Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optical control of nanobody-mediated protein activity modulation with a photocleavable fluorescent protein†
Mizuki
Endo
 ,
Saki
Tomizawa
,
Qiaoyue
Kuang
and
Takeaki
Ozawa
,
Saki
Tomizawa
,
Qiaoyue
Kuang
and
Takeaki
Ozawa
 *
*
Department of Chemistry, School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. E-mail: ozawa@chem.s.u-tokyo.ac.jp; Tel: +81 3-5841-4351
First published on 25th June 2024
Abstract
Antibodies are crucial in various biological applications due to their specific binding to target molecules, altering protein function and structure. The advent of single-chain antibodies such as nanobodies has paved the way for broader applicability in both research and therapies due to their small size and efficient tissue penetration. Recently, several approaches have been reported to optically control the antigen-binding affinity of nanobodies. Here, we show an alternative strategy for creating photo-activatable nanobodies. By fusing the photocleavable protein PhoCl with the N-terminus of the nanobody (named optoNb60), we successfully demonstrated light-dependent restoration of the antigen-binding ability and the following modulation of the activity of a target protein, the beta-2 adrenergic receptor. Moreover, the activation of optoNb60 was monitored by the fluorescence changes upon photoconversion. The compatibility of the uncaging design with the previously reported optogenetic molecules using nanobodies will contribute to the further optimization of the response capabilities of existing optogenetic tools, thereby expanding their applicability.
Introduction
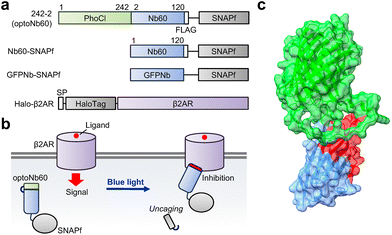
The unique ability of antibodies to specifically bind to target molecules with high affinity is indispensable in various biological applications, including protein activity modulation.1,2 By selectively binding to specific regions of target proteins, antibodies activate or inactivate protein functions by altering protein conformations and blocking active sites. In contrast to conventional antibodies, which typically exist as tetrameric structures with large molecular weights, the emerging use of single-chain antibodies, such as nanobodies,3 has recently gained significant attention.4–9 The remarkably small size of nanobodies enables exceptional tissue-penetrating capabilities, which allow them to access target molecules within tissues more effectively than traditional antibodies. Nanobodies are also used for the modulation of protein functions; for instance, a nanobody called Nb60 binds to the beta-2 adrenergic receptor (β2AR) and inhibits ligand-dependent β2AR-mediated signaling.10 The versatility of nanobodies as modulators for protein functions has led to their widespread use in research and therapeutics, underscoring their importance in understanding and manipulating biological processes at the molecular level.Various strategies have been developed to regulate the antigen-binding affinities of nanobodies. One such approach involves employing chemogenetic control by integrating circularly permutated bacterial dihydrofolate reductase (cpDHFR)11 into complementary-determining region 3 (CDR3) of the nanobody, named ligand-modulated antibody fragments (LAMAs).12 This allows for the modulation of antigen binding by administration of the DHFR inhibitor trimethoprim. Alternatively, chemical induction of protein–protein interactions has been utilized to reassemble split nanobody fragments.13 Furthermore, a split-protein system has been combined with a light-inducible protein–protein interaction system, known as an optobody, utilizing optical dimerization tools called Magnets.14 Another optogenetic strategy akin to LAMAs involves the incorporation of a light-responsive LOV domain into the non-CDR loop, which enables reversible modulation of the affinity.15 Although these design strategies enable the artificial control of antigen binding, the activation of modified antibodies cannot be confirmed until the analysis of target protein functions.
In the present study, we aimed to develop a new design for generating photo-activatable caged nanobodies by genetically fusing a photocleavable protein called PhoCl16 with the N-terminal side of the nanobody locating an antigen-binding region. PhoCl undergoes photoconversion upon 405 nm illumination and then irreversibly cleaves into two fragments. By fusing PhoCl with the N-terminus of Nb60 (optoNb60), light-dependent uncaging of the fused nanobody was achieved, which enabled the subsequent modulation of β2AR function. Moreover, the activation of the caged nanobody can be monitored using the photoconversion of fluorescence originating from the fused PhoCl. The present uncaging design will contribute to the precise control of target protein functions by tracking the photoconversion fluorescence, thereby expanding their applicability such as quantitative control of biological phenomena.
Materials and methods
Plasmid construction
The cDNAs of SNAPf tag, Halo tag, and PhoCl were amplified from pSNAPf-ADRB2 (NEB), p53-HaloTag (Promega), and PC-cadherin,17 respectively. The cDNA of Nb60 was generated by PCR. The PCR products were subcloned into the pcDNA4/myc-His B vector (Thermo Fisher Scientific, CA, USA).Cell culture and transfection
Human embryonic kidney (HEK) 293 cells (CRL-1573, ATCC) and COS7 (CRL-1651, ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, high glucose; Wako Pure Chemical Industries Ltd, Japan) supplemented with 10% fetal bovine serum (FBS; Gibco, CA, USA), 100 unit mL−1 penicillin and 100 μg mL−1 streptomycin (Gibco) at 37 °C in 5% CO2. HEK293 cells stably expressing β2AR were cultured in DMEM supplemented with 10% FBS, 100 units mL−1 penicillin, 100 μg mL−1 streptomycin (Gibco), 0.8 μg μL−1 G418 sulfate (Millipore Sigma), and 0.05 μg μL−1 Zeocin™ selection reagent (Thermo Fisher Scientific). HEK293 cells and COS7 cells were transiently transfected with expression vectors using the TransIT-LT1 transfection reagent (Mirus) or polyethylenimine “Max” (Mw 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) – high potency linear PEI (Cosmo Bio Co., Ltd), respectively, according to the manufacturer's protocol.
000) – high potency linear PEI (Cosmo Bio Co., Ltd), respectively, according to the manufacturer's protocol.
Western blot analysis
Cells were stimulated with 405 nm LED light (6 mW cm−2 at 405 nm, 30 s min−1, TH-160X120VL405-M1, CCS) for 30 min before cell collection. The cells were collected in lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 50 mM NaF, and 0.5% (v/v) NP-40) and boiled with 5× sample buffer (50 mM Tris-HCl, 2% sodium dodecyl sulfate (SDS), 5% glycerol, 1% 2-mercaptoethanol, and 0.004% bromophenol blue) at 95 °C for 5 min.For immunoprecipitation analysis, samples were lysed with mild lysis buffer (20 mM HEPES, 100 mM NaCl, and 0.5% maltose-neopentyl glycol). Insoluble materials were removed by centrifugation (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min). Halo-β2AR-myc was immunoprecipitated with Myc-Trap® agarose (Proteintech Group).
000g for 5 min). Halo-β2AR-myc was immunoprecipitated with Myc-Trap® agarose (Proteintech Group).
The samples were separated by SDS polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane (GE Healthcare). The membrane was blocked with 5% skim milk in Tris-buffered saline containing Tween-20 (TBS-T; 150 mM NaCl, 0.05% Tween-20, 50 mM Tris-HCl, pH 8.0). The membrane was incubated with the indicated primary antibodies and the appropriate secondary antibodies labelled with horseradish peroxidase (ESI Table S2†). Immunoblot bands were detected using the SuperSignal West Femto substrate (Thermo Scientific, IL, USA) with an image analyzer (LAS-4000 mini; Fuji Photo Film Co. Ltd, Tokyo, Japan).
Microscopy observation
Transiently transfected cells seeded on a 35 mm glass bottom dish (IWAKI) were stained with the HaloTag TMR ligand (Promega) and/or SNAP-Cell 647-SiR (New England Biolabs) according to the manufacturer's protocol. The cells were observed with an observation medium (Dulbecco's modified Eagle's medium (high glucose)) without L-glutamine and phenol red (FUJIFILM Wako Pure Chemical Corp.) and 10% fetal bovine serum under a confocal fluorescence microscope (IX-81, FV-1000D, Olympus). The acquired images were analyzed using ImageJ software (NIH).18Bioluminescence assay
For the cAMP assay, the cells were transiently transfected with the plasmids of the caged nanobody and the pGloSensorTM-22F cAMP plasmid (Promega). The cells were seeded in a 96-well multiplate (Corning). The seeded cells were stimulated with 405 nm LED light (7.2 mW cm−2 at 405 nm, 30 s min−1, TH-160X120VL405-M1, CCS) for 30 min before the assay. The medium was exchanged with a measurement medium (Dulbecco's modified Eagle's medium (high glucose) with L-glutamine and phenol red, 10% fetal bovine serum, 1% penicillin–streptomycin, and 2% GloSensor™ cAMP reagent stock solution (Promega)), and then incubated in the dark for 2 h. The cells were then stimulated with 1 nM isoproterenol. The bioluminescence was measured using a microplate reader TriStar LB941 (Berthold).Results
Photocleavage of the caged nanobody
The complementary-determining region of Nb60, which is the binding interface of the target β2AR, is located on the N-terminal side. To modulate the binding ability of Nb60, candidate molecules were generated by fusing the C-terminus of the photocleavable protein, called PhoCl,17 with Nb60 at Gln2 or Val3, directly or via glycine linkers (Gly, Gly–Gly, Gly–Gly–Gly) (Fig. 1, Fig. S1a†). For the following characterization, the FLAG tag (DYKDDDDK) and SNAPf tag were attached to the C-terminus. Nb60 fused with the SNAPf tag and an anti-GFP nanobody (GFPNb) fused with the SNAPf tag were also prepared as controls. Upon photocleavage of PhoCl by 405 nm light illumination, optoNb60 possibly binds to β2AR and inhibits ligand-induced signalling (Fig. 1b).To examine the photocleavage of PhoCl in the candidate molecules upon blue light illumination, HEK293 cells transiently expressing the molecules were stimulated with blue light (405 nm) for 30 min and then subjected to western blotting (Fig. S1b†). The immunoblotted data clearly showed a light-dependent appearance of the band at around 37 kDa in candidate molecules where PhoCl was fused with Nb60 at Gln2 directly (242-2) or via Gly (242-G-2) or fused at Val3via Gly (242-G-3). The band size corresponded to the estimated molecular weight of Nb60 fused with the FLAG and SNAPf tags. Considering that the position of PhoCl should be close enough to cover the antigen-binding surface, the candidate molecule 242-2, which showed clear photocleavage and the shortest linker length between PhoCl and Nb60, was named optoNb60. Indeed, the antigen-binding surface of optoNb60 was partially hindered by the fused PhoCl according to the 3D structure predicted by AlphaFold2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 (Fig. 1c). The incomplete photocleavage might be improved by future optimization under illumination conditions or using the improved version of PhoCl.20 Based on these results, we conclude that PhoCl fused with Nb60 detached from Nb60 upon illumination, which could be regarded as light-dependent uncaging.
19 (Fig. 1c). The incomplete photocleavage might be improved by future optimization under illumination conditions or using the improved version of PhoCl.20 Based on these results, we conclude that PhoCl fused with Nb60 detached from Nb60 upon illumination, which could be regarded as light-dependent uncaging.
Light-dependent recovery of optoNb60's antigen-binding ability
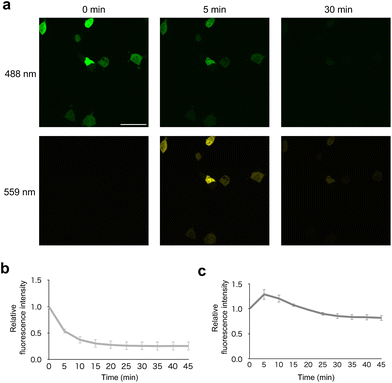
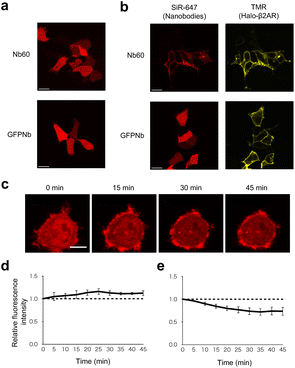
To confirm the photoconversion of PhoCl in optoNb60 in living cells, it was analyzed using fluorescence emitted by excitation light at 488 nm and 559 nm (Fig. 2a). Before photoconversion by illumination at 405 nm, the fluorescence was detected upon 488 nm excitation. Upon illumination at 405 nm, the fluorescence emitted by excitation at 488 nm gradually disappeared within 10 min (Fig. 2b). In contrast, the fluorescence emitted by excitation at 559 nm transiently increased around 5 min after the onset of illumination and slowly decayed within 20 min (Fig. 2c). These results suggest that the photoconversion of PhoCl in optoNb60 occurred within 5 min, followed by dissociation of the cleaved fragments over 20 min under the illumination conditions.To examine where the interaction between Nb60 and β2AR occurs inside the cells, their intracellular localization was investigated. In cells without β2AR expression, both GFPNb and Nb60 labelled with SNAP-Cell 647-SiR showed cytosolic expression (Fig. 3a). In contrast, Nb60 showed localization on the plasma membrane in cells transiently expressing β2AR labelled with HaloTag TMR, which also localized on the plasma membrane (Fig. 3b). Taken together, these results confirmed that the interaction between Nb60 and β2AR occurred on the plasma membrane.
To investigate the antigen-binding ability of optoNb60 before and after 405 nm illumination, the localization of optoNb60 was analyzed by fluorescence from the labelled dye linked via the SNAPf tag (Fig. 3c). Before photoconversion, the caged nanobody was expressed in the cytosol, suggesting the absence of interaction with β2AR located on the plasma membrane. Upon 405 nm illumination, the fluorescence from optoNb60 increased around the plasma membrane and reached a plateau around 25 min after the onset of illumination (Fig. 3d), whereas the fluorescence in the cytosol gradually decreased (Fig. 3e). The interaction between β2AR and photocleaved optoNb60 was also detected by immunoprecipitation (Fig. S2†). These results indicate that the antigen-binding ability of Nb60 was inhibited by the fusion of PhoCl with optoNb60 and was recovered upon photoconversion and subsequent dissociation of PhoCl by 405 nm illumination.
Modulation of β2AR activity by the caged nanobody
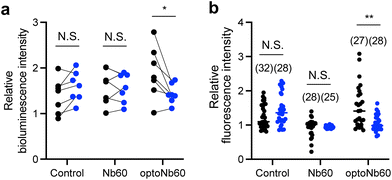
β2AR couples to the stimulatory G protein (Gs), which stimulates the effector adenylyl cyclase to transiently produce cAMP.21,22 To investigate ligand-induced activation of β2AR, cytosolic cAMP levels were monitored using GloSensor-22F.23 HEK293 cells stably expressing β2AR were transiently cotransfected with GloSensor-22F and optoNb60. The transfected cells were illuminated with 405 nm LED light (7.2 mW cm−2, 30 s min−1) or kept in the dark for 30 min, and then stimulated with 10 nM isoproterenol. Ligand-induced changes in cAMP levels were monitored using the bioluminescence of GloSensor-22F (Fig. 4a, Fig. S3†). Considering that the expression level of β2AR was suppressed by the cotransfected proteins (Fig. S3b†), the bioluminescence of Glosensor-22F was normalized by that before stimulation in each condition in independent experiments. In cells expressing optoNb60, the cAMP level increased upon ligand activation by 1.90-fold in the dark compared to that in non-stimulated cells. In contrast, ligand-induced cAMP level changes in the cells stimulated with 405 nm light decreased 1.43-fold over the non-stimulated cells. In cells expressing GloSensor-22F and Nb60-SNAPf or expressing only GloSensor-22F, ligand-induced changes in cAMP levels were not affected by light illumination at 405 nm. Additionally, β2AR mobilizes Ca2+via activation of phospholipase C, which is independent of the cAMP pathway.24 To further confirm the inhibitory effect of optoNb60 on β2AR activity, the intracellular Ca2+ level was monitored by R-GECO.25 HEK293 cells stably expressing β2AR were transiently cotransfected with R-GECO and optoNb60. The cells were stimulated with light and then subjected to Ca2+ level analysis under a confocal microscope (Fig. 4b). In control cells, the intracellular Ca2+ level increased in response to ligand stimulation, which was inhibited by Nb60 expression. In cells expressing optoNb60 in the dark, the Ca2+ level increased upon ligand activation by 1.52-fold compared to that in non-stimulated cells. In contrast, ligand-induced Ca2+ level changes in the cells stimulated with 405 nm light decreased 1.06-fold over the non-stimulated cells. These results demonstrate that optoNb60 inhibits β2AR activity by light-induced uncaging.Conclusions
In this study, we developed a genetically encoded caged nanobody named optoNb60, whose antigen-binding ability against β2AR was optically controlled by light-dependent uncaging of N-terminally fused PhoCl. We confirmed that the fused PhoCl undergoes photoconversion and cleaves upon 405 nm illumination, which results in the recovery of the antigen-binding ability of the nanobody. The optically uncaged nanobody allows binding to β2AR on the plasma membrane and inhibits ligand-dependent activation of downstream signaling. The activation of the caged nanobody can also be monitored by the photoconversion of fused PhoCl, whose fluorescence emitted by excitation at 559 nm transiently increased upon photoconversion. The improvement in the photocleavage efficiency will further enhance the inhibitory effect. The present uncaging design will contribute to the precise control of target protein functions by tracking the photoconversion fluorescence, thereby expanding their applicability, such as the quantitative control of biological phenomena.Author contributions
M. E. and T. O. conceived the project, M. E., Q. K. and S. T. performed the experiments, and M. E. and T. O. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (JP19K05538 (M. E.) and 22H00322 (T. O.)) and the Core-to-Core Program (T. O.).References
- M. L. Chiu, D. R. Goulet, A. Teplyakov and G. L. Gilliland, Antibody Structure and Function: The Basis for Engineering Therapeutics, Antibodies, 2019, 8(4), 55 CrossRef CAS PubMed.
- S. Magaki, S. A. Hojat, B. Wei, A. So and W. H. Yong, An Introduction to the Performance of Immunohistochemistry, Methods Mol. Biol., 2019, 1897, 289–298 CrossRef CAS PubMed.
- C. Hamers-Casterman, T. Atarhouch, S. Muyldermans, G. Robinson, C. Hammers and E. B. Songa, et al., Naturally occurring antibodies devoid of light chains, Nature, 1993, 363(6428), 446–448 CrossRef CAS PubMed.
- G. Bao, M. Tang, J. Zhao and X. Zhu, Nanobody: a promising toolkit for molecular imaging and disease therapy, EJNMMI Res., 2021, 11(1), 6 CrossRef PubMed.
- H. Revets, P. De Baetselier and S. Muyldermans, Nanobodies as novel agents for cancer therapy, Expert Opin. Biol. Ther., 2005, 5(1), 111–124 CrossRef CAS PubMed.
- W. Wolfson, Ablynx Makes Nanobodies from Llama Bodies, Chem. Biol., 2006, 13(12), 1243–1244 CrossRef CAS PubMed.
- B.-k. Jin, S. Odongo, M. Radwanska and S. Magez, Nanobodies: A Review of Generation, Diagnostics and Therapeutics, Int. J. Mol. Sci., 2023, 24(6), 5994 CrossRef CAS PubMed.
- E. Beghein and J. Gettemans, Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein–Protein Interaction Analysis, and Protein Function Exploration, Front. Immunol., 2017, 8, 771 CrossRef PubMed.
- M. Rashidian and H. Ploegh, Nanobodies as non-invasive imaging tools, Immuno-Oncol. Technol., 2020, 7, 2–14 CrossRef CAS PubMed.
- D. P. Staus, L. M. Wingler, R. T. Strachan, S. G. Rasmussen, E. Pardon and S. Ahn, et al., Regulation of β2-adrenergic receptor function by conformationally selective single-domain intrabodies, Mol. Pharmacol., 2014, 85(3), 472–481 CrossRef PubMed.
- M. Iwakura and T. Nakamura, Effects of the length of a glycine linker connecting the N-and C-termini of a circularly permuted dihydrofolate reductase, Protein Eng., Des. Sel., 1998, 11(8), 707–713 CrossRef CAS PubMed.
- H. Farrants, M. Tarnawski, T. G. Müller, S. Otsuka, J. Hiblot and B. Koch, et al., Chemogenetic Control of Nanobodies, Nat. Methods, 2020, 17(3), 279–282 CrossRef CAS PubMed.
- D. Yu, H. Lee, J. Hong, H. Jung, Y. Jo and B.-H. Oh, et al., Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins, Nat. Methods, 2019, 16(11), 1095–1100 CrossRef CAS PubMed.
- F. Kawano, H. Suzuki, A. Furuya and M. Sato, Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins, Nat. Commun., 2015, 6(1), 6256 CrossRef CAS PubMed.
- A. A. Gil, C. Carrasco-López, L. Zhu, E. M. Zhao, P. T. Ravindran and M. Z. Wilson, et al., Optogenetic control of protein binding using light-switchable nanobodies, Nat. Commun., 2020, 11(1), 4044 CrossRef CAS PubMed.
- W. Zhang, A. W. Lohman, Y. Zhuravlova, X. Lu, M. D. Wiens and H. Hoi, et al., Optogenetic control with a photocleavable protein, PhoCl, Nat. Methods, 2017, 14(4), 391–394 CrossRef CAS PubMed.
- M. Endo, T. Iwawaki, H. Yoshimura and T. Ozawa, Photocleavable Cadherin Inhibits Cell-to-Cell Mechanotransduction by Light, ACS Chem. Biol., 2019, 14(10), 2206–2214 CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair and T. Pietzsch, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682 CrossRef CAS PubMed.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, ColabFold: making protein folding accessible to all, Nat. Methods, 2022, 19(6), 679–682 CrossRef CAS PubMed.
- X. Lu, Y. Wen, S. Zhang, W. Zhang, Y. Chen and Y. Shen, et al., Photocleavable proteins that undergo fast and efficient dissociation, Chem. Sci., 2021, 12(28), 9658–9672 RSC.
- S. B. Liggett, Molecular and genetic basis of β2 -adrenergic receptor function, J. Allergy Clin. Immunol., 1999, 104(2), S42–SS6 CrossRef CAS PubMed.
- J. D. Violin, L. M. DiPilato, N. Yildirim, T. C. Elston, J. Zhang and R. J. Lefkowitz, β2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics, J. Biol. Chem., 2008, 283(5), 2949–2961 CrossRef CAS PubMed.
- B. F. Binkowski, F. Fan and K. V. Wood, Luminescent Biosensors for Real-Time Monitoring of Intracellular cAMP, in Signal Transduction Protocols, ed. L. M. Luttrell and S. S. G. Ferguson, Humana Press, Totowa, NJ, 2011, pp. 263–271 Search PubMed.
- M. Galaz-Montoya, S. J. Wright, G. J. Rodriguez, O. Lichtarge and T. G. Wensel, β2-Adrenergic receptor activation mobilizes intracellular calcium via a non-canonical cAMP-independent signaling pathway, J. Biol. Chem., 2017, 292(24), 9967–9974 CrossRef CAS PubMed.
- Y. Zhao, S. Araki, J. Wu, T. Teramoto, Y.-F. Chang and M. Nakano, et al., An Expanded Palette of Genetically Encoded Ca2+ Indicators, Science, 2011, 333(6051), 1888–1891 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00433g |
| This journal is © The Royal Society of Chemistry 2024 |