acids at the pT-2 position,31 or cyclization using a bis-alkylated His residue32 or a novel orthogonally protected Glu residue.33 Plk1 affords an attractive target to develop bivalent inhibitors that simultaneously engage the two intramolecular domains.16,18,22 Although peptide 2 had been one of the highest PBD-binding ligands yet reported, recently, we were able to achieve more than 100-fold affinity enhancement relative to 2 by preparing bivalent ligands that simultaneously target both the KD and PBD.34
 | ||
| Fig. 1 Structures of the parent pentapeptides 1 and 2, and bivalent ligands possessing a BI2536 moiety conjugated from the PBD-binding peptide C-terminus (3 and 4) and N-terminus (5 and 6). | ||
It is noteworthy that a structure of full-length Plk1 has yet to be reported (neither crystal, solution, nor cryo-EM). This may possibly be due to high flexibility of the inter-domain linker (IDL) that connects the KD and PBD. However, the potential relative orientation of these two domains has been informed by a co-crystal structure of the isolated Plk1 KD and PBD from zebrafish (D. rerio), which is a homolog to human Plk1. This structure was stabilized by including the PBD-binding motif of Map205 (Map205PBM) from Drosophila (PDB: 4J7B) as a molecular glue (Fig. 2(a)).35
 | ||
| Fig. 2 (a) Superimposed structure of the isolated zebrafish Plk1 KD and PBD complex (4J7B) with BI2536-bound human Plk1 KD (2RKU) and human Plk1 PBD-bound peptide 2 (3RQ7). The KD and PBD are shown in light blue and yellow ribbons, respectively. Carbon atoms of BI2536 and peptide 2 are shown in magenta and green, respectively. Nitrogen and oxygen atoms are shown in blue and red. The magenta and green meshes represent binding sites of the ligands; (b) and (c) MD simulation results of Plk1 with the KD and PBD arranged to allow binding of the bivalent ligands 4 in (b) and 6 in (c).33 The linkers (Lys) are shown in yellow. The orange and cyan meshes represent binding sites of the bivalent ligands 4 and 6, respectively. (d) Structure of KD complexed with BI2536 (2RKU, human KD, left) and KD complexed with Wortmannin (3D5X, zebrafish KD, right). The carbon atoms of Wortmannin are shown in orange. The magenta and orange meshes represent binding sites of BI2536 and Wortmannin in the KD, respectively. The structure of Wortmannin bound to K68 of the KD is shown. (e) Superimposed structure of human KD-bound BI2536 (2RKU) and zebrafish KD-bound Wortmannin (3D5X). The human KD is shown in transparent light blue ribbon. | ||
We recently reported bivalent constructs 3–6 that employed the known Plk1 KD-targeting inhibitor BI253636 tethered to 2 by PEG linkers. We observed that binding affinities to the Plk1 PBD in full-length Plk1 were not affected by linker length (n = 4 or 0) (Fig. 1).34 We interpreted this to suggest that there are many possible conformations of full-length Plk1 in which its KD and PBD can be stably engaged. This hypothesis is also supported by our MD simulation studies (Fig. 2(b) and (c)).34,37 We were intrigued by the idea that tight binding bivalent ligands might reduce the conformational mobility of full-length Plk1 sufficiently to solve its structure. Our current work was undertaken to develop bivalent ligands that incorporate one component that can bind covalently to the KD.
Results and discussion
Design and synthesis of bivalent ligands that incorporate Wortmannin
Binding of covalent inhibitors to their target proteins can result in highly stable complexes. We envisioned that when a bivalent ligand possesses a covalent component, the simultaneous binding of KD and PBD would function like a door bolt to form a stable complex. Wortmannin is a non-specific covalent phosphoinositide 3-kinases (PI3Ks) inhibitor that is known as an anti-inflammatory agent.38 Wortmannin has been shown to bind to Plk1 in an ATP-competitive manner and to form a covalent bond with the ε-amino group of Lys68 in the zebrafish Plk1 KD (PBD: 3D5X) and Lys82 in the human Plk1 KD (Fig. 2(d) and (e)).39–41 Binding of Wortmannin to the Plk1 KD has been reported to result in Michael addition-type conjugation of the side chain amine group of Lys68 (zebrafish) or Lys82 (human) and the C20 of Wortmannin (Fig. 2(d) and (e)). This position is part of a reactive electrophilic furan ring double bond that forms an α,β-unsaturated lactone and a vinylogous α,β-unsaturated ketone. This nucleophilic attack at the C20 position opens the furan ring (Fig. 2(d)).42,43 Wortmannin initially binds to Plk1 reversibly and subsequently undergoes time-dependent covalent bond formation with the target Lys ε-amino group, resulting irreversible binding.44 The enzymatic inhibitory activity and binding affinity of Wortmannin for Plk1 KD are comparable to those of BI2536.36,39,45 The crystal structure of Wortmannin bound to the isolated Plk1 KD shows that an acetyl group is exposed to solvent, indicating that this portion of Wortmannin could potentially be modified without undue deleterious effects on binding interactions (Fig. 2(d)).We have previously observed that bivalent ligands of BI2536 and 2 conjugated from the N- and C-terminus of 2 (3–6, Fig. 1) exhibit similar Plk1 PBD binding affinities for both full length protein and isolated PBD domain, as well as KD binding affinity for full-length PlK1 and kinase inhibitory activity of full-length Plk1.34 Accordingly, in our current work we examined both C- and N-terminal linkages for our bivalent constructs. We designed and synthesized bivalent ligands 7 and 8 that possess Wortmannin as a KD-directed warhead. Peptide 2 was tethered to Wortmannin by means of a linker composed of succinic acid, a Lys residue, and four mini-PEG units (Scheme 1). Our synthesis began with chain elongation on NovaSyn® TGR resin by general Fmoc-based solid-phase peptide synthesis (Fmoc-SPPS). Coupling of the non-standard amino acids Fmoc-His*-OH,25 Fmoc-Lys(ivDde)-OH, and Fmoc-Thr{PO(OBn)OH}-OH were conducted using 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate (HATU) as a coupling reagent in the presence of N,N-diisopropylethylamine (DIPEA). The ivDde moiety on the Lys ε-amino group was selectively deprotected on-resin by treatment with 2% hydrazine. The resulting free amine was then coupled with succinylated Wortmannin to afford the bivalent constructs 7 and 8.
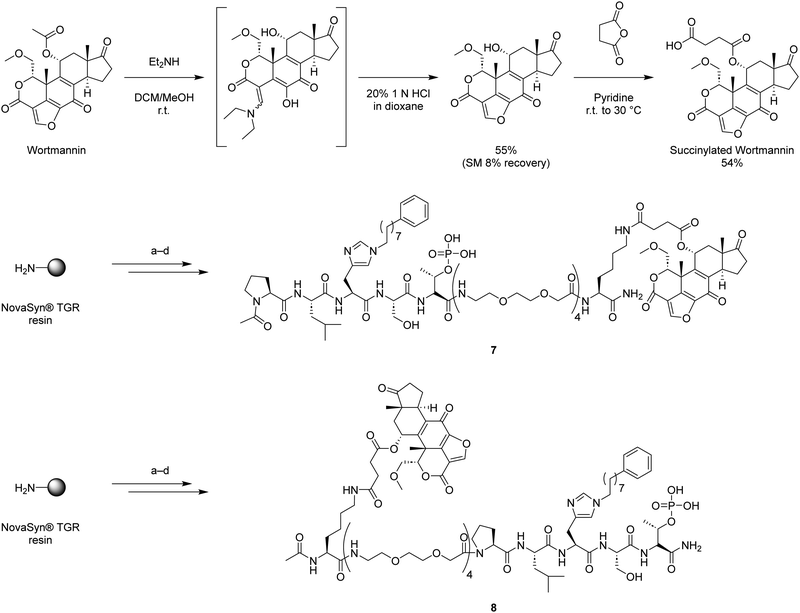 | ||
Scheme 1 Synthesis of Wortmannin-containing bivalent ligands 7 and 8. (a) Fmoc-based solid phase peptide synthesis using Fmoc-His*-OH,25 Fmoc-Lys(ivDde)-OH, and Fmoc-Thr{PO(OBn)OH}-OH with 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate (HATU) and N,N-diisopropylethylamine (DIPEA); (b) 2% H2NNH2 in NMP; (c) succinylated Wortmannin, HATU, DIPEA, NMP; (d) TFA/TIPS/H2O = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5. 2.5. | ||
Biological evaluation of bivalent ligands possessing Wortmannin as a KD-directed warhead
We evaluated the PBD-binding affinities of the synthetic bivalent ligands 7 and 8 using our recently reported fluorescence polarization (FP) assays with full length Plk1 in the presence or absence of excess Wortmannin (Table 1).34 We found that Wortmannin-containing bivalent ligands 7 and 8 exhibited greater affinity enhancement (IC50 = 4.6 nM (Exp. 1)/18 nM (Exp. 2), and IC50 = 4.5 nM (Exp. 1)/20 nM (Exp. 2), respectively) as compared to the parent peptide 2 (IC50 = 180 nM (Exp. 1)/1200 nM (Exp. 2) and 390 nM (Exp. 1)/1900 nM (Exp. 2) in the presence of 10 μM Wortmannin). These affinities are comparable or slightly lower than that of the bivalent ligands 3 and 5 conjugated to BI2536 (IC50 = 3.5 nM (Exp. 1)/2.0 nM (Exp. 2) and 4.9 nM (Exp. 1)/2.4 nM (Exp. 2), respectively).| Ligands | PBD-binding Affinity (nM)a (full-length Plk1) | |
|---|---|---|
| Exp. 1b | Exp. 2 | |
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Exp.: experiment. c N.a.: not applicable. | ||
| 2 | 180 ± 15 | 1200 ± 84 |
| Wortmannin | No inhibition | N.a. |
| 2 + 10 μM of Wortmannin | 390 ± 11 | 1900 ± 140 |
| BI2536 | No inhibition | N.a. |
| 3 | 3.5 ± 0.18 | 2.0 ± 0.040 |
| 5 | 4.9 ± 0.72 | 2.4 ± 0.072 |
| 3 + 100 μM of Wortmannin | N.a.c | 3.5 ± 0.55 |
| 5 + 100 μM of Wortmannin | N.a. | 26 ± 17 |
| 7 | 4.6 ± 0.27 | 18 ± 1.4 |
| 8 | 4.5 ± 0.24 | 20 ± 0.68 |
| 7 + 100 μM of Wortmannin | N.a. | 140 ± 16 |
| 8 + 100 μM of Wortmannin | N.a. | 82 ± 4.7 |
Next, we conducted FP binding assays using Plk1 pre-treated with Wortmannin to confirm the bivalent binding of 7 and 8 as well as 3 and 5 (Table 2). Since Wortmannin can form a covalent bond with the Plk1 KD, we removed free Wortmannin by dialysis using a 10 kDa filter. This permitted us to examine the effect of Wortmannin bound to the catalytic cleft (Table 2). However, as previously reported, irreversible binding of Wortmannin to Plk1 occurs in a time-dependent manner.44 Therefore, over the time course of dialysis, covalent binding of Wortmannin may be incomplete and free Wortmannin released from the KD could be removed. The fact that dialysis did not completely reverse the effect of Wortmannin is consistent with partial irreversible binding to Plk1 KD resulting incomplete occupation of the Plk1 KD.
| PBD-binding affinity (nM)a (full-length Plk1) | ||||
|---|---|---|---|---|
| Exp. 1b | Exp. 2 | |||
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Exp.: experiment. c Plk1 was treated with Wortmannin (10 eq.) at 4 °C for 2 h, and then the protein solution was dialyzed with assay buffer for 5 times using 10 kDa filter. d Plk1 solution was dialyzed with assay buffer for 5 times using 10 kDa filter. e N.a.: not applicable. | ||||
| Ligands | Wortmannin (+)c | Wortmannin (−)d | Wortmannin (+) | Wortmannin (−) |
| 2 | 1000 ± 210 | 340 ± 54 | 1000 ± 6.6 | 830 ± 380 |
| 3 | 2.8 ± 0.48 | 0.36 ± 0.26 | 3.8 ± 0.38 | 1.7 ± 0.083 |
| 5 | N.a.e | N.a. | 4.6 ± 1.1 | 2.3 ± 0.30 |
| 7 | 40 ± 16 | 5.1 ± 1.7 | 83 ± 7.7 | 30 ± 4.7 |
| 8 | N.a. | N.a. | 82 ± 10 | 44 ± 7.3 |
As anticipated, Wortmannin treatment attenuated ligand affinities for full length Plk1. Peptide 2 showed attenuation of its PBD-binding affinity by treatment with Wortmannin (IC50 values are 1000 nM (Exp. 1)/1000 nM (Exp. 2) for Wortmannin (+) and 340 nM (Exp. 1)/830 nM (Exp. 2) for Wortmannin (−)). This suggests that binding of Wortmannin to the KD attenuates autoinhibition (intramolecular association of the KD and PBD),35 and therefore, the probe (FITC-labeled 2) is allowed to more easily access to the PBD-binding cleft. The higher affinity of the FP probe results in decreased IC50 values for peptide 2 following Wortmannin treatment. This is consistent with previous reports suggesting that ATP competitive compounds increase the affinity of the PBD for PBD-binding phosphopeptides.46,47 The bivalent compounds 3, 5, 7, and 8 showed attenuated PBD-binding affinities by treatment with Wortmannin (IC50 values for Wortmannin (+) are as follows; 3: 2.8 nM (Exp. 1)/3.8 nM (Exp. 2), 5: 4.6 nM (Exp. 2), 7: 40 nM (Exp. 1)/83 nM (Exp. 2), and 8: 82 nM (Exp. 2), and IC50 values for Wortmannin (−) are as follows; 3: 0.36 nM (Exp. 1)/1.7 nM (Exp. 2), 5: 2.3 nM (Exp. 2), 7: 5.1 nM (Exp. 1)/30 nM (Exp. 2), and 8: 44 nM (Exp. 2)). This could potentially be caused by suppression of bivalent binding. These results suggest that binding of the KD components (Wortmannin or BI2536) enhance the PBD binding of the peptide 2 moiety in the bivalent constructs.
Because Wortmannin constructs 7 and 8 exhibited similar affinities, we arbitrarily decided to proceed using the bivalent ligand 7. We employed our in-house fluorescence recovery assays to evaluate the KD binding affinities of the bivalent ligand 7 (Fig. 3).45 We observed that the binding curve of the bivalent compound 7 showed a steep Hill slope, which was consistent with stoichiometric binding of 7 to the target protein. Signal enhancement in the presence of 1 μM of peptide 2 may reflect peptide 2 binding to PBD, which could cancel autoinhibition (association of intramolecular KD and PBD).35 We then conducted Plk1 KD binding assays of BI2536 and Wortmannin to confirm the time-dependent KD binding of Wortmannin using in-house fluorescence recovery assays (Table 3). We observed that Wortmannin inhibited probe binding in a time-dependent fashion. We found that 4 h incubation at both room temperature and at 4 °C resulted in effective binding to Plk1. We conducted dialysis experiments to wash out the non-covalent Plk1 binders (Fig. 4). We found by fluorescence recovery assays, that Wortmannin bound to Plk1 more tightly than BI2536 (Fig. 4(a)). However, bivalent ligand with BI2536 3 showed extremely tight binding to Plk1 that was comparable to the Wortmannin-containing bivalent ligand 7 even after dialysis using 50 kDa filter (Fig. 4(b), results of the same experiments of Fig. 4(b) using 10 kDa filter is also shown in Fig. S1, ESI†). These findings are consistent with the results of FP-based PBD binding assays in Tables 1 and 2.
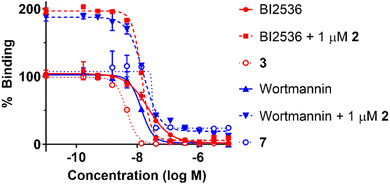 | ||
| Fig. 3 Results from fluorescence recovery assays to evaluate Plk1 KD affinity using FITC-miniPEG-BI2536 as a probe.45 The IC50 values of each compound are BI2536 (23 ± 1.4 nM), BI2536 + 1 μM 2 (14 ± 0.27 nM), 3 (4.5 ± 0.13 nM), Wortmannin (13 ± 0.48 nM), Wortmannin + 1 μM 2 (16 ± 0.30 nM), and 7 (32 ± 3.4 nM). Numbers are from three independent experiments and fit using non-linear regression in GraphPad Prism 10. | ||
| Ligands | KD-binding affinity (nM)a (full-length Plk1) | ||
|---|---|---|---|
| Incubation time | |||
| 0 h | 1 h | 4 h | |
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments; Plk1 was incubated with various concentrations of BI2536 or Wortmannin at r.t. or 4 °C for 0, 1, or 4 h, and then added the probe solution. | |||
| BI2536 (r.t.) | 45 ± 4.5 | 41 ± 2.4 | 38 ± 2.5 |
| BI2536 (4 °C) | 45 ± 1.2 | 22 ± 0.17 | 21 ± 0.46 |
| Wortmannin (r.t.) | 130 ± 4.9 | 51 ± 1.3 | 29 ± 1.5 |
| Wortmannin (4 °C) | 84 ± 2.0 | 27 ± 1.4 | 9.5 ± 0.49 |
Finally, we performed cell growth assays using HeLa cells (Fig. S2, ESI†). As we previously reported,25,34 peptide 2 does not show cellular inhibitory activity below 400 μM. We did not observe Wortmannin cytotoxicity at the tested concentrations (0 to 20 μM). This was consistent with previous reports.48,49 In spite of the fact that the bivalent ligands 7 and 8 showed single-digit nanomolar affinity to Plk1, they did not inhibit HeLa cell proliferation within the tested concentration range (0 to 200 μM). However, at the highest concentration of 7 and 8 (200 μM), the compounds did exhibit slight inhibition of HeLa cell proliferation. These results may reflect low cell membrane penetration of the bivalent ligands. They could also result from Plk1 inhibition by the bivalent ligands possessing Plk1 PBD inhibitor 2 moiety.
Conclusions
We have previously reported several high-affinity monomeric PBD-binding ligands based on the pentapeptide 2. These ligands demonstrate remarkable selectivity for the PBD of Plk1 versus the PBDs of Plk2 and Plk3 PBDs. However, affinities of these variants for Plk1 represent modest improvements relative to the parent peptide 2. In further studies we have found that the bivalent compounds 3–6 exhibit two-orders of magnitude affinity enhancement relative to 2 and that they display good selectivity when examined against a panel of kinases. They also display modest cytotoxicity against HeLa cells (Fig. 1). Based on these findings, we undertook the current work to extend our bivalent approach to ligands that incorporate as a KD-binding warhead, the non-selective covalent kinase inhibitor, Wortmannin. We found that tethering Wortmannin to 2 from either the C- or N-terminus provided the bivalent ligands 7 and 8, which showed binding affinity enhancements for the Plk1 PBD that were similar to the bivalent ligands 3 and 5, which have BI2536 as a KD-binder (Table 1). We observed using fluorescence recovery assays that the binding curve of compound 7 showed a steep Hill slope (Fig. 3). This is consistent with stoichiometric binding to the target protein, however further evaluation will be necessary to confirm the binding mode. Dialysis experiments indicated that Wortmannin bound to Plk1 more tightly than BI2536 (Fig. 4(a)) Interestingly, we also found that bivalent ligand 3 having BI2536 as the KD-binding component, showed extremely tight Plk1 binding to Plk1 compared to Wortmannin-containing 7 following dialysis with a 50 kDa filter (Fig. 4(b) and Fig. S1, ESI†). Despite their high target affinities, bivalent ligands 7 and 8, showed only modest cytotoxicity at 200 μM concentration (Fig. S2, ESI†). These results indicate that regardless of extremely high Plk1-binding affinities, the cellular efficacies of peptide-based Plk1 PBD inhibitors can be significantly attenuated, possibly due to their low cell membrane permeability. We have previously shown that attaching a CPP to a PBD-binding monomeric peptide can partially overcome this issue.50 In closing, our current work reports bivalent constructs employing a non-specific covalent kinase inhibitor, Wortmannin. We found that these exhibit extremely high Plk1 PBD-binding affinities that are similar to affinities of bivalent ligands that employ the Plk1 selective inhibitor, BI2536. Dialysis experiments support tight binding of bivalent constructs to Plk1. Although MD simulation models suggest the possibility of stable conformations for intramolecular binding of the bivalent construct to a single Plk1 molecule (Fig. 2(b) and (c)),34 it is possible that the bivalent constructs could engage two Plk1 proteins in an intermolecular fashion, with one end of the bivalent ligand binding to the KD-domain of one protein and the other through the PBD-binding of a second protein. Future studies will be required to resolve these possible modes of binding. In any event, the current bivalent constructs represent useful chemical biology probes that may shed light on how Plk1 is regulated and also into the conformational transitions it undergoes during activation. They may also be useful in stabilizing specific conformations of Plk1 and in so doing, facilitate its structural determination. Our findings provide new insights that should facilitate further development of Plk1 inhibitors.Author contributions
KT: conceptualization, funding acquisition, investigation, project administration, writing – original draft; HT: funding acquisition, supervision, writing – review & editing; TB: conceptualization, funding acquisition, project administration, supervision, writing – review & editing.Conflicts of interest
Aspects of the work presented are contained within one or more patent applications.Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (Z01-BC 006198, TB), by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at the NIH (KT), by JSPS KAKENHI grant numbers 22K15243 (KT), and by AMED under grant number JP23ama121043 (Platform Project for Supporting Drug Discovery and Life Science Research, BINDS) (HT). This research is based on a collaboration between the NIH and the Research Center for Biomedical Engineering. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. Appreciation is expressed to Dr. David Hymel for helpful discussion.References
- C. Chittasupho, Ther. Delivery, 2012, 3, 1171–1187 CrossRef CAS PubMed.
- B. J. Mayer, Methods Mol. Biol., 2006, 332, 79–99 CAS.
- T. Pawson and P. Nash, in Handbook of Cell Signaling, ed. R. A. Bradshaw and E. A. Dennis, Academic Press, San Diego, 2nd edn, 2010, pp. 399–411 DOI:10.1016/B978-0-12-374145-5.00057-7.
- T. Hunter, Cell, 2000, 100, 113–127 CrossRef CAS PubMed.
- C. M. Gower, M. E. K. Chang and D. J. Maly, Crit. Rev. Biochem. Mol. Biol., 2014, 49, 102–115 CrossRef CAS PubMed.
- D. Lavogina, E. Enkvist and A. Uri, ChemMedChem, 2010, 5, 23–34 CrossRef CAS PubMed.
- K. J. Cox, C. D. Shomin and I. Ghosh, Future Med. Chem., 2011, 3, 29–43 CrossRef CAS PubMed.
- S. Lee, J. Kim, J. Jo, J. W. Chang, J. Sim and H. Yun, Eur. J. Med. Chem., 2021, 216, 113318 CrossRef CAS PubMed.
- J. Kalous and D. Aleshkina, Cells, 2023, 12, 187 CrossRef CAS PubMed.
- K. Strebhardt and A. Ullrich, Nat. Rev. Cancer, 2006, 6, 321–330 CrossRef CAS.
- J.-M. Schvartzman, R. Sotillo and R. Benezra, Nat. Rev. Cancer, 2010, 10, 102–115 CrossRef CAS PubMed.
- Z. Liu, Q. Sun and X. Wang, Transl. Oncol, 2017, 10, 22–32 CrossRef.
- W. Weichert, M. Schmidt, J. Jacob, V. Gekeler, J. Langrehr, P. Neuhaus, M. Bahra, C. Denkert, M. Dietel and G. Kristiansen, Pancreatology, 2005, 5, 259–265 CrossRef CAS PubMed.
- Q. N. Hassan II, L. Alinari and J. C. Byrd, J. Clin. Invest., 2018, 128, 5206–5208 CrossRef.
- S. B. Jeong, J. H. Im, J.-H. Yoon, Q. T. Bui, S. C. Lim, J. M. Song, Y. Shim, J. Yun, J. Hong and K. W. Kang, Mol. Cancer Ther., 2018, 17, 825–837 CrossRef CAS.
- K. S. Lee, T. R. Burke, Jr., J.-E. Park, J. K. Bang and E. Lee, Trends Pharmacol. Sci., 2015, 36, 858–877 CrossRef CAS.
- R. E. A. Gutteridge, M. A. Ndiaye, X. Liu and N. Ahmad, Mol. Cancer Ther., 2016, 15, 1427–1435 CrossRef CAS.
- J. E. Park, D. Hymel, T. R. Burke, Jr. and K. S. Lee, F1000 Res., 2017, 6, 1024, DOI:1010.12688/f11000research.11398.12681.
- V. Archambault, G. Lépine and D. Kachaner, Oncogene, 2015, 34, 4799–4807 CrossRef CAS PubMed.
- G. Manning, D. B. Whyte, R. Martinez, T. Hunter and S. Sudarsanam, Science, 2002, 298, 1912–1934 CrossRef CAS PubMed.
- S. Zitouni, C. Nabais, S. C. Jana, A. Guerrero and M. Bettencourt-Dias, Nat. Rev. Mol. Cell Biol., 2014, 15, 433–452 CrossRef CAS PubMed.
- A. Berg and T. Berg, ChemBioChem, 2016, 17, 650–656 CrossRef CAS PubMed.
- J. M. Stafford, M. D. Wyatt and C. McInnes, Expert Opin. Drug Delivery, 2023, 18, 65–81 CrossRef CAS.
- S.-M. Yun, T. Moulaei, D. Lim, J. K. Bang, J.-E. Park, S. R. Shenoy, F. Liu, Y. H. Kang, C. Liao, N.-K. Soung, S. Lee, D.-Y. Yoon, Y. Lim, D.-H. Lee, A. Otaka, E. Appella, J. B. McMahon, M. C. Nicklaus, T. R. Burke, Jr., M. B. Yaffe, A. Wlodawer and K. S. Lee, Nat. Struct. Mol. Biol., 2009, 16, 876–882 CrossRef CAS.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, M. Graber, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., Nat. Chem. Biol., 2011, 7, 595–601 CrossRef CAS PubMed.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, A. Scharow, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., ACS Chem. Biol., 2012, 7, 805–810 CrossRef CAS PubMed.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, A. Scharow, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., ChemBioChem, 2012, 13, 1291–1296 CrossRef CAS PubMed.
- X. Z. Zhao, D. Hymel and T. R. Burke, Jr., Bioorg. Med. Chem. Lett., 2016, 26, 5009–5012 CrossRef CAS.
- X. Z. Zhao, D. Hymel and T. R. Burke, Jr., Bioorg. Med. Chem., 2017, 25, 5041–5049 CrossRef CAS.
- X. Z. Zhao, K. Tsuji, D. Hymel and T. R. Burke, Jr., Molecules, 2019, 24, 1488 CrossRef CAS PubMed.
- W.-J. Qian, J.-E. Park, K. S. Lee and T. R. Burke, Jr., Bioorg. Med. Chem. Lett., 2012, 22, 7306–7308 CrossRef CAS PubMed.
- D. Hymel, R. A. Grant, K. Tsuji, M. B. Yaffe and T. R. Burke, Jr., Bioorg. Med. Chem. Lett., 2018, 28, 3202–3205 CrossRef CAS.
- D. Hymel, K. Tsuji, R. A. Grant, R. M. Chingle, D. L. Kunciw, M. B. Yaffe and T. R. Burke, Jr., Org. Biomol. Chem., 2021, 19, 7843–7854 RSC.
- K. Tsuji, D. Hymel, B. Ma, H. Tamamura, R. Nussinov and T. R. Burke, Jr., RSC Chem. Biol., 2022, 3, 1111–1120 RSC.
- J. Xu, C. Shen, T. Wang and J. Quan, Nat. Struct. Mol. Biol., 2013, 20, 1047–1053 CrossRef CAS PubMed.
- M. Steegmaier, M. Hoffmann, A. Baum, P. Lenart, M. Petronczki, M. Krssak, U. Guertler, P. Garin-Chesa, S. Lieb, J. Quant, M. Grauert, G. R. Adolf, N. Kraut, J.-M. Peters and W. J. Rettig, Curr. Biol., 2007, 17, 316–322 CrossRef CAS PubMed.
- H. Ruan, J. Kiselar, W. Zhang, S. Li, R. Xiong, Y. Liu, S. Yang and L. Lai, Phys. Chem. Chem. Phys., 2020, 22, 27581–27589 RSC.
- T. Okada, L. Sakuma, Y. Fukui, O. Hazeki and M. Ui, J. Biol. Chem., 1994, 269, 3563–3567 CrossRef CAS PubMed.
- Y. Liu, K. R. Shreder, W. Gai, S. Corral, D. K. Ferris and J. S. Rosenblum, Chem. Biol., 2005, 12, 99–107 CrossRef CAS PubMed.
- Y. Liu, N. Jiang, J. Wu, W. Dai and J. S. Rosenblum, J. Biol. Chem., 2007, 282, 2505–2511 CrossRef CAS PubMed.
- R. A. Elling, R. V. Fucini and M. J. Romanowski, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 909–918 CrossRef CAS PubMed.
- H. Yuan, K. R. Barnes, R. Weissleder, L. Cantley and L. Josephson, Chem. Biol., 2007, 14, 321–328 CrossRef CAS PubMed.
- M. H. Johansson, Mini-Rev. Med. Chem., 2012, 12, 1330–1344 CAS.
- E. F. Johnson, K. D. Stewart, K. W. Woods, V. L. Giranda and Y. Luo, Biochemistry, 2007, 46, 9551–9563 CrossRef CAS PubMed.
- K. Tsuji, D. Hymel and T. R. Burke, Jr., Anal. Methods, 2020, 12, 4418–4421 RSC.
- M. Raab, M. Sanhaji, L. Pietsch, I. Béquignon, A. K. Herbrand, E. Süβ, S. L. Gande, B. Caspar, D. Kudlinzki, K. Saxena, S. Sreeramulu, H. Schwalbe, K. Strebhardt and R. M. Biondi, ACS Chem. Biol., 2018, 13, 1921–1931 CrossRef CAS PubMed.
- D. Chapagai, G. Merhej, C. McInnes and M. D. Wyatt, ACS Chem. Biol., 2023, 18, 1642–1652 CrossRef CAS.
- H. Hou, Y. Zhang, Y. Huang, Q. Yi, L. Lv, T. Zhang, D. Chen, Q. Hao and Q. Shi, PLoS One, 2012, 7, e35665 CrossRef CAS PubMed.
- M. Hayashi, A. Kawana, D. Endoh and T. Okui, J. Vet. Med. Sci., 2000, 62, 191–194 CrossRef CAS PubMed.
- S. E. Miller, K. Tsuji, R. P. M. Abrams, T. R. Burke, Jr. and J. P. Schneider, J. Am. Chem. Soc., 2020, 142, 19950–19955 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00031e |
| This journal is © The Royal Society of Chemistry 2024 |

