Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A (RP)UHPLC/UV analytical method to quantify dsRNA during the mRNA vaccine manufacturing process†
Sara Sousa
Rosa‡
abc,
Shuran
Zhang‡
a,
Yustika
Sari
a and
Marco P. C.
Marques
 *a
*a
aDepartment of Biochemical Engineering, University College London, Gordon Street, London, WC1E 6BT, UK. E-mail: marco.marques@ucl.ac.uk
bDepartment of Bioengineering, iBB—Institute for Bioengineering and Biosciences, Instituto Superior Técnico, Universidade de Lisboa, Lisboa, Portugal
cAssociate Laboratory i4HB—Institute for Health and Bioeconomy, Instituto Superior Técnico, Universidade de Lisboa, Lisboa, Portugal
First published on 8th July 2024
Abstract
dsRNA is a product related impurity produced during the mRNA manufacturing process. The established immuno-based detection methods lack the flexibility and speed required to be applied throughout the manufacturing process. The RP-HPLC method developed outperforms these in terms of precision, broader detection range, LOD and LOQ, as well as in output variance. Using this method, dsRNA can be quantified in under 30 min for a single sample.
Introduction
There has been widespread interest in mRNA vaccines within both academia and industry since the COVID-19 pandemic. Today, there are over 60 ongoing clinical trials that use mRNA vaccines for a variety of treatments that include prophylactic and cancer treatments, protein replacement and gene editing.1 When compared with traditional vaccine platforms, mRNA vaccines present a number of advantages, namely the fast production, the safety profile, and the vaccine effectiveness. From a clinical and manufacturing point-of-view, consistency of high-quality products is required with little batch-to-batch variability. This can only be achieved with a precise product characterisation and a tight manufacturing process control. Therefore, analytical procedures that rigorously characterise the mRNA and impurities (process- and product-related impurities)2 throughout the manufacturing process are required.3mRNA vaccines are usually produced in a cell-free system where a linearised DNA strand is in vitro transcribed (IVT) into a mRNA strand. RNA polymerases (e.g. T7 RNA polymerase) transcribe the DNA with nucleotides as co-substrates to produce grams per litre of mRNA.4–7 Other reaction components include co-factors, enhancers and additional enzymes, with temperature and pH as critical reaction parameters. The exploration of different reaction conditions coupled with a tight production characterisation8 led already to an improvement in reaction yields, with reported production in the range of 12 g L−1.7,9 However, despite the tight control, there are impurities produced throughout the IVT process. These can be classified into process related (enzymes, residual NTPs, or the DNA template) and product related impurities (malformed mRNAs).2,7 During the IVT reaction, the T7 RNA polymerase can release truncated mRNA molecules, or produce complementary RNA strands that can hybridise and produce double-stranded mRNA (dsRNA). The presence of this particular impurity in the final product must be avoided since it can impact translation and trigger a strong immune response,10 which ultimately can lead to an uncontrolled immune-inflammatory reaction.11 In the absence of dsRNA, protein expression within cells can be increased by 10–1000 fold.12 Therefore, optimised reaction conditions or new purification methods are necessary to eliminate dsRNA as well as precise analytical procedures to quantify this impurity. Nevertheless, there is still a lack of defined concentration limits that are established by the regulatory agencies for dsRNA.3
From a reaction optimisation perspective, multiple approaches have been followed to reduce dsRNA such as engineering T7 RNA polymerase,13,14 or blocking the 3′ end with complementary oligonucleotides to avoid overextension.15 New chromatographic modalities can be applied, exploring physico-chemical differences between mRNA and dsRNA.16 Nonetheless, dsRNA needs to be carefully monitored during the manufacturing process itself and in the final product.3 Analytically, dsRNA can be detected and characterised by several methods, including electrophoresis,17,18 immunoassays that use anti-double-stranded RNA antibodies such as dot blot, ELISA or lateral flow strip assay (LFSA),19 and asymmetric flow field flow fractionation (A4F),20 or even chromatographic methods.8,21–23 Nevertheless, there is a lack of well-established methods that can be applied in the manufacturing process which will quantify dsRNA that meet the regulatory requirements,24 are quick, specific and minimise impurity interference, and can be easily adapted to the different process stages, from the production to the different purification steps, and to the different modalities of the mRNA process (batch, fed-batch, or continuous).
In this communication, we explore a method to quantify dsRNA in the different stages of the manufacturing process. A previously established reverse-phase HPLC method that quantifies total mRNA was coupled to an enzymatic digestion step that digests ssRNA, allowing us to directly measure dsRNA. The developed RP-HPLC method achieved lower limit of detection (LOD) and limit of quantification (LOQ), 1.41 × 10−2 ± 1.78 × 10−3 g L−1, and 4.27 × 10−2 ± 5.4 × 10−3 g L−1, respectively, compared to the current golden standard, the dot blot. Furthermore, the decision limit (CCα, 9.95 × 10−2 ± 1.26 × 10−3 g L−1) and detection capability (CCβ, 1.70 × 10−2 ± 2.14 × 10−3 g L−1) are low, and obtained for a single sample under 30 min when compared with standard methods (e.g. ELISA and dot blot assay).3 HPLC outperformed these and proved to be precise, and less prone to operational errors, even in spiking studies with process-related impurities (NTPs and DNA) where minimal variance in quantification is observed. The implemented RP-HPLC is robust to use during the mRNA manufacturing process and that can be adapted for Process Analytical Technology (PAT) purposes in the future.
Materials and methods
Unless otherwise stated, all chemicals and reagents were purchased from Thermo Fisher Scientific (UK).Template plasmid DNA production
Template design and plasmid production was performed as previously described.7 Briefly, GFP gene (GenBank Accession # AAB02572.1) is flanked by 5′UTR containing the T7 RNA polymerase promoter, eukaryotic translation factor binding site and a Kozak consensus sequence,25 and by a 3′UTR composed of two β-globin tandem repeats followed by a 120 bp poly-A tail segmented with a 6 bp.26 A pUC7 containing a kanamycin resistance is used as a plasmid vector with plasmid propagation performed in e. coli NEB 10-beta (New England Biolabs, UK). The pDNA is obtained by performing an overnight culture in LB media at 37 °C and purified using the GeneJET Plasmid Miniprep Kit.Template production by touchdown polymerase chain reaction
The forward and reverse primers were previously described.7 A T7 RNA polymerase promoter sequence was added to the 5′ end of the complementary strand using the reverse primer. The DNA template for mRNA IVT is produced by touchdown PCR (Applied Biosystems™ Veriti™ 96-Well Thermal Cycler, Thermo Fisher Scientific, UK). Briefly, the reaction mixture comprises 250 ng mL−1 of template plasmid, 0.4 μM of forward and reverse primer, 1× VeriFi™ Buffer, 1× VeriMax Enhancer, and 0.02 U μL−1 high-fidelity VeriFi™ DNA polymerase (PCR Biosystems, UK). The PCR conditions for the denaturation step are 98 °C for 30 s, 20 cycles with a denaturation step at 94 °C for 15 s, an annealing step at 65–55 °C for 30 s and an extension step at 72 °C for 45 s. This is followed by 20 cycles of an annealing step at 55 °C for 30 s and an extension step at 72 °C. The final extension is performed at 72 °C for 2 min. The obtained PCR product is purified and concentrated using a GeneJET PCR Purification Kit. Final concentrations are quantified on a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, UK).dsRNA in vitro transcription reactions
dsRNA is produced in an IVT reaction using previously described reaction conditions.7 Briefly, 89 nM DNA template is mixed with 7.7 mM NTPs, 5.3 mM dithiothreitol, 50 mM magnesium acetate, 40 mM Tris–HCl pH 6.8, 2.3 mM spermidine, 8 U mL−1 inorganic pyrophosphatase and 7750 U mL−1 of T7 RNA polymerase. To this mix, 1500 U mL−1 RNase inhibitor is added to avoid degradation. The mixture is incubated at 43 °C for two hours on an Applied Biosystems™ Veriti™ 96-Well Thermal Cycler (Thermo Fisher Scientific, UK). After incubation, the produced strands of RNA are annealed to form dsRNA by diluting the reaction mixture twice with WFI and incubating in a decreasing temperature gradient (from 85 °C to 30 °C) with a 2 min step at each temperature.dsRNA purification
The DNA template and the remaining mRNA are digested using TURBO™ DNase, and RNase T1, respectively. The enzymes are added to the IVT samples to a final concentration of 0.04 U μL−1 (TURBO™ DNase), and 20 U μL−1 (RNase T1). The samples are incubated at 37 °C for 30 min and purified afterwards using a MEGAclear™ Transcription Clean-Up kit. After purification, the dsRNA is precipitated overnight at −20 °C by adding 500 mM pH 5 ammonium acetate and 2.5 volumes of ethanol. The samples are centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min and the supernatant was discarded. The pellet is air dried and WFI water is added to resuspend the pellet to the desired final concentration. Concentration is determined on a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, UK) using a conversion factor of 47 μg mL−1 Abs260−1.27
000 × g for 15 min and the supernatant was discarded. The pellet is air dried and WFI water is added to resuspend the pellet to the desired final concentration. Concentration is determined on a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, UK) using a conversion factor of 47 μg mL−1 Abs260−1.27
Analytical methodologies
The samples were blotted onto a 0.45 μm nitrocellulose membrane using a pipette tip. The membrane was dried and incubated with 3% (w/v) of BSA (Sigma-Aldrich, USA) in 1× PBS–Tween buffer, containing 1× PBS (10 mM sodium phosphate, 2.68 mM potassium chloride, and 140 mM calcium chloride) and 0.05% (v/v) of Tween-20 (Sigma-Aldrich, USA), for an hour at 22 °C. The membrane was subsequently incubated for an hour with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500 dilution of SCICONS™ J2 mouse anti-dsRNA IgG2a monoclonal antibody (Nordic-MUbio, Netherlands) as primary antibody using 3% (w/v) BSA in 1× PBS–Tween buffer. The membrane was washed by incubating for 5 min in the PBS–Tween buffer and repeated three times. Afterwards, the membrane was incubated with 1
1500 dilution of SCICONS™ J2 mouse anti-dsRNA IgG2a monoclonal antibody (Nordic-MUbio, Netherlands) as primary antibody using 3% (w/v) BSA in 1× PBS–Tween buffer. The membrane was washed by incubating for 5 min in the PBS–Tween buffer and repeated three times. Afterwards, the membrane was incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution of secondary antibody, mouse IgG horseradish peroxidase (HRP)-conjugated antibody (R&D Systems, UK) for an hour, followed by three washing steps. Prior to the chemiluminescence exposure, the membrane was treated with the Pierce™ ECL western blotting substrate and incubated for 3 min. All the incubation steps were performed at 22 °C. The visualisation was performed through chemiluminescence exposure using an Amersham™ Imager 600 (GE Healthcare, UK).
2000 dilution of secondary antibody, mouse IgG horseradish peroxidase (HRP)-conjugated antibody (R&D Systems, UK) for an hour, followed by three washing steps. Prior to the chemiluminescence exposure, the membrane was treated with the Pierce™ ECL western blotting substrate and incubated for 3 min. All the incubation steps were performed at 22 °C. The visualisation was performed through chemiluminescence exposure using an Amersham™ Imager 600 (GE Healthcare, UK).
| Species | Concentration | |
|---|---|---|
| Min. | Max. | |
| RNA (g L−1) | 0.1 | 0.4 |
| DNA (nM) | 10 | 90 |
| NTPs (mM) | 2 | 7.5 |
| LOD = 3.3σ/S | (1) |
| LOQ = 10σ/S | (2) |
| CCα = 2.33σ/S | (3) |
| CCβ = CCα + 1.64σ/S | (4) |
Results and discussion
We have explored reverse phase chromatography (RP-HPLC) for the detection of dsRNA based on the physico-chemical properties of mRNA, namely the hydrophobicity.31 Preparative chromatography, namely ion-pair (IP-RP-HPLC), has already been used to separate ssRNA from dsRNA.12,32 The ion-pairs added to the mobile phase will interact with the negatively charged backbone of oligonucleotides and will allow separation according to the number of backbones available.33 In this work, the mobile phase is composed of Tris buffer and EDTA, instead of an ion-pair and thus the separation is achieved by the hydrophobic portions present in the mRNA (e.g. poly(A) tail).28 To facilitate detection, it is necessary to ensure that solely dsRNA is present in samples. To achieve this, an enzymatic digestion prior to injection is performed using RNase T1.7,13 The RNase T1 is an endoribonuclease that cleaves single-stranded RNA at the guanosine residues. When added to an IVT reaction, RNase T1 will digest all the ssRNA, and the dsRNA can be directly quantified by RNA quantification methods. The semi-quantitative assays such as the dot blot or ELISA can be performed without the use of this additional enzymatic digestion since they use specific antibodies (e.g. dsRNA antibody j2).34 However, with these assays, no direct read out of the samples is possible and multiple assay preparation steps are often required, which can potentially increase the associated operator error.The RP-HPLC separates molecules according to its hydrophobicity (Fig. 1a). Small size impurities (e.g. reaction components) do not usually bind to the stationary phase and are eluted during the wash phase (first peak in the chromatograms), while mRNA is eluted during the gradient phase. This is achieved by increasing the organic solvent concentration in the mobile phase. After digestion with the RNase T1 enzyme, the digested ssRNA is eluted during the wash phase while the dsRNA is bound to the stationary phase and requires organic solvent to be eluted. After digestion, there is a shift in the retention time of the RNA peak. This can be attributed to the difference in the molecular weight between dsRNA and ssRNA (Fig. 1b). The dsRNA produced during IVT will have single stranded regions, possible interacting with the stationary phase, while after digestion, only double stranded regions are present.
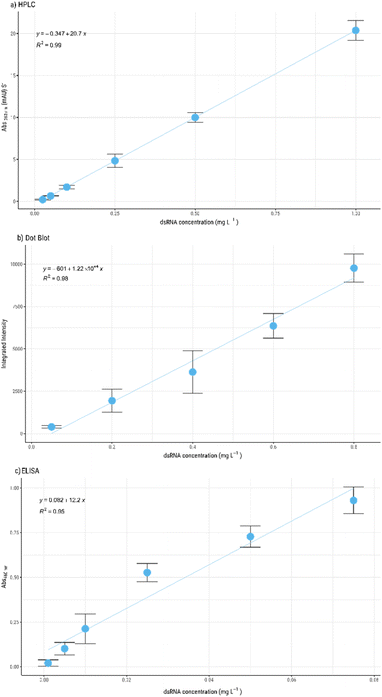
We have validated the HPLC method by analysing the different regulatory requirements (e.g. specificity, range, accuracy, precision, and/or robustness24) and compare it to dot blot and commercially available dsRNA detection ELISA kits (Fig. 2). To achieve this, 6-point calibration curves were prepared with dsRNA concentrations ranging from 0.025 to 1 g L−1, 0.05 to 1 g L−1 and 0.005 to 0.075 mg L−1, for the HPLC, dot blot and ELISA, respectively. The standard curves were evaluated using three independent samples. In the HPLC method, linearity is observed throughout the range of sample concentrations used with a high correlation coefficient, R2 > 0.99 (Fig. 2a). In contrast, the dot blot only presents a linear relationship at concentrations below 0.8 g L−1 with R2 > 0.98 (Fig. 2b). However, the residual evaluation data show that there is homoscedasticity (ESI, Fig. S1.b†), which means that although the R2 is lower, the variance between samples is similar. The ELISA method is most specific of the methods evaluated, as it can detect the smallest concentration of dsRNA. However, this assay has a very narrow range of detection. Linearity is only achieved at concentration below 0.07 mg L−1 (Fig. 2c) with a low correlation coefficient, R2 > 0.95. The lower correlation can be attributed to the high sensitivity of the method which makes the process more prone to operator errors. The residual analysis (ESI, Fig. S1†) and the respective model coefficient (p-value) evaluation show that there is a strong correlation between the variables for the three methods, confirming the goodness of fit of the resulting models.
The limit of detection (LOD) and limit of quantification (LOQ) were calculated for the three methods (Table 2). ELISA presents the lowest LOD and LOQ, 1.51 × 10−5 ± 3.21 × 10−6 and 4.56 × 10−5 ± 9.73 × 10−6 g L−1, respectively. The dot blot presents the highest LOD (0.201 ± 0.06 g L−1) and LOQ (0.609 ± 0.181 g L−1) whilst the HPLC presents 14 times lower LOD and LOQ (0.014 ± 0.002, 0.043 ± 0.005 g L−1), when compared with dot blot assay.
| Parameter | Analytical method | |||||
|---|---|---|---|---|---|---|
| HPLC | Dot blot | ELISA | ||||
![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) (g L−1) (g L−1) |
σ (g L−1) |
![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) (g L−1) (g L−1) |
σ (g L−1) |
![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) (g L−1) (g L−1) |
σ (g L−1) | |
| LOD | 1.41 × 10−2 | 1.78 × 10−3 | 2.01 × 10−1 | 5.96 × 10−2 | 1.51 × 10−5 | 3.21 × 10−6 |
| LOQ | 4.27 × 10−2 | 5.40 × 10−3 | 6.09 × 10−1 | 1.81 × 10−1 | 4.56 × 10−5 | 9.73 × 10−6 |
| CCα | 9.95 × 10−3 | 1.26 × 10−3 | 1.42 × 10−1 | 4.21 × 10−2 | 1.06 × 10−5 | 2.27 × 10−6 |
| CCβ | 1.70 × 10−2 | 2.14 × 10−3 | 2.42 × 10−1 | 7.17 × 10−2 | 1.81 × 10−5 | 3.86 × 10−6 |
The decision limit (CCα) and detection capability (CCβ) were evaluated for all methods. CCα represents the lowest concentration obtained which is reliable (1% of false positive risk), while CCβ is the lowest concentration possible to measure with an error probability of 5% (false negative risk).
ELISA assay presents the lowest CCα and CCβ values (1.06 × 10−5 ± 2.27 × 10−6 and 1.81 × 10−5 ± 3.86 × 10−6 g L−1) while in contrast, dot blot presents the highest values (0.142 ± 0.042 and 0.242 ± 0.07 g L−1). Since ELISA presents the lowest LOD and LOQ evaluated, it can be an ideal method to use with highly pure samples with residual amounts of impurities, namely dsRNA. Currently approved vaccines control the dsRNA levels to be as low as possible throughout the manufacturing process35,36 with no defined value. Currently, the dot blot assay is the method recommended to characterise dsRNA throughout the mRNA manufacturing process3 and evaluation values obtained support this. Nevertheless, the developed HPLC method shows a larger detection range and limits, presenting itself as a more versatile quantification method. Additionally, these ranges can be extended as the maximum concentration of dsRNA that can be quantified by HPLC is contingent on the saturation capacity of the column itself, which can be adapted by selecting columns of different diameters and lengths.
To evaluate further the accuracy and precision of the HPLC method implemented, a spiking study was performed. Pure dsRNA samples were spiked with two process related impurities that typically can be present in process samples, namely NTPs and the DNA template (Table 1). The concentrations of the impurities tested are in the range commonly encountered in the manufacturing process of mRNA vaccines. Additional controls with the spiking reagents (DNA and NTPs) and their mixtures with different concentrations evaluated were also performed. The HPLC precision performance was compared with that of dot blot assay as a similar range can be used in both methods. HPLC analysis shows that the values measured have a more uniform distribution when compared with the dot blot assay (Fig. 2). Statistical analysis performed shows that a significant difference is observed in the dot blot performed with a high concentration of dsRNA (Fig. 3d, 0.4 g L−1), and in the presence of a high concentration of NTPs (ESI, Tables S1–S4†). This may be due to the occurrence of unspecific binding of the antibody used to perform the dot blot assay.
No significant differences in the dsRNA measured concentrations were found when using HPLC, indicating that the method is more accurate (ESI, Tables S1–S4†). Comparing the concentrations obtained for high and low range of dsRNA concentrations, the results show that there are no significant differences between both methods. For high concentration of dsRNA (0.4 g L−1), HPLC measured 0.41 ± 0.05 g L−1, while with dot blot assay, a concentration of 0.45 ± 0.151 g L−1 was obtained. For low dsRNA concentrations (0.1 g L−1), 0.09 ± 0.016 g L−1 and 0.13 ± 0.017 g L−1 were obtained for the HPLC and dot blot assay, respectively.
Overall, the HPLC method presents a better precision and range, as it presented a better correlation between the concentration range (0.025 to 1 g L−1) evaluated when compared with dot blot assay, but ELISA presents the lowest detection range (0.005 to 0.075 mg L−1). Nevertheless, when comparing analytical methods, it is also required to look into the overall analytical performance (Table 3). Although ELISA is a method that can accurately measure the smallest concentration of dsRNA present in samples, the results are solely obtained after 12 hours due to the multiple incubation steps involved. Additionally, the multiple steps involved in this assay also increase operator error occurrence. In terms of time-to-result, the dot blot is the faster method among the immunoassays (three hours). Nonetheless, similar to ELISA, it requires multiple incubation steps and reagent additions. The HPLC is the fastest for single sample, requiring minimum sample preparation and operator input to run the method. However, the process time can increase if an additional digestion step with RNase T1 is required. Nevertheless, the throughput is limited to HPLC sampler capacity and the method running time. Higher throughputs are obtained with the ELISA and dot blot. On the other hand, the HPLC method is a fully quantitative method while dot blot relies on a semi quantitative analysis based on densitometry measurements.
| Method validation | Performance overview | ||||||
|---|---|---|---|---|---|---|---|
| Sensibility | Range | Precision | Assay time | Complexity | Operator error | Detection | |
| ELISA | +++ | + | n/a | >12 h | ++ | +++ | Quantitative |
| Dot blot | ++ | ++ | ++ | >3 h | + | ++ | Semi-quantitative |
| HPLC | ++ | +++ | +++ | <1 h | +++ | + | Quantitative |
Cost can also be a predominant factor when choosing the analytical method to be used. By nature of the assay itself, the immuno-assays are costly. In particular, the dot blot costs are associated with the primary antibodies used. Comparatively, these are up to 10× higher than the RNase T1 enzyme required for HPLC assay, but cost will be reduced with increased throughput. Evidently, HPLC costs are strongly correlated with the stationary phase and equipment used. Finally, at the manufacturing level, HPLC can be used as an at-line method integrated in the process workflow37 allowing data to be obtained faster and consequently enabling a tight process control.
Conclusions
dsRNA is a product related impurity produced during the mRNA vaccine manufacturing. This impurity has a strong impact on mRNA performance within the cells as it decreases the translation rate and increases the inflammatory response, and the presence of this particular impurity in the final product must be quantified and avoided.In this work, the use of HPLC for dsRNA quantification is explored, and compared with established immunoassays, namely the dot blot and ELISA. With the developed RP-HPLC method it was possible to quantify dsRNA in samples in under 30 min. The sensitivity and precision of this method are high, with a broader detection range 0.025 to 1 g L−1 and minimum impurity detection interference. From a regulatory perspective, it achieved the lowest limit of detection (LOD) and limit of quantification (LOQ), 1.41 × 10−2 ± 1.78 × 10−3 g L−1, and 4.27 × 10−2 ± 5.4 × 10−3 g L−1, respectively, compared to the current golden standard, the dot blot. The occurrence of false positive or negatives with this method is also low given the decision limit and detection capability obtained.
Additionally, the implementation of the HPLC method requires minimum operator input and sample handling with throughput achieved by tunning the method running time. This, combined with the ability to use this method at-line and prone to automation makes the HPLC an ideal method to be used to quantify dsRNA throughout the manufacturing process. Precise and reliable analytical assays are of paramount importance to have a well-established manufacturing process that delivers high quality products.
Data availability
The data supporting this article have been included as part of the ESI.† Raw data are available upon request from the authors.Author contributions
Conceptualization, M. P. C. M. and S. S. R.; data curation, formal analysis, investigation and methodology, S. S. R., S. Z., and Y. S.; supervision, M. P. C. M; validation, S. S. R. and Y. S.; visualization, S. S. R.; writing – original draft, S. S. R.; writing – reviewing and editing, M. P. C. M. and S. S. R.; funding acquisition, M. P. C. M.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the support of the Future Biomanufacturing Research Hub [grant EP/S01778X/1], funded by the Engineering and Physical Sciences Research Council (EPSRC) and Biotechnology and Biological Sciences Research Council (BBSRC) as part of UK Research and Innovation. The Fundação para a Ciência e a Tecnologia (FCT, Portugal) is acknowledged for funding Sara Sousa Rosa's [SFRH/BD/148437/2019] PhD studentship.Notes and references
- Y. S. Wang, M. Kumari, G. H. Chen, M. H. Hong, J. P. Y. Yuan and J. L. Tsai, et al., mRNA-based vaccines and therapeutics: an in-depth survey of current and upcoming clinical applications, J. Biomed. Sci., 2023, 30(1), 1–35 CrossRef PubMed.
- J. Grinsted, J. Liddell, E. Bouleghlimat, K. Y. Kwok, G. Taylor and M. P. C. Marques, et al., Purification of therapeutic & prophylactic mRNA by affinity chromatography, Cell Gene Therapy Insights, 2022, 8(2), 335–349 CrossRef.
- Organization WH and others, Evaluation of the Quality, Safety and Efficacy of RNA-Based Prophylactic Vaccines for Infectious Diseases: Regulatory Considerations, retrieved January 2020, 1, 2022 Search PubMed.
- S. Bancel, W. J. Issa, J. G. Aunins and T. Chakraborty, Manufacturing Methods for Production of RNA Transcripts, US Pat., 10138507 2018 Search PubMed.
- J. M. Henderson, A. Ujita, E. Hill, S. Yousif-Rosales, C. Smith and N. Ko, et al., Cap 1 Messenger RNA Synthesis with Co-transcriptional CleanCap® Analog by In Vitro Transcription, Curr. Protoc., 2021, 1(2), e39 CrossRef CAS PubMed.
- A. Wochner, T. Roos and T. Ketterer, Methods and Means for Enhancing Rna Production, US Pat., US20170114378A1, 2017.
- S. S. Rosa, D. Nunes, L. Antunes, D. M. F. Prazeres, M. P. C. Marques and A. M. Azevedo, Maximizing mRNA vaccine production with Bayesian optimization, Biotechnol. Bioeng., 2022, 119(11), 3127–3139 CrossRef CAS PubMed.
- D. Pregeljc, J. Skok, T. Vodopivec, N. Mencin, A. Krušič and J. Ličen, et al., Increasing yield of in vitro transcription reaction with at-line high pressure liquid chromatography monitoring, Biotechnol. Bioeng., 2023, 120(3), 737–747 CrossRef CAS PubMed.
- J. Skok, P. Megušar, T. Vodopivec, D. Pregeljc, N. Mencin and M. Korenč, et al., Gram-Scale mRNA Production Using a 250-mL Single-Use Bioreactor, Chem. Ing. Tech., 2022, 94(12), 1928–1935 CrossRef CAS.
- S. Linares-Fernández, C. Lacroix, J. Y. Exposito and B. Verrier, Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response, Trends Mol. Med., 2020, 26(3), 311–323 CrossRef PubMed.
- G. Milano, J. Gal, A. Creisson and E. Chamorey, Myocarditis and COVID-19 mRNA vaccines: a mechanistic hypothesis involving dsRNA, Future Virol., 2021, 17(3), 191–196 CrossRef PubMed.
- K. Kariko, H. Muramatsu, J. Ludwig and D. Weissman, Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA, Nucleic Acids Res., 2011, 39(21), e142 CrossRef CAS PubMed.
- A. Dousis, K. Ravichandran, E. M. Hobert, M. J. Moore and A. E. Rabideau, An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts, Nat. Biotechnol., 2023, 41(4), 560–568 CrossRef CAS PubMed.
- M. Miller, O. Alvizo, S. Baskerville, A. Chintala, C. Chng, J. Dassie, J. Dorigatti, G. Huisman, S. Jenne, S. Kadam and N. Leatherbury, An Engineered T7 RNA Polymerase for efficient co-transcriptional capping with reduced dsRNA byproducts in mRNA synthesis, Faraday Discuss., 2024 10.1039/d4fd00023d.
- Y. Gholamalipour, A. Karunanayake Mudiyanselage and C. T. Martin, 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses, Nucleic Acids Res., 2018, 46(18), 9253–9263 CrossRef CAS PubMed.
- S. S. Rosa, D. M. F. Prazeres, A. M. Azevedo and M. P. C. Marques, mRNA vaccines manufacturing: challenges and bottlenecks, Vaccine, 2021, 39(16), 2190–2200 CrossRef CAS PubMed.
- A. C. D. Peña, M. Vaduva, N. S. Li, S. Shah, M. B. Frej and A. Tripathi, Enzymatic isolation and microfluidic electrophoresis analysis of residual dsRNA impurities in mRNA vaccines and therapeutics, Analyst, 2024, 149(5), 1509–1517 RSC.
- A. C. D. Peña, N. Li, M. Vaduva, L. Bwanali and A. Tripathi, A microfluidic electrophoretic dual dynamic staining method for the identification and relative quantitation of dsRNA contaminants in mRNA vaccines, Analyst, 2023, 148(16), 3758–3767 RSC.
- D. Luo, Z. Wu, D. Wang, J. Zhang, F. Shao and S. Wang, et al., Lateral flow immunoassay for rapid and sensitive detection of dsRNA contaminants in in vitro-transcribed mRNA products, Mol. Ther.--Nucleic Acids, 2023, 32, 445–453 CrossRef CAS PubMed.
- K. Eskelin, M. Lampi, C. Coustau, J. Imani, K. H. Kogel and M. M. Poranen, Analysis and purification of ssRNA and dsRNA molecules using asymmetrical flow field flow fractionation, J. Chromatogr. A, 2022, 1683, 463525 CrossRef PubMed.
- A. O. Nwokeoji, A. W. Kung, P. M. Kilby, D. E. Portwood and M. J. Dickman, Purification and characterisation of dsRNA using ion pair reverse phase chromatography and mass spectrometry, J. Chromatogr. A, 2017, 1484, 14–25 CrossRef CAS PubMed.
- J. De Vos, K. Morreel, P. Alvarez, H. Vanluchene, R. Vankeirsbilck and P. Sandra, et al., Evaluation of size-exclusion chromatography, multi-angle light scattering detection and mass photometry for the characterization of mRNA, J. Chromatogr. A, 2024, 1719, 464756 CrossRef CAS PubMed.
- J. Camperi, S. Lippold, L. Ayalew, B. Roper, S. Shao and E. Freund, et al., Comprehensive Impurity Profiling of mRNA: Evaluating Current Technologies and Advanced Analytical Techniques, Anal. Chem., 2024, 96(9), 3886–3897 CrossRef CAS PubMed.
- Guideline, I. C. H. H. Validation of Analytical Procedures Q2 (R2), ICH, Geneva, Switzerland, 2022 Search PubMed.
- M. Tusup, L. E. French, M. De Matos, D. Gatfield, T. Kundig and S. Pascolo, Design of in vitro Transcribed mRNA Vectors for Research and Therapy, Chimia, 2019, 73(5), 391–394 CrossRef CAS PubMed.
- Z. Trepotec, J. Geiger, C. Plank, M. K. Aneja and C. Rudolph, Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life, RNA, 2019, 25(4), 507–518 CrossRef CAS PubMed.
- A. O. Nwokeoji, P. M. Kilby, D. E. Portwood and M. J. Dickman, Accurate Quantification of Nucleic Acids Using Hypochromicity Measurements in Conjunction with UV Spectrophotometry, Anal. Chem., 2017, 89(24), 13567–13574 CrossRef CAS PubMed.
- M. WilliamIssa, Methods for Hplc Analysis, US Pat., US20210163919A1, 2021.
- Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on in vitro diagnostic medical devices and repealing Directive 98/79/EC and Commission Decision 2010/227/EU (Text with EEA relevance.), internet, OJ L, 32017R0746 May 5, 2017, available from: https://data.europa.eu/eli/reg/2017/746/oj/eng Search PubMed.
- J. Van Loco, A. Jànosi, S. Impens, S. Fraselle, V. Cornet and J. M. Degroodt, Calculation of the decision limit (CCα) and the detection capability (CCβ) for banned substances: the imperfect marriage between the quantitative and the qualitative criteria, Anal. Chim. Acta, 2007, 586(1), 8–12 CrossRef CAS PubMed.
- M. I. Aguilar, Reversed-phase High-performance Liquid Chromatography, HPLC of Peptides and Proteins: Methods and Protocols, 2004, pp. 9–22 Search PubMed.
- D. Weissman, N. Pardi, H. Muramatsu and K. Karikó, HPLC purification of in vitro transcribed long RNA, Synthetic Messenger RNA and Cell Metabolism Modulation: Methods and Protocols, 2013, pp. 43–54 Search PubMed.
- L. Baronti, H. Karlsson, M. Marušič and K. Petzold, A guide to large-scale RNA sample preparation, Anal. Bioanal. Chem., 2018, 410(14), 3239–3252 CrossRef CAS PubMed.
- J. Schonborn, J. Oberstraβ, E. Breyel, J. Tittgen, J. Schumacher and N. Lukacs, Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts, Nucleic Acids Res., 1991, 19(11), 2993–3000 CrossRef CAS PubMed.
- E. Moderna, Assessment Report COVID-19 Vaccine Moderna. Procedure No. EMEA/H/C/005791/0000, European Medicines Agency, Netherlands, 2021 Search PubMed.
- E. Pfizer, Assessment Report Comirnaty. Procedure No. EMEA/H/C/005735/0000, European Medicines Agency, Netherlands, 2021 Search PubMed.
- D. Pregeljc, J. Skok, T. Vodopivec, N. Mencin, A. Krušič and J. Ličen, et al., Increasing yield of in vitro transcription reaction with at-line high pressure liquid chromatography monitoring, Biotechnol. Bioeng., 2023, 120(3), 737–747 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1: residual plots for the calibration curves used to calculate dsRNA. Fig. S2: residual plots for the dsRNA spiking studies. Table S1: dot blot Tukey test results for high dsRNA concentration. Table S2: dot blot Tukey test results for low dsRNA concentration. Table S3: HPLC Tukey test results for high dsRNA concentration. Table S4: HPLC Tukey test results for low dsRNA concentration. See DOI: https://doi.org/10.1039/d4ay00560k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |