Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Determining monolignol oxifunctionalization by direct infusion electrospray ionization tandem mass spectrometry†
Rannei
Skaali
 *a,
Hanne
Devle
*a,
Hanne
Devle
 a,
Katharina
Ebner
b,
Dag
Ekeberg
a,
Katharina
Ebner
b,
Dag
Ekeberg
 a and
Morten
Sørlie
a
a and
Morten
Sørlie
a
aFaculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, Christian Magnus Falsens vei 18, 1433, Ås, Norway. E-mail: rannei.skaali@nmbu.no
bBisy GmbH, Wünschendorf 292, 8200 Hofstätten an der Raab, Austria
First published on 8th May 2024
Abstract
We have successfully developed a validated high-throughput analysis method for the identification and quantification of native and oxifunctionalized monolignols using direct infusion electrospray ionization tandem mass spectrometry (DI-ESI-MS/MS). Oxifunctionalized monolignols generated through unspecific peroxygenase catalysis present a sustainable alternative to fossil aromatic hydrocarbons. This study emphasizes a sustainable analytical approach for these renewable biocatalytic precursors, addressing challenges such as matrix effects, accuracy, precision, and sensitivity of the method. Our findings demonstrate the potential of overcoming quantification difficulties using DI-ESI-MS. Notably, this analytical methodology represents a novel utilization of DI-ESI-MS/MS in examining monolignols and their functionalization, thereby advancing the exploration of lignin as a valuable and sustainable bioresource.
Introduction
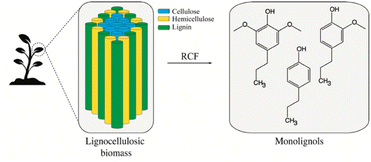
Lignin is the most abundant aromatic polymer in nature, serving as an essential element of protection and structural support in plant cell walls. Accordingly, lignin holds the potential to serve as a sustainable substitute for fossil aromatic hydrocarbons. However, the utilization of this biopolymer is currently limited due to the high cost and energy requirement for lignin depolymerization as a consequence of lignin's resistance to decomposition.1 Sustainable lignin valorization relies on effective biorefinery of intricate mixtures of lignin-derived compounds, into valuable and reactive depolymerized lignin (Fig. 1). One example is reductive catalytic fractionating (RCF) of lignocellulose, a promising strategy in lignin-first biorefinery. RCF yields extensively depolymerized lignin and, depending on the conditions, nearly theoretical quantities of lignin monomers such as 4-propylphenol (4PP), 4-propylguaiacol (4PG), and 4-propylsyringol (4PS), here termed monolignols.2 The value of such lignin monomers is enhanced by oxifunctionalization, which involves the introduction of functional groups through, i.e., selective hydroxylation.3 Functional groups such as hydroxyl and carbonyl groups serve as versatile intermediates for target-oriented synthesis. Oxifunctionalized monolignols hence serve as alternative intermediates for a sustainable green industry (Fig. 2A). Targets for oxifunctionalization include the aromatic ring and the propyl chain of monolignols. An oxifunctionalized benzene ring is used to produce epoxy thermoplastics,4–9 agrochemicals,10 pharmaceuticals,10 and cosmetics.11 The oxidation of the monolignol propyl chain facilitates aromatic ring opening12 as well as ketone production. Such ketones find application in pharmaceuticals12 and cosmetics.13 An oxifunctionalized propyl chain is also attractive as an intermediate in industries such as pharmaceuticals14,15 and bio-based benzoxazine production.14,15 Achieving the targeted oxifunctionalization of organic compounds stands as a challenge within synthetic chemistry, where the utilization of chemical catalysts raises environmental concerns and substantial expenses. Moreover, especially for propyl chain hydroxylation, control over regioselectivity is a concern. To address these challenges and expand the scope of versatile intermediates, our attention shifted toward biotransformations using fungal unspecific peroxygenases (UPO, E.C. 1.11.2). UPOs (Fig. 2B) are biocatalysts that hold a potential for catalyzing the oxifunctionalization of the monolignol structure. A strong indication for this was given by the O-demethylation and cleavage of various non-phenolic lignin model compounds catalyzed by AaeUPO using H2O2.16 UPOs represent a rather young class of iron-dependent oxidative enzymes first described by Ullrich et al. in 2004.17 Similar to cytochrome P450 monooxygenases (P450s), these enzymes are capable of complex oxyfunctionalization chemistry catalyzing regio- and stereoselective oxidization of C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and C–C bonds. However, in contrast to the complex cytochrome P450 system, UPOs are self-sufficient, require only H2O2 as a co-substrate, and are naturally secreted soluble proteins.18 Additionally, they are known for their outstanding substrate spectrum (over 400 different substrates have been tested) and ability to catalyze one-as well as two-electron transfers. This catalytic peculiarity suggests that they are related to both cytochrome P450 monooxygenases and chloroperoxidases and represent the “missing link” between these two classes of enzymes.19 Phylogenetically, there are two classes of UPOs, “short” and “long” family UPOs. They differ not only in their average molecular mass – around 29 kDa for short and 44 kDa for long type UPOs – but also their conserved protein motifs and catalytic residues, with -PCP-EHD-E- and a histidine or -PCP-EGD-R-E- and an arginine as the charge stabilizer in “short” and “long” UPOs, respectively.20 The broad substrate scope, catalytic versatility, and fungal origin of UPOs suggest involvement in various environmental detoxification reactions e.g. plant phytoalexins, microbial toxins, and xenobiotics, as well as in lignin and humus degradation. However, due to their structural characteristics – narrow substrate channel and internal active center – UPOs are presumed to be active on small lignin fragments rather than on the lignin polymer.18,21
C, and C–C bonds. However, in contrast to the complex cytochrome P450 system, UPOs are self-sufficient, require only H2O2 as a co-substrate, and are naturally secreted soluble proteins.18 Additionally, they are known for their outstanding substrate spectrum (over 400 different substrates have been tested) and ability to catalyze one-as well as two-electron transfers. This catalytic peculiarity suggests that they are related to both cytochrome P450 monooxygenases and chloroperoxidases and represent the “missing link” between these two classes of enzymes.19 Phylogenetically, there are two classes of UPOs, “short” and “long” family UPOs. They differ not only in their average molecular mass – around 29 kDa for short and 44 kDa for long type UPOs – but also their conserved protein motifs and catalytic residues, with -PCP-EHD-E- and a histidine or -PCP-EGD-R-E- and an arginine as the charge stabilizer in “short” and “long” UPOs, respectively.20 The broad substrate scope, catalytic versatility, and fungal origin of UPOs suggest involvement in various environmental detoxification reactions e.g. plant phytoalexins, microbial toxins, and xenobiotics, as well as in lignin and humus degradation. However, due to their structural characteristics – narrow substrate channel and internal active center – UPOs are presumed to be active on small lignin fragments rather than on the lignin polymer.18,21
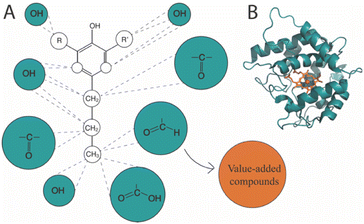 | ||
| Fig. 2 Oxifunctionalization of the monolignol propyl chain. (A) Oxifunctionalization of the monolignol structure produces versatile intermediates suitable for generating value-added compounds. Proposed precursor monolignols are 4-propylphenol (R = R′ = H), 4-propylguaiacol (R = H, R′ = –OCH3), and 4-propylsyringol (R = R′ = –OCH3). (B) Unspecific peroxygenases have demonstrated catalytic activity towards aromatic substrates. We propose leveraging this property for the monolignol activation in the context of the proposed DI-ESI-MS/MS method. The presented crystalline structure is HspUPO (PDB: 7O2G). | ||
We consider an optimized enzymatically catalyzed oxifunctionalization of the monolignols to be a specific, possibly cost-effective, and environmentally favorable process for monolignol functionalization and would represent a breakthrough in lignin valorization. To achieve this, it is crucial to establish a viable analytical method for efficient, comprehensive, and sustainable screening concerning the nature and quantity of the derived products. Commonly used screening techniques include colorimetric and spectrophotometric assays, and more specific and comprehensive analytics such as high-performance liquid chromatography (HPLC) and gas chromatography (GC) in combination with mass spectrometry (MS). Both analytics come with their own set of advantages and disadvantages. Colorimetric and spectrophotometric assays are regarded as straightforward and fast. Still, information on the regioselectivity of the oxifunctionalization is often elusive. The use of HPLC-MS and GC-MS provides qualitative and quantitative information about the products, even for beyond-model substrates. GC-MS has been recognized as the golden standard for structure elucidation. The downside is the time consumed for analysis, specialized equipment, and extensive sample preparation. However, the use of liquid tandem mass spectrometry (LC-MS/MS) has increased as an analytical tool as this technique requires less analytical time and sample preparation compared with conventional GC-MS. LC-MS/MS holds the potential for structure elucidation of a broad range of compounds especially due to the soft ionization techniques such as atmospheric pressure ionization mass spectrometry (API-MS) and electrospray ionization mass spectrometry (ESI-MS). Examples using LC-MS/MS include the structure elucidation of lignin oligomers.22 ESI-MS exhibits selectivity towards analytes that are either acidic or basic making this an interesting ionization technique for the weak acidic phenols found in the monolignols. ESI has been utilized for the characterization of analogous molecules; e.g. model compounds of processed lignin described by Haupert et al.23 However, so far ESI has not been a traditional choice of ionization for the structural elucidation of lignin monomers. Lignin monomers are traditionally ionized by electron ionization (EI) or chemical ionization (CI), where the lignin monomers are identified based on pyrolysis- or reductive cleavage GC.24,25
This paper introduces a method that aims to strike a balance between the straightforwardness of a plate assay and the precision and diversity of the time-consuming chromatography. We present a flow injection analysis (FIA) method designed for high-throughput analysis of both native and oxifunctionalized monolignols using direct infusion electrospray ionization tandem mass spectrometry (DI-ESI-MS/MS), offering advantages such as minimal reagent usage, straightforward sample preparation, and a rapid analytical time without compromising the analytical accuracy. Our focus is on the validation and application of DI-ESI analytics for the identification and quantification of 4PP, 4PG, 4PS, and various oxifunctionalized monolignols. Moreover, we demonstrate the utility of DI-ESI-MS for qualitative and quantitative analyses of oxifunctionalized monolignols using two separate UPO-based catalysts, one “short” (HspUPO) and one “long” (CmaUPO-I). To our knowledge, this analytical method marks the first instance of the identification and quantification of monolignols using DI-ESI-MS/MS.
Results and discussion
Identification of native and oxifunctionalized monolignols
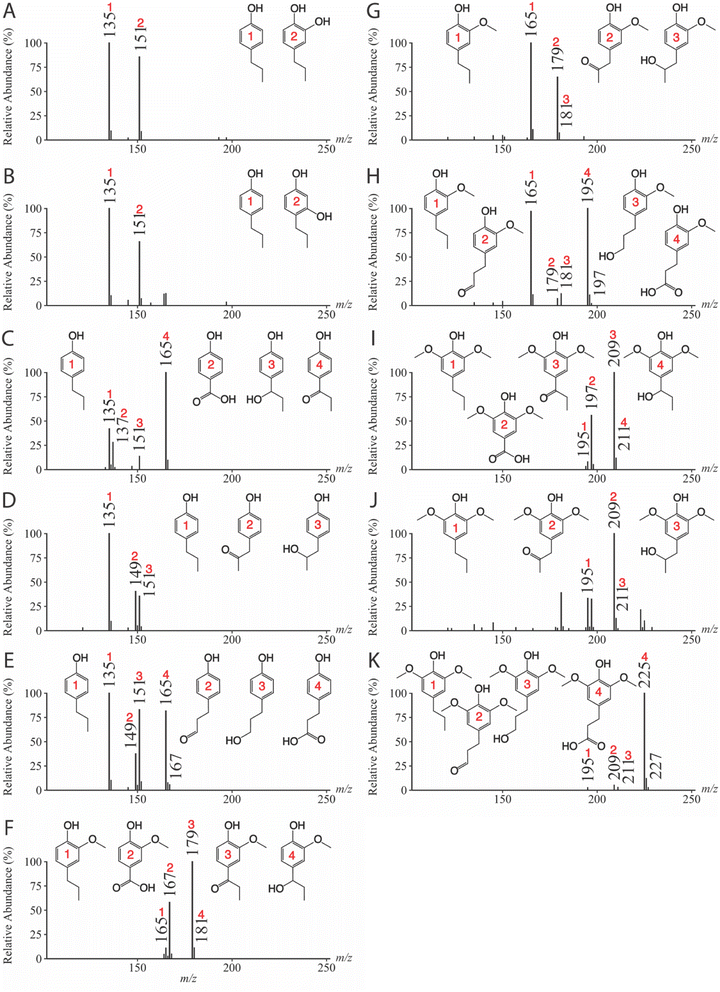
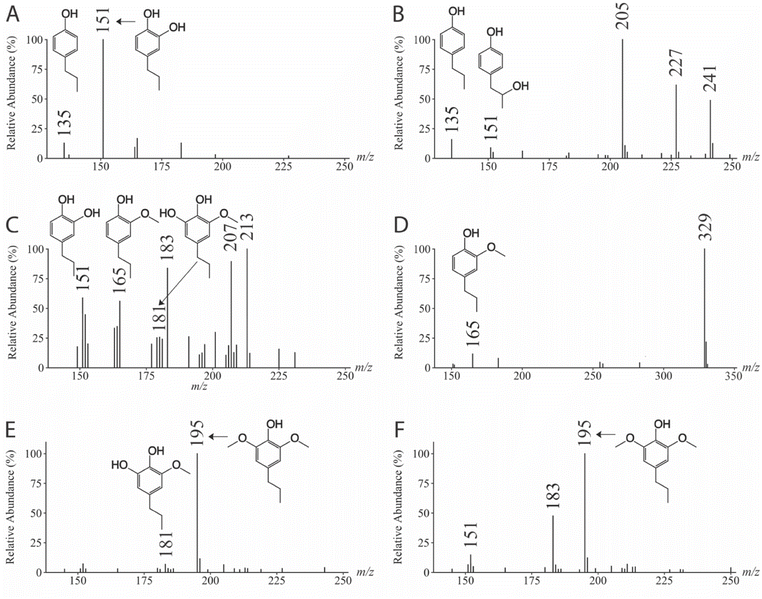
| Monolignol | CE (eV) | Precursor ion (m/z) | Product ions (m/z) | Reference |
|---|---|---|---|---|
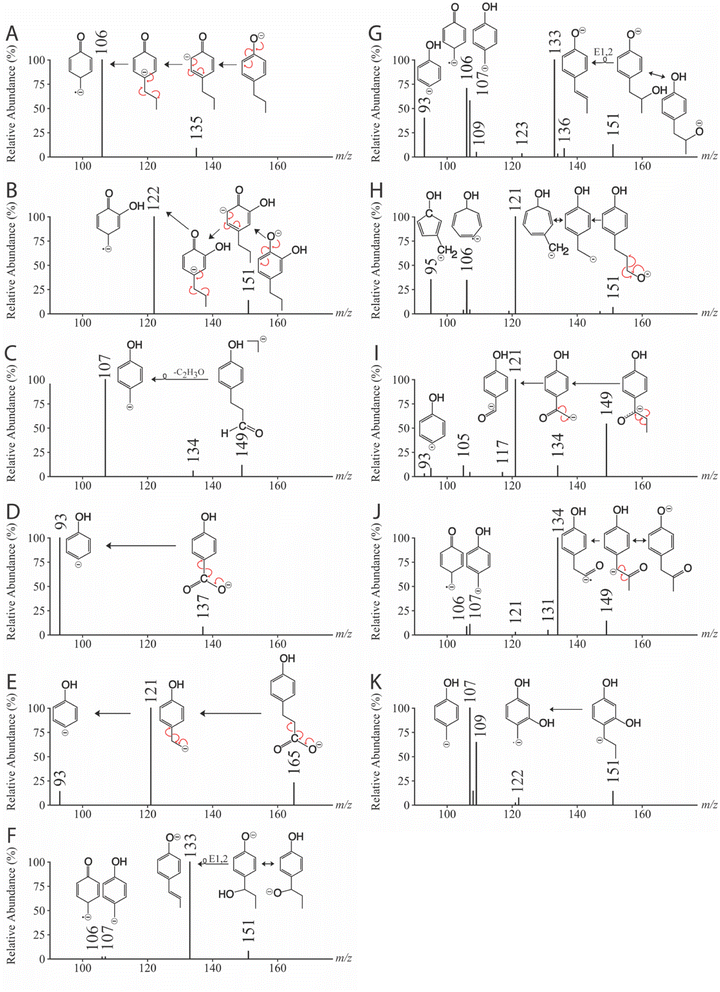
| 4-Propylphenol | 34 | 135 | 106 | Fig. 4A |
| 4-Propylbenzene-1,2-diol | 34 | 151 | 122 | Fig. 4B |
| 3-(4-Hydroxyphenyl)propanal | 27 | 149 | 107 | Fig. 4C |
| 4-Hydroxybenzoic acid | 28 | 137 | 93 | Fig. 4D |
| 3-(4-Hydroxyphenyl)propanoic acid | 26 | 165 | 93, 121 | Fig. 4E |
| 4-(1-Hydroxypropyl)phenol | 30 | 151 | 107, 122 | Fig. 4F |
| 4-(2-Hydroxypropyl)phenol | 36 | 151 | 107, 133 | Fig. 4G |
| 4-(3-Hydroxypropyl)phenol | 34 | 151 | 95, 106, 121 | Fig. 4H |
| 3-(4-Hydroxyphenyl)propane-1-one | 27 | 149 | 93, 95, 105, 107, 117, 121, 134 | Fig. 4I |
| 3-(4-Hydroxyphenyl)propane-2-one | 31 | 149 | 106, 107, 121, 131, 134 | Fig. 4J |
| 4-Propylbenzene-1,3-diol | 34 | 151 | 107, 108, 109, 121, 122 | Fig. 4K |
| 4-Propylguaiacol | 26 | 165 | 150 | Fig. S3A |
| 1-(4-Hydroxy-3-methoxyphenyl)propane-1-one | 24 | 179 | 164 | Fig. S3B |
| 4-Hydroxy-3-methoxybenzoic acid | 24 | 167 | 123, 152 | Fig. S3C |
| 1-(4-Hydroxy-3-methoxyphenyl)propane-2-one | 25 | 179 | 164 | Fig. S3D |
| 4-(3-Hydroxypropyl)-2-methoxyphenol | 24 | 181 | 166 | Fig. S3E |
| 3-(4-Hydroxy-3-methoxyphenyl)propanal | 27 | 179 | 137, 164 | Fig. S3F |
| 3-(4-Hydroxy-3-methoxyphenyl)propanoic acid | 23 | 195 | 119, 123, 136, 151, 177, 180 | Fig. S3G |
| 4-Propylsyringol | 24 | 195 | 180 | Fig. S3H |
| 1-(4-Hydroxy-3,5-methoxyphenyl)propane-1-one | 23 | 209 | 194 | Fig. S3I |
| 4-Hydroxy-3,5-dimethoxybenzoic acid | 22 | 197 | 120, 153, 182 | Fig. S3J |
| 1-(4-Hydroxy-3,5-methoxyphenyl)propane-2-one | 24 | 209 | 194 | Fig. S3K |
| 4-(3-Hydroxypropyl)-2,6-dimethoxyphenol | 22 | 211 | 196 | Fig. S3L |
| 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanal | 25 | 209 | 167, 194 | Fig. S3M |
| 3-(4-Hydroxy-3,5-dimethoxyphenyl propanoic acid) | 23 | 225 | 149, 153, 166, 181, 207, 210 | Fig. S3N |
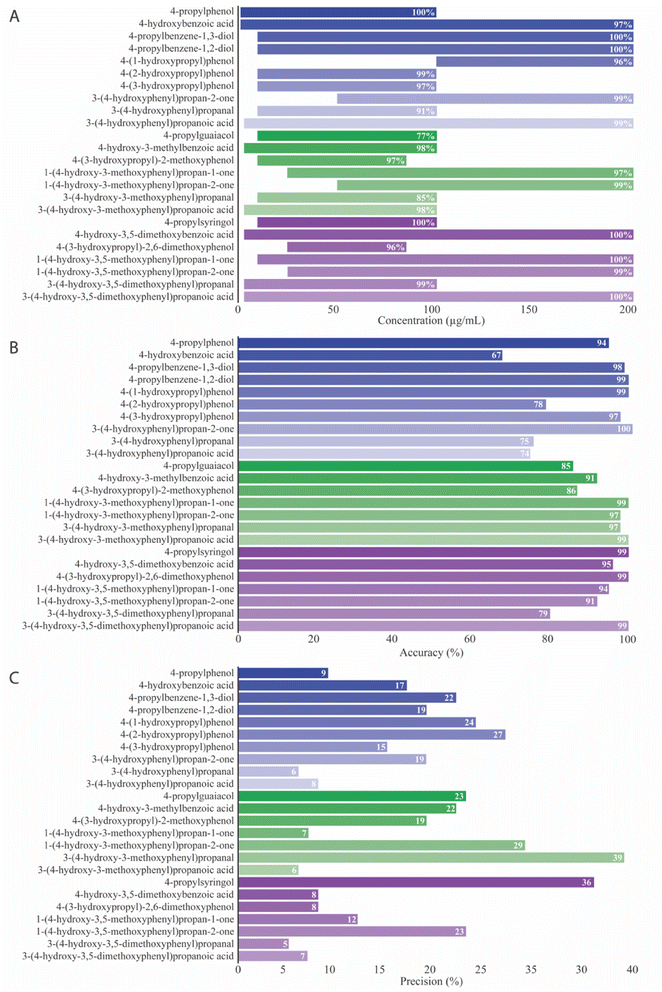
Quantification of native and oxidized monolignols
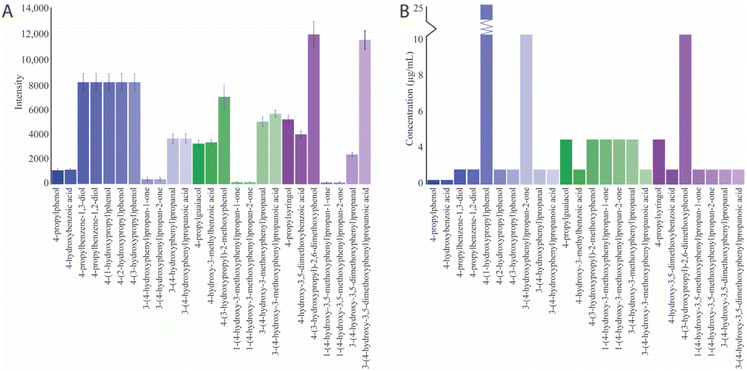
Selected ion monitoring (SIM) mode measured the intensity of the deprotonated molecular ions for quantification purposes within a run time of 24 seconds. This targeted approach is particularly advantageous for achieving high sensitivity and selectivity and minimizing interference from background noise in complex sample matrices.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840 ± 1006 for the different monolignols (Fig. 7A). Compounds exhibiting the least acidity seem to achieve the lowest LOB. LOD, the lowest quantifiable analyte concentration discernible from background noise, was found to range from 0.3–25 μg mL−1 (Fig. 7B). Compounds such as 4-(1-hydroxypropyl)phenol (25 μg mL−1), 3-(4-hydroxyphenyl)propan-2-one (10 μg mL−1), and 4-(3-hydroxypropyl)-2,6-dimethoxyphenol (10 μg mL−1) exhibited the highest LOD and were the compounds with lowest acidity.
840 ± 1006 for the different monolignols (Fig. 7A). Compounds exhibiting the least acidity seem to achieve the lowest LOB. LOD, the lowest quantifiable analyte concentration discernible from background noise, was found to range from 0.3–25 μg mL−1 (Fig. 7B). Compounds such as 4-(1-hydroxypropyl)phenol (25 μg mL−1), 3-(4-hydroxyphenyl)propan-2-one (10 μg mL−1), and 4-(3-hydroxypropyl)-2,6-dimethoxyphenol (10 μg mL−1) exhibited the highest LOD and were the compounds with lowest acidity.
Detection of monolignols generated by UPO-catalyzed biotransformation
Having established the DI-ESI-MS method, we turned to the nature (Fig. 8) and quantity of products (Table 2) resulting from the oxifunctionalization of monolignols catalyzed by UPOs. Incubating 4PP, 4PG, and 4PS with HspUPO and CmaUPO-I in the presence of H2O2 revealed varied oxidation specificity, as evidenced by comparing the established monolignol fragmentation patterns with CID scans from the UPO-catalyzed reactions (Fig. S4†). These variations are attributed to differences in the enzyme composition and the methoxy groups of the monolignols, potentially influencing the interaction within the enzyme–substrate complex and variations in the transition state energy.| UPO | Native monolignol | Oxifunctionalized monolignols | ||||
|---|---|---|---|---|---|---|
| Substrate | Concentration prior to reaction (μg mL−1) | Concentration after reaction (μg mL−1) | Products | Concentration (μg mL−1) | Reference | |
| a The absence of the standard introduces uncertainty, and the assumption made is a tentative inference relying on the observed CID spectra, mass calculations, and reaction pathways. Hence, the final concentration remains non-estimated. | ||||||
| HspUPO | 4-Propylphenol | 1362 ± 100 | 341 ± 46 | 4-Propylbenzene-1,2-diol | 1081 ± 123 | Fig. 8A |
| 4-Propylguaiacol | 1662 ± 278 | 83 ± 45 | 4-Propylbenzene-1,2-diol 3-methoxy-5-propylbenzene-1,2-diola | 307 ± 49 | Fig. 8C | |
| — | ||||||
| 4-Propylsyringol | 1962 ± 230 | 357 ± 141 | 3-Methoxy-5-propylbenzene-1,2-diola | — | Fig. 8E | |
| CmaUPO-I | 4-Propylphenol | 1362 ± 99 | 5 ± 1 | 4-(2-Hydroxypropyl)phenol | 66 ± 1 | Fig. 8B |
| 4-Propylguaiacol | 1938 ± 336 | 433 ± 160 | Dimerizationa | — | Fig. 8D | |
| 4-Propylsyringol | 1962 ± 230 | 568 ± 170 | Unknowna | — | Fig. 8F | |
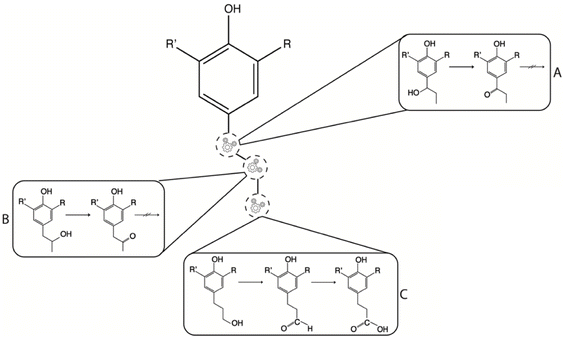
For the enzymatically oxyfunctionalization of 4PP (start concentration 1362 ± 99 μg mL−1), HspUPO catalyzed an electrophilic attack on the π system to yield 4-propylbenzene-1,2-diol (1081 ± 123 μg mL−1, m/z 151, Fig. 8A) as the main product. At the end of the reaction, 341 ± 46 μg mL−1 4PP remained after biotransformation, which is not unexpected as there is a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio between 4PP and H2O2. In contrast, CmaUPO-I targeted mainly the Cβ position of the propyl chain, producing 4-(2-hydroxypropyl)phenol (66 ± 1 μg mL−1, m/z 151, Fig. 8B). This reaction is likely the result of the hydroxylation of Cβ–H bond by compound I. Upon completion of the reaction, 5 ± 1 μg mL−1 of 4PP remained. Given that there is a discrepancy between the amount of substrate and the hydroxylated product, there is likely a competition between a peroxygenase and a peroxidase mechanism. The latter will likely have a dimerization product formation initially (see more below). Moreover, inspection of the mass spectrum shows products with m/z values deviating what is expected of what is obtained by only insertion of one oxygen in a C–H bond. Poor chemo selectivity in alkane oxidation is a known problem leading to low yields of the desired products.28 B3LYP-D3 calculations indicate the relative activation energies of the phenol H, Cα–H, Cβ–H, and Cγ–H of 4PP to be 20.5, 20.5, 27.2, and 41.8 kJ mol−1, respectively. Similar trends were observed for 4-propylguaiacol and 4-propylsyringol. This shows that, kinetically, the Cγ–H is the most difficult to oxifunctionalize. Still, the key parameter for using enzymes for regioselectivity is the positioning of the substrate in the active site concerning which C–H will be in the closest proximity to the activated oxygen species.
1 molar ratio between 4PP and H2O2. In contrast, CmaUPO-I targeted mainly the Cβ position of the propyl chain, producing 4-(2-hydroxypropyl)phenol (66 ± 1 μg mL−1, m/z 151, Fig. 8B). This reaction is likely the result of the hydroxylation of Cβ–H bond by compound I. Upon completion of the reaction, 5 ± 1 μg mL−1 of 4PP remained. Given that there is a discrepancy between the amount of substrate and the hydroxylated product, there is likely a competition between a peroxygenase and a peroxidase mechanism. The latter will likely have a dimerization product formation initially (see more below). Moreover, inspection of the mass spectrum shows products with m/z values deviating what is expected of what is obtained by only insertion of one oxygen in a C–H bond. Poor chemo selectivity in alkane oxidation is a known problem leading to low yields of the desired products.28 B3LYP-D3 calculations indicate the relative activation energies of the phenol H, Cα–H, Cβ–H, and Cγ–H of 4PP to be 20.5, 20.5, 27.2, and 41.8 kJ mol−1, respectively. Similar trends were observed for 4-propylguaiacol and 4-propylsyringol. This shows that, kinetically, the Cγ–H is the most difficult to oxifunctionalize. Still, the key parameter for using enzymes for regioselectivity is the positioning of the substrate in the active site concerning which C–H will be in the closest proximity to the activated oxygen species.
When 4PG (start concentration 1662 ± 278 μg mL−1) was the substrate for HspUPO, 83 ± 45 μg mL−1 remained upon completion of the reaction. Again, hydroxylation of the aromatic ring (m/z 181, Fig. 8C) was observed. Interestingly, HspUPO catalyzed a demethylation reaction to yield 4-propylbenzene-1,2-diol (307 ± 49 μg mL−1, m/z 151, Fig. 8C) as the primary product. The addition of ascorbate enhanced the yield of reaction products by preventing phenol radical formation. CmaUPO-I catalyzed mainly a peroxidase reaction resulting in the potential dimerization of 4PG (m/z 329, Fig. 8D). Here, the 4PG concentration decreased to 433 ± 160 μg mL−1. The same biotransformation was tested in the presence of ascorbate to see if this would favor the peroxygenase reaction. Now, the dimerization of 4PG was restricted, yet there was no significant substrate degradation.
HspUPO catalyzed demethylation of 4PS (start concentration 1962 ± 230 μg mL−1) that produced ions at m/z 181 (Fig. 8E). Upon completion of the reaction, the 4PS concentration was 357 ± 141 μg mL−1. Although the proposed identity of 3-methoxy-5-propylbenzene-1,2-diol could not be verified due to a lack of a suitable standard, the fact that the same peak with comparable fragmentation patterns was observed in the HspUPO-catalyzed biotransformation of 4PG, we express confidence that the compounds at m/z 181 produced from 4PG and 4PS are the same, likely 3-methoxy-5-propylbenzene-1,2-diol. CmaUPO-I catalyzed the biotransformation of 4PS resulting in substrate degradation to a concentration of 568 ± 170 μg mL−1. However, the nature of the products could not be determined (Fig. 8F).
It is essential to emphasize that although all experiments were conducted under identical conditions, altering parameters such as temperature, component concentrations, or buffers could significantly impact the biotransformations. We repeated the biotransformation of 4PS catalyzed by CmaUPO-I at 37 °C to explore potential product formation, as the nature of the products was unclear at 25 °C. Subsequently, we identified a formed product at m/z 211, indicating a potential isomeric mixture of hydroxylated 4PS (Fig. S5†). Additionally, a product emerged at m/z 209, indicating second-step oxidation of the Cα, Cβ, and Cγ of the hydroxylated products. Notably, this resulted in an isomeric mixture of ketones and an aldehyde (Fig. S5†). Quantification of the isomeric mixtures was not attempted; nevertheless, the intensities of the deprotonated molecular ions at m/z 209 and m/z 211 were significantly higher compared to the controls.
Materials and methods
Standard solution preparation
Monolignol standard stock solutions (1.0 mg mL−1) were prepared separately in 50% ACN (LC-MS grade) and 50% Milli-Q water with 150 mM NH4OH and filtered (0.2 μm, Pall Corporation, Port Washington, New York, USA). The stock solutions comprised the following compounds, each purchased from their respective suppliers: 4-propylphenol, 4-hydroxy benzoic acid, 4-(3-hydroxypropyl)phenol, 3-(4 hydroxyphenyl) propanoic acid, 4-propylguaiacol, 4-hydroxy-3-methoxy-benzoic acid, 3-(4-hydroxy-3-methoxyphenyl)propanoic, 4-hydroxy-3,5-methoxy-benzoic acid, and 3-(4-hydroxy-3-methoxyphenyl)propanal (Sigma-Aldrich, Burlington, Massachusettes, USA), 4-(3-hydroxypropyl)-2-methoxy phenol, 3-(4-hydroxyphenyl) propanal, 1-(4-hydroxyphenyl)propane-1-one, 1-(4-hydroxy-3-methoxy-phenol)propane-1-one, 1-(4-hydroxy-3-methoxy-phenol)propane-2-one, and 1-(4-hydroxy-3,5-dimethoxy-phenol)propane-2-one (BLD Pharmatech Ltd, Shanghai, China), 4-propylsyringol, 4-(2-hydroxypropyl) phenol, 4-(3-hydroxypropyl)-2,6-dimethoxy phenol, and 1-(4-hydroxy-3,5-dimethoxy-phenol)propane-1-one (Enamine, Kyiv, Ukraine), 3-(4-hydroxy-3,5-dimethoxyphenyl) propanal (ChemSpace, Riga, Latvia), 3-(4-hydroxy-3,5-dimethoxyphenyl) propanoic acid (Accela, San Diego, California, USA), 4-(1-hydroxypropyl) phenol (LGC Standards Ltd, Teddington, Middlesex, UK), and 1-(4-hydroxyphenyl)propane-2-one (Tokyo Chemical Industry, Tokyo, Japan).Instrumentation and conditions
MS analyses were performed using a Dionex Ultimate 3000 RS autosampler (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a linear ion trap (LTQ XL Thermo Fisher Scientific, Waltham, MA, USA) equipped with an ESI source. Instrument control, data acquisition, and processing were carried out using LTQ Tune software and Xcalibur 2.2 SP1.48 (Thermo Fisher Scientific, Waltham, MA, USA). All injections were performed as three replicates of 10 μL with three injection parallels. The transfer capillary was set at 275 °C and comprised the sheath, aux, and sweep gas flow of 12, 2, and 0 arb. units, respectively. 50% ACN with NH4OH (pH 10) was the mobile phase that directed the monolignol standard solutions to the ESI source with a flow of 0.3 mL min−1. The MS operated in negative mode with an electrospray voltage of 3.0 kV, providing optimal ionization conditions for these phenolic compounds.Identification of native and oxidized monolignols
Quantification of native and oxidized monolignols
Unspecific peroxygenase activity assays and analyzation of monolignol oxifunctionalization
Conclusions
This study addresses the challenges associated with lignin valorization by focusing on the oxifunctionalization of lignin monomers, particularly 4-propylphenol, 4-propylguaiacol, and 4-propylsyringol. We emphasize the potential of lignin monomers as valuable intermediates for a sustainable green industry. Oxifunctionalization enhances the value of the monomers that can be achieved by using UPO catalysts. Our research introduces a novel, validated, and efficient method using DI-ESI-MS in full scan, SIM, and MS/MS mode for efficient and sustainable identification and quantification of native and oxifunctionalized monolignols. Each analysis segment is completed within a concise run time of 24 seconds. The proposed fragmentation mechanisms provide valuable insights into the deprotonation and identification of these compounds. Additionally, we present the quantification of monolignols, addressing challenges associated with matrix effects, accuracy, precision, and sensitivity. Our findings demonstrate the potential of overcoming quantification difficulties using DI-ESI-MS. This screening method overcomes the limitations of existing techniques by providing a balance between the straightforwardness of plate assays and the precision of chromatography and represents an analytical breakthrough. We illustrate the method's utility in both qualitative and quantitative analyses of oxifunctionalized monolignol products derived from different UPO catalyzes. The varied oxidation specificity observed with different UPOs highlights the potential for tailoring enzymatic processes for specific applications. Notably, this analytical method represents a versatile application of DI-ESI-MS/MS for the analysis of monolignols, contributing to the exploration of lignin as a valuable and sustainable bioresource.Author contributions
K. E. expressed HspUPO and CmaUPO-I. R. S. purified HspUPO and CmaUPO-I, performed the formal analysis and investigation, ensured experiment reproducibility, and project administration, and authored the manuscript including visualization and data presentation. D. E. and H. D. provided supervision, investigation, and manuscript review. M. S. was responsible for conceptualization, resources, and funding acquisition and contributed to manuscript review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge funding from the European Innovation Council (EIC) Pathfinder program under grant agreement no. 101046815. We also acknowledge Professor Tomasz Borowski for the computation of the activation parameters of the monolignols and Dr Gabriela Schröder for helpful discussions. The PhD thesis of K. E. was funded by the Eureka project PUMLA (FFG Basisprogramm).Notes and references
- A. J. Ragauskas, G. T. Beckham, M. J. Biddy, R. Chandra, F. Chen, M. F. Davis, B. H. Davison, R. A. Dixon, P. Gilna, M. Keller, P. Langan, A. K. Naskar, J. N. Saddler, T. J. Tschaplinski, G. A. Tuskan and C. E. Wyman, Science, 2014, 344, 1246843 CrossRef PubMed
.
- S. Van den Bosch, W. Schutyser, R. Vanholme, T. Driessen, S. F. Koelewijn, T. Renders, B. De Meester, W. J. J. Huijgen, W. Dehaen, C. M. Courtin, B. Lagrain, W. Boerjan and B. F. Sels, Energy Environ. Sci., 2015, 8, 1748–1763 RSC
.
- Y. Y. Wang, X. Meng, Y. Pu and A. J. Ragauskas, Polymers, 2020, 12(10), 2277 CrossRef CAS PubMed
.
- W. Schutyser, T. Renders, S. Van den Bosch, S. F. Koelewijn, G. T. Beckham and B. F. Sels, Chem. Soc. Rev., 2018, 47, 852–908 RSC
.
- J. F. Zhang, Y. Jiang, L. F. Easterling, A. Anstner, W. R. Li, K. Z. Alzarieni, X. M. Dong, J. Bozell and H. I. Kenttamaa, Green Chem., 2021, 23, 983–1000 RSC
.
- C. K. Zhao, Z. H. Hu, L. L. Shi, C. Wang, F. X. Yue, S. X. Li, H. Zhang and F. C. Lu, Green Chem., 2020, 22, 7366–7375 RSC
.
- A. De Santi, M. V. Galkin, C. W. Lahive, P. J. Deuss and K. Barta, Chemsuschem, 2020, 13, 4468–4477 CrossRef CAS PubMed
.
- A. Kumar and B. Thallada, Sustainable Energy Fuels, 2021, 5, 3802–3817 RSC
.
- J. Zhu, C. Yan, X. Zhang, C. Yang, M. Jiang and X. Zhang, Prog. Energy Combust. Sci., 2020, 76, 100788 CrossRef
.
- J. Bomon, M. Bal, T. K. Achar, S. Sergeyev, X. Wu, B. Wambacq, F. Lemière, B. F. Sels and B. U. W. Maes, Green Chem., 2021, 23, 1995–2009 RSC
.
- J.-M. Gaudin and J.-Y. de Saint Laumer, Eur. J. Org Chem., 2015, 2015, 1437–1447 CrossRef CAS
.
- E. Erickson, A. Bleem, E. Kuatsjah, A. Werner, J. Dubois, J. McGeehan, L. Eltis and G. Beckham, Nat. Catal., 2022, 5, 86–98 CrossRef CAS
.
- J. Beekwilder, I. M. van der Meer, O. Sibbesen, M. Broekgaarden, I. Qvist, J. D. Mikkelsen and R. D. Hall, Biotechnol. J., 2007, 2, 1270–1279 CrossRef CAS PubMed
.
- A. Piazzon, U. Vrhovsek, D. Masuero, F. Mattivi, F. Mandoj and M. Nardini, J. Agric. Food Chem., 2012, 60, 12312–12323 CrossRef CAS PubMed
.
- M. W. Hackl, M. Lakemeyer, M. Dahmen, M. Glaser, A. Pahl, K. Lorenz-Baath, T. Menzel, S. Sievers, T. Böttcher, I. Antes, H. Waldmann and S. A. Sieber, J. Am. Chem. Soc., 2015, 137, 8475–8483 CrossRef CAS
.
- M. Kinne, M. Poraj-Kobielska, S. A. Ralph, R. Ullrich, M. Hofrichter and K. E. Hammel, J. Biol. Chem., 2009, 284, 29343–29349 CrossRef CAS PubMed
.
- R. Ullrich, J. Nüske, K. Scheibner, J. Spantzel and M. Hofrichter, Appl. Environ. Microbiol., 2004, 70, 4575–4581 CrossRef CAS PubMed
.
-
M. Hofrichter, H. Kellner, M. J. Pecyna and R. Ullrich, Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450, 2015, vol. 851, pp. 341–368 Search PubMed
.
-
M. Hofrichter, H. Kellner, R. Herzog, A. Karich, C. Liers, K. Scheibner, V. W. Kimani and R. Ullrich, in Grand Challenges in Fungal Biotechnology, ed. H. Nevalainen, Springer International Publishing, Cham, 2020, pp. 369–403, DOI: DOI:10.1007/978-3-030-29541-7_14
.
- M. Faiza, S. Huang, D. Lan and Y. Wang, BMC Evol. Biol., 2019, 19, 76 CrossRef PubMed
.
- M. Hofrichter, H. Kellner, R. Herzog, A. Karich, J. Kiebist, K. Scheibner and R. Ullrich, Antioxidants, 2022, 11, 163 CrossRef CAS PubMed
.
- R. Seraglia, L. Molin, I. Isak and P. Traldi, Eur. J. Mass Spectrom., 2012, 18, 195–203 CrossRef CAS PubMed
.
- L. J. Haupert, B. C. Owen, C. L. Marcum, T. M. Jarrell, C. J. Pulliam, L. M. Amundson, P. Narra, M. S. Aqueel, T. H. Parsell, M. M. Abu-Omar and H. I. Kenttämaa, Fuel, 2012, 95, 634–641 CrossRef CAS
.
- J. MacKay and D. Dimmel, J. Wood Chem. Technol., 2001, 21, 1–17 CrossRef
.
- F. Lu and J. Ralph, J. Agric. Food Chem., 1997, 45, 2590–2592 CrossRef CAS
.
- B. C. Owen, L. J. Haupert, T. M. Jarrell, C. L. Marcum, T. H. Parsell, M. M. Abu-Omar, J. J. Bozell, S. K. Black and H. I. Kenttämaa, Anal. Chem., 2012, 84, 6000–6007 CrossRef CAS
.
- M. Mattonai, E. Parri, D. Querci, I. Degano and E. Ribechini, Microchem. J., 2016, 126, 220–229 CrossRef CAS
.
-
J. A. Davies, Selective Hydrocarbon Activation: Principles and Progress, VCH Publishers, 1990 Search PubMed
.
- L. Rotilio, A. Swoboda, K. Ebner, C. Rinnofner, A. Glieder, W. Kroutil and A. Mattevi, ACS Catal., 2021, 11, 11511–11525 CrossRef CAS PubMed
.
- K. Ebner, L. J. Pfeifenberger, C. Rinnofner, V. Schusterbauer, A. Glieder and M. Winkler, Catalysts, 2023, 13, 206 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay00403e |
| This journal is © The Royal Society of Chemistry 2024 |