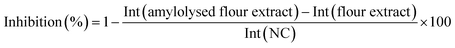Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Screening of α-amylase/trypsin inhibitor activity in wheat, spelt and einkorn by high-performance thin-layer chromatography†
Isabel
Müller
 ,
Bianca
Schmid
,
Loredana
Bosa
and
Gertrud Elisabeth
Morlock
,
Bianca
Schmid
,
Loredana
Bosa
and
Gertrud Elisabeth
Morlock
 *
*
Chair of Food Science, Institute of Nutritional Science, Interdisciplinary Research Centre for Biosystems, Land Use and Nutrition, Justus Liebig University Giessen, Heinrich-Buff-Ring 26-32, 35392 Giessen, Germany. E-mail: gertrud.morlock@uni-giessen.de; Fax: +49-641-99-39149; Tel: +49-641-9939141
First published on 30th April 2024
Abstract
α-Amylase/trypsin inhibitor proteins (ATI) are discussed as possible triggers for non-celiac gluten sensitivity. The potential of high-performance thin-layer chromatography (HPTLC) was studied for the first time to analyse the inhibitory properties of ATIs from flour of wheat, spelt, and einkorn. Inhibition by each flour of the digestive enzymes trypsin or α-amylase was determined by the reduction of released metabolisation products in comparison to non-digested flour, and positive (acarbose) and negative (water) controls. Firstly, amylolysis was carried out in miniaturized form on the HPTLC surface (HPTLC-nanoGIT) after in-vial pre-incubation of the amylase with the inhibitors from flour. α-Amylase inhibition was evident via the reduction of released saccharides, as analysed by normal phase HPTLC. A strong influence of the flour matrix on the assay results (individual saccharides) was evident, caused by an increased amylolysis of further polysaccharides present, making HPTLC analysis more reliable than currently used spectrophotometric sum value assays. The detection and visualization of such matrix influence helps to understand the problems associated with spectrophotometric assays. Only maltotriose was identified as a reliable marker of the amylolysis. The highest α-amylase inhibition and thus the lowest saccharide response was detected for maltotriose in refined spelt, whereas the lowest α-amylase inhibition and thus the highest saccharide response was detected for maltotriose in refined wheat. A comparison of refined and whole grain flours showed no clear trend in the responses. Secondly, trypsin inhibition and proteolysis were performed in-vial, and any inhibition was evident via the reduction of released peptides, analysed by reversed-phase HPTLC. Based on the product pattern of the proteolysis, einkorn and whole wheat showed the highest trypsin inhibition, whereas refined wheat and refined spelt showed the lowest inhibition. Advantageously, HPTLC analysis provided important information on changes in individual saccharides or peptides, which was more reliable and sustainable than spectrophotometric in-vial assays (only sum value) or liquid column chromatography analysis (targeting only the ATI proteins).
Introduction
Digestive problems concerning the consumption of especially wheat (Triticum aestivum) as a carbohydrate diet source have increased in recent decades.1 Studies have shown that complicated dietary adjustments are needed, if possible, to distinguish gastrointestinal disorders,2 such as celiac and inflammatory bowel disease, irritable bowel syndrome, wheat allergy, and non-celiac gluten sensitivity (NCGS). Among these, NCGS is the least studied and has the highest but also varying prevalence (0.5–15%).2–4 To ensure a safe and conclusive diagnosis, analysts tend to identify causative compounds. The ongoing discussion on NCGS involves identifying the specific compound classes that serve as triggers, and multiple approaches have been explored to narrow down and comprehend a diverse range of triggers. Under suspicion are gluten-related gliadins, α-amylase/trypsin inhibitors (ATIs) and/or fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAPs).5 Gliadins and ATIs are cereal-derived compounds, whereas FODMAPs are widely distributed among foods. Since gluten and its components have been well studied, the focus has shifted to the evaluation of the exact role of ATIs and FODMAPs in the diagnosis of NCGS.2,3The ATI proteins with two different binding sites act as bifunctional inhibitors to the digestive enzymes trypsin and α-amylase.6 Originally, this function represents their role in the plant defence mechanism to protect the grain from parasites and pests by inhibiting only their digestive enzymes and not those of the cereals themselves.7 This inhibitory ability could play an important role considering the potential accumulation of digestion-resistant gluten peptides, which are the cause of an immunogenic reaction in patients with gastrointestinal disorders.6,8 Another possible mechanism of action of these inhibitors is the activation of Toll-like receptor 4 complex (TLR4–MD2–CD14) and with it, the release of proinflammatory cytokines in cells.9,10 At least 19 ATIs in their monomeric, dimeric or tetrameric form were reported, which account for 2–4% of the total proteins in wheat.11,12 These low molecular weight proteins (12–15 kDa) are part of the water-soluble fraction and can be extracted according to the Osborne fractionation with the aid of aqueous salt buffers.2 They are known to be heat resistant and thus survive food processing like baking, which implies they still could be bioactive in processed food like pasta, bread or biscuits.5,13 However, there are also contrary studies that mainly focus on α-amylase activity, suggesting a decrease in inhibition after heat treatment.14,15 Such contradictory studies highlight the difficulties in understanding such complex diseases and the problems encountered in interpreting results from in vitro experiments, not to mention the relevance of in vivo data.
Two approaches were pursued for the evaluation of ATI. One was the tandem mass spectrometric analysis of known ATIs to identify/quantify their amounts in cereal products.16–25 The other approach was the screening of the ATI inhibitory potency via enzymatic assays followed by non-selective spectrophotometric detection.7,23,24,26 However, mass spectrometry and enzymatic assays were rarely combined for an ideal matching, difficult to interpret,24 and recently no correlation between ATI amount and inhibition potential was found.26
Because there is only a choice between expensive tandem mass spectrometry (targeting only the ATI proteins) and cheap, non-selective spectrophotometric assay screening (providing only a sum value), this study exploited the potential of high-performance thin-layer chromatography (HPTLC). HPTLC has the advantage of reducing any interfering matrix effects via its planar separation, thus detecting individual saccharides or peptides. The miniaturised, sustainable nano gastrointestinal tract (nanoGIT) HPTLC methods dealing with digestive products of amylolysis27,28 and proteolysis,27 as well as TLC/HPTLC methods for α-amylase29,30 and trypsin31 inhibition, served as motivation. It was hypothesised that changes in enzyme activity caused by ATIs and, thus, in the saccharide or peptide profiles can be measured. It was expected that fewer digestion products would be released due to the inhibition of amylolysis or proteolysis. As samples, ATI-containing flour extracts from wheat (refined and whole), spelt (refined and whole) and einkorn were selected.
Materials and methods
Chemicals and materials
Acarbose (≥95%), D-(+)-glucose (≥99.5%, Glc), maltotriose hydrate (97%, Mal3), 4-aminobenzoic acid (≥99%), D-(+)-maltose monohydrate (≥99%, Mal), trypsin inhibitor from Glycine max (90%), triethylamine (>99.5%), casein from bovine milk (technical grade), aniline (≥99.5%), diphenylamine (≥99%), pyridine (≥99%), ammonium hydroxide solution (25%, p. a.), peptone/tryptone from casein (tryptic digested, suitable for microbiology), α-amylase from hog pancreas (45.5 U mg−1) and trypsin from bovine pancreas (97%; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 BAEE U per mg protein) were purchased from Sigma Aldrich Fluka (Steinheim, Germany). o-Phosphoric acid (85%, purissimum) and dichloromethane (≥99.9%) were from Th. Geyer (Renningen, Germany). 2-Propanol (≥99.8%), calcium chloride dihydrate (≥98%), formic acid (≥98.0%), sodium dihydrogen phosphate monohydrate (≥98%), disodium hydrogen phosphate (≥99%), albumin fraction V (BSA, ≥98%), hydrogen chloride (37%, p. a.), sodium hydroxide (≥99%) and citric acid monohydrate (≥99.5%) were from Carl Roth (Karlsruhe, Germany). Acetic acid (99–100%), acetonitrile (≥99.9%), starch soluble (analytical grade) and potassium iodide (Puriss, p. a. grade) were from Riedel-de Haen (Seelze, Germany). Acetone (≥99.9%) and sodium chloride (≥99%) were purchased from VWR International (Darmstadt, Germany). n-Hexane (≥96%), iodine (p. a.), ninhydrin (analytical grade), HPTLC plates silica gel 60 (NP) and silica gel 60 RP-18 W (both 20 × 10 cm), and 3 kDa Amicon Ultra-0.5 mL centrifugal filters (low-binding regenerated cellulose) were purchased from Millipore (Merck, Darmstadt, Germany). Fluorescamine was supplied by abcr (Karlsruhe, Germany). 2-Butanol (99%) was purchased from Alfa Aesar (Kandel, Germany). Ethanol (≥99.8%) was supplied by Thermo Fisher Scientific (Geel, Belgium). Bi-distilled water was produced using Heraeus Destamat B-18E (Thermo Fisher Scientific, Dreieich, Germany).
000 BAEE U per mg protein) were purchased from Sigma Aldrich Fluka (Steinheim, Germany). o-Phosphoric acid (85%, purissimum) and dichloromethane (≥99.9%) were from Th. Geyer (Renningen, Germany). 2-Propanol (≥99.8%), calcium chloride dihydrate (≥98%), formic acid (≥98.0%), sodium dihydrogen phosphate monohydrate (≥98%), disodium hydrogen phosphate (≥99%), albumin fraction V (BSA, ≥98%), hydrogen chloride (37%, p. a.), sodium hydroxide (≥99%) and citric acid monohydrate (≥99.5%) were from Carl Roth (Karlsruhe, Germany). Acetic acid (99–100%), acetonitrile (≥99.9%), starch soluble (analytical grade) and potassium iodide (Puriss, p. a. grade) were from Riedel-de Haen (Seelze, Germany). Acetone (≥99.9%) and sodium chloride (≥99%) were purchased from VWR International (Darmstadt, Germany). n-Hexane (≥96%), iodine (p. a.), ninhydrin (analytical grade), HPTLC plates silica gel 60 (NP) and silica gel 60 RP-18 W (both 20 × 10 cm), and 3 kDa Amicon Ultra-0.5 mL centrifugal filters (low-binding regenerated cellulose) were purchased from Millipore (Merck, Darmstadt, Germany). Fluorescamine was supplied by abcr (Karlsruhe, Germany). 2-Butanol (99%) was purchased from Alfa Aesar (Kandel, Germany). Ethanol (≥99.8%) was supplied by Thermo Fisher Scientific (Geel, Belgium). Bi-distilled water was produced using Heraeus Destamat B-18E (Thermo Fisher Scientific, Dreieich, Germany).
Five flour samples were purchased from supermarkets. Refined, but not bleached wheat flour type 405 (Lidl Belbake) and organic whole wheat flour (Rewe Bio), both produced by Friesinger Mühle, Bad Wimpfen, Germany, refined, but not bleached spelt flour type 630 (Gut und Günstig, Edeka Zentrale, Hamburg, Germany), organic whole spelt flour (Rewe Bio), produced by BioKorn, Aalen, Germany, and organic whole einkorn flour (Spielberger, Brackenheim, Germany).
Defatting and buffered extraction of ATI
According to a protocol for defatting and ATI extraction,24 flour (1 g) was weighed into a 15 mL reaction tube, defatted twice using 10 mL n-hexane and in-between it was centrifuged at 3000×g for 1 min. Excessive n-hexane was evaporated using a flow of nitrogen (TH 26, HLC BioTech, Bovenden, Germany). Extraction buffer (5 mL, 150 mM sodium chloride in 1.3 mM phosphate buffer pH 7) was added to each defatted flour sample, which was then vortexed for 10 min using a multi-tube holder (Vortex Genie 2, Scientific Industries, New York City, NY, USA). The supernatant was collected after centrifugation at 3000×g for 10 min. The buffer extraction was repeated, both turbid supernatants were combined, centrifuged for 30 s, and the resulting supernatant was filtered through a sterile 0.45 μm cellulose acetate membrane syringe filter (VWR International), resulting in clear ATI-containing flour extracts (i.e., 0.1 g flour per mL buffer). These should be analysed within few weeks, as it was observed that turbidity occurred and increased for longer storage.Removal of saccharides by centrifugal membrane filtration
Each ATI-containing flour extract (50 μL) was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with extraction buffer pre-cooled to 4 °C (to reduce potential protein denaturation during long centrifugation periods in an uncooled centrifuge). The 500 μL aliquot was filtered twice via the 3 kDa Amicon filter by centrifugation at 14
10 with extraction buffer pre-cooled to 4 °C (to reduce potential protein denaturation during long centrifugation periods in an uncooled centrifuge). The 500 μL aliquot was filtered twice via the 3 kDa Amicon filter by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 30 min, whereby the first remaining 48 μL concentrate (remaining inside the filter insert) was filled with 452 μL cooled extraction buffer and filter-centrifuged again. To collect the clear twice-membrane-filtered concentrate (48 μL), the filter was inserted upside down into a clean tube and centrifuged at 1000×g for 2 min. Both saccharide-containing filtrates were discarded.
000×g for 30 min, whereby the first remaining 48 μL concentrate (remaining inside the filter insert) was filled with 452 μL cooled extraction buffer and filter-centrifuged again. To collect the clear twice-membrane-filtered concentrate (48 μL), the filter was inserted upside down into a clean tube and centrifuged at 1000×g for 2 min. Both saccharide-containing filtrates were discarded.
HPTLC-nanoGIT (amylolysis)-FLD/Vis screening of saccharides after in-vial α-amylase inhibition by the flour extracts
The in-vial pre-incubation at 37 °C without rotation for 30 min was performed in 1.5 mL sample vials with 0.2 mL conical inserts. In each insert, 5 μL (2.5 μg, 114 mU) of the α-amylase solution (0.5 mg mL−1, 22.8 U mL−1 in water) was pipetted and mixed with 6 μL either membrane-filtered flour extract (equivalent to 600 μg flour) or bi-distilled water as negative control (NC) or aqueous acarbose solution as positive controls (PC1 10 ng μL−1: 60 ng; PC2 5 ng μL−1: 30 ng).If not stated otherwise, HPTLC instrumentation (CAMAG, Muttenz, Switzerland) was operated under VisionCATS software version 3.1. NP-HPTLC plates were pre-washed by front elution up to 90 mm with acetonitrile, followed by drying for 10 min. The entire pre-incubated enzyme–inhibitor mixture (11 μL per band) was applied onto the NP-HPTLC plate and oversprayed with 2 μL per band soluble starch substrate solution (1 mg mL−1) using the following ATS 4 settings: band length 7 mm, track distance 11 mm, distance from lower and left edge 10 mm, dosage speed 50 nL s−1, filling speed 8 μL s−1, filling vacuum time 0 s, syringe volume 25 μL and the option fill only programmed volume.
As references, the corresponding membrane-filtered flour extracts (6 μL per band) as well as saccharide standards (each 1 mg mL−1) of glucose (Glc, 0.5 μL per band), maltose (Mal, 1 μL per band), and maltotriose (Mal3, 1 μL per band), soluble starch (2 μL per band, 1 mg mL−1), α-amylase (5 μL per band, 0.5 mg mL−1, 22.8 U mL−1) and acarbose (6 μL per band, 10 ng μL−1) were applied.
The plate was instantly wetted with 2.5 mL sodium chloride solution (0.1 M) by piezoelectrical spraying (yellow nozzle, level 6, Derivatizer), whereby only the application zone was wetted because the residual adsorbent area was covered by an NP-HPTLC plate cut to 8.5 cm × 20 cm, layer faced upwards.28 The still-covered plate was incubated for 60 min at 37 °C in a humid plastic box (26.5 × 16 × 10 cm, ABM, Wolframs-Eschenbach, Germany) lined with wet filter paper and filled with 70 mL water. The enzymatic on-surface reaction was stopped by heating the plate (120 °C, 10 min, TLC Plate Heater).
After chamber saturation for 5 min, the plate was developed with 8 mL acetonitrile/water/2-propanol/acetone 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (V/V/V/V) up to 70 mm (Twin-Trough Chamber), and sprayed piezoelectrically as mentioned with the p-aminobenzoic acid reagent (4 mL, 2 g p-aminobenzoic acid in 252 mL glacial acetic acid/water/acetone/o-phosphoric acid 1
0.5 (V/V/V/V) up to 70 mm (Twin-Trough Chamber), and sprayed piezoelectrically as mentioned with the p-aminobenzoic acid reagent (4 mL, 2 g p-aminobenzoic acid in 252 mL glacial acetic acid/water/acetone/o-phosphoric acid 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.04, V/V/V/V), followed by heating (140 °C, 5 min) and detection at 366 nm (TLC Visualizer 2). To perform a reagent sequence on the same plate, the latter was automated dipped (immersion time 2 s, immersion speed 3 cm s−1, Chromatogram Immersion Device 3) in the diphenylamine aniline o-phosphoric acid reagent (diphenylamine/aniline/2-propanol/water/o-phosphoric acid 0.1
0.04, V/V/V/V), followed by heating (140 °C, 5 min) and detection at 366 nm (TLC Visualizer 2). To perform a reagent sequence on the same plate, the latter was automated dipped (immersion time 2 s, immersion speed 3 cm s−1, Chromatogram Immersion Device 3) in the diphenylamine aniline o-phosphoric acid reagent (diphenylamine/aniline/2-propanol/water/o-phosphoric acid 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1
0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 : 1
8 : 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/V/V/V/V), followed by heating at 120 °C for 5–10 min, documentation under white light illumination (TLC Visualizer 2), and densitometric detection at 370 nm (absorbance measurement, deuterium/tungsten lamp, slit 5.0 mm × 0.2 mm, TLC Scanner 4).
1, w/V/V/V/V), followed by heating at 120 °C for 5–10 min, documentation under white light illumination (TLC Visualizer 2), and densitometric detection at 370 nm (absorbance measurement, deuterium/tungsten lamp, slit 5.0 mm × 0.2 mm, TLC Scanner 4).
Calculation of the relative α-amylase inhibition
After densitometric measurement, the signal height intensity (Int) of an individual saccharide in the flour extract was subtracted from the signal in the corresponding amylolysed flour extract and divided by the signal of the NC (bi-distilled water; amylolysis without inhibitor). This corrected relative signal was subtracted from 1 and multiplied by 100 for calculation of the percentage inhibition as follows:RP-HPTLC-FLD screening of peptides after in-vial trypsin inhibition by the flour extracts
The pre-incubation and proteolysis were performed in 1.5 mL sample vials with 0.2 mL conical inserts. An aliquot of 200 μL trypsin solution (0.33 mg mL−1, 3300 BAEE U mL−1 in 5 M calcium chloride monohydrate solution, pH 4)24 was deacidified by adding 2 μL sodium hydroxide solution (1 M) and then adjusted to pH 8 before use. For the enzyme–inhibitor reaction, 3 μL (1 μg) trypsin solution (0.33 mg mL−1, pH 8) was mixed in each vial with either 7 or 17 μL unfiltered flour extract (equivalent to 0.7 or 1.7 mg flour, 100 mg mL−1) or 3 μL (0.9 μg) trypsin inhibitor solution (0.3 mg mL−1) used as PC or 17 μL bi-distilled water used as NC. As references, 17 μL of the corresponding unfiltered flour extract was filled in vials (without trypsin, no proteolysis). All vials were filled to 20 μL with bi-distilled water and incubated at 37 °C without rotation for 30 min. For the enzyme–substrate reaction, 100 μL (100 μg) of casein substrate solution (1 mg mL−1) was added, followed by further incubation for 120 min. The final assay volume was 120 μL. The enzyme-to-substrate ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100.
100.
The HPTLC RP-18 W plate was dipped (immersion time 8 s, immersion speed 3 cm s−1, Chromatogram Immersion Device 3) in phosphate–citrate buffer (6 g per L citric acid and 10 g per L di-sodium hydrogen phosphate, adjusted to pH 12 by sodium hydroxide powder). After plate drying (5 min, 100 °C), an aliquot of 10 μL of each trypsin assay solution (resulting in 80 ng trypsin, ATI-containing extract from 60 or 140 μg flour, 80 ng trypsin inhibitor, and 8 μg casein per band) was applied (with the ATS 4 settings as mentioned, but band length 8 mm and distance from left edge 9 mm) on the buffered plate, which was developed with 8 mL 2-propanol/formic acid/acetonitrile 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (V/V/V) up to 60 mm (Twin-Trough Chamber).
5 (V/V/V) up to 60 mm (Twin-Trough Chamber).
For derivatisation, the plate was dipped (immersion time 2 s, immersion speed 3 cm s−1, Chromatogram Immersion Device 3) in fluorescamine reagent (10% w/V in acetone), followed by plate drying (1 min). For fluorescence enhancement/stabilisation, the plate was dipped subsequently in a triethylamine solution (10% V/V in dichloromethane), followed by plate drying (5 min at 20 °C, room temperature). The fluorescent peptide signals were detected at FLD 366 nm in enhanced mode (TLC Visualizer 2). Alternatively, the plate was derivatised with the ninhydrin reagent (ninhydrin/ethanol/glacial acetic acid 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 92
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, w/V/V).
8, w/V/V).
Results and discussion
After flour defatting, the ATIs were extracted with a sodium chloride-containing phosphate buffer according to Call et al.24 and the inhibitory potential towards α-amylase and trypsin was evaluated. Two approaches were pursued for the evaluation of the flour extracts. The first was the HPTLC α-amylase inhibition assay, in which inhibition by ATIs was detected at the end of analysis after separation. However, this approach was critical, as it probably caused the denaturation of ATI proteins by organic solvents during chromatographic separation, and thus loss of the ability to inhibit α-amylase in the final assay detection. The second approach was the HPTLC-nanoGIT (amylolysis)-FLD/Vis method. Amylolysis was performed in the starting zone prior to chromatographic separation. This approach seemed promising as a combined in-vial/on-surface incubation method. Both approaches were discussed in detail as follows.HPTLC-α-amylase inhibition assay (caused denaturation of ATI)
Initial experiments followed the HPTLC-α-amylase inhibition assay.29–31 The assay was performed after separation to detect ATIs, here α-amylase inhibitors, in the flour extracts. The unfiltered ATI extracts were applied onto an NP-HPTLC plate and separated with 2-butanol/pyridine/ammonia/water 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (V/V/V/V) up to 65 mm. The optimised mobile phase mixture (Fig. S1†) differed in the solvent ratios compared to the peptide separations by Pasilis et al.32 (39
6 (V/V/V/V) up to 65 mm. The optimised mobile phase mixture (Fig. S1†) differed in the solvent ratios compared to the peptide separations by Pasilis et al.32 (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26, V/V/V/V) and Biller et al.33 (39
26, V/V/V/V) and Biller et al.33 (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31, V/V/V/V). The α-amylase inhibition assay was performed via piezoelectrical spraying.34 First, the amylase solution, followed by 30 min incubation, then the soluble starch substrate solution, followed by 20 min incubation, and finally, for the detection, the derivatisation reagent iodine/potassium iodide solution30 was sprayed, or iodine vapour29 was used (Fig. S2†). The α-amylase-inhibiting zones should be visible as deep-purple zones on a less purple background. However, no purple inhibitor zones were revealed. Only an unstable white zone (vanished with longer incubation time) was observed and characterised as a saccharide via selective derivatisation with the diphenylamine aniline reagent (Fig. S2†). However, ATIs are proteins, and selective derivatisation with the ninhydrin reagent showed several zones in the separated ATI extract, these zones did not show any inhibition response, as mentioned. The assumption was that the ATIs were denatured during separation with the highly organic mobile phase and were no longer active. Therefore, purple inhibitor zones were not observed.
31, V/V/V/V). The α-amylase inhibition assay was performed via piezoelectrical spraying.34 First, the amylase solution, followed by 30 min incubation, then the soluble starch substrate solution, followed by 20 min incubation, and finally, for the detection, the derivatisation reagent iodine/potassium iodide solution30 was sprayed, or iodine vapour29 was used (Fig. S2†). The α-amylase-inhibiting zones should be visible as deep-purple zones on a less purple background. However, no purple inhibitor zones were revealed. Only an unstable white zone (vanished with longer incubation time) was observed and characterised as a saccharide via selective derivatisation with the diphenylamine aniline reagent (Fig. S2†). However, ATIs are proteins, and selective derivatisation with the ninhydrin reagent showed several zones in the separated ATI extract, these zones did not show any inhibition response, as mentioned. The assumption was that the ATIs were denatured during separation with the highly organic mobile phase and were no longer active. Therefore, purple inhibitor zones were not observed.
Removal of initial saccharides in flour by membrane filtration and NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis method development
The next experiments studied the on-surface metabolization in the start zone (nanoGIT), followed by the analysis of the released metabolization products on the same surface, as developed by Morlock et al.27 As predominant amylolysis products, the released saccharides glucose (Glc), maltose (Mal) and maltotriose (Mal3) were separated, previously shown to be detected quantitatively.28 Nevertheless, the five flour extract samples contained a high amount of native saccharides (Fig. S3†), which had to be removed before the analysis of the reduction in saccharide release. Therefore, each flour extract was cleaned via a 3 kDa centrifugal membrane filter. This ensured no loss of ATIs (mainly > 15 kDa), but the passage of saccharides (≤0.5 kDa), much smaller than the membrane cut-off. Simultaneous with centrifugal membrane filtration, the ATI extract was 10-fold concentrated (from 500 μL to 48 μL). To avoid concentration of the extract and subsequent protein precipitation, the sample was previously diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to maintain a consistent concentration. Two 30 min centrifugation steps were sufficient to remove all native saccharides (Fig. S3†).
10 to maintain a consistent concentration. Two 30 min centrifugation steps were sufficient to remove all native saccharides (Fig. S3†).
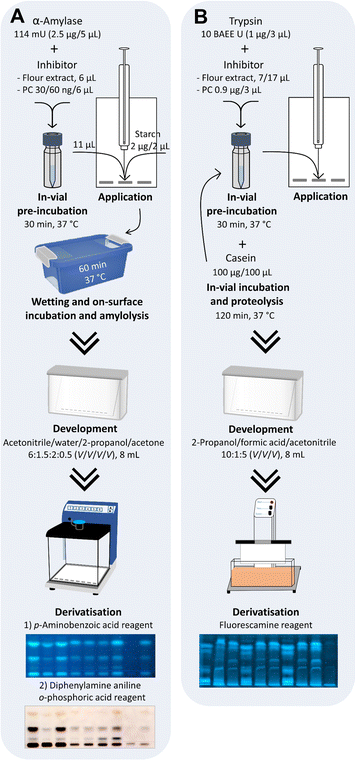
First, the inhibition of α-amylase by ATI and the substrate reaction with soluble starch were performed in the same start zone. The design of experiment included two separate on-surface incubations: a 30 min pre-incubation of the enzyme with ATI-containing flour extract (to guarantee proper interaction), followed by a 60 min substrate incubation.24,27 The known α-amylase inhibitor acarbose was used as PC for proof of the inhibition of amylolysis and bi-distilled water as NC. However, the approach to perform all steps on the same HPTLC surface failed because of the two incubation steps and the associated diffusion of the inhibitor, enzyme, and substrate on the start zone (Fig. S4†). To reduce the number of incubations on the HPTLC plate, a 30 min pre-incubation was performed in a sample vial (in-vial pre-incubation), whereas the following enzyme–substrate reaction remained on the NP-HPTLC plate surface (on-surface incubation/metabolization, Fig. 1A).
The in-vial pre-incubation volume was selected as small as possible. Enzyme–inhibitor mass ratios (E/I) of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 124
100 and 124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 25 μL were evaluated, whereof only 11 μL was applied onto the plate. The enzyme–substrate mass ratio (E/S) for on-surface incubation was set to 1
100 in 25 μL were evaluated, whereof only 11 μL was applied onto the plate. The enzyme–substrate mass ratio (E/S) for on-surface incubation was set to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Co-application of the NC (bi-distilled water) proved that the α-amylase activity was retained after pre-incubation, whereas co-application of the PC (acarbose) inhibited α-amylase (Fig. S5†). The combination of in-vial pre-incubation (for enzyme–inhibitor reaction) and on-surface incubation (for enzyme–substrate reaction) was successful. The nanoGIT chromatograms showed total α-amylase inhibition at a higher E/I of 25
2. Co-application of the NC (bi-distilled water) proved that the α-amylase activity was retained after pre-incubation, whereas co-application of the PC (acarbose) inhibited α-amylase (Fig. S5†). The combination of in-vial pre-incubation (for enzyme–inhibitor reaction) and on-surface incubation (for enzyme–substrate reaction) was successful. The nanoGIT chromatograms showed total α-amylase inhibition at a higher E/I of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 124
100 and 124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. Total inhibition was not of interest, as the signals should be in a dynamic response range. In addition, no signals could not exclude a failed enzymatic reaction. Consequently, the E/I was optimised to 83
100. Total inhibition was not of interest, as the signals should be in a dynamic response range. In addition, no signals could not exclude a failed enzymatic reaction. Consequently, the E/I was optimised to 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the total pre-incubation volume was reduced to 11 μL and entirely applied onto the HPTLC plate. The E/S (1
1, and the total pre-incubation volume was reduced to 11 μL and entirely applied onto the HPTLC plate. The E/S (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) was changed to 2.5
2) was changed to 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Thus, for the PC acarbose, the favoured moderate inhibition was detected, being strongest for Mal3, weakest for Mal, and absent for Glc (Fig. S5†). Derivatisation with the p-aminobenzoic acid reagent detected monosaccharides more sensitively than polysaccharides at FLD 366 nm.28,34 To improve polysaccharide detection, a second derivatisation with the diphenylamine aniline o-phosphoric acid reagent, followed by detection under white light illumination, was performed as a reagent sequence on the same plate. The final amounts and ratios used for in-vial incubation were 2.5 μg α-amylase (114 mU) and ATI-containing extract from 600 μg flour or PC acarbose (60 ng as PC1, 30 ng as PC2). For subsequent on-surface amylolysis, 2 μg soluble starch substrate was applied. Calculated for the PC, this corresponded to an E/I of 42
2. Thus, for the PC acarbose, the favoured moderate inhibition was detected, being strongest for Mal3, weakest for Mal, and absent for Glc (Fig. S5†). Derivatisation with the p-aminobenzoic acid reagent detected monosaccharides more sensitively than polysaccharides at FLD 366 nm.28,34 To improve polysaccharide detection, a second derivatisation with the diphenylamine aniline o-phosphoric acid reagent, followed by detection under white light illumination, was performed as a reagent sequence on the same plate. The final amounts and ratios used for in-vial incubation were 2.5 μg α-amylase (114 mU) and ATI-containing extract from 600 μg flour or PC acarbose (60 ng as PC1, 30 ng as PC2). For subsequent on-surface amylolysis, 2 μg soluble starch substrate was applied. Calculated for the PC, this corresponded to an E/I of 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–83
1–83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and an E/S of 2.5
1 and an E/S of 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Unfortunately, these conditions could not be compared with published studies, since either α-amylase activity was not reported, a common inaccuracy in studies on enzymatic activities, or other substrates (synthesised chromogenic starch-like substances) were used.
2. Unfortunately, these conditions could not be compared with published studies, since either α-amylase activity was not reported, a common inaccuracy in studies on enzymatic activities, or other substrates (synthesised chromogenic starch-like substances) were used.
NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis screening of α-amylase inhibitor activity of flours
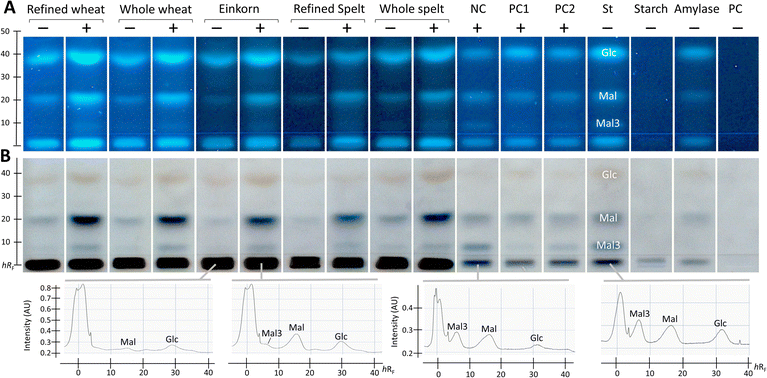
After successful demonstration of α-amylase inhibition using the PC acarbose, the developed NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis method was used for screening and comparing the five membrane-filtered ATI extracts from flours of wheat (refined and whole), einkorn, and spelt (refined and whole). The resulting nanoGIT chromatograms after derivatisation with the p-aminobenzoic acid reagent (Fig. 2A), followed by the diphenylamine aniline o-phosphoric acid reagent (Fig. 2B), showed amylolytic products. The absorbance of each saccharide was measured using densitometry after the second reagent, which detected the Mal3 zones more sensitively (Fig. 2B). A great advantage of the developed NP-HPTLC-nanoGIT-FLD/Vis method was the selective detection of individual saccharides. This allowed to investigate the correlation between the inhibition of amylolysis and the release of individual saccharides.In contrast, current in vial assays detect only the overall saccharide release as a sum value, which can be influenced by matrix effects as discussed in the following. A disproportionate increase in Mal was revealed for the flour extracts. This was explained by the long-chain polysaccharides (visible at the start zone), which were retained due to membrane filtration in the clear extracts and were co-metabolised by α-amylase in addition to the starch substrate. A cheaper but not time-saving alternative would be dialysis, which was investigated in a separate study.35 Such individual increases in released saccharides would not be detected in spectrophotometric (sum value) assays. When subtracting the respective not amylolysed flour extract, such unexpected co-metabolization would result in underestimated relative inhibition or negative absorbance (as the inhibitory reaction released a higher amount of detected product).
As expected, and as a successful proof, the NC (bi-distilled water) revealed three amylolysis products. However, saccharide impurities (Glc and Mal) in the purchased enzyme, which was co-applied as α-amylase reference, were detected. Such impurities did not influence the calculation of the relative inhibition but were still problematic regarding the detector limits, which can be exceeded more quickly and falsify the results of the inhibition assay. For this reason, the α-amylase impurities were not subtracted, as they were present in both the flour extract and the NC.
Additionally, the flour extracts contained saccharide impurities despite prior purification (membrane filtration) of the extracts. Hence, the absorbance of individual saccharides in the amylolysed flour extract was corrected by subtracting the absorbance of the flour extract and then calculating the relative α-amylase inhibition with reference to the NC (Fig. 3 and Table S1†).
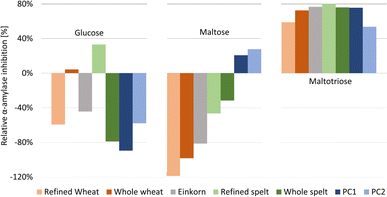 | ||
| Fig. 3 Relative α-amylase inhibition by the flour extracts and positive control acarbose at two different amounts (PC1 60 ng per band, PC2 30 ng per band) calculated for the three saccharides released (full dataset in Table S1†). | ||
The relative inhibition was only single determined as first proof of the quantification of α-amylase inhibition and for comparison with other methods.
For Glc and Mal, the relative inhibition values were mainly negative for the flour extracts, that is, more Glc and Mal products were released and detected in the amylolysed flour extracts than in the NC without any inhibitor. As mentioned, the higher absorbance values for Mal were explained by the co-metabolization of the matrix during amylolysis, which could also be the case for Glc. Nevertheless, the whole wheat (4%) and refined spelt extract (33%) showed positive inhibition values regarding Glc (Fig. 3), where whole wheat released more Glc than refined spelt, resulting in lower inhibition. No trend regarding their whole grain or refined counterparts was observed. Thus, the evaluation of the release of Mal and Glc for the calculation of the relative inhibition was classified as critical without any further purification of the extracts. However, the reduction in Mal3 release led to positive relative inhibition values for all amylolysed flour extracts, with the strongest inhibition observed for refined spelt and the lowest inhibition observed for refined wheat.
According to Geisslitz et al.,19 einkorn contained the lowest amount of ATIs compared to wheat, spelt, and emmer, resulting in the lowest α-amylase inhibition, as confirmed by Simonetti et al.36 Our results did not support this hypothesis. The flour extract of einkorn showed no significantly lower inhibition than all other investigated flours, keeping in mind the possible presence of other α-amylase inhibitors, the affinity of the enzyme regarding its origin (human or animal),26 the different analytical approaches, and the natural variety of the flour composition. Other studies on α-amylase inhibition26,36 have also reported moderate to high standard deviations. Calculating the overall inhibition (Table S1†) as a simulation of values obtained by spectrophotometric assays, refined spelt (15%) showed the strongest inhibition, and refined wheat the lowest (−40%). Interestingly, the inhibition was only positive for refined and whole spelt, whereas the other extracts resulted in negative inhibition (not evaluable in a spectrophotometric assay). Repeatedly, the investigation of the Mal3 release evaluated the same trend as the overall inhibition, but with consistently positive inhibition values and thus more reliable results. In conclusion, the influence of the flour matrix plays an important but often neglected role in determining the inhibitory potential, which was uncovered with the developed NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis method. The comparison of refined and whole grain flours regarding Mal3-release showed contrary results; whole wheat showed a higher and whole spelt a lower inhibition than its corresponding refined types. ATIs are predominantly present in the starchy endosperm of the grain,37,38 and to a lesser extent in the outer aleurone layer. The latter is removed in refined flour together with the cereal bran. In contrast, whole grain flour contains an aleurone layer and other components that influence the weight distribution. Consequently, the ATI amount was expected to be enriched in refined flour.
Saccharide release should be reduced by increasing the amount of the α-amylase inhibitor. However, the addition of more PC (PC1 60 ng acarbose > PC2 30 ng acarbose) led to unexpected increases in Glc and Mal release (Fig. 2, Table S1†). Especially, Glc was released in a higher amount than in the NC, resulting in negative inhibition (Fig. 3). The acarbose reference standard showed no impurities, but acarbose is a pseudo-tetrasaccharide that may fit into the active centre of α-amylase and become partially digested, which explains the additional and thus higher Glc release. In conclusion, acarbose may not be the first choice as PC, which was further investigated in another study.35 All in all, only a decrease in Mal3 was observed with increasing acarbose amount, confirming the reduction in the Mal3 release as a suitable mechanism for the screening of the inhibitory potential of ATI in flours.
Comparing the overall inhibition (Table S1†) of PC1 and PC2, they revealed the same inhibitory potential (25% and 26%, respectively, despite differing amounts) as consequence of the higher Glc release with higher acarbose amounts masking the lower Mal3 release. Hence, in a spectrophotometric assay, doubling the amount of PC would result in an underestimated inhibition for acarbose. This highlights the better reliability of the developed NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis method in contrast to the spectrophotometric (sum value) assays.
Method development to measure the trypsin inhibition by ATI
ATIs are bi-functional inhibitors for α-amylase and trypsin. After successful inhibition of α-amylase, it was necessary to also prove trypsin inhibition. First, the nanoGIT on-surface incubation/metabolization was attempted. The zone diffusion on the NP-HPTLC plate was expected to be less (compared to the α-amylase inhibition study) because of the higher molecular weights of trypsin (24 kDa) and casein (25 kDa). First, the enzyme–substrate reaction was optimised with trypsin and tryptone (tryptic-digested casein) by evaluating different E/S from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 to 1
100 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S6†). The in-vial proteolytic products were separated with 2-butanol/ammonia/pyridine/water 19
1 (Fig. S6†). The in-vial proteolytic products were separated with 2-butanol/ammonia/pyridine/water 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 (V/V/V/V) and detected under white light illumination after derivatisation with the ninhydrin reagent. The resulting NP-HPTLC-nanoGIT (proteolysis)-Vis chromatogram showed that the separation was increasingly impaired (i.e., peptides were pushed upwards) the more trypsin was applied on the plate. The interferences were assumed to be caused by the associated high-molar (5 M) calcium chloride solution. Only the lowest E/S of 2
13 (V/V/V/V) and detected under white light illumination after derivatisation with the ninhydrin reagent. The resulting NP-HPTLC-nanoGIT (proteolysis)-Vis chromatogram showed that the separation was increasingly impaired (i.e., peptides were pushed upwards) the more trypsin was applied on the plate. The interferences were assumed to be caused by the associated high-molar (5 M) calcium chloride solution. Only the lowest E/S of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 showed no interferences but also no peptide increase, so the amounts were considered too low.
100 showed no interferences but also no peptide increase, so the amounts were considered too low.
Next, the stationary phase was switched from polar NP to more apolar RP-18 W plates. The water-wettable RP-18 W adsorbent is more polar than RP-18 but rather apolar in contrast to NP.39 Due to its acidic pH (pH ≈ 4), the RP-18 W plate was buffered with citrate–phosphate buffer (pH 12) to neutral pH. A mobile phase for peptide separation was developed, resulting in 2-propanol/formic acid/acetonitrile 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (V/V/V). Peptide separation worked better on buffered versus non-buffered plates, as observed in the RP-HPTLC-nanoGIT (proteolysis)-Vis chromatograms (Fig. S7†). As expected, the acidic mobile phase caused an acid gradient with the lowest pH at the bottom and the highest pH at the top, which was confirmed via planar pH electrode measurement and by the background colour gradient formed after derivatisation with the ninhydrin reagent. By replacing formic acid in the mobile phase with ammonia (25%), no gradient in the coloured background was observed, but the separation was too low in elution strength and would require further optimisation. Proteolysis on RP-18 W plates still showed no increase in peptide zones (Fig. S7†); as tryptone is a tryptically pre-digested casein, further proteolytic products could be marginal and thus undetectable. Hence, the in-vial proteolysis was performed with casein to ensure sufficient enzymatic degradation. The fluorescamine derivatisation reagent was tested as an alternative to the ninhydrin reagent (Fig. 1B). Proteolysis products were detected more sensitively as blue fluorescent zones in the RP-HPTLC chromatogram at FLD 366 nm. An incubation time of 2 h with an E/S of 1
5 (V/V/V). Peptide separation worked better on buffered versus non-buffered plates, as observed in the RP-HPTLC-nanoGIT (proteolysis)-Vis chromatograms (Fig. S7†). As expected, the acidic mobile phase caused an acid gradient with the lowest pH at the bottom and the highest pH at the top, which was confirmed via planar pH electrode measurement and by the background colour gradient formed after derivatisation with the ninhydrin reagent. By replacing formic acid in the mobile phase with ammonia (25%), no gradient in the coloured background was observed, but the separation was too low in elution strength and would require further optimisation. Proteolysis on RP-18 W plates still showed no increase in peptide zones (Fig. S7†); as tryptone is a tryptically pre-digested casein, further proteolytic products could be marginal and thus undetectable. Hence, the in-vial proteolysis was performed with casein to ensure sufficient enzymatic degradation. The fluorescamine derivatisation reagent was tested as an alternative to the ninhydrin reagent (Fig. 1B). Proteolysis products were detected more sensitively as blue fluorescent zones in the RP-HPTLC chromatogram at FLD 366 nm. An incubation time of 2 h with an E/S of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and an application volume of 10 μL was considered sufficient (Fig. S8†). Again, a higher E/S interfered with the separation, caused by trypsin in its high-molar calcium chloride solution. To prove the inhibition of proteolysis, the trypsin inhibitor from Glycine max was used as PC and bi-distilled water as NC. Different E/I and pre-incubation periods were evaluated for the PC and the flour extract of refined wheat (Fig. S9†). For the PC, an E/I of 11
100 and an application volume of 10 μL was considered sufficient (Fig. S8†). Again, a higher E/S interfered with the separation, caused by trypsin in its high-molar calcium chloride solution. To prove the inhibition of proteolysis, the trypsin inhibitor from Glycine max was used as PC and bi-distilled water as NC. Different E/I and pre-incubation periods were evaluated for the PC and the flour extract of refined wheat (Fig. S9†). For the PC, an E/I of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 showed already a complete inhibition after 15 min. A lower E/I was preferred to prevent false-positive results if the enzyme assay did not work. A pre-incubation time of 30 min was chosen for the enzyme–inhibitor reaction. For wheat, the best results were achieved by adding 7 μL (60 μg flour) or 17 μL (140 μg flour) of the flour extract (Fig. S9†), whereas higher volumes (27 μL) resulted in excessive inhibition, potentially leading to false-positive results that are more challenging to recognise.
10 showed already a complete inhibition after 15 min. A lower E/I was preferred to prevent false-positive results if the enzyme assay did not work. A pre-incubation time of 30 min was chosen for the enzyme–inhibitor reaction. For wheat, the best results were achieved by adding 7 μL (60 μg flour) or 17 μL (140 μg flour) of the flour extract (Fig. S9†), whereas higher volumes (27 μL) resulted in excessive inhibition, potentially leading to false-positive results that are more challenging to recognise.
RP-HPTLC screening of trypsin inhibitor activity of flours
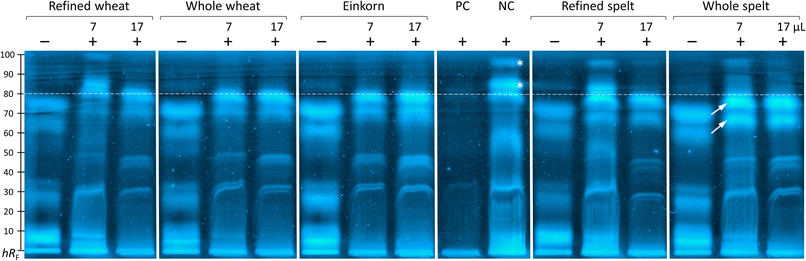
The inhibition of the proteolysis was effective for all samples. The PC (Glycine max trypsin inhibitor) showed strong inhibition, whereas the NC (bi-distilled water) did not (Fig. 4). Screening of the five unfiltered flour extracts of wheat (refined and whole), einkorn, and spelt (refined and whole) showed satisfactory trypsin inhibition in the chromatogram in comparison to the NC and their non-proteolyzed counterparts. In particular, the zones with a retardation factor times 100 (hRF) ≥ 80 (Fig. 4, marked *) were well-suited for evaluating the inhibition of the proteolysis. Zones in the proteolyzed unfiltered flour extracts containing trypsin in 5 M calcium chloride solution (Fig. 4, marked +) were eluted and pushed together towards a higher hRF (Fig. 4, marked by arrows) owing to the comparatively higher elution power caused by the calcium chloride load on the start zone. Only the extracts of 60 μg refined flours of wheat and spelt suggested at hRF 81 and 91 peptide zone responses equivalent to the NC. Indicating, that nearly all extracts inhibited zones ≥ hRF 80 and showed trypsin inhibitory properties. With comparable intensity, the extracts from the flours of einkorn and whole wheat inhibited trypsin most strongly.These results were consistent with the findings of Simonetti et al.,36 which evaluated the highest trypsin inhibition for einkorn samples; however, contrary to the results of Geisslitz et al.,19 where einkorn had the lowest ATI amount. Unfortunately, the assay conditions were not comparable to those of other studies24,36 since they used chromogenic substrates, which have a different affinity to trypsin compared to the used casein.
Qualitative screening and visual evaluation of the chromatogram was performed. The current separation was not suited for densitometric measurement and calculation of the relative trypsin inhibition. In future, firstly, the elution strength of the mobile phase could be reduced to better separate the upper chromatogram part, where clear differences were observed. Secondly, the salt load could be reduced to overcome the interference caused by high trypsin-associated calcium chloride load on the start zone. Thirdly, the transfer from in-vial to on-surface incubation with shorter incubation periods (to reduce the diffusion of reactants in the start zone) could be tested.
Conclusions
Two screening methods were successfully developed to evaluate the α-amylase and trypsin inhibitory properties of extracts from five different flours. Analysis of the α-amylase inhibitory potential consisted of in-vial pre-incubation, followed by NP-HPTLC-nanoGIT (amylolysis)-FLD/Vis, whereas analysis of the trypsin inhibition used in-vial pre-incubation and proteolysis, followed by RP-HPTLC-FLD. In contrast to spectrophotometric assays, the developed HPTLC screening methods detected individual saccharides or peptides, and the inhibitory potential of flours was determined via the reduction of released saccharides during amylolysis or proteolysis, respectively. A strong influence of the flour matrix on the assay results (individual saccharides) was observed, explained by an increased amylolysis of further polysaccharides. The visualization of such matrix influence made HPTLC analysis a more reliable and information-rich method than currently used spectrophotometric sum value assays and helped to understand the problems associated with spectrophotometric assays (matrix effects depending on the chromogenic substrate used). HPTLC screening was not only more reliable and sustainable than conventional in-vial assays but also than liquid column chromatography analysis targeting only the ATI proteins. In comparison, up to 17 samples can be analysed all at once in HPTLC, consuming far less materials. Organic solvents were needed in the HPTLC workflow for conclusive results, but there is potential to use solvents of renewable sources for improvements regarding green chemistry.In the future, the workflow of both methods could be optimised using a full on-surface digestion strategy. Further purification of the flour extracts via fractionation and ensuring their stability over time could improve the accuracy. Additional effect-directed detection on the planar surface could provide new information about further non-proteinogenic inhibitors of the flour extracts. The results of both the α-amylase and trypsin inhibition assays need to be validated and verified by an independent method. Additionally, an investigation of consumer products such as bread would reveal the remaining activity of ATIs after exposure to high temperatures and provide more comprehensive diet information.
Author contributions
Isabel Müller: conceptualization, methodology, experimental analysis, data analysis, writing – original draft. Loredana Bosa: experimental analysis, data analysis. Bianca Schmid: experimental analysis, data analysis. Gertrud E. Morlock: conceptualization, methodology, supervision, writing – original draft, writing – review and editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this study.Acknowledgements
Instrumentation was partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – INST 162/536-1 FUGG and INST 162/471-1 FUGG.References
- A. Sapone, J. C. Bai, C. Ciacci, J. Dolinsek, P. H. R. Green, M. Hadjivassiliou, K. Kaukinen, K. Rostami, D. S. Sanders, M. Schumann, R. Ullrich, D. Villalta, U. Volta, C. Catassi and A. Fasano, Spectrum of gluten-related disorders: consensus on new nomenclature and classification, BMC Med., 2012, 10, 13 CrossRef PubMed.
- S. Geisslitz, P. Shewry, F. Brouns, A. H. P. America, G. P. I. Caio, M. Daly, S. D’Amico, R. de Giorgio, L. Gilissen, H. Grausgruber, X. Huang, D. Jonkers, D. Keszthelyi, C. Larré, S. Masci, C. Mills, M. S. Møller, M. E. Sorrells, B. Svensson, V. F. Zevallos and P. L. Weegels, Wheat ATIs: Characteristics and Role in Human Disease, Front. Nutr., 2021, 8, 667370 CrossRef PubMed.
- F. I. Cárdenas-Torres, F. Cabrera-Chávez, O. G. Figueroa-Salcido and N. Ontiveros, Non-Celiac Gluten Sensitivity: An Update, Medicina, 2021, 57(6), 526 CrossRef PubMed.
- C. Catassi, J. C. Bai, B. Bonaz, G. Bouma, A. Calabrò, A. Carroccio, G. Castillejo, C. Ciacci, F. Cristofori, J. Dolinsek, R. Francavilla, L. Elli, P. Green, W. Holtmeier, P. Koehler, S. Koletzko, C. Meinhold, D. Sanders, M. Schumann, D. Schuppan, R. Ullrich, A. Vécsei, U. Volta, V. Zevallos, A. Sapone and A. Fasano, Non-Celiac Gluten sensitivity: the new frontier of gluten related disorders, Nutrients, 2013, 5, 3839–3853 CrossRef PubMed.
- M. G. Mumolo, F. Rettura, S. Melissari, F. Costa, A. Ricchiuti, L. Ceccarelli, N. de Bortoli, S. Marchi and M. Bellini, Is Gluten the Only Culprit for Non-Celiac Gluten/Wheat Sensitivity?, Nutrients, 2020, 12, 3785 CrossRef CAS PubMed.
- M. Cuccioloni, M. Mozzicafreddo, I. Ali, L. Bonfili, V. Cecarini, A. M. Eleuteri and M. Angeletti, Interaction between wheat alpha-amylase/trypsin bi-functional inhibitor and mammalian digestive enzymes: Kinetic, equilibrium and structural characterization of binding, Food Chem., 2016, 213, 571–578 CrossRef CAS PubMed.
- S. Priya, S. Kumar, N. Kaur and A. K. Gupta, Specificity of α-amylase and trypsin inhibitor proteins in wheat against insect pests, N. Z. J. Crop Hortic. Sci., 2013, 41, 49–56 CrossRef CAS.
- L. Shan, Ø. Molberg, I. Parrot, F. Hausch, F. Filiz, G. M. Gray, L. M. Sollid and C. Khosla, Structural basis for gluten intolerance in celiac sprue, Science, 2002, 297, 2275–2279 CrossRef CAS PubMed.
- Y. Junker, S. Zeissig, S.-J. Kim, D. Barisani, H. Wieser, D. A. Leffler, V. Zevallos, T. A. Libermann, S. Dillon, T. L. Freitag, C. P. Kelly and D. Schuppan, Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4, J. Exp. Med., 2012, 209, 2395–2408 CrossRef CAS PubMed.
- V. F. Zevallos, V. Raker, S. Tenzer, C. Jimenez-Calvente, M. Ashfaq-Khan, N. Rüssel, G. Pickert, H. Schild, K. Steinbrink and D. Schuppan, Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells, Gastroenterology, 2017, 152, 1100–1113.e12 CrossRef CAS PubMed.
- S. B. Altenbach, W. H. Vensel and F. M. Dupont, The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86, BMC Res. Notes, 2011, 4, 242 CrossRef CAS PubMed.
- F. M. Dupont, W. H. Vensel, C. K. Tanaka, W. J. Hurkman and S. B. Altenbach, Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry, Proteome Sci., 2011, 9, 10 CrossRef CAS PubMed.
- M. Kostekli and S. Karakaya, Protease inhibitors in various flours and breads: Effect of fermentation, baking and in vitro digestion on trypsin and chymotrypsin inhibitory activities, Food Chem., 2017, 224, 62–68 CrossRef CAS PubMed.
- P. E. Granum, Studies on α-amylase inhibitors in foods, Food Chem., 1979, 4, 173–178 CrossRef CAS.
- P. Gélinas, C. McKinnon and F. Gagnon, Inhibitory activity towards human α-amylase in cereal foods, LWT–Food Sci. Technol., 2018, 93, 268–273 CrossRef.
- U. Bose, K. Byrne, C. A. Howitt and M. L. Colgrave, Targeted proteomics to monitor the extraction efficiency and levels of barley α-amylase trypsin inhibitors that are implicated in non-coeliac gluten sensitivity, J. Chromatogr. A, 2019, 1600, 55–64 CrossRef CAS PubMed.
- U. Bose, A. Juhász, J. A. Broadbent, K. Byrne, C. A. Howitt and M. L. Colgrave, Identification and Quantitation of Amylase Trypsin Inhibitors Across Cultivars Representing the Diversity of Bread Wheat, J. Proteome Res., 2020, 19, 2136–2148 CrossRef CAS PubMed.
- S. Geisslitz, C. F. H. Longin, P. Koehler and K. A. Scherf, Comparative quantitative LC-MS/MS analysis of 13 amylase/trypsin inhibitors in ancient and modern Triticum species, Sci. Rep., 2020, 10, 14570 CrossRef CAS PubMed.
- S. Geisslitz, C. Ludwig, K. A. Scherf and P. Koehler, Targeted LC-MS/MS Reveals Similar Contents of α-Amylase/Trypsin-Inhibitors as Putative Triggers of Nonceliac Gluten Sensitivity in All Wheat Species except Einkorn, J. Agric. Food Chem., 2018, 66, 12395–12403 CrossRef CAS PubMed.
- S. Tchewonpi Sagu, E. Landgräber, M. Rackiewicz, G. Huschek and H. Rawel, Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars, Molecules, 2020, 25, 5982 CrossRef PubMed.
- S. Tchewonpi Sagu, L. Zimmermann, E. Landgräber, T. Homann, G. Huschek, H. Özpinar, F. J. Schweigert and H. M. Rawel, Comprehensive Characterization and Relative Quantification of α-Amylase/Trypsin Inhibitors from Wheat Cultivars by Targeted HPLC-MS/MS, Foods, 2020, 9(10), 1448 CrossRef PubMed.
- B. Prandi, A. Faccini, T. Tedeschi, G. Galaverna and S. Sforza, LC/MS analysis of proteolytic peptides in wheat extracts for determining the content of the allergen amylase/trypsin inhibitor CM3: influence of growing area and variety, Food Chem., 2013, 140, 141–146 CrossRef CAS PubMed.
- S. Tchewonpi Sagu, G. Huschek, J. Bönick, T. Homann and H. M. Rawel, A New Approach of Extraction of α-Amylase/trypsin Inhibitors from Wheat (Triticum aestivum L.), Based on Optimization Using Plackett-Burman and Box-Behnken Designs, Molecules, 2019, 24(19), 3589 CrossRef PubMed.
- L. Call, E. V. Reiter, G. Wenger-Oehn, I. Strnad, H. Grausgruber, R. Schoenlechner and S. D'Amico, Development of an enzymatic assay for the quantitative determination of trypsin inhibitory activity in wheat, Food Chem., 2019, 299, 125038 CrossRef CAS PubMed.
- L. Call, E. Haider, S. D'Amico, E. Reiter and H. Grausgruber, Synthesis and accumulation of amylase-trypsin inhibitors and changes in carbohydrate profile during grain development of bread wheat (Triticum aestivum L.), BMC Plant Biol., 2021, 21, 113 CrossRef CAS PubMed.
- N. Jahn, C. F. H. Longin, K. A. Scherf and S. Geisslitz, No correlation between amylase/trypsin-inhibitor content and amylase inhibitory activity in hexaploid and tetraploid wheat species, Curr. Res. Food Sci., 2023, 7, 100542 CrossRef CAS PubMed.
- G. E. Morlock, L. Drotleff and S. Brinkmann, Miniaturized all-in-one nanoGIT+active system for on-surface metabolization, separation and effect imaging, Anal. Chim. Acta, 2021, 1154, 338307 CrossRef CAS PubMed.
- I. Müller and G. E. Morlock, Quantitative saccharide release of hydrothermally treated flours by validated salivary/pancreatic on-surface amylolysis (nanoGIT) and high-performance thin-layer chromatography, Food Chem., 2023, 432, 137145 CrossRef PubMed.
- V. Sonkamble, G. Zore and L. Kamble, A simple method to screen amylase inhibitors using thin layer chromatography, Sci. Res. Rep., 2014, 4, 85–88 Search PubMed.
- S. Agatonovic-Kustrin and D. W. Morton, HPTLC – bioautographic methods for selective detection of the antioxidant and α-amylase inhibitory activity in plant extracts, MethodsX, 2018, 5, 797–802 CrossRef PubMed.
- P. Houghton, M. Simmonds and S. Larssen, Detection of trypsin inhibition and antioxidant effects on TLC, Planta Med., 2010, 76(12) DOI:10.1055/s-0030-1264254.
- S. P. Pasilis, V. Kertesz, G. J. van Berkel, M. Schulz and S. Schorcht, Using HPTLC/DESI-MS for peptide identification in 1D separations of tryptic protein digests, Anal. Bioanal. Chem., 2008, 391, 317–324 CrossRef CAS PubMed.
- J. Biller, L. Morschheuser, M. Riedner and S. Rohn, Development of optimized mobile phases for protein separation by high performance thin layer chromatography, J. Chromatogr. A, 2015, 1415, 146–154 CrossRef CAS PubMed.
- G. E. Morlock, L. P. Morlock and C. Lemo, Streamlined analysis of lactose-free dairy products, J. Chromatogr. A, 2014, 1324, 215–223 CrossRef CAS PubMed.
- I. Müller, I. Scheibelhut and G. E. Morlock, Study of the quantitative α-amylase or trypsin inhibition by refined and whole wheat and einkorn using high-performance thin-layer chromatography–nanoGIT versus conventional spectrophotometry, in submission.
- E. Simonetti, S. Bosi, L. Negri and G. Dinelli, Amylase Trypsin Inhibitors (ATIs) in a Selection of Ancient and Modern Wheat: Effect of Genotype and Growing Environment on Inhibitory Activities, Plants, 2022, 11(23), 3268 CrossRef CAS PubMed.
- C. Finnie, S. Melchior, P. Roepstorff and B. Svensson, Proteome analysis of grain filling and seed maturation in barley, Plant Physiol., 2002, 129, 1308–1319 CrossRef CAS PubMed.
- G. Guo, D. Lv, X. Yan, S. Subburaj, P. Ge, X. Li, Y. Hu and Y. Yan, Proteome characterization of developing grains in bread wheat cultivars (Triticum aestivum L.), BMC Plant Biol., 2012, 12, 147 CrossRef CAS PubMed.
- I. Klingelhöfer and G. E. Morlock, Sharp-bounded zones link to the effect in planar chromatography-bioassay-mass spectrometry, J. Chromatogr. A, 2014, 1360, 288–295 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay00402g |
| This journal is © The Royal Society of Chemistry 2024 |