Supramolecular pyrrole radical cations for bacterial theranostics†
Yue
Han‡
a,
Jing
Li‡
b,
Lihua
Zheng‡
a,
Yunhua
Chen
 a,
Yan
Yang
*c,
Kai
Liu
*d,
Yiyue
Zhang
a,
Yan
Yang
*c,
Kai
Liu
*d,
Yiyue
Zhang
 *b and
Meng
Gao
*b and
Meng
Gao
 *a
*a
aNational Engineering Research Center for Tissue Restoration and Reconstruction, Key Laboratory of Biomedical Engineering of Guangdong Province, Key Laboratory of Biomedical Materials and Engineering of the Ministry of Education, Innovation Center for Tissue Restoration and Reconstruction, School of Materials Science and Engineering, South China University of Technology, Guangzhou 510006, China. E-mail: msmgao@scut.edu.cn
bSchool of Medicine, South China University of Technology, Guangzhou 510006, China. E-mail: mczhangyy@scut.edu.cn
cFoshan University, Foshan 528225, China. E-mail: 87852182yan@163.com
dInstitute of Marine Drugs, Guangxi University of Chinese Medicine, Nanning 530200, China. E-mail: kailiu@gxtcmu.edu.cn
First published on 20th November 2023
Abstract
Bacterial infections with emerging resistance to antibiotics require urgent development of antibacterial agents with new core skeletons. Recently, a series of antibacterial agents have been reported based on positively charged organic groups, such as ammonium, guanidine, and phosphonium groups, which can selectively bind and destroy negatively charged bacterial membranes. To achieve imaging-guided precise antibacterial therapy, these positively charged organic groups usually require further decoration with imaging modalities, such as fluorescence. However, most fluorophores with electron-closed shell structures usually suffer from tedious synthetic procedures for preparation. We herein prepare a series of positively charged and deep-red fluorescent supramolecular pyrrole radical cations (P˙+–CB[7]) based on the simple mixing of pyrroles and CB[7] in water under air. The readily available deep-red fluorescent P˙+–CB[7] can not only be used for selective imaging and killing of live Gram-positive bacteria with excellent biocompatibility, but also for imaging of dead Gram-negative bacteria killed by drugs and in vivo monitoring of phagocytosis of bacteria by innate immune cells in zebrafish. It is believed that the deep-red fluorescent pyrrole radical cations as a new core skeleton are promising in bacterial theranostics.
Introduction
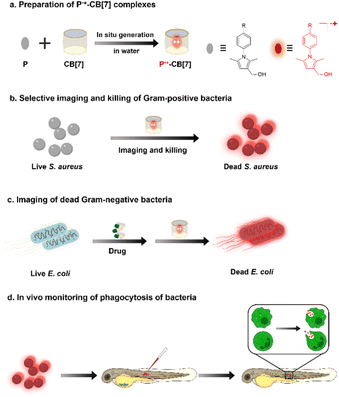
Bacterial infections are becoming serious health threats due to emerging resistance to antibiotics.1 It is thus highly desirable to develop antibacterial agents with new core skeletons.2,3 Recently, a series of antibacterial agents have been developed based on positively charged organic groups (e.g., ammonium, aza-aromatic, guanidine, sulfonium and phosphonium), which can selectively recognize and destroy negatively charged bacterial membranes through electrostatic interactions.4–10 However, to achieve imaging-guided precise killing of bacteria, positively charged groups usually require further decoration with imaging modalities, such as positron emission tomography (PET),11 magnetic resonance imaging (MRI),12 photoacoustic13 and fluorescence.14–16 Among these imaging modalities, red-to-near-infrared (NIR) fluorescence imaging has significant advantages in terms of low autofluorescence interference and high spatial resolution.17 However, conventional organic red-to-NIR fluorophores with electron-closed shell structures, such as D–π–A conjugated cyanine and squaraine, suffer from tedious synthetic procedures for preparation.18–20 Compared with fluorophores with electron-closed shell structures, radical cations usually feature red-to-NIR absorption and have been used for photothermal treatment of bacterial infections.21–25However, as far as we know, red-to-NIR fluorescent radical cations have not been reported for bacterial theranostics. Recently, we found that weak acids (pH ∼ 2–3) can induce the generation of pyrrole radical cations (P˙+), which are stable in a water solution containing supramolecular host CB[7].26–28 Inspired by the fact that CB[7] also features an acidic cavity and its pKa can downshift up to 4.5 units,29 herein, we systematically investigate the substituent effect of pyrroles for the direct preparation of the corresponding radical cations in a water solution by simple mixing of pyrroles and CB[7]. Furthermore, based on the positively charged structure of pyrrole radical cations with deep-red fluorescence emission, we investigate their promising applications for bacterial theranostics, including selective imaging and killing of Gram-positive bacteria, imaging of dead Gram-negative bacteria killed by drugs, and in vivo monitoring of phagocytosis of bacteria by innate immune cells in zebrafish (Scheme 1).
Results and discussion
Preparation and characterization of fluorescent P˙+–CB[7]
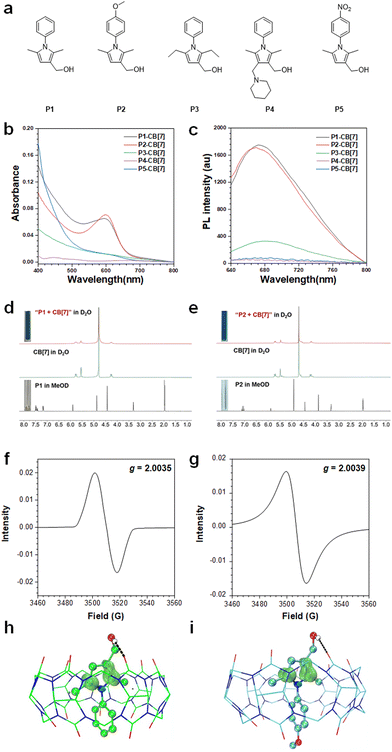
Pyrrole compounds (P1–P5) are prepared according to our previously reported methods (Scheme S1 and Fig. S1–S4, ESI†).26,27 As shown in Fig. 1a–c, after mixing pyrrole compounds with CB[7] at an optimized molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in water under air (Fig. S5, ESI†), pyrroles P1 and P2 decorated with a 2,5-dimethyl-3-hydroxylmethyl group and an N-phenyl or N-(4-methoxyphenyl) group can readily afford the corresponding radical cations P1˙+ and P2˙+ with λabs ∼ 600 nm and λem ∼ 670 nm (Table S1 and Fig. S6, ESI†). The dynamic color changing process from colorless to red and then to blue was also in situ monitored (Video S1, ESI†), which is similar to the acid-induced generation of the corresponding radical cations at pH = 2,26 indicating that the acidic cavity of CB[7] can efficiently induce the generation of pyrrole radical cations in the water solution (Fig. S7, ESI†). The generation of pyrrole radical cations P1˙+ and P2˙+ is verified by the disappearance of the hydrogen signals of pyrroles in the 1H NMR spectra after mixing P1 or P2 with CB[7] in D2O (Fig. 1d and e). After lyophilization of the water solution of P1˙+–CB[7] and P2˙+–CB[7], a g value of ∼2.004 was observed in their electron paramagnetic resonance (EPR) spectra (Fig. 1f and g). The theoretical calculations of the supramolecular complexes of P1˙+–CB[7] and P2˙+–CB[7] show that the electron spin is mainly located at the C-2 and C-5 positions of the pyrrole ring, which indicates that the 2,5-dimethyl group is critical for stabilizing the pyrrole radical cations by inhibiting radical coupling reactions (Fig. 1h, i and Table S2, ESI†). Compared with compounds P1 and P2, compound P3 decorated with a 2,5-diethyl group shows a much lower efficiency in generating the corresponding radical cation P3˙+, which may be because the 2,5-diethyl group with higher steric hindrance restricts the encapsulation of compound P3 by CB[7]. For compounds P4 and P5 decorated with a 4-(piperidin-1-ylmethyl) group and an N-PhNO2 group, the corresponding radical cations were not observed, which can be ascribed to their steric hindrance effect and electron-withdrawing properties, respectively.
1 in water under air (Fig. S5, ESI†), pyrroles P1 and P2 decorated with a 2,5-dimethyl-3-hydroxylmethyl group and an N-phenyl or N-(4-methoxyphenyl) group can readily afford the corresponding radical cations P1˙+ and P2˙+ with λabs ∼ 600 nm and λem ∼ 670 nm (Table S1 and Fig. S6, ESI†). The dynamic color changing process from colorless to red and then to blue was also in situ monitored (Video S1, ESI†), which is similar to the acid-induced generation of the corresponding radical cations at pH = 2,26 indicating that the acidic cavity of CB[7] can efficiently induce the generation of pyrrole radical cations in the water solution (Fig. S7, ESI†). The generation of pyrrole radical cations P1˙+ and P2˙+ is verified by the disappearance of the hydrogen signals of pyrroles in the 1H NMR spectra after mixing P1 or P2 with CB[7] in D2O (Fig. 1d and e). After lyophilization of the water solution of P1˙+–CB[7] and P2˙+–CB[7], a g value of ∼2.004 was observed in their electron paramagnetic resonance (EPR) spectra (Fig. 1f and g). The theoretical calculations of the supramolecular complexes of P1˙+–CB[7] and P2˙+–CB[7] show that the electron spin is mainly located at the C-2 and C-5 positions of the pyrrole ring, which indicates that the 2,5-dimethyl group is critical for stabilizing the pyrrole radical cations by inhibiting radical coupling reactions (Fig. 1h, i and Table S2, ESI†). Compared with compounds P1 and P2, compound P3 decorated with a 2,5-diethyl group shows a much lower efficiency in generating the corresponding radical cation P3˙+, which may be because the 2,5-diethyl group with higher steric hindrance restricts the encapsulation of compound P3 by CB[7]. For compounds P4 and P5 decorated with a 4-(piperidin-1-ylmethyl) group and an N-PhNO2 group, the corresponding radical cations were not observed, which can be ascribed to their steric hindrance effect and electron-withdrawing properties, respectively.
Selective imaging and killing of Gram-positive bacteria
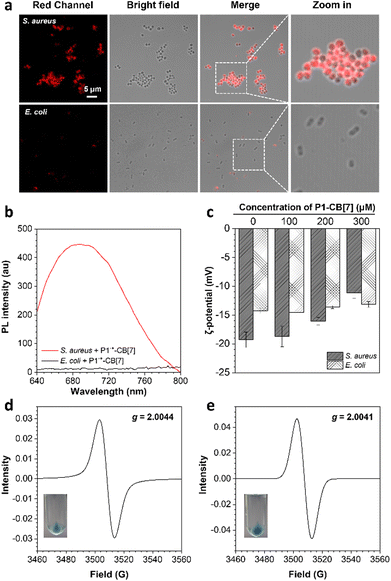
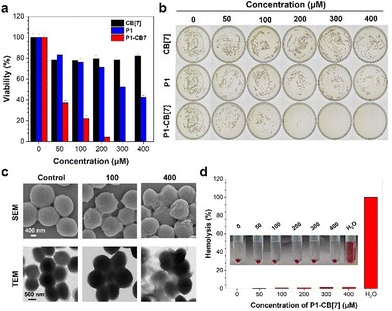
We then investigated the bacterial imaging and killing abilities of P˙+–CB[7]. Confocal laser scanning microscopy (CLSM) images show that Gram-positive S. aureus can be stained with P1˙+–CB[7] with a strong deep-red fluorescence signal, whereas almost no fluorescence signal was observed for live Gram-negative E. coli (Fig. 2a and Fig. S8, ESI†). The selective staining of live Gram-positive S. aureus by P1˙+–CB[7] was further verified by PL spectrum measurement, and a maximum emission wavelength at 685 nm was observed (Fig. 2b). Moreover, a gradually increasing zeta potential (ζ) was observed for Gram-positive S. aureus from −19.30 to −11.17 mV with an increased concentration of P1–CB[7] from 0 to 300 μM, whereas the zeta potential (ζ) almost did not change for Gram-negative E. coli (∼−13.40 mV) (Fig. 2c). A similar phenomenon was observed for P2˙+–CB[7], which can also selectively stain live Gram-positive bacteria with deep-red emission (Fig. S9, ESI†). The selective staining of live Gram-positive S. aureus by P1˙+–CB[7] and P2˙+–CB[7] was further verified by EPR spectrum measurement with g ∼ 2.004 (Fig. 2d and e).Based on the selective staining of live Gram-positive bacteria by P˙+–CB[7], we further investigated their bacterial killing abilities against Gram-positive bacteria with S. aureus as an example. The bacterial viability of Gram-positive S. aureus decreased to almost zero with increasing concentrations of P1–CB[7] or P2–CB[7] from 0 to 300 μM (Fig. 3a, b and Fig. S10, ESI†). To gain insight into the bacterial killing mechanism of P˙+–CB[7], the bacterial morphology changes were observed by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). After treatment with P1˙+–CB[7], a rough bacterial membrane was observed in SEM images (Fig. 3c), whereas bacterial lysis was not observed in TEM images. With DiSC3(5) dye as the membrane potential indicator, an obviously increased bacterial membrane depolarization was observed for Gram-positive S. aureus with increasing concentrations of P1˙+–CB[7] and P2˙+–CB[7], which suggests that the positively charged P1˙+–CB[7] can efficiently bind with the cytoplasmic membrane of S. aureus and induce membrane depolarization (Fig. S11, ESI†). This unique antibacterial mode of P1˙+–CB[7] without causing bacterial lysis of Gram-positive bacteria was further verified by co-staining with the nucleic acid dye SYTOX Green, which has poor penetration abilities toward intact bacterial membranes (Fig. S12, ESI†).30,31 Therefore, the bacterial killing ability of P˙+–CB[7] towards Gram-positive bacteria can be ascribed to their membrane depolarization ability.32–35 Moreover, both P1˙+–CB[7] and P2˙+–CB[7] can efficiently kill Gram-positive methicillin-resistant S. aureus (MRSA) (Fig. S13, ESI†), which suggests that they are also promising for the treatment of drug-resistant Gram-positive bacteria. We further investigated the biocompatibility of P1˙+–CB[7] and P2˙+–CB[7], which showed a low hemolytic activity towards rabbit red blood cells and low cytotoxicity for mammalian L929 cells, suggesting they have excellent biocompatibility (Fig. 3d and Fig. S14, S15, ESI†).
Imaging of dead Gram-negative bacteria killed by drugs
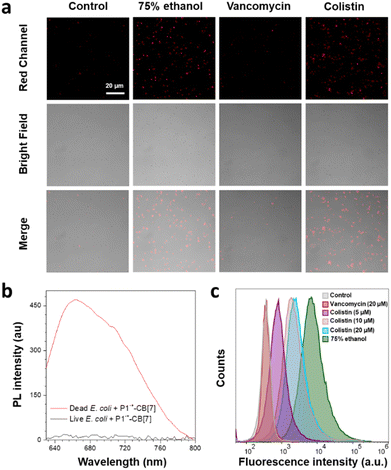
Although P1˙+–CB[7] cannot stain and kill live Gram-negative E. coli with a dense outer lipopolysaccharide layer (Fig. S16, ESI†),36,37 both CLSM images and PL spectrum measurements show that it can stain dead E. coli treated with 75% ethanol (Fig. 4a and b). Inspired by the turn-on staining of dead Gram-negative E. coli, we then investigated whether P1˙+–CB[7] can be used for the evaluation of the bacterial killing ability of antibiotics with vancomycin and colistin sulfate as examples. The CLSM images show that P1˙+–CB[7] only stains E. coli treated with colistin sulfate, which can efficiently kill Gram-negative bacteria. Meanwhile, no fluorescence signal was observed for E. coli treated with vancomycin, which can only kill Gram-positive bacteria. Moreover, an increasing concentration of colistin sulfate (0, 5, 10, and 20 μM) led to an increased fluorescence signal of P1˙+–CB[7], which was also verified by flow cytometry measurement (Fig. 4c). These results suggest that P1˙+–CB[7] is promising for the evaluation of the antibacterial efficacy of antibiotics against Gram-negative bacteria.In situ monitoring of phagocytosis of bacteria
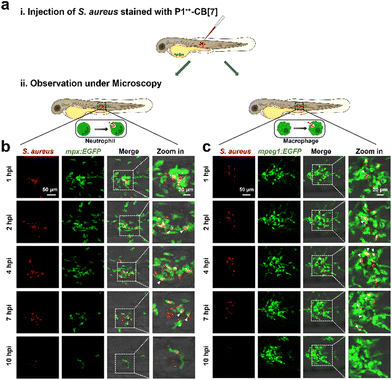
Considering that innate immune responses are critical for clearance of in vivo bacterial infections, we then investigated whether P1˙+–CB[7] can be used for in situ monitoring of phagocytosis of bacteria by innate immune cells, such as neutrophils and macrophages. Because a long-term traceability of bacteria over different infection stages is required for phagocytosis monitoring, we first investigated the long-term bacterial tracing ability of P1˙+–CB[7]. Both CLSM images and flow cytometry analysis show that P1˙+–CB[7] has an excellent long-term retention ability in S. aureus for at least 12 h without the loss of fluorescence signals (Fig. S17, ESI†), which suggests that it is promising for in situ monitoring of phagocytosis of bacteria. We then used P1˙+–CB[7] for in situ monitoring of phagocytosis of Gram-positive S. aureus by ex vivo macrophage 264.7 cells and the time-dependent 2D and 3D CLSM images show that S. aureus can be phagocytosed by macrophage cells within 1.5 h (Fig. S18, ESI†).38We then investigated the in vivo antibacterial immune response with zebrafish as a model because the vertebrate innate immune system is highly conserved.39–42 We subcutaneously injected P˙+–CB[7]-labeled S. aureus over one somite at 3 days post fertilization (dpf) into Tg(mpx:EGFP) and Tg(mpeg1:EGFP) larvae, in which EGFP was expressed specifically in neutrophils and macrophages, respectively. The phagocytosis of bacteria by neutrophils and macrophages was then in situ monitored by confocal microscopy (Fig. 5 and Videos S2, S3, ESI†). At 1 h postinjection (hpi), both neutrophils and macrophages migrated to the bacterial infection site from all directions to phagocytose bacteria, and most bacteria were cleared after 7 h. These results suggest that P˙+–CB[7] can be conveniently used for in vivo monitoring of phagocytosis of bacteria.
Experimental
Synthesis and characterization of the P˙+–CB[7] complex
The P˙+–CB[7] complex was prepared by dissolving pyrrole compounds and CB[7] at an optimized molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in a DMSO/H2O mixture (v
1 in a DMSO/H2O mixture (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v = 1
v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 99) under sonication (KQ-600KDE, 23 °C, 40 kHz, 600 W) for 10 min.
99) under sonication (KQ-600KDE, 23 °C, 40 kHz, 600 W) for 10 min.
CSLM imaging of bacteria
200 μL bacterial suspensions (OD600 = 1.0) in a 2 mL centrifuge tube were harvested by centrifugation (8000 rpm, 3 min), and then the supernatant was removed. 1 mL of PBS solution containing P˙+–CB[7] ([P1] = 100 μM, [CB[7]] = 33.3 μM) was added and the solution was incubated on a shaking incubator (220 rpm) at 37 °C for 2 h. After centrifugation for 3 min at 8000 rpm, the supernatant solution was discarded, and the bacteria were resuspended in 50 μL of PBS. 2 μL of the stained bacterial solution were transferred to a glass slide and then covered by a coverslip for CLSM imaging.Assessment of bacterial killing ability
250 μL bacterial suspensions (OD600 = 0.1) were treated with different concentrations of P˙+–CB[7] for 2 h on a shaking incubator (220 rpm) at 37 °C. After further gradient elution 1000 times, the bacterial suspensions (10 μL) were plated on agar plates and dispersed with an L-shaped cell spreader. Finally, these agar plates were incubated for 18 h at 37 °C. The antibacterial viability was assessed by counting the number of bacteria on agar plates and comparing them with the control.Imaging of dead Gram-negative bacteria killed by drugs
200 μL of E. coli (OD600 = 1.0) in a 2 mL centrifuge tube was harvested by centrifugation (8000 rpm, 3 min), and then the supernatant was removed. After washing with PBS twice, antibiotics (colistin sulfate and vancomycin) were added to PBS at different concentrations and the mixture was then incubated on a shaking incubator (220 rpm) for 3 h at 37 °C. After centrifugation for 3 min at 8000 rpm, the supernatant solution was discarded, 1 mL of P˙+–CB[7] ([P1] = 100 μM, [CB[7]] = 33.3 μM) in PBS was then added, and the solution was incubated for 2 h on a shaking incubator (220 rpm) at 37 °C. After centrifugation for 3 min at 8000 rpm, the supernatant solution was discarded, and the bacteria were resuspended in 100 μL PBS. The fluorescence signals of the bacteria were analyzed by CLSM and flow cytometry.In vivo monitoring of phagocytosis of bacteria
All studies involving zebrafish were performed in accordance with the Guiding Opinions on the Treatment of Experimental Animals and the Guangdong Province Laboratory Animal Management Regulation and were approved by the Animal Ethics Committee of the South China University of Technology. 200 μL of S. aureus (OD600 = 1.0) in a 2 mL centrifuge tube was harvested by centrifugation (8000 rpm, 3 min), and then the supernatant was removed. 1 mL of P˙+–CB[7] ([P1] = 100 μM, [CB[7]] = 33.3 μM) in PBS was added, and the solution was incubated at room temperature for 2 h on a shaking incubator (180 rpm) at 37 °C. After centrifugation for 3 min at 8000 rpm, the supernatant solution was discarded, and the bacteria were resuspended in 50 μL of PBS. Zebrafish embryos (3 dpf) were anesthetized with 0.02% tricaine and then injected subcutaneously over a somite with ∼0.5 nL of the above bacterial suspension using a Milli-Pulse Pressure Injector.Conclusions
In conclusion, deep-red fluorescent supramolecular complexes of P˙+–CB[7] can be readily prepared by simple mixing of pyrroles and CB[7] in water under air. The substituent effect of pyrrole compounds for the generation of P˙+ in the presence of CB[7] is systematically investigated. P˙+–CB[7] can not only discriminate between live Gram-positive and Gram-negative bacteria, but also efficiently kill Gram-positive bacteria with excellent biocompatibility. Moreover, P˙+–CB[7] can be used for imaging of dead Gram-negative bacteria killed by drugs and long-term in vivo monitoring of phagocytosis of bacteria by neutrophils and macrophages in zebrafish. We believe that the deep-red fluorescent supramolecular complexes of P˙+–CB[7] as a new core skeleton are promising for bacterial theranostics.Author contributions
Y. Yang, K. Liu, Y. Zhang and M. Gao conceived the original idea for investigation and wrote the manuscript. Y. Han and L. Zheng synthesized the compounds. Y. Han, J. Li, Y. Chen and L. Zheng measured the photophysical properties of the compounds and performed the bacterial imaging and therapy experiments. K. Liu performed the quantum chemical calculations. All authors discussed the progress of research and reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key R&D Program of China (2022YFC2410104), the National Natural Science Foundation of China (22175065) and the Natural Science Foundation of Guangdong Province (2020B1515020010 and 2020A1515110746). K. L. thanks the Special Fund for the Hundred Talents Program for Universities in Guangxi and the GuiPai Traditional Chinese Medicine Inheritance and Innovation Team Project of the Guangxi University of Chinese Medicine (2022A007).Notes and references
- M. O. A. Sommer, C. Munck, R. V. Toft-Kehler and D. I. Andersson, Prediction of antibiotic resistance: time for a new preclinical paradigm?, Nat. Rev. Microbiol., 2017, 15, 688–695 Search PubMed.
- L. D. Blackman, T. D. Sutherland, P. J. De Barro, H. Thissen and K. E. S. Locock, Addressing a future pandemic: how can non-biological complex drugs prepare us for antimicrobial resistance threats?, Mater. Horiz., 2022, 9, 2076–2096 RSC.
- E. D. Brown and G. D. Wright, Antibacterial drug discovery in the resistance era, Nature, 2016, 529, 336–343 CrossRef CAS.
- Y. Qian, S. Deng, Z. Cong, H. Zhang, Z. Lu, N. Shao, S. A. Bhatti, C. Zhou, J. Cheng, S. H. Gellman and R. Liu, Secondary Amine Pendant beta-Peptide Polymers Displaying Potent Antibacterial Activity and Promising Therapeutic Potential in Treating MRSA-Induced Wound Infections and Keratitis, J. Am. Chem. Soc., 2022, 144, 1690–1699 CrossRef CAS.
- H. Chen, S. Li, M. Wu, K. Z. Huang, C. S. Lee and B. Liu, Membrane-Anchoring Photosensitizer with Aggregation-Induced Emission Characteristics for Combating Multidrug-Resistant Bacteria, Angew. Chem., Int. Ed., 2020, 59, 632–636 CrossRef CAS PubMed.
- X. Y. Wang, Q. L. Cui, C. Yao, S. L. Li, P. B. Zhang, H. Sun, F. T. Lv, L. B. Liu, L. D. Li and S. Wang, Conjugated Polyelectrolyte-Silver Nanostructure Pair for Detection and Killing of Bacteria, Adv. Mater. Technol., 2017, 2, 1700033 CrossRef.
- I. Roy, D. Shetty, R. Hota, K. Back, J. Kim, C. Kim, S. Kappert and K. Kim, A Multifunctional Subphthalocyanine Nanosphere for Targeting, Labeling, and Killing of Antibiotic-Resistant Bacteria, Angew. Chem., Int. Ed., 2015, 54, 15152–15155 CrossRef CAS.
- Q. Kong, G. Li, F. Zhang, T. Yu, X. Chen, Q. Jiang and Y. Wang, N-Arylimidazoliums as Highly Selective Biomimetic Antimicrobial Agents, J. Med. Chem., 2022, 65, 11309–11321 CrossRef CAS PubMed.
- M. Xiong, M. W. Lee, R. A. Mansbach, Z. Song, Y. Bao, R. M. Peek, Jr., C. Yao, L.-F. Chen, A. L. Ferguson, G. C. L. Wong and J. Cheng, Helical antimicrobial polypeptides with radial amphiphilicity, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 13155–13160 CrossRef CAS.
- X. Chen, L. Huang, Y. Jia, R. Hu, M. Gao, L. Ren and B. Z. Tang, AIE-Based Theranostic Probe for Sequential Imaging and Killing of Bacteria and Cancer Cells, Adv. Opt. Mater., 2020, 8, 1902191 CrossRef CAS.
- K. Sachin, E.-M. Kim, S.-J. Cheong, H.-J. Jeong, S. T. Lim, M.-H. Sohn and D. W. Kim, Synthesis of N4′-[18F]Fluoroalkylated Ciprofloxacin as a Potential Bacterial Infection Imaging Agent for PET Study, Bioconjugate Chem., 2010, 21, 2282–2288 CrossRef CAS.
- I. Kim, S.-M. Jin, E. H. Han, E. Ko, M. Ahn, W.-Y. Bang, J.-K. Bang and E. Lee, Structure-Dependent Antimicrobial Theranostic Functions of Self-Assembled Short Peptide Nanoagents, Biomacromolecules, 2017, 18, 3600–3610 CrossRef CAS.
- L. Xie, X. Pang, X. Yan, Q. Dai, H. Lin, J. Ye, Y. Cheng, Q. Zhao, X. Ma, X. Zhang, G. Liu and X. Chen, Photoacoustic Imaging-Trackable Magnetic Microswimmers for Pathogenic Bacterial Infection Treatment, ACS Nano, 2020, 14, 2880–2893 CrossRef CAS.
- S. Cao, J. Shao, L. K. E. A. Abdelmohsen and J. C. M. Hest, Amphiphilic AIEgen-polymer aggregates: Design, self-assembly and biomedical applications, Aggregate, 2021, 3, e128 CrossRef.
- J. J. Hu, W. Jiang, L. Yuan, C. Duan, Q. Yuan, Z. Long, X. Lou and F. Xia, Recent advances in stimuli-responsive theranostic systems with aggregation-induced emission characteristics, Aggregate, 2021, 2, 48–65 CrossRef CAS.
- C. Zhou, W. Xu, P. Zhang, M. Jiang, Y. Chen, R. T. K. Kwok, M. M. S. Lee, G. Shan, R. Qi, X. Zhou, J. W. Y. Lam, S. Wang and B. Z. Tang, Engineering Sensor Arrays Using Aggregation-Induced Emission Luminogens for Pathogen Identification, Adv. Funct. Mater., 2019, 29, 1805986 CrossRef.
- S. Lu, J. Ke, X. Li, D. Tu and X. Chen, Luminescent nano-bioprobes based on NIR dye/lanthanide nanoparticle composites, Aggregate, 2021, 2, e59 CrossRef CAS.
- Q. Li, Y. Li, T. Min, J. Gong, L. Du, D. L. Phillips, J. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, R. T. K. Kwok, C. L. Ho, K. Li, J. Wang and B. Z. Tang, Time-Dependent Photodynamic Therapy for Multiple Targets: A Highly Efficient AIE-Active Photosensitizer for Selective Bacterial Elimination and Cancer Cell Ablation, Angew. Chem., Int. Ed., 2020, 59, 9470–9477 CrossRef CAS PubMed.
- M. Kang, C. Zhou, S. Wu, B. Yu, Z. Zhang, N. Song, M. M. S. Lee, W. Xu, F.-J. Xu, D. Wang, L. Wang and B. Z. Tang, Evaluation of Structure–Function Relationships of Aggregation-Induced Emission Luminogens for Simultaneous Dual Applications of Specific Discrimination and Efficient Photodynamic Killing of Gram-Positive Bacteria, J. Am. Chem. Soc., 2019, 141, 16781–16789 CrossRef CAS PubMed.
- W. Qiao, T. Ma, S. Wang, L. Li, M. Liu, H. Jiang, Y. Wu, J. Zhu and Z. Li, Designing Squaraine Dyes with Bright Deep-Red Aggregation-Induced Emission for Specific and Ratiometric Fluorescent Detection of Hypochlorite, Adv. Funct. Mater., 2021, 31, 2105452 CrossRef CAS.
- H. Wang, K. F. Xue, Y. Yang, H. Hu, J. F. Xu and X. Zhang, In Situ Hypoxia-Induced Supramolecular Perylene Diimide Radical Anions in Tumors for Photothermal Therapy with Improved Specificity, J. Am. Chem. Soc., 2022, 144, 2360–2367 CrossRef CAS PubMed.
- Y. Yang, P. He, Y. Wang, H. Bai, S. Wang, J.-F. Xu and X. Zhang, Supramolecular Radical Anions Triggered by Bacteria In Situ for Selective Photothermal Therapy, Angew. Chem., Int. Ed., 2017, 56, 16239–16242 CrossRef CAS PubMed.
- W. Z. Li, H. Chen, M. N. Shen, Z. Yang, Z. Fan, J. Xiao, J. Chen, H. Zhang, Z. Wang and X. Q. Wang, Chaotropic Effect Stabilized Radical-Containing Supramolecular Organic Frameworks for Photothermal Therapy, Small, 2022, 18, e2108055 CrossRef PubMed.
- X. Li, H. Bai, Y. Yang, J. Yoon, S. Wang and X. Zhang, Supramolecular Antibacterial Materials for Combatting Antibiotic Resistance, Adv. Mater., 2019, 31, e1805092 CrossRef PubMed.
- Z. Mi, P. Yang, R. Wang, J. Unruangsri, W. Yang, C. Wang and J. Guo, Stable Radical Cation-Containing Covalent Organic Frameworks Exhibiting Remarkable Structure-Enhanced Photothermal Conversion, J. Am. Chem. Soc., 2019, 141, 14433–14442 CrossRef CAS PubMed.
- S. Dai, M. Tao, Y. Zhong, Z. Li, J. Liang, D. Chen, K. Liu, B. Wei, B. Situ, M. Gao and B. Z. Tang, In Situ Generation of Red-to-NIR Emissive Radical Cations in Stomach for Gastrointestinal Imaging, Adv. Mater., 2023, e2209940 CrossRef PubMed.
- L. Zheng, W. Zhu, Z. Zhou, K. Liu, M. Gao and B. Z. Tang, Red-to-NIR emissive radical cations derived from simple pyrroles, Mater. Horiz., 2021, 8, 3082–3087 RSC.
- Z. Zhou, J. Qian, K. Liu, Y. Zhang, M. Gao and B. Z. Tang, A Water-Stable and Red-Emissive Radical Cation for Mutp53 Cancer Therapy, Angew. Chem., Int. Ed., 2022, 61, e202212671 CrossRef CAS.
- A. Praetorius, D. M. Bailey, T. Schwarzlose and W. M. Nau, Design of a Fluorescent Dye for Indicator Displacement from Cucurbiturils: A Macrocycle-Responsive Fluorescent Switch Operating through a pKa Shift, Org. Lett., 2008, 10, 4089–4092 CrossRef CAS PubMed.
- W. Kim, W. Zhu, G. L. Hendricks, D. Van Tyne, A. D. Steele, C. E. Keohane, N. Fricke, A. L. Conery, S. Shen, W. Pan, K. Lee, R. Rajamuthiah, B. B. Fuchs, P. M. Vlahovska, W. M. Wuest, M. S. Gilmore, H. Gao, F. M. Ausubel and E. Mylonakis, A new class of synthetic retinoid antibiotics effective against bacterial persisters, Nature, 2018, 556, 103–107 CrossRef CAS.
- J. T. Trevors, Fluorescent probes for bacterial cytoplasmic membrane research, J. Biochem. Biophys. Methods, 2003, 57, 87–103 CrossRef CAS PubMed.
- D. J. Phillips, J. Harrison, S. J. Richards, D. E. Mitchell, E. Tichauer, A. T. M. Hubbard, C. Guy, I. Hands-Portman, E. Fullam and M. I. Gibson, Evaluation of the Antimicrobial Activity of Cationic Polymers against Mycobacteria: Toward Antitubercular Macromolecules, Biomacromolecules, 2017, 18, 1592–1599 CrossRef CAS.
- M. Toyofuku, N. Nomura and L. Eberl, Types and origins of bacterial membrane vesicles, Nat. Rev. Microbiol., 2019, 17, 13–24 CrossRef CAS PubMed.
- L. Zheng, Y. Zhu, Y. Sun, S. Xia, S. Duan, B. Yu, J. Li and F. J. Xu, Flexible Modulation of Cellular Activities with Cationic Photosensitizers: Insights of Alkyl Chain Length on Reactive Oxygen Species Antimicrobial Mechanisms, Adv. Mater., 2023, 35, 2302943 CrossRef CAS.
- L. Zheng, J. Li, M. Yu, W. Jia, S. Duan, D. Cao, X. Ding, B. Yu, X. Zhang and F.-J. Xu, Molecular Sizes and Antibacterial Performance Relationships of Flexible Ionic Liquid Derivatives, J. Am. Chem. Soc., 2020, 142, 20257–20269 CrossRef CAS.
- R. Hu, F. Zhou, T. Zhou, J. Shen, Z. Wang, Z. Zhao, A. Qin and B. Z. Tang, Specific discrimination of gram-positive bacteria and direct visualization of its infection towards mammalian cells by a DPAN-based AIEgen, Biomaterials, 2018, 187, 47–54 CrossRef CAS.
- Z. Ye, W. He, Z. Zhang, Z. Qiu, Z. Zhao and B. Z. Tang, AIEgens for microorganism-related visualization and therapy, Interdiscip. Med., 2023, 1, e20220011 CrossRef.
- M. Gu, Z. Zeng, M. Y. Wu, J. K. Leung, E. Zhao, S. Wang and S. Chen, Imaging Macrophage Phagocytosis Using AIE Luminogen-Labeled E. coli, Chem. – Asian J., 2019, 14, 775–780 CrossRef CAS.
- G. J. Lieschke and N. S. Trede, Fish immunology, Curr. Biol., 2009, 19, R678–R682 CrossRef CAS.
- D. M. Tobin, R. C. May and R. T. Wheeler, Zebrafish: A See-Through Host and a Fluorescent Toolbox to Probe Host-Pathogen Interaction, PLoS Pathog., 2012, 8, e1002349 CrossRef CAS.
- P. Herbomel, B. Thisse and C. Thisse, Ontogeny and behaviour of early macrophages in the zebrafish embryo, Development, 1999, 126, 3735–3745 CrossRef CAS.
- G. J. Lieschke, A. C. Oates, M. O. Crowhurst, A. C. Ward and J. E. Layton, Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish, Blood, 2001, 98, 3087–3096 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Equipment and methods, photophysical characterization, CLSM images and bacterial viability data. See DOI: https://doi.org/10.1039/d3bm01472j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |