Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessing CaDPA levels, metabolic activity, and spore detection through deuterium labeling†
Rasmus
Öberg‡
 ab,
Timir Baran
Sil‡
ab,
Timir Baran
Sil‡
 a,
André
Ohlin
a,
André
Ohlin
 c,
Magnus
Andersson
c,
Magnus
Andersson
 *a and
Dmitry
Malyshev
*a and
Dmitry
Malyshev
 *a
*a
aDepartment of Physics, Umeå University, 901 87, Umeå, Sweden. E-mail: magnus.andersson@umu.se; dmitry.malyshev@umu.se; Tel: +46 90 786 6336
bSwedish Defence Research Agency (FOI), Cementvägen 20, 906 21 Umeå, Sweden
cDepartment of Chemistry, Umeå University, 901 87, Umeå, Sweden
First published on 1st February 2024
Abstract
Many strains among spore-forming bacteria species are associated with food spoilage, foodborne disease, and hospital-acquired infections. Understanding the impact of environmental conditions and decontamination techniques on the metabolic activity, viability, and biomarkers of these spores is crucial for combatting them. To distinguish and track spores and to understand metabolic mechanisms, spores must be labeled. Staining or genetic modification are current methods for this, however, these methods can be time-consuming, and affect the viability and function of spore samples. In this work, we investigate the use of heavy water for permanent isotope labeling of spores and Raman spectroscopy for tracking sporulation/germination mechanisms. We also discuss the potential of this method in observing decontamination. We find that steady-state deuterium levels in the spore are achieved after only ∼48 h of incubation with 30% D2O-infused broth and sporulation, generating Raman peaks at cell silent region of 2200 and 2300 cm−1. These deuterium levels then decrease rapidly upon spore germination in non-deuterated media. We further find that unlike live spores, spores inactivated using various methods do not lose these Raman peaks upon incubation in growth media, suggesting these peaks may be used to indicate the viability of a spore sample. We further observe several Raman peaks exclusive to deuterated DPA, a spore-specific chemical biomarker, at e.g. 988 and 2300 cm−1, which can be used to track underlying changes in spores involving DPA. In conclusion, permanent spore labeling using deuterium offers a robust and non-invasive way of labeling bacterial spores for marking, viability determination, and characterising spore activity.
1 Introduction
Bacterial spores are dormant forms of certain bacteria that can cause food poisoning, hospital-acquired infections, and be used as biological warfare agents.1,2 When exposed to favorable conditions, they can become active again. Spores are highly resistant to conventional decontamination methods such as heat, chemicals, and electromagnetic radiation.3 The robustness of spores and variations in resilience among strains have motivated research into understanding the factors behind spore resilience.When studying bacterial spores, it is important to distinguish spores based on factors like different strains and genetic mutants. Consequently, developing techniques to mark or stain spores is vital for investigating distinctions among spore populations. However, current techniques such as dye staining or fluorescent gene labeling have drawbacks. Dye staining can be time-consuming, while the dyes themselves can be light sensitive and toxic to humans.4,5 In addition, fluorophores and fluorescent gene labels can undergo photobleaching, impairing their effectiveness.6 Furthermore, in a sample, there is the possibility of non-specific binding of dyes or fluorophores, which can create background noise, and there may also be limitations in the penetration of a dye stain into the sample.7,8 Similarly, genetic modification to include fluorophores demands significant time, cannot be used with field samples, and is associated with notable variability from one experiment to another.9
To avoid the problems caused by dyes and fluorophores in bacterial samples, one can use isotope labeling with deuterium. The key advantages of deuterium-labeling are the non-toxic nature of the label, the stability and longevity of the label, and the ability to use it without any genetic modification to the cells. By incubating bacteria in deuterated water (heavy water, D2O) enriched media, bacteria will incorporate carbon–deuterium (C–D) bonds instead of carbon–hydrogen (C–H) bonds. The increased mass of deuterium atoms causes a change in the vibrational frequencies of bonds formed with deuterium, in contrast to those involving hydrogen.10 During cell growth, metabolic reactions, like NADP + redox processes, introduce deuterium into the cell, leading to H–D exchange in activities like Acetyl-CoA fatty acid synthesis. Consequently, deuterium accumulates in proteins and lipids, replacing C–H bonds with C–D bonds.11 These C–D bonds produce a distinct Raman peak around 2200 cm−1, which indicates metabolic activity in a cell.12 This method has been used to track the metabolic activity of bacteria,11,13,14 to test disinfectants on bacteria15 as well as growth and differentiation of different metabolic activity in fungi.16 It has also been used as a label to track the metabolic activity in spore germination and outgrowth of bacterial cells.17 Tracking the metabolism-induced C–D Raman peak around 2200 cm−1 in germinating spores is a more effective measurement of viability than other Raman peaks in the fingerprint region 600–1800 cm−1 such as the prevalent calcium dipicolinate (CaDPA) peak at 1016 cm−1, as changes in these peaks can be caused by other factors than germination, such as inactivation.18
In this study we enhance this labeling method by evaluating its applicability to bacterial spores. We assess the growth and stability of the characteristic Raman peak at 2200 cm−1 during bacterial growth, division, and subsequent sporulation within deuterium-enriched growth media. Additionally, we explore the persistence of this peak during germination in non-deuterated media. Moreover, we analyze how different decontaminants impact the spectral fingerprint of deuterium-labeled spores, demonstrating the method's potential in determining spore viability. To comprehend the underlying mechanism behind deuterium-labeling of bacterial spores, we evaluate the effect of deuteration on the wider spore Raman spectrum 600–2400 cm−1 using mutant strains as well as density functional theory, a computational tool to calculate molecular vibrations. We also investigate the effect of deuteration on dipicolinic acid (DPA), a prevalent substance in the spore core responsible for many of the spores’ resisting qualities.19 Thus, we present a new, versatile, non-invasive spectroscopic spore labeling method for use in a lab environment. This method has several applications, such as distinguishing between spore strains in a mixture, investigating metabolic activity post-decontamination procedures, and evaluating DPA content in spores.
2 Methods
2.1 Strains and origin
We evaluated our deuterium-labeling technique on B. paranthracis (also referred to in literature as B. cereus) NVH 0075/95 spores, with the strain provided by Marina Aspholm, Norwegian University of Life Sciences.B. subtilis str. 168 wild-type20 and the DPA-deficient mutant strains21 (ΔspoVAC::lox72 ΔgerA::CmΔ sleB::erm) were used to verify the Raman identity of deuterium-shifted DPA spectrum. The strains were provided by David Rudner, Harvard University.
2.2 Spore stock preparation
The spore stock was prepared on TSA agar (Bacto™, BD) at 37 °C for one week, long enough to cause sporulation. After growing the sample, spores were harvested from the plate, suspended in autoclaved distilled water and washed three times through centrifugation (4500 rpm, 5 min) in distilled water. The stock was then stored in autoclaved distilled water at 4 °C until use. Ahead of use, the spore suspension concentration was adjusted to 106 spores per ml, and vortexed at 3000 rpm (VM3 Vortex, M. Zipperer GmBH) for 15 seconds.2.3 D2O-substituted growth media preparation and spore labeling
TSA agar was prepared using pre-made TSA powder (Bacto™, BD). D2O-enriched agar plates were prepared by mixing TSA powder with 15% or 30% of the water replaced with D2O (99.8% deuterium oxide, 11327088, Fisher Scientific). Previous studies have shown these concentrations of D2O do not significantly inhibit spore germination and bacterial growth, while higher concentrations may inhibit bacterial growth.17,22To prepare deuterium-labeled spore stocks, spores from the appropriate stock were spread on the D2O-enriched agar plates with an inoculating loop and incubated at 37 °C for one week, and processed as detailed above.
2.4 Raman spectroscopy system
To acquire Raman spectra from single spores, we use a custom-built laser tweezer Raman spectroscopy (LTRS) setup built around an inverted microscope (IX71, Olympus), described in greater detail in earlier studies.23,24 Such an LTRS setup allows us to achieve more repeatable and high signal-to-noise ratio Raman spectra compared to regular Raman setups. The LTRS system uses a 785 nm Gaussian laser beam with a maximum power of 120 mW (08-NLD, Cobolt), coupled into the microscope system using a dichroic shortpass mirror with cut-off wavelength 650 nm (DMSP650, Thorlabs). The beam is focused onto the sample using a 60× water immersion objective with numerical aperture 1.2 (UPlanSApo, Olympus) and a working distance 0.28 mm. The Raman signal is collected from backscattered light, with Rayleigh scattered light filtered out using a 785 nm notch filter (NF785-33, Thorlabs). The resulting light is then filtered using a 150 μm diameter pinhole in the focal point of the beam telescope before it is coupled into a spectrometer (Model 207, McPherson) and dispersed using a 600 grooves per mm holographic grating with 800 nm blaze. Lastly, the spectral signal is captured using a Peltier-cooled CCD detector (Newton 920N-BR-DDXW-RECR, Andor) operated at −95 °C. The sample is placed on a temperature controlled sample holder and the temperature of the sample is maintained at 25 °C to avoid temperature-related intensity fluctuation during measurement. The spectral resolution of our instrument is <3 cm−1 and the Solis (Solis v4.30, Andor) software is used to control the detector and acquire spectra.2.5 Sample preparation
We prepare a sample for Raman measurement by diluting 1 μl of deuterized spore stock in 9 μl deionised water to reduce the background signal from the broth. The 10 μl spore suspension is then placed on a 24 × 60 mm glass coverslip (no. 1, Paul Marienfeld GmbH & Co., Lauda-Königshofen, Germany), after which the sample is closed by placing a vacuum grease ring around the drop and a 20 × 20 mm coverslip (no. 1, Paul Marienfeld GmbH & Co., Lauda-Königshofen, Germany) on top. This seal keeps the sample in place and prevents evaporation. Once placed in the LTRS system, Raman spectra are recorded by trapping single spores or cells using laser tweezers with laser power 70 mW acting on the sample. In this work, we gathered 10 accumulations of 10 seconds to acquire spectra on at least 15 spores in 2 separate solutions for each experiment. Each experiment thus used 30 technical replicates and 2 biological replicates. Displayed spectra show an average of these 30 replicates. The total energy applied on the spore during laser exposure is ∼6 J, well below the 20 J shown to cause spore damage.25,26 To avoid variation in Raman shift and intensity over time, the system is calibrated before each measurement by recording Raman spectra of polystyrene beads. Each measurement was performed with the spore 100 μm above the glass surface to maintain a consistent background signal without interference from the glass coverslip. Lastly, we acquired 3 technical replicates of background spectra from the background solution for spectral background subtraction.2.6 Decontamination protocol
The decontamination process for each sample proceeded as outlined below. 90 μl of spore suspension in water at a concentration of 106 spores per ml was mixed with 10 μl of biocide stock. The biocide stocks used were 5% sodium hypochlorite (NaOCl), in the form of a commercial bleach (Klorin Original, Colgate-Palmolive), with its pH adjusted to pH 8 using hydrochloric acid. We also used 1500 ppm chlorine dioxide (ClO2) from a commercial stock (DK-DOX 1500, Dr Küke GmbH). Thus the actual concentration of biocides were 0.5% NaOCl and 150 ppm (0.015%) ClO2.Spore suspensions were incubated in the biocide for 10 minutes. The biocide was then neutralised with 1% sodium thiosulphate, by mixing the spore suspension with a required volume of 10% sodium thiosulphate. The spores were then washed with deionised water by centrifuging and discarding the supernatant twice to remove reacted chemicals. A negative control was prepared using the same method, substituting the biocide for deionised water.
We also disinfected spores with 253.7 nm ultraviolet light. For this method, we placed 1 ml 106 spores per ml spore suspension into 3 ml quartz glass cuvettes (10 mm, Agilent). The cuvettes were exposed to ultraviolet light from a mercury lamp (EVG65-80 W/1,5A-PH, ZED), with the full emission profile seen in Fig. S1.† The cuvettes were placed under the mercury lamp at a distance 20 cm from the lamp, exposing the sample to UV-radiation at 100 mW for 10 min.
Neutralised samples were incubated in TSA for 4 hours at 37 °C. The incubated suspensions were diluted in deionised water and Raman spectra were acquired as detailed above.
2.7 Data analysis
To process our Raman spectra, we use an open-source Matlab (Matlab2018b, Mathworks) script provided by the Vibrational Spectroscopy Core Facility at Umeå University.27 Background subtraction and baseline correction of Raman spectra are done using an asymmetrical least squares algorithm28 and with the algorithm parameters set to λ = 106 and p = 10−3. For those spectra where we apply smoothing, we use Savitzky–Golay filtering29 with polynomial order 1 and frame length 33 for peak area calculations. We use polynomial order 1 and a frame rate of 5 for other spectra showing the 2200 cm−1 Raman peak.We evaluate the peak intensity of the broad 2200 cm−1 Raman peak by calculating the area under the curve between 2060 and 2260 cm−1 for the main peak and between 2265 and 2348 cm−1 for the 2300 cm−1 smaller peak. Peak intensities of sharper Raman peaks in the 600–1800 cm−1 region are evaluated using maximum peak height.
2.8 Statistical analysis
We performed statistical analysis on our Raman data using Prism 10 (Prism 10.0.2, GraphPad Software). We used two-way ANOVA with Šidák's multiple comparisons tests, comparing peak areas over time and in response to different chemicals. When comparing the 2300 cm−1 peak, we used the Kruskal–Wallis test with multiple comparisons to assess the difference in Raman peak intensity compared to the control set. We also used the Shapiro–Wilk test to assess if the data had a normal distribution.The box plots show the 1st and 3rd quartiles (box limits), mean values (squares), median values (solid line), and 1.5 interquartile range (whiskers).
2.9 Density functional theory
Raman spectra were computed using density functional theory at the B3LYP30–33/cc-pvtz34,35 level of theory, as implemented in Gaussian 16 ver. C.01,36 with an ultra-fine integration grid and implicit solvation in water using the polarizable continuum model.37 Spectra for deuterated species were computed using the ReadIsotopes keyword. Raman frequencies were scaled by 0.955, 0.963 and 0.953 in the regions 0–1000, 1001–2000, and >2000 cm−1 respectively.383 Results and discussion
3.1 Producing permanently labeled spore samples through deuterium incorporation
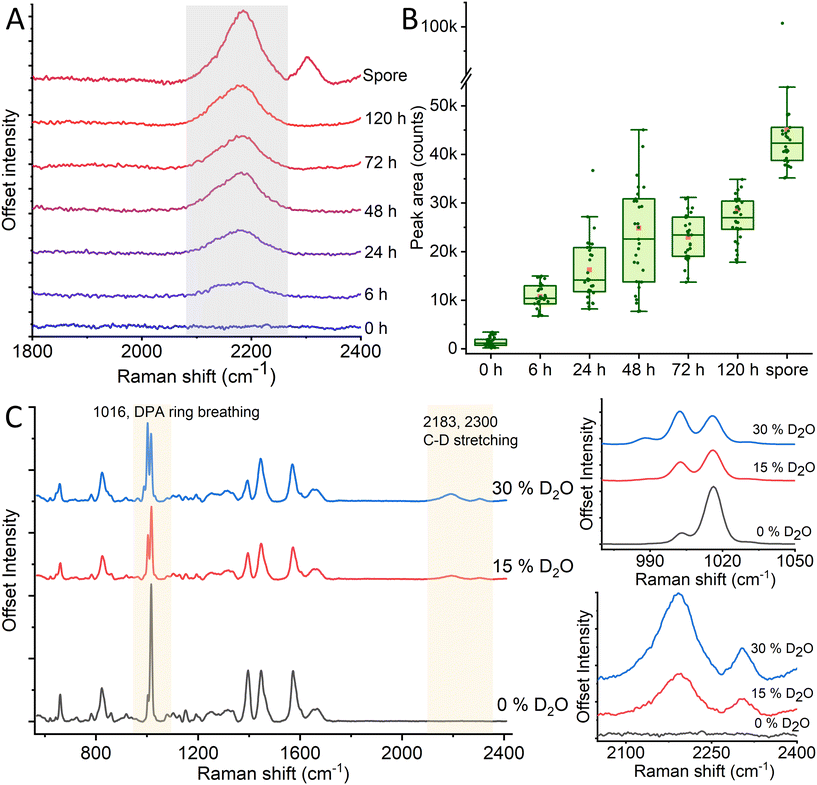
Labeling any biological sample requires balancing the method's complexity, the stability of the labeling agent, and its toxicity to the sample. In this context, deuterium-labeling is a method that combines several desirable traits for a labeling system. It is a stable, non-toxic (up to concentration as high as 70%), and metabolism-driven method that does not require any genetic modification.11 Deuteration is used as a label since it has no significant effect on the chemical reactions in metabolism.39 At the same time, the shift in molecular mass distribution will create unique vibrations that can be detected using Raman spectroscopy.To prepare a deuterium-labeled spore sample, we grow bacteria to form spores in 30% heavy water (D2O)-enriched medium. The relative concentration of D2O will determine the deuteration level of the spore and, thus, the intensity of the deuterium-induced Raman peaks around 2200 cm−1 (Fig. 1). The deuterium level will also depend on the sample's incubation time, as seen in Fig. 1A, with an increased incubation time permitting the bacteria to metabolize for longer and thus incorporate more deuterium into their structure. While incubating a sample for 120 h, we observe that the bacteria reach a steady-state in their deuterium-induced C–D peak after ∼48 h, as seen in Fig. 1B. Once having reached this steady-state, the bacteria will retain the C–D bond through sporulation. Upon spore germination in non-deuterated media, the reverse happens and the peak rapidly decreases as the spores grow and divide (Fig. S2†). Another notable feature is that the peak area of a spore is significantly higher than that for non-sporulated bacteria, even after 120 hours of incubation. This could be due to the varying sizes and densities of non-water substances present in vegetative cells and spores. Vegetative cells are elongated and can reach up to 5 μm in length and 1 μm in width, which is much larger than the laser beam width (400 nm waist). Thereby, cell parts outside the beam focus do not contribute to the Raman spectra. By contrast, spores are only ∼1 μm long and ∼0.5 wide, meaning a much larger percentage of the spore falls into the trap. Vegetative cells also have higher hydration than spores, and since water does not have C–D bonds, the signal from proteins and lipids in vegetative cells is lower. Furthermore, after sporulation, a secondary Raman peak appears at 2300 cm−1. This peak does not appear in the bacterial form and must thus stem from a spore-specific structure (for example CaDPA) originating during sporulation, as will be discussed in section 3.2.
Although the “quiet” Raman region around 2200 cm−1 displays the most notable D2O-induced peak in the spore, the changed chemical bond within the spores will affect the rest of the Raman spectrum within the fingerprint region ∼600–1600 cm−1 as well. To observe the effects of different deuteration levels on the spores, we record Raman spectra of spores prepared in 0, 15, and 30% heavy water induced broth, respectively. As previously discussed, deuterated spores display wide Raman peaks at ∼2200 and 2300 cm−1 which grow in intensity with higher levels of D2O as seen in Fig. 1C. However, we also observe the changes in the peak intensities of characteristic DPA peak established in previous works18,40 around 1016 cm−1 in D2O-labeled spores compared to non-deuterated ones. The intensity of the 1016 cm−1 peak decreases and the intensity of the 1003 cm−1 peak increases due to increased deuterium concentration. Additionally, a new peak arises at 988 cm−1, and its intensity increases with enhanced deuterium concentration (Fig. S3†). Within the 1200–1800 cm−1 range, we observe prominent peaks at 1395, 1448, and 1572 cm−1. Within these, we observe a relative decline in the 1395 and 1572 peaks compared to the central 1448 peak. Previous studies have assigned these two former peaks to DPA, while the latter has been assigned to a combination of DPA and non-specific CH2 vibrations (δCH2).40–42 From this, we can infer that the DPA is affected by the deuteration process to a larger degree than other molecular structures. This may partly be due to the relatively high concentration of DPA in bacterial spores and thus the high initial intensity of the peaks, such as the 1016 cm−1 peak. In addition, other prominent Raman peaks such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O from protein, lipid and DNA O–P–O will not be significantly affected by deuteration since hydrogen is not part of the bond. By contrast, the ring breathing mode that causes the 1016 cm−1 peak will be affected by deuteration, as there are three C–H bonds on the ring itself, which during sporulation could be substituted for C–D.
O from protein, lipid and DNA O–P–O will not be significantly affected by deuteration since hydrogen is not part of the bond. By contrast, the ring breathing mode that causes the 1016 cm−1 peak will be affected by deuteration, as there are three C–H bonds on the ring itself, which during sporulation could be substituted for C–D.
The dormant state of spores implies that the deuterium label remains stable and unchanged during storage, even when the spores are kept in water that does not contain deuterium. However, once allowed to germinate in non-deuterated growth media, the bacteria will increase in size and gradually incorporate regular hydrogen into its structure, decreasing the C–D Raman peak. We observe this decrease soon after starting incubation, with a 60% loss in the peak area of the 2200 cm−1 after 2 h of incubation, and a near complete peak loss after 3 h (Fig. S2†). The C–D Raman peak of deuterated spores can be used as a reliable and stable label for a spore sample, and the disappearance of the peak can be used to detect germination in a sample rapidly.
3.2 CaDPA-deficient mutants highlight the effect of deuteration on CaDPA Raman biomarker in spores
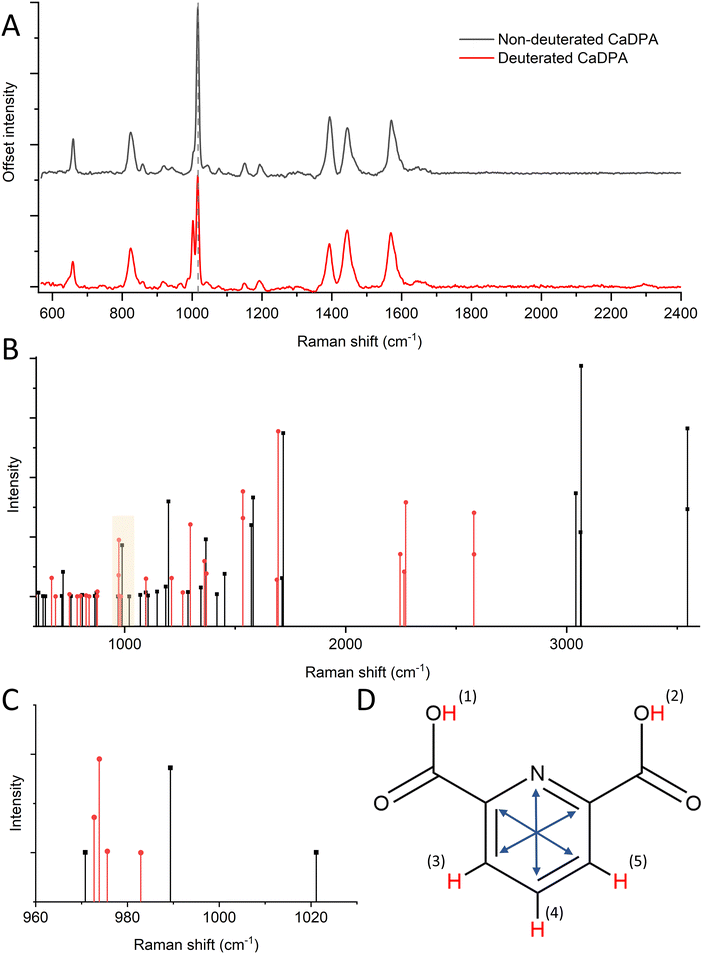
To determine the role of CaDPA in the Raman spectra of deuterated spores, we grew CaDPA-deficient B. subtilis spores alongside WT and measured their Raman signal. The ΔspoVAC mutant cannot take up CaDPA from the mother cell during sporulation due to a non-functional transport channel, while the ΔgerA ΔsleB mutation prevents unwanted germination of the spores.21 Thus, the combined mutant results in spores that have no internal CaDPA but are otherwise structurally and chemically the same as the wild-type, giving us a way to directly assess the effect of deuteration on DPA in the spore.Similar to the WT spores containing CaDPA, we observe the 2200 cm−1 peak in the deuterated CaDPA-deficient spores as seen in Fig. 2 and Fig. S4A.† However, unlike the WT, the deuterated CaDPA-deficient spores do not display a peak at 2300 cm−1, confirming that this peak directly correlates to the deuterated DPA as previously hypothesized. We think that both 2200 cm−1 and 2300 cm−1 peak originates from C–D vibration; however, the former corresponds to aliphatic C–D and the latter represents aromatic C–D vibration. As DPA is the major aromatic component of the spore, 2300 cm−1 peak intensity can be correlated with C–D vibration of DPA and a marker of deuterated DPA concentration.
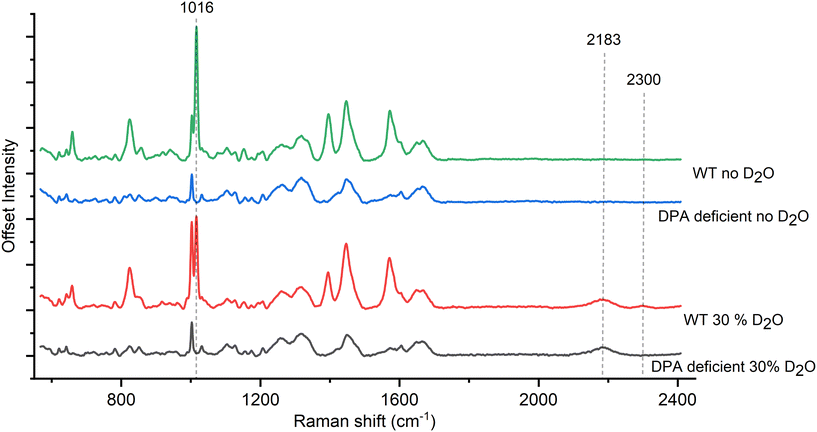 | ||
| Fig. 2 (A) Comparison of spore Raman spectra between WT and DPA-deficient mutants of B. subtilis. The absence of 2300 cm−1 peak in the DPA-deficient mutant confirms its association with DPA. | ||
To better observe the effects of deuterated CaDPA on the spore Raman spectrum, we subtract Raman spectra of CaDPA-deficient mutants from the WT of both deuterated and non-deuterated spores, with the differential spectra seen in Fig. 3A and Fig. S4B.† These difference spectra effectively correspond to regular and deuterated forms of CaDPA, in the pristine spore environment. This is important, as synthesised CaDPA spectra have differences depending on whether it is in solution, powder or crystalline form.43 In addition to the previously mentioned peak at 2300 cm−1, we also observe significant shifts around 1000 cm−1. The characteristic CaDPA peak at 1016 cm−1 has partly diminished in favour of two new peaks; a larger one around 1003 cm−1, and a smaller one around 988 cm−1. The larger one is largely unobservable within the context of a full spore organism due to the presence of the phenylalanine Raman peak at 1001 cm−1, however, the smaller one can be observed as a shoulder to the left of the main Raman peaks around 1000 cm−1. Additional shifts include a ratio shift between the peaks at 1395, 1448, and 1572 cm−1 as described in the previous section, as well as the formation of a shoulder to the left of the 658 cm−1 peak around 646 cm−1. Thus, these measurements highlight the deuteration-related spectral changes of CaDPA in the spores and provide Raman spectra of CaDPA and deuterated CaDPA in their pure form.
3.3 DFT reveals the role of H–D substitution in the Raman spectrum of DPA
To better understand the role of H–D substitution in Raman spectra of the DPA biomarker, we computed the spectra of relevant DPA isotopologues (molecules with an identical chemical formula but different isotopes) using DFT. These calculations were performed on non-chelated DPA because the arrangement of Ca2+ ions in relation to DPA in the spore core is not well characterized. DPA contains five unique hydrogen atoms that can be replaced with deuterium during sporulation, though individual substitutions in two pairs of H atoms are indistinguishable due to molecule symmetry. By replacing all the H atoms in DPA with D atoms, we observed in the DFT calculations a shifting trend towards lower relative Raman shifts (Fig. 3B). This trend is expected as higher atomic mass results in lower vibrational frequencies. The trend towards lower relative Raman shifts for deuterated DPA can be seen most clearly at higher Raman shifts (>2000 cm−1), where we observe the major C–H peak at around 3050 cm−1 (not shown in our Raman spectra due to the range limit of our system), while the C–D peak discussed in this manuscript appears around 2250 cm−1. Looking at another area of interest, around 1000 cm−1, we observed a similar pattern, where the aromatic ring breathing mode around 1004 cm−1 (here 990 cm−1) shifts lower toward 975 cm−1 (Fig. 3C). This is in line with Raman shifts predicted using DFT for benzene rings.44 Also, benzene rings are structurally similar to DPA and have similar ring breathing modes in the 1000 cm−1 region. This is further backed up by experiments as we see the appearance of a Raman peak around 988 cm−1 in deuterated CaDPA (Fig. 3A).Looking more closely at the effect of individual H–D substitutions, we can divide the substitutions into two categories; substitutions in the carboxyl groups (hydrogen 1–2), and substitutions in the pyridine ring (hydrogen 3–5), as seen in Fig. 3D. Substitutions in the carboxyl groups yielded a peak at 2600 cm−1, suggesting these substitutions are not responsible for the characteristic C–D vibrations seen at 2200 cm−1. These vibrations can instead be attributed to substitutions in the H atoms making up the pyridine ring. Substituting one or several of these atoms resulted in the appearance of several Raman vibrations around 2250 cm−1. Similarly, substitutions in the pyridine ring are, as expected, the main contributing factor to shifts in the characteristic ring breathing mode (marked with blue arrows, Fig. 3D) of DPA around 1000 cm−1. More specifically, substitutions in the 3 and 5 H atoms result in a larger shift from the 990 cm−1 peak down to 970 cm−1, while substitutions of only the 4 H-atom results in a lower shift down to 980 cm−1. This suggests these substitutions are responsible for the experimental shift towards lower Raman shifts observed around 1000 cm−1. Substitutions in the carboxyl group also result in slight shifts in the ring breathing mode, however, these measure less intense and similarly low shifts at ∼980 cm−1. Overall, the shifts observed from substitutions in the 3–5 hydrogen atoms appear more significant to the overall Raman activity and match more closely to experimental data. This further appears likely as the population of hydrogens in the carboxyl group would be largely non-present in CaDPA, and is generally prone to H–D exchange in an aqueous environment. A full list of DFT computed vibrations can be found in Table S1.† Thus, we concluded that substitutions in the pyridine ring of DPA are the major cause of the shifts in the Raman signal induced during deuteration.
3.4 The 2200 cm−1 Raman peak in deuterium-labeled spores acts as viability indicator
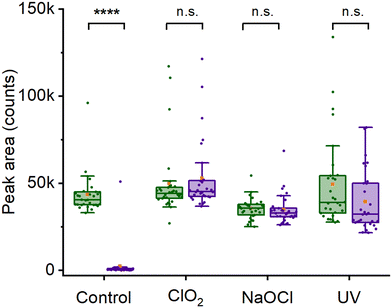
Another application for heavy water labeling is viability assessment. Chemical agents can neutralize spores, which is crucial for sanitizing surfaces in the food industry and medical settings. However, effectively neutralizing spores can prove difficult, especially when certain strains exhibit increased resistance to disinfectants like sodium hypochlorite45 and UV light.3 Although these spores might seem neutralized, they could still remain viable and eventually develop into fully-grown cells, albeit with a delay.46 In this context, the utilization of deuterium-labeling might offer a means to verify the inactivity of decontaminated spores on a metabolic level. This approach could then aid in confirming the effectiveness of the decontamination procedure.To illustrate this, we inactivated spore samples using different decontamination procedures and followed their C–D Raman peak evolution when incubated in growth media. As a control, we used a reference sample not exposed to any decontamination procedure, which indicated an almost complete disappearance of the 2200 cm−1 Raman peak after 4 hours of incubation, see Fig. 4 and Fig. S5.† This shows that deuterium-labeled spores incubated in regular TSA broth germinate and divide normally, leading to the complete loss of the C–D peak in 4 hours (p ≤ 0.0001). By contrast, spores decontaminated in 150 ppm chlorine dioxide do not divide and the C–D peak remains largely unchanged after 4 hours of incubation (p = 0.92) (Fig. 4 and Fig. S6A and B†). Spores treated with 0.5% hypochlorite (pH 8) appear visually changed, with the exosporium missing.18 However, similar to the chlorine dioxide treated sample, the C–D also did not significantly change in intensity over 4 hours (p ≥ 0.99) (Fig. 4 and Fig. S6C and D†). However, it should be noted that chlorine dioxide and hypochlorite-treated spores showed many spectral outliers, both directly after treatment and after 4 hours (Fig. S6C and D†). Apart from chemical decontamination, we also performed decontamination of spores using UV-C radiation. However, similar to the chemical decontamination, the intensity of the 2200 cm−1 Raman peak does not differ significantly between immediately after decontamination and subsequent growth over 4 hours (Fig. S6E and F†). Thus, we concluded that the viability of deuterium-labeled spores can be rapidly evaluated using their 2200 cm−1 Raman peak.
3.5 Evaluating the mode of action of different decontamination procedures using the 2300 cm−1 Raman peak in deuterium-labeled spores
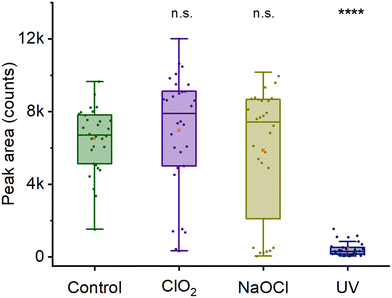
As discussed in the previous section, the 2300 cm−1 Raman peak is characterized as a CaDPA marker in its deuterated form. Hence, the change in intensity of 2300 cm−1 peak after decontamination can be used to evaluate spores’ DPA content after different modes of decontamination. Fig. 5 and Fig. S6† show the intensity of the 2300 cm−1 Raman peak after decontamination of spore using 0.015% chlorine dioxide, 0.5% hypochlorite (pH 8) and 100 mW UV-C radiation for 10 min. The intensity of the chlorine dioxide treated spores is similar to the control (p = 0.42), indicating no loss of DPA upon decontamination. For hypochlorite, there is no overall difference in peak intensity (p > 0.99). However, unlike the control and chlorine dioxide-treated spores, spores treated with hypochlorite follow a bimodal distribution indicating some spores lose their DPA, while others retain it. This is likely due to spores being degraded by hypochlorite, leading to loss of core material.47 The normal distribution of the control (p = 0.83) and chlorine dioxide-treated (p = 0.49) and the non-normal distribution of hypochlorite-treated spores (p = 0.0014) is confirmed by the Shapiro–Wilk test. In addition, for UV-treated spores, we observe the absence of the 2300 cm−1 peak in most of the spores, indicating a loss of DPA. In all samples, the low peak area values (≤2000 counts) do, in fact, correspond to a complete absence of the peak. The non-zero values are an artifact of the curve integration procedure. There is no peak in the actual spectra, as shown in Fig. S6E and F.† Thus, these observations suggest a possible mechanism of action of different decontaminants. For example, chlorine dioxide acts rapidly and does not change the Raman signal and its core structure, as suggested earlier in Malyshev et al.18 For UV mediated decontamination, unlike chlorine dioxide, decontamination leads to the release of DPA from the core of spores as observed in Öberg et al.,42 suggesting a different mode of action. Additionally, unlike the 1016 cm−1 peak, a widely used marker of DPA content, 2300 cm−1 peak appears in the ‘quiet’ region of the spores’ Raman spectra. Furthermore, partial overlap between 1016 cm−1 peak and 1003 cm−1 peak requires prior deconvolution of the spectra to measure the area under the curve for DPA concentration measurement, especially in deuterated condition (Fig. 1C). Hence, the intensity of 2300 cm−1 Raman peak can act as a better marker of DPA content in its deuterated form and changes in the intensity of this peak upon decontamination provide useful information about the mode of action of different decontaminant.4 Conclusion
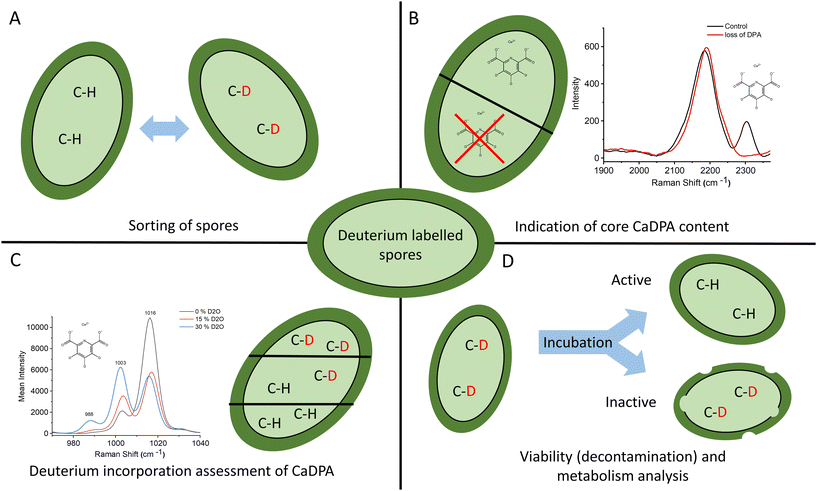
Bacterial spores are problematic within, among other areas, the food industry and healthcare. Due to the problems they can cause, a lot of research is done on spores to mitigate their hazards, and this requires robust techniques to label, track, and monitor spore activity. Herein, we present a method to label spores permanently using heavy water. By incubating a spore sample in D2O for 48 h, and then letting it re-sporulate, the spores are permanently labeled with a characteristic Raman fingerprint. This fingerprint is most notable as a pair of peaks at 2200 and 2300 cm−1, an otherwise quiet Raman region, allowing for easy labeling, sorting, and tracking of spores (Fig. 6A). The 2300 cm−1 peak specifically correlates to the presence of deuterated CaDPA within the spores making it an excellent indicator of core CaDPA content (Fig. 6B). Additional spectral changes in the more conventional Raman fingerprint region include shifts in peak ratios in the 1380–1600 cm−1 range and other small peaks appearing correlating with deuterated CaDPA at 988 and 646 cm−1. Furthermore, shifts in ratios between deuterated and non-deuterated CaDPA around the ∼1000 cm−1 Raman region allow for quantitative evaluation of deuterium uptake (Fig. 6C). DFT calculations suggest these Raman peaks correlate with deuterations, primarily in the central pyridine ring of DPA. Accordingly, it is possible to use D2O-labeling to track CaDPA in spores. Lastly, thanks to the permanence of the D2O-label within the spore form, it is possible to use deuterated spores to indicate spore viability by whether or not the spores keep their characteristic C–D Raman peak during germination (Fig. 6D). In conclusion, this paper demonstrates a method of labeling spores permanently with D2O and provides examples of its application in bacterial spore research.Author contributions
DM and RÖ conceived the experiments. MA secured funding and guided the project. DM prepared the deuterium-labeled spores. RÖ and TS carried out the Raman measurements. AO and RÖ performed and analyzed the DFT calculations. RÖ, TS and DM analyzed the data. RÖ, TS, DM and MA wrote the manuscript. All authors reviewed and revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Swedish Research Council (2019-04016); the Umeå University Industrial Doctoral School (IDS); Kempestiftelserna (JCK-1916.2); and Swedish Department of Defence, Project No. 470-A400823. The authors acknowledge the facilities and technical assistance of the Umeå Core Facility for Electron Microscopy (UCEM) at the Chemical Biological Centre (KBC), Umeå University, a part of the National Microscopy Infrastructure NMI (VR-RFI 2019-00217). We thank Marina Aspholm for providing the B. cereus spores. We thank David Rudner for providing the B. subtilis wild-type and DPA-deficient mutant spores.References
- A. Andersson, U. Rönner and P. E. Granum, Int. J. Food Microbiol., 1995, 28, 145–155 CrossRef CAS PubMed.
- A. K. Goel, World J. Clin. Cases, 2015, 3, 20 CrossRef PubMed.
- P. Setlow, J. Appl. Microbiol., 2006, 101, 514–525 CrossRef CAS PubMed.
- P. L. Choyke, R. Alford, H. M. Simpson, J. Duberman, G. Craig Hill, M. Ogawa, C. Regino and H. Kobayashi, Mol. Imaging, 2009, 8, 341–354 Search PubMed.
- A. Ali, K. Gaurav, N. Senthilnathan, T. P. Radhakrishnan, C. Sasikala and C. V. Ramana, J. Microbiol. Methods, 2020, 177, 106019 CrossRef PubMed.
- C. P. Toseland, J. Chem. Biol., 2013, 6, 85–95 CrossRef PubMed.
- B. A. Kairdolf, M. C. Mancini, A. M. Smith and S. Nie, Anal. Chem., 2008, 80, 3029–3034 CrossRef CAS PubMed.
- E. A. Te Velde, T. Veerman, V. Subramaniam and T. Ruers, Eur. J. Surg. Oncol., 2010, 36, 6–15 CrossRef CAS PubMed.
- V. Gupta, A. Cherkassky, P. Chatis, R. Joseph, A. L. Johnson, J. Broadbent, T. Erickson and J. DiMeo, Nucleic Acids Res., 2003, 31, e13 CrossRef PubMed.
- G. E. Walrafen, J. Chem. Phys., 1964, 40, 3249–3256 CrossRef CAS.
- M. Zhang, W. Hong, N. S. Abutaleb, J. Li, P. T. Dong, C. Zong, P. Wang, M. N. Seleem and J. X. Cheng, Adv. Sci., 2020, 7, 1–14 Search PubMed.
- J. Xu, D. Zhu, A. D. Ibrahim, C. C. Allen, C. M. Gibson, P. W. Fowler, Y. Song and W. E. Huang, Anal. Chem., 2017, 89, 13305–13312 CrossRef CAS PubMed.
- D. Berry, E. Mader, T. K. Lee, D. Woebken, Y. Wang, D. Zhu, M. Palatinszky, A. Schintlmeister, M. C. Schmid, B. T. Hanson, N. Shterzer, I. Mizrahi, I. Rauch, T. Decker, T. Bocklitz, J. Popp, C. M. Gibson, P. W. Fowler, W. E. Huang and M. Wagner, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E194–E203 CAS.
- K. Yang, H. Z. Li, X. Zhu, J. Q. Su, B. Ren, Y. G. Zhu and L. Cui, Anal. Chem., 2019, 91, 6296–6303 CrossRef CAS PubMed.
- Y. Liu, Y. Ma, L. Zhang, X. Sun, J. Yang, X. Li, F. Li, R. Chen, P. Zhu, J. Xu and F. Yang, J. Raman Spectrosc., 2022, 53, 902–910 CrossRef CAS.
- M. Yasuda, N. Takeshita and S. Shigeto, Sci. Rep., 2021, 11, 1279 CrossRef CAS PubMed.
- R. Öberg, T. Dahlberg, D. Malyshev and M. Andersson, Analyst, 2023, 148, 2141–2148 RSC.
- D. Malyshev, T. Dahlberg, K. Wiklund, P. O. Andersson, S. Henriksson and M. Andersson, Anal. Chem., 2021, 93, 3146–3153 CrossRef CAS PubMed.
- B. Setlow, S. Atluri, R. Kitchel, K. Koziol-Dube and P. Setlow, J. Bacteriol., 2006, 188, 3740–3747 CrossRef CAS PubMed.
- D. R. Zeigler, Z. Prágai, S. Rodriguez, B. Chevreux, A. Muffler, T. Albert, R. Bai, M. Wyss and J. B. Perkins, J. Bacteriol., 2008, 190, 6983–6995 CrossRef CAS PubMed.
- Y. Gao, R. D. C. Barajas-Ornelas, J. D. Amon, F. H. Ramírez-Guadiana, A. Alon, K. P. Brock, D. S. Marks, A. C. Kruse and D. Z. Rudner, Genes Dev., 2022, 36, 634646 CrossRef PubMed.
- M. F. Lu, Y. J. Zhang and H. Y. Zhang, Afr. J. Microbiol. Res., 2013, 7, 604–611 CAS.
- T. Stangner, T. Dahlberg, P. Svenmarker, J. Zakrisson, K. Wiklund, L. B. Oddershede and M. Andersson, Opt. Lett., 2018, 43, 1990 CrossRef PubMed.
- T. Dahlberg, D. Malyshev, P. O. Andersson and M. Andersson, Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XXI, 2020, p. 28 Search PubMed.
- D. Malyshev, R. Öberg, T. Dahlberg, K. Wiklund, L. Landström, P. O. Andersson and M. Andersson, Spectrochim. Acta, Part A, 2022, 265, 120381 CrossRef CAS PubMed.
- D. Malyshev, N. F. Robinson, R. Öberg, T. Dahlberg and M. Andersson, J. Biophotonics, 2022, 15, 1–7 CrossRef PubMed.
- Vibrational spectroscopy core facility, Umeå Unversity, accessed 22-10-04, 2022, https://www.umu.se/en/research/infrastructure/visp/downloads/.
- P. H. C. Eilers, Anal. Chem., 2004, 76, 404–411 CrossRef CAS PubMed.
- M. J. E. Savitzky and A. Golay, Anal. Chem., 1964, 36, 1627–1639 CrossRef.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- B. Miehlich, A. Savin, H. Stoll and H. Preuss, Chem. Phys. Lett., 1989, 157, 200–206 CrossRef CAS.
- S. H. Vosko, L. Wilk and M. Nusair, Can. J. Phys., 1980, 58, 1200–1211 CrossRef CAS.
- R. A. Kendall, T. H. Dunning and R. J. Harrison, J. Chem. Phys., 1992, 96, 6796–6806 CrossRef CAS.
- E. R. Davidson, Chem. Phys. Lett., 1996, 260, 514–518 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian ∼ 16 Revision C.01, Gaussian Inc, Wallingford CT, 2016 Search PubMed.
- G. Scalmani and M. J. Frisch, J. Chem. Phys., 2010, 132, 114110 CrossRef PubMed.
- J. C. Zapata Trujillo and L. K. McKemmish, J. Phys. Chem. A, 2023, 127, 1715–1735 CrossRef CAS PubMed.
- S. L. Harbeson and R. D. Tung, Annual Reports in Medicinal Chemistry, Academic Press, 2011, pp. 403–417 Search PubMed.
- S. S. Huang, D. Chen, P. L. Pelczar, V. R. Vepachedu, P. Setlow and Y. Q. Li, J. Bacteriol., 2007, 189, 4681–4687 CrossRef CAS PubMed.
- E. Kočišová and M. Procházka, J. Raman Spectrosc., 2018, 49, 2050–2052 CrossRef.
- R. Öberg, T. B. Sil, A. Johansson, D. Malyshev, L. Landström, S. Johansson, M. Andersson and P. O. Andersson, J. Phys. Chem. B, 2024 Search PubMed , Advance online publication, https://doi.org/10.1021/acs.jpcb.3c07062.
- L. Kong, P. Setlow and Y. Q. Li, Analyst, 2012, 137, 3683 RSC.
- A. Sakamoto and M. Tasumi, J. Raman Spectrosc., 2021, 52, 2282–2291 CrossRef CAS.
- A. N. Edwards, S. T. Karim, R. A. Pascual, L. M. Jowhar, S. E. Anderson and S. M. McBride, Front. Microbiol., 2016, 7, 1–13 Search PubMed.
- B. Setlow, C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow and P. Setlow, J. Appl. Microbiol., 2002, 92, 362–375 CrossRef CAS PubMed.
- D. Malyshev, I. A. Jones, M. McKracken, R. Öberg, G. M. Harper, L. T. Joshi and M. Andersson, BMC Microbiol., 2023, 23, 59 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Emission spectrum of the used UV source as well as all the computed DFT peaks for different H–D substitutions in CaDPA; additional figures. See DOI: https://doi.org/10.1039/d3an02162a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |