Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Distinct DNA conformations during forward and backward translocations through a conical nanopore†
Fei
Zheng
 *a and
Quan
Han
b
*a and
Quan
Han
b
aCavendish Laboratory, University of Cambridge, CB3 0HE, Cambridge, UK. E-mail: fz284@cam.ac.uk
bSchool of Mechanical Engineering, Nanjing Forest University, 211100, Nanjing, China
First published on 27th August 2024
Abstract
DNA conformations, which encompass the three-dimensional structures of the DNA strand, play a crucial role in genome regulation. During DNA translocation in a nanopore, various conformations occur due to interactions among force fields, the fluidic environment, and polymer features. The most common conformation is folding, where DNA moves through the nanopore in a two-strand or multi-strand manner, influencing the current signature. Factors such as hydrodynamic drag, ionic environments, and DNA length significantly affect these conformations. Notably, conical nanopores, with their asymmetrical geometry, impose unique constraints on DNA translocation. Our findings reveal that during forward translocation, from the narrow (cis) end to the wide (trans) end, DNA experiences less resistance, resulting in shorter translocation times and higher blockade currents. Conversely, backward translocation, from the wide (trans) end to the narrow (cis) end, leads to longer translocation times and more complex conformations due to increased hydrodynamic drag and geometric constraints. This study employs molecular ping-pong methods to confine DNA, further highlighting the intricate dynamics of DNA folding within nanopores. These insights enhance the understanding of DNA behavior in confined environments, contributing to advancements in nanopore-based sensing and sequencing technologies, with implications for genome regulation and biomedical applications.
Introduction
DNA conformations, which encompass the three-dimensional structures of the DNA strand, play a critical role in various biological activities related to genome regulation.1–9 For instance, during the intricate process of genome packaging, DNA undergoes looping, folding, and coiling, ultimately resulting in the formation of highly condensed structures known as chromosomes.10 This dynamic reorganization is essential for the efficient compaction of the genome within the nucleus and for the regulation of gene expression, as different conformation states can influence the accessibility of DNA to transcription factors and other regulatory proteins.11 Analogous to genome packaging where long DNA polymers are threaded through a small aperture into a constricted volume, DNA translocation in a nanopore also presents various conformations due to the intricate interactions among the force field, fluidic environment, and polymer features.12–14 The most common conformation during DNA translocation is the folding type, where DNA moves through the nanopore in a two-strand or multi-strand way, which, in the current signature, induces a deeper blockade current with the amplitude proportional to the number of folded strands.15–17DNA folding in the nanopore is influenced by several factors.18–24 First, before entering the nanopore, DNA moves from an open space to a confined area and, under the influence of an external driving force, adjusts its conformation by bending into a fold.13 The fluidic environment is another important factor. Hydrodynamic drag resulting from a high-viscosity solution disturbs the local force equilibrium of the DNA polymer, causing it to deform into complex conformations.25 Additionally, certain ion types, such as lithium, can more effectively screen the charges of the DNA backbone, indirectly reducing the electric force by which DNA is stretched and maintains a linear conformation.26 Furthermore, the probability of folding is proportional to DNA length, with a greater possibility of longer DNA existing in a folded conformation. Nevertheless, how nanopore shape, especially a conical nanopore with confined space on one side, affects the DNA conformation during the translocation has not been fully discussed.
DNA translocation in a conical nanopore exhibits distinct velocity behaviors in the forward and backward directions.27–29 In this context, forward translocation refers to movement from the tip (cis side) to the back (trans side) of the nanopore, while backward translocation is in the opposite direction. During backward translocation, the DNA has a longer effective length, which is dragged by the flow, thus slowing down its movement. In contrast, during forward translocation, the DNA buckles within the nanopore, resulting in a shorter effective length and less drag force.27 Due to the distinct force profiles and steric hindrance, DNA translocation in the two directions should exhibit different conformational behaviors.
Herein, we investigated the DNA conformations during forward and backward translocations in a conical nanopore. We show the distinct translocation features of the forward and backward translocation, including the translocation time and the current blockade. All current point histograms show that DNA exhibits more complex conformations in the backward translocation than the forward one. We also employed the molecular ping-pong method to restrict the DNA molecule in a more confined space, where the DNA exists with even more complicated conformations. This work has implications for the DNA polymer dynamics in a confined nanoscale space, and potentially contributes to understanding genome regulation.
Results and discussion
Fig. 1 illustrates how we define a forward and backward translocation in a conical nanopore. In all experiments, we utilized asymmetric, conical glass nanopores characterized previously,30,31 having diameters of 14 ± 3 nm (mean ± standard deviation) and a cone semi-angle of 0.05 ± 0.01 radians (ESI Fig. S2†). As shown in Fig. 1a, a forward translocation in a conical nanopore refers to the movement of a DNA molecule through the nanopore from the narrow (cis) end to the wide (trans) end, which is typically driven by an electric field applied at the trans side, causing the molecule to pass through the constricted channel. In contrast, backward translocation involves the passage of molecules from the wide (trans) end to the narrow (cis) end, moving in the opposite direction of forward translocation (Fig. 1b). In this process, a positive voltage is applied to the cis side. Due to the limited volume on the trans side, the diffusion of DNA molecules, particularly longer ones, is significantly restricted, making it challenging for them to diffuse from the wide region to the nanopore before being captured by the electric field. To enable backward translocation, we initially performed forward translocation for 1–2 hours and subsequently reversed the voltage polarity, and thus we could obtain a desired translocation frequency of DNA molecules.Fig. 2 illustrates the current signals of forward and backward translocations in a conical nanopore. Fig. 2a depicts three forward translocation signals, while Fig. 2b presents a single backward translocation signal. Notably, the translocation time for backward translocation is approximately three times that for forward translocation (∼9 ms for forward translocation and ∼27 ms for backward translocation). A DNA buckling theory has been proposed to explain this discrepancy.27 Due to the asymmetric electric field distribution, during forward translocation, the DNA molecule encounters less electric force as it enters the deeper region of the nanopore, leading to buckling and reduced hydrodynamic resistance. In contrast, during backward translocation, the electric field strength increases from the back to the tip of the nanopore, stretching the DNA molecule and increasing hydrodynamic drag, thereby prolonging translocation time.
Several additional factors contribute to the extended backward translocation time. First, in a conical nanopore, the opening sizes differ, with one end being wider and the other narrower, causing forward translocation (narrow to wide) to encounter less resistance and benefit from a stronger electric field driving the molecule through the pore. Conversely, backward translocation (wide to narrow) faces increased resistance and moves against an increasing electric field, slowing its progress. Due to this gradient of electric field strength inside the nanopore, the DNA translocation speed varies during the process instead of remaining constant. In addition, the molecular conformation and entropic state are influenced by the nanopore geometry, with forward translocation allowing the molecule to stretch and align more easily, reducing translocation time. Backward translocation requires the molecule to adopt a more compact, entropically unfavorable conformation, increasing the time needed for translocation, which we will show in detail later.
Another intriguing phenomenon is the observed difference in blockade current between forward and backward translocations, as shown in Fig. 2a and b, and the scatter distribution in Fig. 2c, where each scatter represents a single translocation event. The blockade current during backward translocation is smaller than that during forward translocation (∼0.15 nA for the backward event and ∼0.1 nA for the forward event). One possible explanation is that the stronger hydrodynamic force in the backward translocation compacts the DNA strand, reducing the number of blocked ions and resulting in a smaller blockade current. Additionally, during forward translocation, the rapidly moving DNA molecule, with its substantial negative charge density, accelerates counter-ions in its electric double layer, likely enhancing the blockade current.
Fig. 2d presents the frequency distribution of translocation times for both forward and backward translocations. Consistent with the linear events in Fig. 2a and b, the translocation time for backward translocations is roughly three times that for forward translocations (the average translocation times are 5.8 ms for forward translocation and 17.2 ms for backward translocation). As illustrated in Fig. 2c and d, the feature distribution of individual backward translocation events is more divergent compared to forward translocations. We attribute this divergence to the more diverse conformations that the DNA molecule adopts during backward translocation.
During nanopore translocation, DNA molecules can adopt various conformations that significantly influence its behavior and the resulting signal. The linear conformation is the simplest form, where the DNA molecule passes through the nanopore in an extended, straight manner, resembling a stretched thread. This conformation is often observed due to the applied electric field that drives the DNA through the nanopore, aligning it along the pore axis and providing a straightforward signal with only one strand blocking the pore (Fig. 3a and b). In contrast, the folded conformation occurs when the DNA molecule folds back on itself, creating loops or multiple segments entering the pore simultaneously. This can happen due to initial configurations or interactions with the nanopore environment, resulting in more complex and varied signal patterns that can reveal information about the number of strands blocking the nanopore concurrently.
We show how we derive the DNA conformations from the nanopore translocation signals, as shown in Fig. 3. First, the current blockade of DNA translocation is proportional to the number of strands within the nanopore. Basically, each DNA strand blocks approximately the same current by occupying the same amount of space of moving ions inside the nanopore. Hence, the amplitude of the current blockade indicates the conformations of DNA. In Fig. 3a, the translocation signal consists of a constant current blockade, indicating that only one DNA strand passes through the nanopore in a linear manner (Fig. 3b). In contrast, the signal in Fig. 3c includes a deeper current blockade at the beginning where the amplitude is 2 times that of the linear blockade in Fig. 3a. That is, the DNA adopts a folded conformation where two DNA strands move through the nanopore concurrently (Fig. 3d). DNA can evolve into more complex conformations during translocation. As shown in Fig. 3e, a deeper current blockade appears at the middle of the translocation signal, of which the amplitude is 3 times of the linear blockade. This is likely attributed to an ‘S’ fold conformation (Fig. 3f) as it translocates the nanopore with three strands simultaneously. A knot conformation5 or plectoneme32 also can induce the similar current signal; however, they feature the same 3-strand conformation. Fig. 3g shows that the current blockade of the signal increases by a factor of 3 compared to that of the linear translocation. This indicates that the DNA translocation adopts a more complex ‘S’ fold conformation within the nanopore.
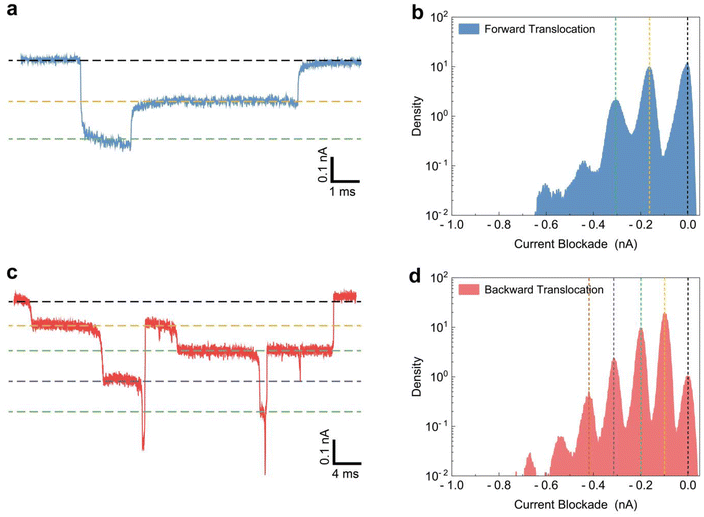
Fig. 4a and c illustrate typical folded event signals for forward and backward translocations, respectively. In the forward translocation signal (Fig. 4a), two distinct levels of current blockade are observed: the linear blockade level (indicated by the yellow dashed line) and the folded blockade level (indicated by the green dashed line), with the black dashed line representing the baseline current where no DNA molecule blocks the nanopore. Conversely, the backward translocation event (Fig. 4c) exhibits multiple folded levels, signifying greater DNA compaction within the constricted nanopore region. The number of current blockade levels during backward translocation is markedly higher compared to that during forward translocation. To elucidate the current blockade levels of folded events, we used an all current point histogram plotted from 300 individual translocation events. This histogram graphically represents the distribution of ionic current levels as the DNA translocates through the nanopore, with the x-axis denoting current blockade levels and the y-axis indicating their frequency. The peaks in the histogram correspond to frequently occurring blockades, reflecting the specific number of DNA strands and allowing inference of the DNA conformation. Fig. 4b and d present the all current point histograms for forward and backward translocation events, respectively. Notably, aside from the baseline peak (0 nA), Fig. 4b displays two peaks at a frequency density over 10−1, whereas Fig. 4d shows four peaks, indicating that DNA molecules adopt more compacted folded conformations during backward translocation. This increased folding during backward translocation through a conical nanopore is primarily due to the asymmetrical geometry of the nanopore, featuring a narrow tip and wider base. As DNA moves from the wider base towards the narrow tip, the decreasing cross-sectional area forces the molecule to become compact and fold. Electrostatic interactions between the negatively charged DNA and the charged nanopore surfaces enhance this compaction. Furthermore, the energy barriers associated with the translocation process are higher in the backward direction, favoring configurations where the DNA is more folded to minimize free energy. Hydrodynamic drag forces also play a role, as the resistance encountered during backward movement prompts the DNA to adopt a more compact and folded state. These combined factors underscore the more compacted and folded state of DNA molecules during backward translocation through a conical nanopore.
To investigate how geometric constriction affects the folded conformation of DNA, we employed the molecular ping-pong method to immediately recapture the DNA molecule after its forward translocation (see Materials and methods for details). Compared to the previous approach, where the DNA molecule was driven backwards after a 1 hour forward translocation, the ping-pong method ensures that the DNA molecule remains in the highly constricted region near the nanopore, as it has less than 20 ms to diffuse into a wider space. As illustrated in Fig. 5a, the ping-pong method involves reversing the voltage polarity once the DNA molecule completes its forward translocation, observable from the current trace. Following the voltage switch, the DNA molecule undergoes immediate backward translocation from the more constricted space.
Fig. 5b shows that the all-current point histogram for the backward translocation in the ping-pong experiment exhibits seven peaks with frequency densities exceeding 10−1, significantly more than that observed with the previous method. This suggests that the more constricted the conical nanopore, the more folded the DNA molecule becomes. Fig. 5 presents a typical backward event signal in a ping-pong experiment, displaying more than ten levels of current blockade. While determining the specific structure of the DNA molecule from this signal is challenging, these blockade levels quantify the complexity of the DNA's folded conformation. This method could potentially be applied to investigate how capsid size affects the complex conformation of DNA molecules during genome packaging.
Conclusion
In this study, we have meticulously explored the distinct DNA conformations during forward and backward translocations through a conical nanopore. By utilizing precise measurements and the molecular ping-pong technique, we demonstrated that DNA exhibits markedly different behaviors depending on the direction of translocation. Our findings highlight that during backward translocation, DNA adopts more complex and compact conformations due to the increased hydrodynamic drag and geometric constraints imposed by the nanopore's asymmetrical structure. These conformational differences significantly affect the translocation time and current blockade signals, providing deeper insights into the DNA polymer dynamics in confined nanoscale environments.The results presented here have important implications for understanding genome regulation and the behavior of DNA under constrained conditions, akin to those found in cellular environments. By revealing how DNA's structural adaptations influence its translocation through nanopores, this work contributes to the broader field of nanopore-based sensing and sequencing technologies. Future research could build upon these findings to further elucidate the interplay between the DNA conformation and nanopore geometry, potentially leading to advancements in the design of nanopore devices for biomedical applications. Our study underscores the critical role of the nanopore shape in determining DNA behavior, paving the way for more sophisticated manipulation and analysis of genetic materials at the nanoscale.
Materials and methods
Nanopore fabrication
Quartz glass capillaries (0.2 mm inner diameter and 0.5 mm outer diameter) were sourced from Sutter Instruments (California, USA). These capillaries were then pulled to specific nanometer sizes using a laser-heated pipette puller (P-2000, Sutter Instrument, California, USA). Most experiments involved fabricated nanopores with a diameter of 14 ± 3 nm (mean ± standard deviation) and a cone semi-angle of 0.05 ± 0.01 radians (mean ± standard deviation) based on previous characterization. The pulling parameters were HEAT = 460, FIL = 0, VEL = 25, DEL = 170, and PULL = 225, detailed in the P2000 user guide. Afterward, the capillary was cut to the desired length and positioned in a custom PDMS microfluidic chip chamber. The PDMS used was the Sylgard 184 silicone elastomer kit from The Dow Chemical Company (MI, USA). Eight capillaries were assembled into the chip using the same method. The chip was then treated with plasma (Femto, Diener Electronic, Germany) for 20 seconds and bonded to a glass slide. Liquid PDMS sealed the capillaries to the chip, followed by heating at 100 °C for 2 hours. The chip underwent another plasma treatment for 5 minutes to make the nanopore surface hydrophilic. Finally, 4 M LiCl solution (1× TE buffer, pH 9.0) was used for wetting and allowing flow through the nanopores.Nanopore measurements
We performed nanopore measurements using a patch clamp amplifier (Axopatch 200B, Molecular Devices, CA, USA), with current signals collected at a 1 MHz sampling rate. A PCI-6251 data card (National Instruments) digitized and transmitted the data, while a Model 900CT low-pass Bessel filter (Frequency Devices) filtered the signal at 50 kHz. Two Ag/AgCl electrodes, prepared by oxidizing Ag wires in a 10% NaClO solution, were placed in the central (cis) and outer (trans) reservoirs to create an electric circuit across the nanopore. The chamber openings were sealed with tape to prevent water evaporation and subsequent LiCl concentration changes. Prior to sample measurements, we scanned the current–voltage characteristic of nanopores from −600 mV to 600 mV to determine the pore diameter. All experiments maintained a root-mean-square (RMS) noise below 6.5 pA for the baseline current. We used a custom LabVIEW GUI to monitor and record current signals, and Python scripts for data analysis. The number of translocation events was automatically counted, and a ‘kick-out’ (voltage polarity reversal) was triggered when a molecule got stuck in the pore.To prevent self-circularization of Lambda DNA, which has two sticky ends with complementary sequences, samples were heated at 70 °C for 10 minutes before measurements. Throughout the process, the Lambda DNA samples were handled gently using large-diameter, RNase-free pipette tips (200 μL size, Catalog number AM12650, Thermo Fisher Scientific, Waltham, USA) without vigorous centrifugation to avoid fragmentation. DNA samples were prepared at a concentration of 0.5 to 1 nM by mixing varying volumes of DNA, 8 M LiCl, and 4 M LiCl to achieve a final 4 M LiCl solution, depending on the original DNA concentration and molecular weight. During typical measurements, the translocation frequency of Lambda DNA was about 1 event per second, which is significantly longer than its Zimm relaxation time (67 ms) and translocation time (∼10 ms). This ensured that co-translocation of two DNA molecules was highly unlikely, minimizing any interference from other molecules during translocation.
Forward and backward experiments
Forward translocations were enabled by applying the positive voltage at the trans side, with the electrode immersed at the cis side grounded. After the forward experiment was conducted for 1 hour, the voltage polarity was reversed for both electrodes. In this situation, the post-translocation DNA molecules at the trans side would undergo backward translocation to the cis side.Ping-pong experiments
Voltage polarity switching was achieved using custom LabVIEW algorithms. The LabVIEW script continuously reads the current trace in cyclic loops, each loop lasting 4 ms, with a sampling frequency of 250 kHz and 1000 data points per loop. The script calculates the max–min value (difference between maximum and minimum currents) for each loop. A max–min value below 0.1 nA indicates no blockade, while a value between 0.1 nA and 3 nA signifies a blockade event. Upon detecting a blockade, the script initiates a waiting period defined by Tdelay without modifying the voltage. The number of loops for this waiting period is determined by dividing Tdelay by the loop duration (4 ms). After this period, the voltage is reversed. This ensured that the next forward translocation did not overlap with the current relaxation phase and prevented molecule trapping inside the nanopore. The ping-pong data were extracted and analyzed using a custom Python script. We calculated the ECD value for all translocation events to ensure these events are from the same DNA molecule (ESI Fig. S3†).Materials
The experiments utilized the following commercial reagents: nuclease-free water (Ambion, AM9937), 100 × Tris-EDTA buffer solution (Sigma-Aldrich, T9285), ≥99% pure lithium chloride for molecular biology (Sigma-Aldrich, L9650), and Tris-HCl BioPerformance certified with ≥99% purity (Sigma-Aldrich, T5941). Nanopore-measurement solutions and buffers made from these reagents were filtered twice using 0.22 μm Millipore syringe filter units (MF-Merck Millipore™, GSWP04700). Lambda DNA was sourced from NEB (500 μg ml−1, N3011S).Author contributions
Fei Zheng: Conceptualization, formal analysis, writing – original draft, validation, methodology, investigation. Quan Han: Investigation, funding acquisition, and writing – review & editing.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The datasets generated and/or analyzed during the current study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.Conflicts of interest
The authors declare no financial interests/personal relationships which may be considered as potential competing interests.Acknowledgements
F. Z. acknowledges funding from the China Scholarship Council (202106090221). Q. H. is supported by the China Postdoctoral Science Foundation (Certificate Number: 2023 M740593) and the Natural Science Research of Jiangsu Higher Education Institutions of China (22KJB460029).References
- Y.-L. Ying, Z.-L. Hu, S. Zhang, Y. Qing, A. Fragasso, G. Maglia, A. Meller, H. Bayley, C. Dekker and Y.-T. Long, Nat. Nanotechnol., 2022, 17, 1136–1146 CrossRef CAS PubMed.
- S. Pud, S.-H. Chao, M. Belkin, D. Verschueren, T. Huijben, C. Van Engelenburg, C. Dekker and A. Aksimentiev, Nano Lett., 2016, 16, 8021–8028 CrossRef CAS PubMed.
- S. Tetter, N. Terasaka, A. Steinauer, R. J. Bingham, S. Clark, A. J. Scott, N. Patel, M. Leibundgut, E. Wroblewski and N. Ban, Science, 2021, 372, 1220–1224 CrossRef CAS PubMed.
- K. Coshic, C. Maffeo, D. Winogradoff and A. Aksimentiev, Nature, 2024, 627, 905–914 CrossRef CAS PubMed.
- R. Kumar Sharma, I. Agrawal, L. Dai, P. S. Doyle and S. Garaj, Nat. Commun., 2019, 10, 1–9 CrossRef CAS PubMed.
- K. Chen, I. Jou, N. Ermann, M. Muthukumar, U. F. Keyser and N. A. Bell, Nat. Phys., 2021, 17, 1043–1049 Search PubMed.
- M. Mihovilovic, N. Hagerty and D. Stein, Phys. Rev. Lett., 2013, 110, 028102 CrossRef PubMed.
- K. Jeong, K. Luo, M. Lim, J. Jung, J. Yu, K. Kim and Y. Kim, Small, 2018, 14, 1801375 CrossRef PubMed.
- C. Plesa, L. Cornelissen, M. W. Tuijtel and C. Dekker, Nanotechnology, 2013, 24, 475101 CrossRef PubMed.
- R. T. Dame, F.-Z. M. Rashid and D. C. Grainger, Nat. Rev. Genet., 2020, 21, 227–242 CrossRef CAS PubMed.
- P. Mach, P. I. Kos, Y. Zhan, J. Cramard, S. Gaudin, J. Tünnermann, E. Marchi, J. Eglinger, J. Zuin and M. Kryzhanovska, Nat. Genet., 2022, 54, 1907–1918 CrossRef CAS PubMed.
- C.-Y. Lin, R. Fotis, Z. Xia, K. Kavetsky, Y.-C. Chou, D. J. Niedzwiecki, M. Biondi, F. Thei and M. Drndić, Nano Lett., 2022, 22, 8719–8727 CrossRef CAS PubMed.
- V. Sharma, N. Farajpour, L. S. Lastra and K. J. Freedman, Small, 2022, 18, 2106803 CrossRef CAS PubMed.
- F. Zheng, Y. Tao, W. Xu and J. Sha, Phys. Chem. Chem. Phys., 2023, 25, 10440–10446 RSC.
- I. M. Fujinami Tanimoto, J. Zhang, B. Cressiot, B. Le Pioufle, L. Bacri and J. Pelta, Chem. – Asian J., 2022, 17, e202200888 CrossRef PubMed.
- S. Confederat, I. Sandei, G. Mohanan, C. Wälti and P. Actis, Biophys. J., 2022, 121, 4882–4891 CrossRef CAS PubMed.
- X. Liu, P. Zimny, Y. Zhang, A. Rana, R. Nagel, W. Reisner and W. B. Dunbar, Small, 2020, 16, 1905379 CrossRef CAS PubMed.
- I. F. Davidson and J.-M. Peters, Nat. Rev. Mol. Cell Biol., 2021, 22, 445–464 CrossRef CAS PubMed.
- W. Liu, C. Ma, H. Wang and J. Sha, Langmuir, 2024, 40, 9622–9629 CrossRef CAS PubMed.
- Z. Zhang, J. Shen, H. Wang, Q. Wang, J. Zhang, L. Liang, H. Ågren and Y. Tu, J. Phys. Chem. Lett., 2014, 5, 1602–1607 CrossRef CAS PubMed.
- V. Wang, N. Ermann and U. F. Keyser, Nano Lett., 2019, 19, 5661–5666 CrossRef CAS PubMed.
- J. Li, M. Gershow, D. Stein, E. Brandin and J. A. Golovchenko, Nat. Mater., 2003, 2, 611–615 CrossRef CAS PubMed.
- C. Ma, W. Xu, W. Liu, C. Xu, G. Qin, D. Chen and J. Sha, J. Phys. Chem. B, 2024, 128, 2792–2798 CrossRef CAS PubMed.
- C. Ma, F. Zheng, W. Xu, W. Liu, C. Xu, Y. Chen and J. Sha, Small Methods, 2023, 2301485 CrossRef PubMed.
- N. Ermann, N. Hanikel, V. Wang, K. Chen, N. E. Weckman and U. F. Keyser, J. Chem. Phys., 2018, 149, 163311 CrossRef PubMed.
- S. W. Kowalczyk, D. B. Wells, A. Aksimentiev and C. Dekker, Nano Lett., 2012, 12, 1038–1044 CrossRef CAS PubMed.
- N. A. Bell, K. Chen, S. Ghosal, M. Ricci and U. F. Keyser, Nat. Commun., 2017, 8, 1–8 CrossRef CAS PubMed.
- F. Zheng, M. Alawami, J. Zhu, C. M. Platnich, J. Sha, U. F. Keyser and K. Chen, Nano Lett., 2023, 23, 11145–11151 CrossRef CAS PubMed.
- K. Chen, N. A. Bell, J. Kong, Y. Tian and U. F. Keyser, Biophys. J., 2017, 112, 674–682 CrossRef CAS PubMed.
- N. A. Bell and U. F. Keyser, Nat. Nanotechnol., 2016, 11, 645–651 CrossRef CAS PubMed.
- K. Chen, A. Choudhary, S. E. Sandler, C. Maffeo, C. Ducati, A. Aksimentiev and U. F. Keyser, Adv. Mater., 2023, 35, 2207434 CrossRef CAS PubMed.
- F. Zheng, A. Suma, C. Maffeo, K. Chen, M. Alawami, J. Sha, A. Aksimentiev, C. Micheletti and U. F. Keyser, arXiv, 2024, preprint, arXiv:2407.16290, https://arxiv.org/abs/2407.16290.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an01068j |
| This journal is © The Royal Society of Chemistry 2024 |