Open Access Article
Open Access ArticleDynamic video recognition for cell-encapsulating microfluidic droplets†
Yuanhang
Mao‡
,
Xiao
Zhou‡
,
Weiguo
Hu
,
Weiyang
Yang
and
Zhen
Cheng
 *
*
Department of Automation, Tsinghua University, Beijing, 100084, China. E-mail: zcheng@mail.tsinghua.edu.cn
First published on 16th February 2024
Abstract
Droplet microfluidics is a highly sensitive and high-throughput technology extensively utilized in biomedical applications, such as single-cell sequencing and cell screening. However, its performance is highly influenced by the droplet size and single-cell encapsulation rate (following random distribution), thereby creating an urgent need for quality control. Machine learning has the potential to revolutionize droplet microfluidics, but it requires tedious pixel-level annotation for network training. This paper investigates the application software of the weakly supervised cell-counting network (WSCApp) for video recognition of microdroplets. We demonstrated its real-time performance in video processing of microfluidic droplets and further identified the locations of droplets and encapsulated cells. We verified our methods on droplets encapsulating six types of cells/beads, which were collected from various microfluidic structures. Quantitative experimental results showed that our approach can not only accurately distinguish droplet encapsulations (micro-F1 score > 0.94), but also locate each cell without any supervised location information. Furthermore, fine-tuning transfer learning on the pre-trained model also significantly reduced (>80%) annotation. This software provides a user-friendly and assistive annotation platform for the quantitative assessment of cell-encapsulating microfluidic droplets.
Introduction
Microfluidic chips integrate biochemical operating units, regulate the fluid flow through microchannels, and create the lab-on-a-chip or micro total analysis system (μTAS).1 Their vast applications are seen in gene sequencing, in vitro diagnosis, and health monitoring.2 Droplet microfluidics is one of the most sensitive and high-throughput technologies for injecting immiscible fluids into microfluidic chips,3 generating uniform droplets,4 encapsulating biological contents (e.g. nucleic acids, proteins or cells), and subsequently manipulating and detecting droplets.5,6 It has been successfully implemented in many fields, including biomedicine, industrial chemistry, and environmental science.7 Numerous applications have been developed, such as the digital droplet polymerase chain reaction (ddPCR),8 single-cell sequencing,9 digital enzyme-linked immunosorbent assay (dELISA),10 material synthesis,11 drug delivery12 and high-throughput screening.13 Since 2015, various single-cell sequencing techniques, including Drop-seq,14 indrop15 and scRNA-seq,16 have been used to deconstruct cell populations and infer gene relationships by encapsulating a single cell into a microdroplet. Instead of manual analysis and tedious counting, growing advances in droplet microfluidics have created an urgent requirement for quality control of uniform droplets encapsulating a single cell.Passive generation of homogeneous droplets has been proposed using different techniques,17 while most microfluidic droplets are generated in the handcrafted polydimethylsiloxane (PDMS) chip.18 For optimal reproducibility and robustness, flow-focusing structures have been widely used to generate microdroplets and encapsulate single cells. However, the Poisson random distribution limits the number of encapsulated cells per droplet with a theoretical maximum of 36.78% droplets containing only one cell.19 Although several active droplet methods have been proposed to break this limit,17 it is still of great significance for real-time monitoring and control of the single-cell encapsulation rate.
In addition, various dynamic factors related to the generation process can also significantly affect the final performance of microdroplets,20 such as fluid stability (bubbles) which affects the diameter uniformity and solution dispersibility (cell adhesion or sedimentation) which affects the encapsulation rate. For instance, a 3% deviation in diameter leads to an 8.5% deviation in the quantitative results of ddPCR and dELISA.21 Encapsulation rate is essential in determining the delivery efficiency.22 More importantly, the single-cell encapsulation rate or the co-encapsulation rate of single cells and encoding magnetic/gel beads in one microdroplet greatly affects the efficiency of cell/microbial screening and the reliability of single-cell sequencing.19 It is also the reason for developing post-processing algorithms to remove empty droplets23 and double cell droplets24 or designing new structures to improve the encapsulation rate.25 In conclusion, computational and intelligent methods for evaluating dynamic droplet uniformity and quantifying encapsulated cells are highly desired.
To independently distinguish both droplets and encapsulated cells in microscopic images, two-stage object recognition methodologies were preferentially applied.26,27 In the first stage, potential droplet candidates are created by morphological analysis,28e.g. edge-feature extraction with Hough transform (HT),29,30 background models, and connected component analysis,31 to find the circular contours of the droplet foreground and segment their borders by masks. These methods are effective for transparent and separable droplets, but struggle with opaque and adherent droplets. In the second stage, researchers investigated droplets containing particles by measuring the grayscale deviation32 and standard deviation (SD) of the distance between the contour and the gravity center.26 However, these morphological approaches are strongly limited by image quality. Machine learning techniques are also being developed to identify encapsulated cells and categorize droplets (see Table 1). Random forest was implemented to identify beads within droplets.27 To categorize encapsulating droplets, handcrafted features were also fed into a support vector machine (SVM)33 and convolutional neural networks (CNNs).34,35 However, these traditional classifiers and morphological approaches cannot count the cell quantity in each droplet. Influential object detectors, e.g. You Only Look Once (YOLO), have also been applied to classify droplets and detect cells showing significant improvements.36,37 We have noticed that the primary distinction between single-cell and multi-cell encapsulation is the cell quantity rather than the divergence of cell-like properties and have recently developed a weakly supervised cell-counting network (WSCNet) for image recognition.38–41 Different from fully supervised learning (e.g. YOLO) that requires cell-level labels, e.g. cell population or precise location,37,42 WSCNet utilized droplet-level labels (empty, single-cell, and multi-cell encapsulation) to prevent tedious annotation.
| Method | Droplet application | Tested types | Detection output | Cell location | Precision | Ref. |
|---|---|---|---|---|---|---|
| Methodology | Cell sorting | 1 | Droplet category | Yes | 85% | 28 |
| Handcrafted CNN | Cell sorting | 3 | Droplet category | No | 90% | 35 |
| YOLO v4-tiny | Cell sorting | 3 | Droplet category/cell quantity | Yes | 92% | 36 |
| YOLO v3/v5 | Cell encapsulation | 1 | Droplet category/cell quantity | Yes | 91% | 37 |
| WSCApp | Cell encapsulation | 6 | Droplet category/cell quantity | Yes | >94% | This work |
Furthermore, several studies have concentrated on cell-encapsulating droplets utilizing microscope videos captured by a high-speed camera,31,32,43 while most droplets generated in microchannels are transparent and separable. With the dynamic recognition of video frames, real-time control of droplet generation and cell encapsulation can be realized to improve the diameter stability and single-cell rate and finally realize high-throughput droplet sorting.35–37 Therefore, we believe that it is critical to design user-friendly application software with a graphical user interface (GUI) to facilitate the quantitative assessment of both the droplet images and videos. To address this need, we propose a new application software of the weakly supervised cell-counting network (WSCApp), which is designed for video recognition of cell-encapsulating microfluidic droplets. WSCApp integrates our original WSCNet model38 (for droplet classification, cell counting, and cell location) with traditional classifiers (for droplet classification) to enhance detection accuracy. Furthermore, we also designed dark-field and bright-field segmentation algorithms (optional) for different imaging conditions. To evaluate the generalizability of the models, our methods were systematically verified on different droplets encapsulating six types of cells/beads, which were collected from various microfluidic structures. We also summarized the advantages of our methods over previous work, as shown in Table 1 (detailed in ESI Table S1†), with a micro-F1 score > 0.94. Compared to fully supervised learning frameworks, WSCApp's use of pre-trained models and the transfer learning strategy has also improved the labelling efficiency (triple droplet-level labels) and further reduced manual annotation (>80%). Quantitative experimental results on intricate data also indicated that our approach can not only accurately distinguish droplet encapsulations, but also locate each cell.
Working principle
Principle of droplet generation and cell encapsulation
Anna et al. proposed flow-focusing structures to generate microdroplets for the first time.50 In this geometry, the dispersed phase is subjected to both the pressure and shear force of two symmetrically continuous phases and breaks into microdroplets. As the dispersed and two continuous phases continue to enter the intersection, the fracture process occurs periodically. Given its ability to generate small droplets at high frequency and throughput, the flow-focusing structure is currently widely used.One of the most commonly used methods for single-cell droplet encapsulation involves diluting cells into the dispersed phase prior to droplet formation.19 The fundamental idea is to sufficiently dilute the cell suspension so that only one cell appears in a single droplet (ESI Fig. S1†). Assuming that there are λ cells per microdroplet (CPD, cell density divided by droplet volume), the following formula can be used to obtain the Poisson probability of k cells per microdroplet:
 | (1) |
Droplet recognition, classification and cell counting
We have developed comprehensive application software named WSCApp with a GUI for dynamic recognition of cell-encapsulating microfluidic droplets. WSCApp enables objective and automatic statistical analysis of droplet quality and cell quantity. The two-stage object recognition methodology is utilized by WSCApp as shown in Fig. 1. The droplet proposals are first segmented, and then, the proposals are fed to optional detectors to remove false positive droplets, categorize droplets, and further count the cell population.In the first stage, to increase the generalization of detection in both droplet images and videos, we create both dark-field and bright-field droplet recognition algorithms, which are optional on WSCApp, for different illumination scenarios. We have recently developed a novel morphological approach named adaptive scale template matching (ASTM)40 to generate proposals of opaque and adherent droplets for static microscopic images. Considering the video characteristics in which most droplets are clearly separated and transparent, in this study, we designed a bright-field algorithm that utilized contour extraction followed by HT (CEHT).
In the second stage, WSCApp integrates both conventional classifiers and our WSCNet38 for droplet classification, cell counting and location, respectively. Firstly, widely studied CNN-based classifiers are applied as baseline methods for droplet classification. Secondly, WSCNet was used to estimate the number of cells within each droplet to categorize droplets according to the number of encapsulated cells. Additionally, WSCNet offers location prediction for each cell, which is more comprehensible and rational than a traditional classifier. Cell localization is also critical for subsequent verification and analysis of the characteristics of each cell. To enhance the software's real-time performance, the code was reconstructed into the C++ code for arithmetic acceleration. Please refer to ESI Note 1† for further details.
Assistive annotation and transfer learning using interactive WSCApp
Currently, most machine learning algorithms assume that the feature distribution of training and test data is similar, but this is often not feasible for different droplet data collected by different laboratories. Given the large differences between imaging systems, microfluidic chips, experimental conditions, and cell types, the standard procedure for applying algorithms is usually to relabel all data from scratch, which is very time consuming and limited by the quantity and quality of the available data. Finally, networks need to be retrained to ensure accuracy.To further mitigate the burden of data annotation, we investigated pre-trained models and transfer learning strategies in this study, as shown in Fig. 2. First, we trained both classifiers and WSCNet using the static droplet image we had collected experimentally38 and integrated them into WSCApp. Second, we applied WSCApp directly to new droplet data and provided initial annotation. Third, the annotation results could be viewed and modified directly on the GUI for assistive annotation. Fourth, we retrained classifiers or WSCNet models with triple droplet-level labels. If the target domain data were insufficient, transfer learning could be used to transfer knowledge to a new scenario,44 fine-tuning the initial model weights to improve classification and recognition performance. Lastly, we compared and validated the generalization capability of the algorithm on complex data collected from the literature (ESI Table S2†).
Results and discussion
Pre-trained model results on droplet datasets
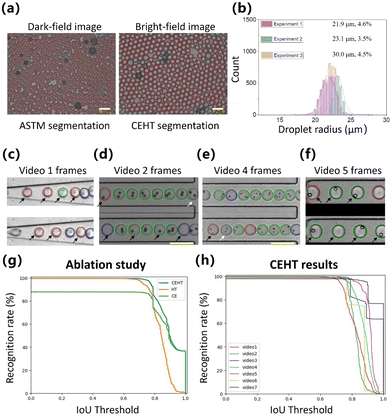
By performing microfluidic experiments and image acquisition, we collected static images of encapsulating droplets for pre-training both the WSCNet model and traditional classifiers. Droplet video datasets were collected for the validation of assistive annotation and transfer learning on WSCApp software. The classifiers and WSCNet model were applied directly to the new video dataset, and preliminary annotations were provided. Representative results of droplet segmentation and classification are shown in Fig. 3.After comparative analysis, the ASTM algorithm has proven to be superior in segmenting highly adherent and opaque droplets in dark-field images. The CEHT method, on the other hand, displays excellent performance in detecting transparent droplets on bright-field images. They showed recall values exceeding 95% and 98% for 21k dark-field and 11k bright-field droplets, respectively, which well demonstrated the detection performance of the two algorithms in their specific scenarios. Conversely, the ASTM algorithm leads to false negative droplets in bright-field images, with less than 10% of droplets being segmented (ESI Fig. S2 and S3†), which is also the motivation behind the design of bright-field algorithms for this study. In addition to good performance on bright-field images, CEHT can also be applied to dark-field images to some extent, achieving 95% recall and 93% accuracy and indicating a low number of false positives and negatives. The main distinction between the two processing algorithms lies in whether the droplets are connected after binarization and filling and whether each droplet possesses distinct edges. Therefore, CEHT has a wider scope of application and can be used as a preliminary attempt in the case of undemanding recognition rate requirements.
Second, in addition to automatically displaying droplet statistics, such as mean diameter and SD value, on the message window, WSCApp also generates recognized text files that contain the droplet position and category for classifiers and the WSCNet model, or the cell number and position only for the WSCNet model. We collected 4135, 2286 and 3981 droplets from approximately 20 images in three independent experiments. Their diameter distribution is plotted in Fig. 3(b) with mean radius and CV values. The CVs of 3.5–4.6% indicate good monodispersity and uniformity in droplet size. Further parameters of Fig. 3(b) are listed in ESI Table S3.†
Next, the GUI allows immediate review, sequential browsing of images or video frames, and automatic loading of the corresponding annotation files. CEHT's excellent performance further reduces the manual workload on droplet segmentation, as shown in Fig. 3(c–f), while less than 3% of droplets in video datasets need to be remarked with circular contours. To verify the effectiveness of CEHT, ablation studies were conducted comparing CEHT to contour extraction (CE) or HT solely. As shown in Fig. 3(g) and ESI Table S4,† CEHT generally showed a good balance between a high recognition rate (recall ≥98%) and a high IoU threshold, outperforming CE or HT alone. It is further noted that CE is prone to false positives but shows an accurate contour. HT hardly leads to false negatives, but the recognized contour is easily deviated from the true position. Our CEHT algorithm combines the CE's accurate positioning with HT's high recognition rate. Finally, their combination significantly enhances the segmentation performance, as CE shows only 88% recognition rate and HT reduces the average IoU of all droplet proposals by 7%, as shown in ESI Fig. S4 and S5.† Moreover, CEHT could detect more than 89% droplets (except 76% and 79% for video 1 and video 2) at an IoU threshold of 0.80 (relatively strict criterion for object location) in droplet segmentation, as shown in Fig. 3(h) and ESI Fig. S5.† Therefore, we have designed two complementary algorithms, ASTM and CEHT, for droplet segmentation on both bright and dark field images.
Concerning droplet classification, it can be concluded that the pre-trained model obtained on static images shows unsatisfactory accuracy for dynamic video frames, with the representative results shown in Fig. 3(c–f). This phenomenon aligns with our expectations, as it can be caused by different imaging systems, microfluidic chips, and continuous (oil) and dispersed (cell types) phases. Classification models need to be retrained with updated annotations. Consequently, we designed the GUI in WSCApp to enable the visualization of annotation results and reconfirmed droplet labels directly within the software. We have manually modified the video annotations for further training and evaluation. It is worth noting that only three droplet-level labels, including empty, single-cell, and multi-cell encapsulation (0, 1, >1), are adopted in this study to avoid tedious and cell-level annotation. To complete the data modification, we merely selected the droplet category and clicked anywhere in the droplet area that should be changed. We completed the full data annotation on videos 1–7 and only performed approximately 1000 mouse clicks on 1695 video frames with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 584 droplets using assistive annotation. Compared to conventional annotation from scratch, we used a pre-trained model to provide initial annotation, thus saving 97% and ∼40% of annotation workload on droplet segmentation and classification. The unsatisfactory classifiers or WSCNet models are further trained with manually modified labels with weakly supervised information (triple labels) to fit the feature distribution and achieve cell counting and location.
584 droplets using assistive annotation. Compared to conventional annotation from scratch, we used a pre-trained model to provide initial annotation, thus saving 97% and ∼40% of annotation workload on droplet segmentation and classification. The unsatisfactory classifiers or WSCNet models are further trained with manually modified labels with weakly supervised information (triple labels) to fit the feature distribution and achieve cell counting and location.
Qualitative results of retrained models from scratch
Both the CNN-based classifier and the WSCNet model were retrained from scratch, with the data splitting shown in the Materials and methods section, and tested on a GeForce RTX 3060Ti GPU using the Pytorch platform with a 50-epoch patience level for early stopping. The loss function, accuracy, and recall values for the validation set were calculated at each epoch throughout the training.After training, the performance of different models was evaluated on data they had never encountered before. To visualize the WSCApp output, the representative results of droplet classification, cell counting, and cell location are demonstrated in Fig. 4 with droplet data collected from various microfluidic chips. It can be qualitatively concluded that the proposed CEHT segmentation algorithm and the retrained classifier can accurately detect most droplets and provide accurate information about the droplet category, as shown in Fig. 4(a). At the same time, it can be concluded that the previous problem of misclassification of pre-trained models is greatly improved, as models have been retrained to fit the new feature distribution. Except for a few incomplete droplets located at the edge, most droplets can be correctly detected and classified by our software.
Compared to other classification-based algorithms, our WSCNet model can not only distinguish droplet encapsulations but also locate each cell, while the maximum pixel and integral of the density map represent the cell location and amount, respectively. The predicted cells are highlighted with yellow dots in Fig. 4(b). It was observed that most of the cell positions, as well as the number of cells, are accurate when compared to their ground truth. WSCNet learns to recognize cell characteristics from the difference between empty and single-cell droplets and then applies the learned knowledge to multi-cell droplets without any supervised location information. In addition, comparing the WSCNet model with ResNet18 and other classification networks, it was found that the WSCNet model showed superior precision and F1 scores, as it identified fewer false negative and false positive droplets, as shown in ESI Fig. S6.† It is also observed that most existing approaches have achieved comparable performance in recognizing empty droplets, while our counting-based method exhibited better performance in recognizing cells, especially in multi-cell encapsulating droplets.
To further evaluate WSCApp performance on multi-cell encapsulating droplets, we collected a publicly accessible dataset of droplets with encapsulated PC3 cancer cells for validation. Therefore, we have also retrained the WSCNet model on the new dataset. Considering the different categories (this dataset has four categories), we labelled 14k droplets for WSCApp training and testing. Image recognition of 128 independent test images of PC3 cancer cell-encapsulating droplets was finally segmented by CEHT and classified using the WSCNet model, with representative results shown in Fig. 5 and ESI Fig. S7.† It is obvious that after retraining from scratch WSCNet could precisely detect and count most cells, even when there were up to six cells in one droplet, and classify droplets according to cell quantity. Because a multi-cell droplet encapsulation contains at least two cells inside, WSCNet can learn to count the cell population from precise labels (empty and single-cell droplets) and imprecise labels (multi-cell droplets). It is important to note that WSCNet could also locate cells, as indicated by yellow dots in Fig. 5, by searching the local maxima on the density map instead of any location information in the network training. This not only increases the interpretability of our proposed algorithm, but also enables the further use of cell localization for characteristic analysis and monitoring of each cell.
Generalization and quantitative performance of cell recognition on multiple datasets
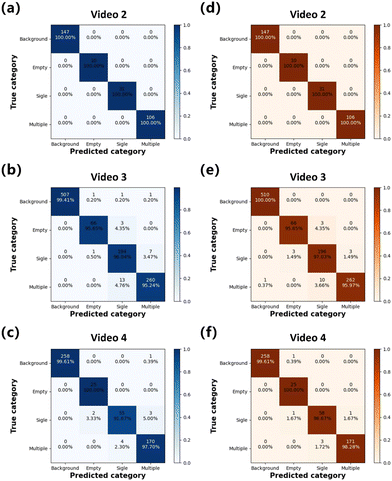
Furthermore, we comprehensively analyzed the recognition results of independent tests, which involved 2335 cancer cell-encapsulating droplets (video 8) and explored the generalization capability of WSCApp software. The performance of a classifier can be thoroughly described by precision, recall, and F1 score. It is concluded that the micro-F1 score of the LeNet-5 classifier on this dataset was 94.4%. The multiclass confusion matrix was also applied as an additional tool to compare model predictions with ground truth labels on the test set for statistical analysis. The confusion matrix in Fig. 6 shows the model performance of both the LeNet-5 classifier and the WSCNet model in predicting the correct class for the segmented droplet candidate. It is seen that both the classifier and WSCNet achieved comparative performance on the background and empty droplets, while WSCNet performed better in the classification of single-cell (98.2% vs. 84.9%) and multi-cell (95% vs. 90%) encapsulating droplets, which is consistent with a previous study.38The confusion matrix in Fig. 6(b) shows the performance of our WSCNet in droplet classification for the different numbers of encapsulated cells. It is shown that only a small number (74) of droplet labels are predicted as the wrong class (see ESI Fig. S8† for model failures). Furthermore, most of the predictions are correctly predicted with the appropriate category. Because the functions of our WSCNet algorithm are basically consistent with the YOLOv3/v5 model, the literature data can be directly used for WSCApp training, validation, and independent testing. The results of the YOLO algorithm, which is 0.97 mean average precision (mAP) @ 0.5 IoU, can be used for qualitative comparison. It is worth noting that the micro-F1 score of the WSCNet model (98.1%) is superior to that reported in the original literature.37 Although the mAP metric is not directly applicable to our region-CNN models, we still observed that our WSCNet model shows better accuracy than the YOLO model used in the previous study37 in the detection of empty (570 vs. 560), single-cell (657 vs. 634) and multi-cell (1027 vs. 988) encapsulations. Therefore, compared to YOLO, our WSCNet model is more light weight with better classification accuracy.
Table 2 summarizes the quantitative classification performance of the WSCNet method and various CNN-based classifiers on eight independent test datasets, including droplets encapsulating mammalian cells, yeast cells, and microspheres. Despite some exceptions, the micro-F1 scores of both CNN-based classifiers and the WSCNet model on these datasets were higher than 0.94, indicating high accuracy in droplet classification. In particular, for the classification of 10 μm microspheres in video 2, all models achieved the ideal performance after CEHT segmentation. Overall, Resnet18 appears to be the preferred choice (of the models we tested) with all micro-F1 scores > 0.94. First, its structure is not complex and its training and inference time (<22 ms) is relatively low, as shown in ESI Table S2.† Second, when the difficulty of the dataset increases, such as cells being difficult to identify due to high transparency, Resnet18's accuracy is much better than other classifiers. The shallow CNN-based classifiers, such as Lenet-5, can also achieve high recognition accuracy (≥0.945) with additional benefit from fast model training and inference, making it particularly useful for large-scale videos. Finally, our WSCNet model achieved >98% accuracy in 5 out of 8 datasets, demonstrating the performance improvement of cell counting networks for droplet classification tasks. Representative results of droplet segmentation and cell classification on the validation and independent test datasets are shown in ESI Fig. S7 and S8.† We show a qualitative evaluation of cell location performance by printing the visualized location of each cell on video frames.
The multiclass confusion matrix was also utilized to analyze the classification performance across multiple datasets, as shown in Fig. 7 and ESI Fig. S9.† If the predicted label matches the true label, it is defined as a true positive and appears on the diagonal of the confusion matrix. As shown in Fig. 7, it can be concluded that the majority of predicted categories, which exceeds 96% in most cases, are true positives, demonstrating the high accuracy of WSCApp in classifying different droplets. The confusion matrixes also showed the comparative performance of the classic ResNet18 classifier and the WSCNet model in predicting the category for the segmented droplet candidates. The classifier and WSCNet displayed a similar performance on the background and empty droplets, while the latter outperformed on all classifications of single-cell and multi-cell encapsulating droplets. Compared to the ResNet18 classifier, the WSCNet model exhibited higher classification accuracy due to its adoption of a weakly supervised learning strategy for counting cells, which is more comprehensible and rational than a conventional classifier. Therefore, it can output the position and number of cells as shown in Fig. 6 and ESI Fig. S8.† The fractions of droplets containing zero, one, or multiple cells determined from WSCNet predictions are in good agreement with hand counting, as shown in ESI Fig. S10.†
However, training classifiers and the WSCNet model from scratch on datasets of unbalanced categories, specifically videos 1, 5, 6, and 7 (see ESI Table S2† for category distribution), also resulted in overfitting the training data and underperformed on a partial class of the test data, as shown in ESI Fig. S9(d) and (g).† This is largely due to the limited quantity and quality of available frames, as videos 2–4 with more frames and balanced categories showed better performance. Notably, videos 5 and 6, containing only empty droplets and single-cell encapsulating droplets, did not yield optimal results with the WSCNet method, likely due to its inability to fully extract cellular localization information from multi-cell encapsulating droplets. This also explains why the performance of the WSCNet model on these two datasets is unremarkable. Despite this, both quantitative experimental results and confusion matrixes confirmed that our approach can not only distinguish droplet encapsulations (micro-F1 score > 0.94) but also locate each cell without any supervised location information.
Transfer learning for dynamic video recognition
To verify the effectiveness of transfer learning and to understand the amount of labeled data required for the algorithm to achieve adequate accuracy, we conducted a control experiment. The results of classifiers trained from scratch with a 100% training sample were first recorded and then compared with those of transfer learning using a 20% training sample while their pre-trained models were previously trained on our static images. As shown in Table 2, the micro-F1 score evaluation results demonstrated the classification performance on all independent tests of video datasets by ResNet18 and MoblieNet classifiers. It can be concluded that through transfer learning, WSCApp software requires only a small number (20%) of training samples to achieve performance that is close (slightly lower) to that of training from scratch with all samples. In particular, the ResNet18 model, after transfer learning, has achieved comparative classification accuracy with other classifiers trained from scratch.Transfer learning involves fine-tuning the pre-trained model weights on a small number of newly annotated datasets and conducting independent tests. We further conducted parameter experiments to explore the impact of varying the number of training video frames on performance. Fig. 8 shows the comparative results of transfer learning and training from scratch by different models. We can clearly draw a conclusion that in the range of a small number of training samples, e.g. 2–10 frames, transfer learning models consistently outperform models trained from scratch because most solid lines are much higher than dashed lines, indicating better performance for transfer learning. However, as the number of training annotations increases, both methods gradually improved the model effectiveness and incorporated artificial experience into the machine learning model. The classification accuracy of the model trained from scratch also gradually improved, approaching that of the transfer learning model, which is also consistent with expectations.
Therefore, when another specific model trained on a similar system is available, e.g. static droplet images, transfer learning has been explicitly verified to be beneficial for improving neural network performance for limited data sets. We have summarized several micro-F1 score results on the comparisons for transfer learning and training from scratch, as shown in Table 3. On three datasets, both methods showed a trend of improving performance on increasing training samples, while transfer learning achieved better accuracy. Notably, on the first two datasets, transfer learning was performed using only 12 frames, and its performance was almost as good as that of training from scratch on all available samples, which are 64 and 586 frames, respectively. This result also suggests that transfer learning fine-tuning on the pre-model can significantly reduce sample annotation workload, e.g. by approximately 80% and 90% for both videos. Overall, this suggests that transfer learning can be an efficient method for improving labelling efficiency. Furthermore, this technique is particularly relevant for the recognition of droplet encapsulation, as the high-level information remains the same regardless of the experimental conditions used to generate droplets or encapsulated cell types.
| Different parameters | Video 1 | Video 3 | Video 8 |
|---|---|---|---|
| 4 labeled frames for training from scratch | 0.682 | 0.395 | 0.502 |
| 4 labeled frames for transfer learning | 0.908 | 0.874 | 0.722 |
| 8 labeled frames for training from scratch | 0.713 | 0.927 | 0.606 |
| 8 labeled frames for transfer learning | 0.922 | 0.945 | 0.769 |
| 12 labeled frames for training from scratch | 0.935 | 0.927 | 0.836 |
| 12 labeled frames for transfer learning | 0.941 | 0.954 | 0.867 |
In addition, transfer learning can be used to transfer knowledge to a new scenario to enhance recognition performance if the target domain data are insufficient. As an illustration of this strategy's utility, we demonstrate its use in the recognition of cell-encapsulating droplets by directly applying the ResNet18 classifier, which was fine-tuned on video 3, to independently test video 2 and video 4 without retraining. In these videos, either the cell type or microfluidic chip parameters are different. Despite the huge variation in content morphology, pre-trained models using transfer learning significantly improved detection and classification accuracy on the test dataset, as seen in Fig. 9. The confusion matrix in Fig. 9(a) indicates that the true positive for empty and single-cell droplets is 100%. Only 2 and 126 multi-cell droplets were missed or misidentified (1425 droplets in total) on video 2. This favorable performance is attributed to the similar distribution of microsphere features in both videos. When the recognized objects change, from microspheres in pre-trained models to algae cells for independent testing, the recognition accuracy decreases, but it is still better than that without transfer training. This is because the micro-F1 score of the fine-tuned ResNet18 classifier for video 4 is 70%, while that without transfer learning falls below 40%.
For new cell-encapsulating droplets that differ significantly in experimental and imaging conditions, large data annotations are usually required for accurate training. However, transfer learning on our pre-trained WSCApp only required a fraction of the initial annotation to achieve comparable accuracy for new droplet data. In summary, we demonstrated the application of WSCApp to various droplet data by using pre-trained models and the transfer learning strategy to improve the labelling efficiency and reduce manual annotation.
Conclusions
In this study, we have illustrated the design of application software as well as the function and performance of recognition algorithms. A novel executable program integrated with different classifiers and weakly supervised algorithms, namely WSCApp, was designed to recognize cell-encapsulating droplets from droplet videos and systematically verified by different datasets. Additionally, we have developed both dark-field and bright-field recognition algorithms, which are optional on WSCApp depending on the illumination scenarios, to enhance accuracy and generalization. Next, we compared and validated the generalization capability of different algorithms on video data collected from the literature to evaluate their classification performance. Finally, assistive annotation and transfer learning strategies were employed to improve the recognition efficiency and reduce manual annotation.WSCApp can not only categorize droplets by encapsulated cell populations, but also locate cells without any supervised location knowledge, showing an advantage over CNN-based classifiers. Cell localization can be applied to subsequent monitoring and detailed analysis of each cell in future studies. It was also proved that the integrated droplet detection and classification software, WSCApp, is feasible for video recognition of single-cell or multi-cell encapsulating droplets. This software is essential to anticipate precise control settings and achieve the desired encapsulation rate. In addition, this video recognition method can allow for a feedback loop so that droplet-based sorting can be achieved according to recognized image results. The entire software is efficient, flexible and user-friendly, making it a comprehensive platform for quantitative evaluation of encapsulating microfluidic droplets.
Materials and methods
CNN-based imaging recognition
We proposed computational application software, namely WSCApp, with different recognition algorithms and CNN-based models to evaluate droplet quality (size and distribution) and further recognize the encapsulated cell (amounts and position). This image and video analysis tool with a user-friendly GUI can fleetly provide dynamic identification of cell-encapsulating microfluidic droplets. WSCApp uses the two-stage object recognition approach, as shown in Fig. 1, to achieve objective and automatic statistical analysis of many droplets and cells. The droplet candidates are first segmented and then fed to CNNs that are used to filter out false positive droplets, categorize droplets, and perform cell counting.For monitoring microfluidic droplets, there are usually two modes: one is video recording of droplet generation in the microfluidic channel, and the other involves capturing static images after droplet generation, as shown in Fig. 1(a). Considering the differences in motion and illumination, the grayscale values inside the droplets are higher (bright-field) or lower (dark-field) than in the dispersed phase. Therefore, as shown in Fig. 1(b), we designed both ASTM and CEHT methods for dark-field and bright-field scenarios, respectively. For adhesive and opaque droplets, ASTM40 adopts a three-step approach to achieve droplet segmentation: (1) adaptive scaling based on the matching response map between the circular template and the image foreground; (2) greedy search to detect droplet candidates; (3) non-maximum suppression filter to remove redundant concentric circles. For separate and transparent droplets, the final segmentation of droplet proposals was produced by CEHT combining the results of two parallel methods: (1) after filtering and thresholding, edge operators are used to detect the droplet edge; (2) HT transforms 2D feature extraction into searching for a point in the high-dimension space determined by the radius and center coordinates of the circle. Both algorithms and key parameters are optional or modifiable in WSCApp, e.g. kernel size and minimum and maximum radius, to improve the accuracy and generalization of droplet segmentation. For a more detailed introduction to the design of the two algorithms, please refer to ESI Note 2 and Fig. S2, S3.†
In the second stage, WSCApp integrates both conventional classifiers and our WSCNet model for droplet classification, cell counting and location, respectively. Firstly, CNN-based classifiers are used as the baseline methods for classifying droplets. We applied a light weight seven-layer LeNet-5 for assistive annotation and then integrated LeNet-5,45 ResNet18,46 and MobileNet47 into the software for comparison. Secondly, WSCNet was used to estimate the number of cells, categorize droplets, and provide position prediction for each cell. The WSCNet consists of classification and counting branches: the former serves as a filter to remove false positive droplet candidates, and the output of the latter branch is a grayscale density map, in which the maximum pixel and the integral of pixel values represent the cell location and amount, as shown in Fig. 1(c), respectively. For a more detailed introduction to the design of CNN-based classifiers and WSCNet models, please refer to ESI Note 3.†
The user interface of WSCApp is developed in C++ programming language and based on Qt, as shown in Fig. 2, enabling fast and iterative design. WSCApp integrated the functions of file selection, arithmetic parameter setting, switching of algorithms and models, automatic droplet/cell detection (invoking recognition algorithms and CNN-based models, respectively), view visual recognition, manual post-recognition correction, performance statistics, and saved modified annotations. Further application details for the proposed software are described in ESI Note 1.†
Dynamic video recognition for cell-encapsulating droplets
WSCApp provides a user-friendly GUI platform for quantitative evaluation of both images and videos of cell-encapsulating droplets, as demonstrated in Fig. 2. Since a video can be considered as a continuous sequence of images, we can apply the image processing methods to video recognition with enhanced real-time performance by code refactoring. VideoCapture and other relevant OpenCV methods were applied to generate frame images corresponding to the video, which would be detected and identified sequentially by WSCApp. Next, the VideoWriter method was used to regenerate a video, which shares the same size and frame rate as the original video, with recognized droplets.In addition, users could select a single video/image or directly load a folder containing multiple videos/images. WSCApp can automatically recognize both static images and dynamic videos with optional segmentation and classification methods. WSCApp directly outputs the recognized image or video, outputs the recognized text file (including frame number, droplet center point coordinates (X, Y), diameter, category; cell number, position), and displays statistical information, such as diameter mean, SD value, single cell encapsulation rate, etc., on the message window. To detect and control droplet encapsulation, the inference time is expected to be fast, preferably in real time, as there are many droplets that need to be detected in real scenarios. Following detection and classification based on optional algorithms and models, the test dataset of droplet frames was run through WSCApp for inference and the consumption time was recorded.
Experimental platform and droplet dataset for pre-training
We have constructed a microfluidic droplet generation platform, comprising microfluidic chips, a multichannel syringe pump, an inverted microscope, a USB camera, and a computer using the proposed algorithms to generate microfluidic droplets and monitor the droplet quality and cell quantity. Three microfluidic chips with different flow-focusing structures were applied to generate droplets, while bacterial cells were simultaneously encapsulated. Microfluidic chips were replicated by standard soft photolithography, fabricated in PDMS, and placed on the stage of an inverted microscope for recording.Syringe pumps injected the dispersed and continuous phases into the corresponding inlets. A mixture of mineral oil (3% w/w EM90 and 0.1% v/v Triton-100 dissolved in M5310 mineral oil) or a mixture of fluorinated oil (1% dSURF surfactant dissolved in Novec 7500 fluorinated oil) was used as the continuous phase to validate the generalization ability of the algorithm on intricate data collected from different geometries and materials. The representative images of cell-encapsulating droplets are demonstrated in Fig. 1(a) and 2(a). Following the same method used in our previous study,38 more than 1245 static images of encapsulating droplets were collected with a resolution of 640 × 480 and a mean of 167 droplets per image. They were further labelled for the pre-training of both the WSCNet model and traditional classifiers in this study.
Microfluidic droplet datasets collected from the literature
We further verified WSCApp with intricate droplet data collected from various microfluidic structures. Instead of generating droplets with encapsulated mammalian cells using our microfluidic system, limited by policy restrictions on medical laboratories, we spared no effort to find related publications that share similar data of cell-encapsulating droplets. The main data supporting the results are available in this article as well as its ESI.† HL60 and K562 cells encapsulated in droplets were collected from ref. 22. The beads and cells co-encapsulating droplets were collected from the ESI of ref. 48. The controlled encapsulation data were collected from the literature.49 The yeast encapsulation data were collected from the Website of Laboratory of BioChemistry at https://www.lbc.espci.fr/home/gallery/microfluidic-movies/. The dataset of PC3 cancer cell encapsulating droplets37 was also evaluated using WSCApp and compared with its YOLO models.The numbers of droplets or frames for different categories in each dataset are summarized in Table 4 and ESI Table S2.† Depending on the dataset quantity, all video frames were randomly divided into a training set, a validation set, and a test set in ratios of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 6
1 or 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. There were four different categories of droplets that needed to be recognized: background, empty, single-cell, and multi-cell droplets. Background samples were collected by randomly selecting areas of the non-droplet backgrounds in equal quantities to the other three categories. The training procedure did not include any cell position or exact quantity of multiple cells; just these three droplet-level labels were provided.
2. There were four different categories of droplets that needed to be recognized: background, empty, single-cell, and multi-cell droplets. Background samples were collected by randomly selecting areas of the non-droplet backgrounds in equal quantities to the other three categories. The training procedure did not include any cell position or exact quantity of multiple cells; just these three droplet-level labels were provided.
| Purpose | |||||
|---|---|---|---|---|---|
| Dataset/content | All | Traininga (droplets/frames) | Inference | Ref. | |
| Train | Validate | Test | |||
| a Distribution of the empty, single-cell and multi-cell encapsulating droplets for network training. The non-droplet background was randomly collected in equal quantities for all categories. b The amount of training data is triple after data augmentation according to the original literature. | |||||
| Images/yeast cells | 208k/1245 | 166k/993 | 21k/126 | 21k/126 | This study |
| Video 1/HL60 and K562 | 537/108 | 317/64 | 109/22 | 111/22 | 22 |
| Video 2/10 μm microspheres | 1425/201 | 1136/160 | 142/20 | 147/21 | 48 |
| Video 3/10 μm microspheres | 5332/733 | 4264/586 | 524/73 | 544/74 | |
| Video 4/mt+ and mt− algae cells | 2540/351 | 2025/280 | 256/35 | 259/36 | |
| Video 5/9.9 μm microspheres | 236/80 | 141/48 | 47/16 | 48/16 | 49 |
| Video 6/HL60 cells | 514/112 | 306/67 | 103/22 | 105/23 | |
| Video 7/yeast cells | 1203/300 | 964/240 | 120/30 | 119/30 | Website |
| Video 8/PC3 cancer cells | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 916/643 916/643 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452/412b 452/412b |
2129/103 | 2335/128 | 37 |
Assistive annotation and transfer learning on new datasets
We demonstrate the application of WSCApp to various droplet data by using pre-trained models and transfer learning strategies to improve labelling efficiency and reduce manual annotation. We first trained both classifiers and WSCNet on the static droplet images we had experimentally collected as base models (pre-trained models), as shown in Fig. 2, and integrated them into WSCApp for assistive annotation to avoid labelling new datasets from scratch.Second, we deployed WSCApp directly to the new droplet dataset and provided preliminary annotation. Next, the GUI allowed immediate review, automatically loading the corresponding annotation files and manually reconfirming the annotation results for retraining. Fourth, classifiers or WSCNet models were trained again with manually modified data using weakly-supervised information (triple droplet-level labels) to fit the feature distribution. In addition, transfer learning could be used to transfer knowledge to a new scenario.44
We applied transfer learning to improve algorithmic accuracy with less annotation. Transfer learning involves fine-tuning the pre-trained model weights on a small number of annotated new datasets and conducting independent tests. The pre-trained models, trained on the original dataset as the relevant source domain, were stored with their structure and weights. The weights of network layers of the training models were fine-tuned to fit the new feature distribution. Both classifiers and WSCNet models (with and without transfer learning) were trained on the training set and their performance was evaluated against the test set.
Finally, we compared and validated the generalization capability of different algorithms on video data collected from the literature to evaluate their recognition and generalization performance. At the end of this study, we also trained final CNN-based classifiers and WSCNet on all the data to allow researchers to fine tune models for new images/videos of cell-encapsulating droplets.
Network implementation and evaluation metrics
In the first stage, we set the ASTM threshold σ, the small constant γ, and the weight ω of the WSCNet model to 0.98, 0.001 and 1, respectively. The Canny edge detector threshold and the center detection threshold of the HoughCircle function in OpenCV are set to 60 and 18, respectively, for CEHT. We use a bounding circle Interaction over Union (IoU) to evaluate the accuracy of droplet positioning, which is derived from the IoU of bounding boxes in object detection. The number of droplets that has an IoU result greater than a threshold θ is illustrated in ESI Fig. S4.† This metric is used to measure the degree of correlation between predicted and actual droplets, and a higher IoU represents a more stringent requirement on the correlation. The diameter deviation of the droplet distribution was calculated by the coefficient of variation (CV). In the second stage, we use the Adam optimizer for training machine learning models.We further evaluate our algorithm using three different metrics. First, widely adopted recall and precision, computed by eqn (2), are used to assess CNN-based classifiers and the WSCNet model in the classification of droplet proposals. Second, the multiclass confusion matrix is used as an additional tool to evaluate the model performance. Third, considering the unbalanced number of categories, the performance of WSCApp in detecting droplets and classifying encapsulated cells is then measured using metric micro F1, pooling per-sample classifications across all classes (background, empty, single-cell, and multi-cell droplets), and computed by eqn (3). To compare different classification-based and counting-based strategies, the F1 score (harmonic mean of precision and recall) and inference time are also considered.
 | (2) |
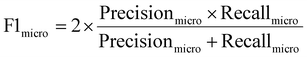 | (3) |
Fourth, the exact number and location of cells in each droplet can be provided by our WSCNet, which can give precise information about encapsulated cells. To demonstrate this function, we print the visualized location of each cell on the original video frames.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The code is available on GitHub at the following link https://github.com/Loyage/WSCNet2 (accessed after 20th Nov., 2023).Author contributions
Conceptualization, Yuanhang Mao and Xiao Zhou; data curation, Weiyang Yang and Xiao Zhou; formal analysis, Yuanhang Mao and Weiguo Hu; funding acquisition, Zhen Cheng and Xiao Zhou; methodology, Xiao Zhou and Zhen Cheng; resources, Zhen Cheng; software, Yuanhang Mao, Weiguo Hu and Xiao Zhou; supervision, Zhen Cheng; validation, Yuanhang Mao and Xiao Zhou; visualization, Zhen Cheng; writing – original draft, Xiao Zhou; writing – review & editing, Zhen Cheng.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The experimental assistance of Juntao Deng and Pengyan Zhang is gratefully acknowledged. This research was funded by the National Natural Science Foundation of China (grant 62103228), supported by Beijing Natural Science Foundation (grant 3242008), and the China Postdoctoral Science Foundation (grant 2023M741850).References
- P. K. Sorger, Microfluidics closes in on point-of-care assays, Nat. Biotechnol., 2008, 26(12), 1345–1346 CrossRef CAS PubMed.
- J. Kim, A. S. Campbell and B. E. F. de Avila, et al., Wearable biosensors for healthcare monitoring, Nat. Biotechnol., 2019, 37(4), 389–406 CrossRef CAS PubMed.
- M. J. Fuerstman, P. Garstecki and G. M. Whitesides, Coding/Decoding and Reversibility of Droplet Trains in Microfluidic Networks, Science, 2007, 315(5813), 828–832 CrossRef CAS PubMed.
- M. Joanicot and A. Ajdari, Droplet control for microfluidics, Science, 2005, 309, 887 CrossRef CAS PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Emerging Droplet Microfluidics, Chem. Rev., 2017, 117(12), 7964–8040 CrossRef CAS PubMed.
- Y. Ding, P. D. Howes and A. J. deMello, Recent Advances in Droplet Microfluidics, Anal. Chem., 2020, 92(1), 132–149 CrossRef CAS PubMed.
- S. Mashaghi, A. Abbaspourrad and D. A. Weitz, et al., Droplet microfluidics: A tool for biology, chemistry and nanotechnology, TrAC, Trends Anal. Chem., 2016, 82, 118–125 CrossRef CAS.
- K. R. Sreejith, C. H. Ooi and J. Jin, et al., Digital polymerase chain reaction technology – recent advances and future perspectives, Lab Chip, 2018, 18(24), 3717–3732 RSC.
- R. Zilionis, J. Nainys and A. Veres, et al., Single-cell barcoding and sequencing using droplet microfluidics, Nat. Protoc., 2017, 12(1), 44–73 CrossRef CAS PubMed.
- L. Cohen, N. Cui and Y. Cai, et al., Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay, ACS Nano, 2020, 14(8), 9491–9501 CrossRef CAS PubMed.
- J. T. Wang, J. Wang and J. J. Han, Fabrication of advanced particles and particle-based materials assisted by droplet-based microfluidics, Small, 2011, 7, 1728 CrossRef CAS PubMed.
- C. X. Zhao, Multiphase flow microfluidics for the production of single or multiple emulsions for drug delivery, Adv. Drug Delivery Rev., 2013, 65, 1420 CrossRef CAS PubMed.
- O. J. Miller, A. E. Harrak and T. Mangeat, et al., High-resolution dose-response screening using droplet-based microfluidics, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 378 CrossRef CAS PubMed.
- E. Z. Macosko, A. Basu and R. Satija, et al., Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets, Cell, 2015, 161(5), 1202–1214 CrossRef CAS PubMed.
- A. M. Klein, L. Mazutis and I. Akartuna, et al., Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells, Cell, 2015, 161, 1187 CrossRef CAS PubMed.
- G. X. Y. Zheng, J. M. Terry and P. Belgrader, et al., Massively parallel digital transcriptional profiling of single cells, Nat. Commun., 2017, 8(1), 14049 CrossRef CAS PubMed.
- P. A. Zhu and L. Q. Wang, Passive and active droplet generation with microfluidics: a review, Lab Chip, 2017, 17(1), 34–75 RSC.
- W. Li, L. Zhang and X. Ge, et al., Microfluidic fabrication of microparticles for biomedical applications, Chem. Soc. Rev., 2018, 47(15), 5646–5683 RSC.
- D. J. Collins, A. Neild and A. deMello, et al., The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation, Lab Chip, 2015, 15(17), 3439–3459 RSC.
- T. M. Ho, A. Razzaghi and A. Ramachandran, et al., Emulsion characterization via microfluidic devices: A review on interfacial tension and stability to coalescence, Adv. Colloid Interface Sci., 2022, 299, 102541 CrossRef CAS PubMed.
- L. B. Pinheiro, V. A. Coleman and C. M. Hindson, et al., Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification, Anal. Chem., 2012, 84(2), 1003–1011 CrossRef CAS PubMed.
- E. W. M. Kemna, R. M. Schoeman and F. Wolbers, et al., High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel, Lab Chip, 2012, 2881–2887 RSC.
- A. T. L. Lun, S. Riesenfeld and T. Andrews, et al., EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data, Genome Biol., 2019, 20(1), 63 CrossRef PubMed.
- N. J. Bernstein, N. L. Fong and I. Lam, et al., Solo: Doublet Identification in Single-Cell RNA-Seq via Semi-Supervised Deep Learning, Cell Syst., 2020, 11(1), 95 CrossRef CAS PubMed.
- X. Yue, X. Fang and T. Sun, et al., Breaking through the Poisson Distribution: A compact high-efficiency droplet microfluidic system for single-bead encapsulation and digital immunoassay detection, Biosens. Bioelectron., 2022, 211, 114384 CrossRef CAS PubMed.
- E. Zang, S. Brandes and M. Tovar, et al., Real-time image processing for label-free enrichment of Actinobacteria cultivated in picolitre droplets, Lab Chip, 2013, 13(18), 3707–3713 RSC.
- C. M. Svensson, O. Shvydkiv and S. Dietrich, et al., Coding of Experimental Conditions in Microfluidic Droplet Assays Using Colored Beads and Machine Learning Supported Image Analysis, Small, 2019, 15(4), 14 Search PubMed.
- M. Sesen and G. Whyte, Image-Based Single Cell Sorting Automation in Droplet Microfluidics, Sci. Rep., 2020, 10, 8736 CrossRef CAS PubMed.
- P. Q. N. Vo, M. C. Husser and F. Ahmadi, et al., Image-based feedback and analysis system for digital microfluidics, Lab Chip, 2017, 17(20), 3437–3446 RSC.
- M. Vaithiyanathan, N. Safa and A. T. Melvin, FluoroCellTrack: An algorithm for automated analysis of high-throughput droplet microfluidic data, PLoS One, 2019, 14(5), 22 CrossRef PubMed.
- J. Jeong, N. J. Frohberg and E. L. Zhou, et al., Accurately tracking single-cell movement trajectories in microfluidic cell sorting devices, PLoS One, 2018, 13(2), 16 CrossRef PubMed.
- A. S. Basu, Droplet morphometry and velocimetry (DMV): a video processing software for time-resolved, label-free tracking of droplet parameters, Lab Chip, 2013, 13(10), 1892–1901 RSC.
- H. F. Zhao, J. Zhou and Y. Y. Gu, et al., Real-Time Computing for Droplet Detection and Recognition, in IEEE International Conference on Real-time Computing and Robotics (IEEE RCAR), IEEE, Kandima, MALDIVES, 2018.
- G. Soldati, F. Del Ben and G. Brisotto, et al., Microfluidic droplets content classification and analysis through convolutional neural networks in a liquid biopsy workflow, Am. J. Transl. Res., 2018, 10(12), 4004–4016 CAS.
- V. Anagnostidis, B. Sherlock and J. Metz, et al., Deep learning guided image-based droplet sorting for on-demand selection and analysis of single cells and 3D cell cultures, Lab Chip, 2020, 20(5), 889–900 RSC.
- L. Howell, V. Anagnostidis and F. Gielen, Multi-object detector YOLOv4-tiny enables high-throughput combinatorial and spatially-resolved sorting of cells in microdroplets, Adv. Mater. Technol., 2022, 7, 2101053 CrossRef CAS.
- K. Gardner, M. M. Uddin and L. Tran, et al., Deep learning detector for high precision monitoring of cell encapsulation statistics in microfluidic droplets, Lab Chip, 2022, 22(21), 4067–4080 RSC.
- X. Zhou, Y. Mao and M. Gu, et al., WSCNet: Biomedical Image Recognition for Cell Encapsulated Microfluidic Droplets, Biosensors, 2023, 13(8), 821 CrossRef PubMed.
- X. Zhou, M. Gu and Z. Cheng, Local Integral Regression Network for Cell Nuclei Detection, Entropy, 2021, 23(10), 1336 CrossRef PubMed.
- X. Zhou, Z. Cheng and F. Chang, N-cell Droplet Encapsulation Recognition via Weakly Supervised Counting Network, in IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 2021, pp. 1424–1428.
- X. Zhou, Z. Cheng and M. Gu, et al., LIRNet: Local Integral Regression Network for Both Strongly and Weakly Supervised Nuclei Detection, in 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, 2020, pp. 945–951.
- K. Sirinukunwattana, S. E. A. Raza and Y. W. Tsang, et al., Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images, IEEE Trans. Med. Imaging, 2016, 35(5), 1196–1206 Search PubMed.
- X. R. Zhu, S. S. Su and B. X. Liu, et al., A real-time cosine similarity algorithm method for continuous monitoring of dynamic droplet generation processes, AIP Adv., 2019, 9(10), 105203 CrossRef.
- S. J. Pan and Q. Yang, A Survey on Transfer Learning, IEEE Trans. Knowl. Data Eng., 2010, 22(10), 1345–1359 Search PubMed.
- Y. LeCun, L. Bottou and Y. Bengio, et al., Gradient-based learning applied to document recognition, Proc. IEEE, 1998, 86(11), 2278–2324 CrossRef.
- K. He, X. Zhang and S. Ren, et al., Deep residual learning for image recognition, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
- A. Howard, M. Sandler and G. Chu, et al., Searching for mobilenetv3, in Proceedings of the IEEE/CVF international conference on computer vision, 2019, pp. 1314–1324.
- T. P. Lagus and J. F. Edd, High-throughput co-encapsulation of self-ordered cell trains: cell pair interactions in microdroplets, RSC Adv., 2013, 3(43), 20512–20522 RSC.
- J. F. Edd, D. Di Carlo and K. J. Humphry, et al., Controlled encapsulation of single-cells into monodisperse picolitre drops, Lab Chip, 2008, 8(8), 1262 RSC.
- S. L. Anna, N. Bontoux and H. A. Stone, Formation of dispersions using “flow focusing” in microchannels, Appl. Phys. Lett., 2003, 82(3), 364–366 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00022f |
| ‡ These authors contributed equally: Yuanhang Mao and Xiao Zhou. |
| This journal is © The Royal Society of Chemistry 2024 |