Open Access Article
Open Access ArticleGetting the SMILES right: identifying inconsistent chemical identities in the ECHA database, PubChem and the CompTox Chemicals Dashboard†
Juliane
Glüge
 *ab,
Kristopher
McNeill
*ab,
Kristopher
McNeill
 a and
Martin
Scheringer
a and
Martin
Scheringer
 a
a
aInstitute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 Zürich, Switzerland. E-mail: juliane.gluege@usys.ethz.ch
bDepartment of Cell Toxicology, UFZ Leipzig-Halle, 04318 Leipzig, Germany
First published on 9th March 2023
Abstract
Chemical databases containing information on substances and their identities are important and useful tools, used in many areas of chemistry and cheminformatics. Errors or inconsistencies in the identities of substances in the databases are a major problem, as they can make QSAR predictions inaccurate, make chemical hazard and risk assessments erroneous, and cause problems for the ordering of chemicals and analytical standards. In the present study, we checked the entries of all mono-constituent organic substances registered under REACH (more than 8500 substances) in the database of the European Chemicals Agency (ECHA), PubChem and the CompTox Chemicals Dashboard and flagged compounds with inconsistent chemical identifiers. In total 736 inconsistent entries, and 48 additional entries where the substance identity was not clear, were identified. This shows that data curation activities are still not sufficient in the databases and that more work needs to be done. Additionally, the identified inconsistent entries were analyzed to understand what kind of mismatches have been introduced in the databases and to avoid these mismatches in the future. Data gathering and processing is described in detail in the current study so that further studies can continue with this work for additional substances and databases. In this way, the study makes an important contribution towards improved and more trustworthy databases.
Environmental significanceThe substances fully registered under REACH are all manufactured in and/or imported to the European Economic Area at more than 1 tonne per year. In terms of tonnage, these are the most important industrial chemicals in Europe, and it is therefore crucial that their chemical identities are correctly listed. However, the analysis of more than 8500 registered mono-constituent organic substances has shown that CAS Registry Numbers and structures of 346 entries did not match in the database of the European Chemical Agency. Inconsistent entries were also found in other databases for these substances. This shows that current data curation activities are still not sufficient in chemical databases and that more work is urgently needed in this area. |
Introduction
Chemical hazard and risk assessment for the thousands of chemicals that are currently on the market or are intended to be put on the market faces various challenges, starting from basic information about chemical identity and chemical properties.1,2 Traditionally, experimental studies have been used to determine chemical property data, but data from read-across and quantitative structure–activity relationships (QSARs) are increasingly used as well. The fourth report on the use of alternatives to testing on animals for the Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) in Europe showed that in 2019 on average 27% of the data in the registrations submitted to the European Chemicals Agency (ECHA) under REACH came from experimental studies, 25% from read-across, 2.6% from QSARs, and 3.7% from weight of evidence. The rest were either data waivers, test proposals or had no information.3 Especially for read-across and QSARs, it is critical that the information about the substances that is used as input to these estimation methods is correct. This includes the line notation of the chemical structure, e.g., as Simplified Molecular-Input Line-Entry System (SMILES) or International Chemical Identifier (InChI) string, the Chemical Abstracts Service Registry Number™ (CAS RN™) and the chemical name and applies to the reference substances in read-across as well as to the substances of interest. Young et al. (2008)4 looked into various databases and found that 0.1% to 3.4% of the chemical structures in the databases were incorrect. Several scientific articles have addressed the importance of chemical structure curation in cheminformatics and QSAR modelling since then,5–10 but there are still two important open issues. First, most articles have looked at data curation workflows, i.e., how data retrieved from a database can be curated to make them suitable for QSAR development. Only few studies really identified and published the incorrect data in the original databases. For example, Waldman et al. (2015)7 mentioned that the errors they discovered have been corrected. However, they did not search systematically for errors in larger databases. Gadaleta et al. (2018)6 investigated large datasets and also reported the rejected substances. However, they did not report the original database, i.e., the sources, of the incorrect entries and it is unlikely that these entries have been corrected in the original databases. Similarly, Young et al. (2008)4 identified incorrect structures but did not publish the structures for each database explicitly. Second, new substances are added to the databases regularly, so curating the databases is an ongoing process.Based on these examples, it can be assumed that there is still a relevant number of incorrect or inconsistent entries in chemical databases. One of the most important databases for chemicals in Europe is the ECHA database, which contains information on substances registered under the REACH Regulation and are manufactured in and/or imported to the European Economic Area at more than 1 tonne per year. The information in the ECHA database originates from the registrants themselves and is supposed to be updated regularly.11 Investigations by ECHA itself but also by the German Environment Agency revealed that quite a high percentage of the data submitted under REACH are not compliant with the REACH regulations.12–15 When we started to look into the database in more detail, we also found a range of inconsistencies in the chemical identities. The aim of the present study was therefore to systematically identify inconsistent chemical identities of organic substances in the ECHA database16 as well as in PubChem17 and the CompTox Chemicals Dashboard.18 Inorganic and organometallic compounds have additional complications with regard to line notation representations and will be treated in a separate study. PubChem and the CompTox Chemicals Dashboard database were chosen as they are two of the largest publicly available databases and because it was possible to retrieve information from these two databases for a large number of substances (semi-)automatically. The commercial database SciFindern![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 that is operated by the Chemical Abstract Service (CAS) was used to cross-check cases where the chemical identities in the ECHA database, PubChem and/or the CompTox Chemicals Dashboard did not agree.
19 that is operated by the Chemical Abstract Service (CAS) was used to cross-check cases where the chemical identities in the ECHA database, PubChem and/or the CompTox Chemicals Dashboard did not agree.
Methods
Selection of substances
The substances investigated in this study are those that have been fully registered under REACH (as of April 2022) and that are organic and mono-constituent. NONS (for “notification of new substances”, which are substances notified before REACH entered into force) were also included in the assessment; intermediates were excluded. Substances whose production has been ceased or whose registration dossier is no longer valid were included if the information on the substances was still available on the ECHA website (and thus publicly accessible). Table 1 shows the number of substances for the entire database and the different subgroups. The majority of the registration dossiers from REACH contained information on the “origin” of the substance (organic/inorganic/organo-metallic/petroleum product) as well as on the composition (mono-constituent/multi-constituent/UVCB). However, for around 19% of the substances, this information was missing and there was no entry for origin and/or composition. In order not to overlook substances, we deselected the unsuitable substances instead of selecting the targeted ones (see Table 1). Remaining substances that were not organic and mono-constituent were subsequently excluded manually. Table 1 shows that 37% of all registered substances (excluding those that are only intermediates) are mono-constituent and organic and do not have the word “reaction” in their name. This set of 8590 substances was included in the present study.| Number of substances | Percentage compared to all | |
|---|---|---|
| a Products such as gasoline, kerosene (jet-fuel), diesel fuel, lubricants, paraffin wax and bitumen that are manufactured from crude oil using a range of refining processes. | ||
| ECHA database – all | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 184 184 |
100% |
| Not inorganic, not an element, not a petroleum producta, not organo-metallic | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 702 702 |
89% |
| Additionally, not UVCB, not multi-constituent | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 156 156 |
70% |
| Additionally, with full registration or NONS | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753 753 |
46% |
| Additionally, without ‘reaction’ in the name | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 529 |
45% |
| Additionally, without those removed manually that had no entry under origin and/or composition but were not organic and mono-constituent | 8590 | 37% |
Chemical identifier
Table 2 gives an overview of, and provides some details on, the chemical identifiers mentioned in this study. To systematically identify inconsistent chemical identities in a database, at least two chemical identifiers are needed: one that represents the chemical structure and one that can be used to verify the entry in another database. In the present study, we used the SMILES for the structural representation and the CAS RN™ for the verification. The chemical names were not used/verified systematically, because many names are provided in the ECHA database as non-IUPAC names and cannot automatically be converted into a structure. The ECHA guidance document for the identification and naming of substances under REACH and CLP20 states that the IUPAC name should be used for the registration of a substance under REACH. However, this is not always respected, and some substances are registered with trade names (e.g., JASMONITRILE) or other non-IUPAC names. We checked the chemical names therefore only for those substances where CAS RN™ and SMILES were not consistent in the ECHA database. Inconsistencies between the name and the given structure are also possible for substances where CAS RN™ and SMILES match. However, this was not checked further.| Chemical identifier | Abbreviation in full | Description |
|---|---|---|
| a In the PubChem terminology, a substance is a chemical sample description provided by a single source and a compound is a normalized chemical structure representation found in one or more contributed substances. b Canonical SMILES represent a unique representation for a particular molecule. However, the original procedure of Weininger et al. (1988)21 did not include a treatment of stereochemistry. Various algorithms have therefore been developed for generating canonical SMILES all of which differed from each other.22 | ||
| IUPAC name | International Union of Pure and Applied Chemistry (IUPAC) name | Name of the substance based on IUPAC nomenclature rules |
| EC number | European Community (EC) number | Unique seven-digit identifier that was assigned to substances for regulatory purposes within the EU |
| CAS RN™ | Chemical Abstracts Service (CAS) Registry Number™ | Unique numerical identifier assigned by the CAS to every chemical substance or compound whose existence has been proven |
| DSSTox substance ID | Distributed structure-search-able (DSS) toxicity substance identifier (DTXSID) | Unique identifier for substances; used mainly in the CompTox Chemicals Dashboard. Substances here are single chemicals, mixtures or polymers |
| DSSTox compound ID | DSS toxicity chemical identifier (DTXCID) | Unique identifier for chemical structures; used mainly in the CompTox Chemicals Dashboard |
| SID | PubChem Substance ID | Unique identifier for substancesa; used mainly in PubChem |
| CID | PubChem Compound ID | Unique identifier for compoundsa; used mainly in PubChem |
| Molecular formula | — | Numbers of each chemical element in a molecule; the Hill notation was used for a uniform order of the elements; does not contain structural information |
| Isomeric SMILES | Isomeric simplified molecular-input line-entry system | Line notation for describing the structure of chemicals; ‘isomeric’ means that it contains isotopic and chiral specifications; one structure can be described by more than one (isomeric) SMILES string even if the canonical SMILES is usedb |
| InChI string | IUPAC International Chemical Identifier string | Standardized way to encode molecular information; uses layers of information; is a unique representation |
| InChIKey | IUPAC International Chemical Identifier Key | Hashed version of the full InChI string that has always 27 characters and allows for easy web searches |
| 2D structure | 2-Dimensional structure of the molecule | Structure created from the SMILES e.g., via Smi2Depict (https://re.edugen.wiley.com/cgibin/Smi2DepictWeb.py); unique representation |
Data gathering
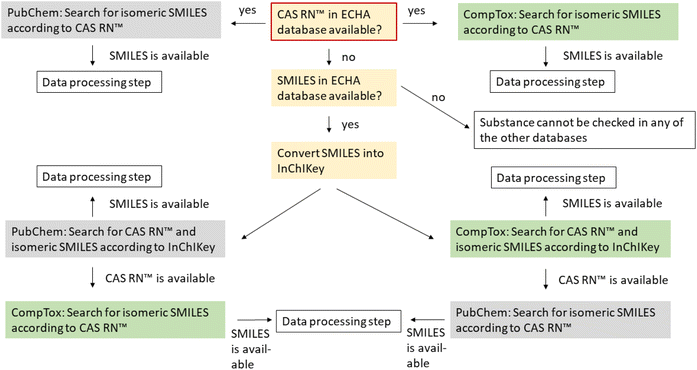
Systematic automated data collection activities (including scraping, data mining, and extraction and re-utilization) of the whole or a substantial part of the ECHA website and the ECHA databases are prohibited. It is therefore not possible to retrieve the registered data directly form the ECHA website. General information on the substances including their CAS RN™, registered tonnage band, composition and origin can be downloaded from https://echa.europa.eu/information-on-chemicals/registered-substances. However, the SMILES is not available via this website and also not available via the study results, which can be downloaded in IUCLID format.23 We therefore contacted ECHA and obtained a list of the registered substances with the available IUPAC names, EC numbers, CAS RNs™, molecular formulas and SMILES notations in January 2021. Additional SMILES codes were obtained manually from the website in February and May 2022, after which SMILES were available for 94% of all organic mono-constituent organic substances. Almost all substances could be identified by an EC number (99.5% of the 8590 substances used in the present study). However, in the dataset provided by ECHA in January 2021, only 73% of the substances had CAS RNs™. After cross-checking with the other databases, different manual checks and updates in the ECHA database, we were able to allocate CAS RNs™ to 88% of the finally investigated substances.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 chemicals which can be accessed online in batch mode. The data in the CompTox Chemicals Dashboard have been manually curated to identify conflicts between identifiers.24 Possible input parameters for the batch search are the chemical name, CAS RN™, InChIKey, DTXSID or DTXCID. Possible outputs are CAS RN™, InChIKey, IUPAC name and SMILES.
000 chemicals which can be accessed online in batch mode. The data in the CompTox Chemicals Dashboard have been manually curated to identify conflicts between identifiers.24 Possible input parameters for the batch search are the chemical name, CAS RN™, InChIKey, DTXSID or DTXCID. Possible outputs are CAS RN™, InChIKey, IUPAC name and SMILES.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S(
S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)(O)C(F)(F)C(F)(F)C(F)(F)C(F)(F)F instead of [K+].[O–]S(
O)(O)C(F)(F)C(F)(F)C(F)(F)C(F)(F)F instead of [K+].[O–]S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)(
O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C(F)(F)C(F)(F)C(F)(F)C(F)(F)F. Despite these (for our case unfavorable) forms of representation, SciFindern is one of the best databases available, mainly because the data are extensively manually curated.
O)C(F)(F)C(F)(F)C(F)(F)C(F)(F)F. Despite these (for our case unfavorable) forms of representation, SciFindern is one of the best databases available, mainly because the data are extensively manually curated.
Searching for the InChIKey in the CompTox Chemicals Dashboard and PubChem resulted in CAS RNs™ that are not included in REACH. These CAS RNs™ are marked in green in the ESI-1† to show that they were not provided by the registrants.
Data processing
If more than two of the three databases had an entry for a substance, the isomeric SMILES obtained from the databases were converted into standard InChI strings and standard InChIKeys using Open Babel version 3.1.1 (ref. 25 and 26) and afterwards compared to each other. For substances where the InChI strings did not match, the SMILES were manually inspected via Smi2Depict27 and grouped afterwards. For mismatching structures or disagreeing stereochemistry, SciFindern was checked additionally to find the ‘correct’ structures for the respective CAS RN™. Since the Chemical Abstract Service that operates SciFindern also assigns CAS RNs™ to structures and is the only authoritative source of CAS RNs™, we assumed that the CAS RN™ and the structural formula are correctly assigned in SciFindern, even if the SMILES strings themselves have some unfavorable representation or are even sometimes incorrect (see the points above).It is important to note that the check for inconsistencies for the compounds in the ECHA database was only possible if the CAS RN™ was available in the ECHA database. Without CAS RN™, it was only possible to check whether or not the SMILES string itself was internally consistent and whether or not it belonged to a valid chemical structure (if a corresponding structure was found in PubChem or the CompTox Chemicals Dashboard).
For substances with CAS RN™ but no SMILES in any of the databases or a SMILES in just one of the databases, the isomeric SMILES was obtained and/or verified via SciFindern. To obtain the correct isomeric SMILES in SciFindern, the given SMILES in SciFindern was taken and manually adjusted for substances with more than one component, taking the stoichiometry and charges in the structural formula and the name into account.
Substances without CAS RN™ and for which the search according to the InChIKey gave no result in PubChem or the CompTox Chemicals Dashboard could not be verified. Here, it was only possible to check whether or not the SMILES string itself was internally consistent and had, e.g., no unusual valence.
Substances where the InChI strings did not match between the databases were assigned to five different groups: (1) inconsistent information on the molecular structure where the assignment could be checked with SciFindern; (2) inconsistent information on the molecular structure in at least two databases and no verification possible via SciFindern; (3) missing cis/trans-isomer information for alkenes; (4) ‘Omitted undefined stereo’ warning in Open Babel, cis/trans isomers are defined for alkenes; (5) tautomers. Tautomers with identical InChI string were marked whenever they were detected in the manual data curation.
For substances in group 1, it was further specified which aspect is inconsistent. The seven different possible specifications were: inconsistency related to the molecular structure itself; no stereochemistry information in the ECHA database while SciFindern has some for the corresponding CAS RN™; deviating stereochemistry; available stereochemistry information in the ECHA database while SciFindern has none for the corresponding CAS RN™; inconsistency related to the cis/trans isomer; substances with unusual valence e.g., unpaired electrons; and substances registered as polymer while the SMILES represents a monomer.
For substances in the ECHA database that belonged to group 1, the so-called ‘brief profiles’ of the ECHA database were checked again manually in June 2022 to find out whether or not the brief profiles of the substances had been updated in the meantime.
An important point for the ECHA database and substances in group 1 is that only the registrants know which substance they intended to register. It is possible that they included a correct SMILES (and maybe name) in the registration but a wrong CAS RN™, but it is also possible that they included the correct CAS RN™ but wrong SMILES. We therefore also converted the names for the substances in group 1 from the ECHA database into structures (using Marvin Sketch 22.18) and compared the structural formulas converted from the names to the structures obtained via the CAS RN™ and those given in the ECHA database directly.
Results & discussion
Identified entries in the various groups
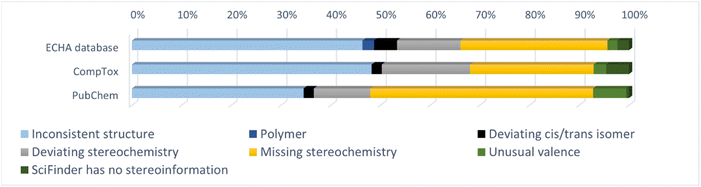
Table 3 gives an overview of the investigated substances and the number of substances in the different groups. In total 8590 substances were checked. Of these, 346 (4.3%), 197 (3.0%) and 193 (2.8%) substances had inconsistent chemical identifiers in the ECHA database, the CompTox Chemicals Dashboard and PubChem, respectively (group 1). In addition, 21, 9, and 18 substances had differing identifiers in the databases, but it was not possible to check the identities in SciFindern (group 2). Table 3 lists additionally the number of substances with missing cis/trans-isomer information for alkenes in each database (group 3) and the number of substances where Open Babel gave the warning ‘Omitted undefined stereo’ that was not due to missing cis/trans-isomer information for alkenes (group 4).| ECHA database | CompTox Chemicals Dashboard | PubChem | ||||
|---|---|---|---|---|---|---|
| Number of substances with SMILES | Percentage | Number of substances with SMILES | Percentage | Number of substances with SMILES | Percentage | |
| Total | 8109 | 6564 | 6989 | |||
| Group 1 | 346 | 4.3% | 197 | 3.0% | 193 | 2.8% |
| Group 2 | 21 | 0.3% | 9 | 0.1% | 18 | 0.3% |
| Group 3 | 115 | 1.4% | 141 | 2.1% | 68 | 1.0% |
| Group 4 | 1722 | 21% | 2112 | 32% | 1532 | 22% |
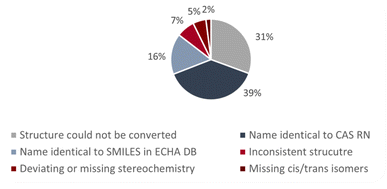
Fig. 2 shows the percentages of the different types of errors and inconsistencies in group 1. In PubChem, most of the inconsistent entries in group 1 were due to missing stereochemistry whereas in the CompTox Chemicals Dashboard and the ECHA database most of the entries in group 1 were due to inconsistent structures. There are also some errors/inconsistencies in group 1 that occurred for many substances. These included (a) substances where the net charge was not zero; (b) substances with LiH, NaH, KH, MgH2 or CaH2 as additional components (instead of counterions) and (c) substances with two or more components while the CAS RN™ only corresponded to one of these components (only in the ECHA database). The investigated substances with their chemical identifiers are provided in the ESI-1,† divided into single substances and substances with multiple components (e.g., salts). The ESI-2† shows the inconsistent structures from the ECHA database in 2D in order to make it easier to correct the entries.
 | ||
| Fig. 2 Percentages of the different types of errors and inconsistencies for the substances in group 1. | ||
Crosschecking of the substance identify in the ECHA database via the chemical name
Fig. 3 shows the results for the name-to-structure conversion for the substances in group 1 of the ECHA database. For 31% of the substances, the name could not be converted into a structure. In most of the cases this was due to errors in the chemical name. In a few cases, the trade name was given and no structure at all could be generated from the trade name. For 39% of the substances, the name corresponded to the CAS RN™ and for 16% of the substances to the SMILES code. For the remaining 14% of the substances, the structures generated from the name did not correspond to the CAS RN™ nor to the SMILES.Discussion on the applied method
Errors such as unusual valence, net charge is not zero, polymer with monomer SMILES and missing cis/trans isomers can be detected independently of other databases and it has been requested in the past that database operators should check for those errors.10 Other errors such as inconsistent or missing stereochemistry and inconsistent structures are harder to detect. In the present study, comparing the entries in two or three databases has been proven very useful. It only fails if all the databases have the same (inconsistent) entry. Unfortunately, the option to check all substances against SciFindern only works to a limited extent, even though we currently consider SciFindern to be the most reliable reference database. A direct comparison is however complicated by the fact that SciFindern represents substances with disconnected structures differently than the other databases.The prerequisite for the comparison of the structures between the databases is in any case a second identifier that is independent of the structure. In the present study, the CAS RN™ was used as second identifier. For substances without a CAS RN™ in the ECHA database, the comparison was less meaningful. In these cases, it could only be checked if the InChIKey from the ECHA database existed in one of the other databases. However, this was not a confirmation that the structure in the ECHA database is correct. The chemical name was only checked for a few substances because many names could not be converted to structures.
Completeness of the check
The semi-automated review of the SMILES in the three databases revealed quite some errors and inaccuracies. However, all three databases are regularly updated, which means that some of the errors might have been corrected since we obtained the data, while others may have been newly introduced. At the time of the submission of this article, the SMILES from PubChem and the CompTox Chemicals Dashboard were quite current as they were retrieved in February 2022. The SMILES from the ECHA database were retrieved more than two years before the publication of this article. In order to at least partially take into account the updates in the ECHA database, all SMILES from the ECHA database for substances in group 1 were manually rechecked (and corrected if necessary) in June 2022. Surprisingly, for 28 substances the inconsistent SMILES had been removed from the ECHA database. In six other cases, however, the SMILES had been adjusted.Significance of the various errors
We divided the identified inconsistencies in the three databases into four groups (see Table 3). The first group contains substances with inconsistent structures as well as substances with inconsistent or missing information on their stereochemistry. SMILES where either single atoms, parts of the molecule or the whole substance are incorrect can lead to significant errors in QSAR predictions. Young et al.4 showed for example that using an O atom instead of the correct S atom in CAS RN™ 34643-46-4, a substance where Young et al. found an inconsistent SMILES/CAS RN™ assignment, resulted in the octanol–water partition coefficient (KOW) to be off by 75%. Similar, the omission of a cyano group from CAS RN™ 15301-48-1 leads to a 48% difference in the mean absolute error for the KOW and an extra carbon in one of the rings of CAS RN™ 60207-93-4 to a difference of 91%. Furthermore, using incorrect SMILES in the training sets of QSARs can lead to less accurate predictions and thus weaker QSARs.4 It has also been shown that organic salts can have different toxicity values compared to their neutral forms.4 The omission or addition of counterions can therefore have an important effect on the evaluation of a substance. Incorrect CAS RNs™, in turn, can lead to errors in the purchasing and using of chemicals and standards. It is therefore of great importance that CAS RNs™ and structures are correct in the databases.The first group also includes substances with deviating or missing information on their stereochemistry. Stereoisomers are mainly important in biological systems and are therefore studied in great detail in clinical pharmacology and pharmacotherapeutics.28 However, this does not mean that they can be ignored for industrial chemicals as also these substances interact with biological systems. For example, bioaccumulation but also toxicity and degradation may differ between enantiomers.29–32 Enantiomers are therefore also not regarded equal under REACH.20 It is therefore important to also pay attention to the stereochemical information when substances are registered and when information is transferred between databases.
Substances that are registered as polymers but have the SMILES of the monomer are mainly an issue in the ECHA database. So far, polymers are exempted from the provisions on registration of Title II of REACH (Article 2(9)),33 however the monomer substances of the polymer have to be registered under REACH. For the substances identified in this work, it seems that the monomers were correctly registered, but under the name (and partly also the CAS RN™) of the polymer.
The second group of incorrect substances contains those substances where we were not able to identify the correct structure. Also, for this group, it would be important to recheck these substances and either correct the structures or the CAS RNs™. The chemical name should also be consistent with the other two identifiers.
The third group contains substances with missing cis/trans-isomer information for alkenes. Similar to the inconsistent or missing stereoisomers in group 1, the cis/trans isomerism might be important in biological systems. Missing information on cis/trans alkene isomers occurred in percentages from 1.0% (PubChem) to 2.1% (CompTox Chemicals Dashboard), showing that the problem is not huge, but still relevant.
The missing stereochemistry information in the fourth group had various reasons. In some cases, these are missing cis/trans-isomer information for N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N or N
N or N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds which are shown as E/Z but in most cases are non-specific and could be represented as crossed bonds. In other cases, chirality for specific stereocenters was not defined in the original SMILES/InChI strings and Open Babel had to omit the stereochemistry definition even if it correctly identified a stereocenter in the structure. This can be problematic as two chemicals with two different three-dimensional conformations would be represented with the same SMILES/InChI, creating confusion when the databased is queried on the basis of the molecular structure.
C bonds which are shown as E/Z but in most cases are non-specific and could be represented as crossed bonds. In other cases, chirality for specific stereocenters was not defined in the original SMILES/InChI strings and Open Babel had to omit the stereochemistry definition even if it correctly identified a stereocenter in the structure. This can be problematic as two chemicals with two different three-dimensional conformations would be represented with the same SMILES/InChI, creating confusion when the databased is queried on the basis of the molecular structure.
Obstacles with checking the databases
One of the biggest problems in cross-checking substances is that there is no unique chemical identifier that is present in all databases. Almost all substances in the ECHA database have an EC number, but the EC number is only partially available in PubChem and the CompTox Chemicals Dashboard. For the chemical name, on the other hand, it is not always possible to convert the chemical name to a structure if non-IUPAC names are used (Fig. 3). From the other identifiers (Table 2) it is only the CAS RN™ that is at least partially available in all three databases. However, the CAS Registry is a proprietary database, and access to most of the data was possible in the past only via a paid service such as SciFinder or STN®. A subset of 8000 substances has been accessible since 2009 via the CAS Common Chemistry database, which is an open web resource provided by CAS.34 This dataset was expanded in 2021 to 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 chemical substances35 and thus now offers free access to a relatively large number of substances in the CAS Registry®. The continued expansion and updating of the CAS Common Chemistry database are critical to the reliable use of CAS RN™ in regulatory systems in the future.
000 chemical substances35 and thus now offers free access to a relatively large number of substances in the CAS Registry®. The continued expansion and updating of the CAS Common Chemistry database are critical to the reliable use of CAS RN™ in regulatory systems in the future.
Another challenge for the identification of inconsistent chemical structures are tautomers. When comparing standardized InChI strings, tautomers must be manually identified, because the standardized InChI strings can differ for tautomers. InChI strings can be converted to detect the keto–enol tautomerism (e.g., in the tool that the IUPAC provides to generate InChI strings), however this may produce non-standard InChI (i.e., starting with ‘InChI=1/’ instead of ‘InChI=1S/’) and other types of tautomerism may not be addressed and standardized and still appear as mismatching structures.22 A related issue are nitro groups that are sometimes represented in the SMILES and the InChI string with a pentavalent nitrogen and sometimes with charge-separated groups. Moreover, many different formats of SMILES exist (e.g., kekulized, canonical, QSAR-ready) and it is not always clear which is the one reported in the various databases or if a standardization of the molecular structure has been done at all.22,36 It has been pointed out that this issue as well as others could be solved with standardization rules for chemistry databases.10
The biggest obstacle, however, is that one must first understand that databases contain errors and that the structures need to be checked before working with them. Williams and Ekins have pointed out in several articles that there is an urgent need for data curation in public databases.10,37 This is to improve the quality of the databases, but also because errors have been found to proliferate from databases such as PubChem to other databases on the internet when the content is downloaded and reused.37 Here we have made the first step for a part of the substances that are registered under REACH to uncover inconsistencies in the ECHA database as well as in the CompTox Chemicals Dashboard and PubChem. For the latter two databases, it is now up to the database operators to also fix these inconsistencies. For the ECHA database, this might be more complicated as only the registrants know what substance they intended to register and are responsible for most data. For this reason, the registrants would need to correct their dossiers first – either by adjusting the CAS RN™ or structure – before the data can be corrected in the ECHA database. To facilitate this process, we present the inconsistent structures in the ESI-2† to this article and hope that this will help to make the correction process faster. An exception are substances that were already listed in the European Inventory of Existing Commercial Chemical Substances (EINECS), the European List of Notified Chemical Substances (ELINCS) and the No-Longer Polymers (NLP) list.20 These are substances whose EC number begins with 2, 3, 4 or 5. The registrants cannot correct errors in EINECS/ELINCS/NLP (i.e., change the associated name and/or CAS RN) because these are closed inventories.38 To change them, a process would have to be opened at ECHA to issue a new list number with new name, and possibly a CAS RN™ associated.
Beside this, more work is still needed on the identification of inconsistent identifiers. We have only checked 37% of the substances registered under REACH and probably the easier ones. The same check would need to be performed for multi-constituent substances and UVCBs as well as (ECHA internally) for those substances where the SMILES is confidential and not available in the public domain. Also, the chemical names would need to be checked systematically. The CAS Common Chemistry database could be very useful here as it can now also be accessed via an application programming interface (API). However, the same systematic errors that occur in SciFindern also occur in the CAS Common Chemistry database and these would have to be managed appropriately in a fully automatic check. However, we think it is worth the effort because it will help to evaluate chemicals in a better and more trustworthy way.
Conclusions
Young et al.4 reported already in 2008 that in between 0.1% and 3.4% of the chemical structures in chemical databases are incorrect. Unfortunately, this still holds true and although there have been efforts to correct inconsistent entries, there is still a substantial number of errors in chemical databases, even in official ones, and users have to carefully check identifiers and structures before working with them. This also shows that more efforts should be dedicated to finding and correcting inconsistent chemical identifier in chemical databases. This could and should be done by the database operators, but should also be supported by scientists working with these databases. Addressing mistakes in publicly available databases is an iterative process that benefits from the inputs and feedbacks of users that find errors and inconsistencies in the data. There are quite some publications on data curation workflows, but it would also be important to report the identified inconsistent entries back to the database operators. An important conclusion is that finding a way to unequivocally represent the chemical structure is not an easy task and most likely errors and inconsistencies will always be found in chemical databases. Different types of SMILES and InChI notations exist because chemicals can exist in different states and forms. Database operators must provide information as clear and accurate as possible, but the final users have to make sure that the identifiers and the structure representation of the intended chemical are correct and appropriate for the context in which they are operating. Cross checking the information in multiple independent databases is a recommended good practice, but it is important to be aware that different databases may have different standardization rules and practices. For the future, we recommend (a) that database operators check their entries (CAS RN™ and SMILES) against other databases (such as the CAS Common Chemistry database) to identify inconsistent entries; (b) the standard use of name-to-structure conversion tools in databases to check the consistency of chemical names; and (c) that users of databases always double-check information that is important to them in a second database.Author contributions
Method development and data curation were mainly done by JG. MS and KM contributed to the method development and data analysis. All authors contributed to writing and reviewed the final manuscript. Funding was acquired by MS.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Antony Williams from the US Environmental Protection Agency for an initial conversation on how important it might be to check the chemical identities in the databases we are using. We also thank Alessandro Sangion (University of Toronto) for his helpful comments and initial support in the project. JG acknowledges funding from the Swiss Federal Office for the Environment.Notes and references
- G. Stieger, M. Scheringer, C. A. Ng and K. Hungerbühler, Assessing the persistence, bioaccumulation potential and toxicity of brominated flame retardants: Data availability and quality for 36 alternative brominated flame retardants, Chemosphere, 2014, 116, 118–123 CrossRef CAS PubMed.
- S. Strempel, M. Scheringer, C. A. Ng and K. Hungerbühler, Screening for PBT Chemicals among the “Existing” and “New” Chemicals of the EU, Environ. Sci. Technol., 2012, 46(11), 5680–5687 CrossRef CAS PubMed.
- ECHA, The Use of Alternatives to Testing on Animals for REACH - Fourth Report under Article 117(3) of the REACH Regulation, 2020 Search PubMed.
- D. Young, T. Martin, R. Venkatapathy and P. Harten, Are the chemical structures in your QSAR correct?, QSAR Comb. Sci., 2008, 27(11–12), 1337–1345 CrossRef CAS.
- D. Fourches, E. Muratov and A. Tropsha, Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research, J. Chem. Inf. Model., 2010, 50(7), 1189–1204 CrossRef CAS PubMed.
- D. Gadaleta, A. Lombardo, C. Toma and E. Benfenati, A new semi-automated workflow for chemical data retrieval and quality checking for modeling applications, J. Cheminf., 2018, 10(1), 1–13 Search PubMed.
- M. Waldman, R. Fraczkiewicz and R. D. Clark, Tales from the war on error: The art and science of curating QSAR data, J. Comput.-Aided Mol. Des., 2015, 29(9), 897–910 CrossRef CAS PubMed.
- K. Mansouri, C. M. Grulke, A. M. Richard, R. S. Judson and A. J. Williams, An automated curation procedure for addressing chemical errors and inconsistencies in public datasets used in QSAR modelling, SAR QSAR Environ. Res., 2016, 27(11), 939–965 CrossRef CAS PubMed.
- P. Gramatica, S. Cassani, P. P. Roy, S. Kovarich, C. W. Yap and E. Papa, QSAR modeling is not “Push a button and find a correlation”: A case study of toxicity of (Benzo-)triazoles on Algae, Mol. Inf., 2012, 31(11–12), 817–835 CrossRef CAS PubMed.
- A. J. Williams, S. Ekins and V. Tkachenko, Towards a gold standard: Regarding quality in public domain chemistry databases and approaches to improving the situation, Drug Discovery Today, 2012, 17(13–14), 685–701 CrossRef CAS PubMed.
- European Commission, Commission implementing regulation (EU) 2020/1435 of 9 October 2020 on the duties placed on registrants to update their registrations under Regulation (EC) No 1907/2006 of the European Parliament and of the Council concerning the Registration, Evaluation, Off. J. Eur. Union, 2020, L 331/24–L 331/29 Search PubMed.
- UBA, REACH Compliance: Data Availability of REACH Registrations – Part 1: Screening of Chemicals > 1000 tpa (43/2015), 2015 Search PubMed.
- UBA, REACH Compliance: Data Availability in REACH Registrations – Part 2: Evaluation of Data Waiving and Adaptations for Chemicals ≥ 1000 tpa (64/2018), 2018 Search PubMed.
- UBA, REACH Compliance : Data Availability in REACH Registrations – Part 3: Evaluation of 100 to 1000 tpa Substances (39/2020), 2020 Search PubMed.
- A. Scott, ECHA to quadruple number of compliance checks on REACH dossiers. C&EN, 2019, available from: https://cen.acs.org/policy/chemical-regulation/EU-quadruple-REACH-compliance-checks/97/i22 Search PubMed.
- ECHA, ECHA Database, 2022, available from: https://echa.europa.eu/information-on-chemicals Search PubMed.
- NIH, PubChem, 2022, available from: https://pubchem.ncbi.nlm.nih.gov/ Search PubMed.
- USEPA, CompTox Chemicals Dashboard, 2022, available from: https://comptox.epa.gov/dashboard/ Search PubMed.
- CAS, SciFinder-n, 2022, available from: https://scifinder-n.cas.org/ Search PubMed.
- ECHA, Guidance for Identification and Naming of Substances under REACH and CLP. Ver 2.1, 2017, pp. 1–127, available from: https://echa.europa.eu/documents/10162/23036412/substance_id_en.pdf/ee696bad-49f6-4fec-b8b7-2c3706113c7d Search PubMed.
- D. Weininger, A. Weininger and J. L. Weininger, SMILES. 2. Algorithm for Generation of Unique SMILES Notation, J. Chem. Inf. Comput. Sci., 1989, 29(2), 97–101 CrossRef CAS.
- N. M. O'Boyle, Towards a Universal SMILES representation - A standard method to generate canonical SMILES based on the InChI, J. Cheminf., 2012, 4(9), 1–14 Search PubMed.
- ECHA, IUCLID6, 2021, available from: https://iuclid6.echa.europa.eu/reach-study-results Search PubMed.
- C. M. Grulke, A. J. Williams, I. Thillanadarajah and A. M. Richard, EPA's DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research, Comput. Toxicol., 2019, 12, 100096 CrossRef PubMed.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, Open Babel: An Open chemical toolbox, J. Cheminf., 2011, 3(10), 1–14 Search PubMed.
- G. Hutchison, Open Babel Version 3.1.1, GitHub, 2020, available from: https://github.com/openbabel/openbabel Search PubMed.
- UCI, ChemDB Portal - Smi2Depict: Generate 2D Images from Molecule Files, 2021, available from: https://re.edugen.wiley.com/cgibin/Smi2DepictWeb.py Search PubMed.
- N. Chhabra, M. Aseri and D. Padmanabhan, A review of drug isomerism and its significance, Int. J. Appl. Basic Med. Res., 2013, 3(1), 16 CrossRef PubMed.
- T. Liu, J. Diao, S. Di and Z. Zhou, Stereoselective bioaccumulation and metabolite formation of triadimefon in Tubifex tubifex, Environ. Sci. Technol., 2014, 48(12), 6687–6693 CrossRef CAS PubMed.
- S. W. Smith, Chiral toxicology: It's the same thing only different, Toxicol. Sci., 2009, 110(1), 4–30 CrossRef CAS PubMed.
- C. Luo, B. Hu, S. Wang, Y. Wang, Z. Zhao and Y. Wang, et al., Distribution and chiral signatures of polychlorinated biphenyls (PCBs) in soils and vegetables around an E-waste recycling site, J. Agric. Food Chem., 2020, 68(39), 10542–10549 CrossRef CAS PubMed.
- Z. Frková, A. Johansen, L. W. de Jonge, P. Olsen, U. Gosewinkel and K. Bester, Degradation and enantiomeric fractionation of mecoprop in soil previously exposed to phenoxy acid herbicides – New insights for bioremediation, Sci. Total Environ., 2016, 569–570, 1457–1465 CrossRef PubMed.
- ECHA, Guidance for monomers and polymers, 2012, available from: https://op.europa.eu/s/wRAM Search PubMed.
- CAS, CAS Common Chemistry, 2022, available from: https://commonchemistry.cas.org/ Search PubMed.
- A. Jacobs, D. Williams, K. Hickey, N. Patrick, A. J. Williams and S. Chalk, et al., CAS Common Chemistry in 2021: Expanding Access to Trusted Chemical Information for the Scientific Community, J. Chem. Inf. Model., 2022, 62(11), 2737–2743 CrossRef CAS PubMed.
- K. Mansouri, A. Abdelaziz, A. Rybacka, A. Roncaglioni, A. Tropsha and A. Varnek, et al., CERAPP: Collaborative estrogen receptor activity prediction project, Environ. Health Perspect., 2016, 124(7), 1023–1033 CrossRef CAS PubMed.
- A. J. Williams and S. Ekins, A quality alert and call for improved curation of public chemistry databases, Drug Discovery Today, 2011, 16(17–18), 747–750 CrossRef CAS PubMed.
- G. Vollmer, K. Rasmussen, G. Christ, O. Nørager, J. B. Davis and A. Van Der Wielen, et al., Compilation of EINECS: Descriptions and definitions used for substances, impurities and mixtures, Toxicol. Environ. Chem., 1998, 65(1–4), 113–122 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: ESI-1 contains the investigated substances with their chemical identifiers; ESI-2 shows the inconsistent structures from the ECHA database in 2D. See DOI: https://doi.org/10.1039/d2va00225f |
| This journal is © The Royal Society of Chemistry 2023 |