Solid-state emitting twisted π-conjugate as AIE-active DSE-gen: in vitro anticancer properties against FaDu and 4T1 with biocompatibility and bioimaging†
Shouvik
Bhuin
 a,
Pravesh
Sharma
a,
Pravesh
Sharma
 b,
Purbali
Chakraborty
b,
Purbali
Chakraborty
 b,
Onkar Prakash
Kulkarni
b,
Onkar Prakash
Kulkarni
 b and
Manab
Chakravarty
b and
Manab
Chakravarty
 *a
*a
aDepartment of Chemistry, Birla Institute of Technology and Science-Pilani, Hyderabad Campus Jawahar Nagar, Shamirpet, Hyderabad, Telangana 500078, India. E-mail: manab@hyderabad.bits-pilani.ac.in
bDepartment of Pharmacy, Birla Institute of Technology and Science-Pilani, Hyderabad Campus Jawahar Nagar, Shamirpet, Hyderabad, Telangana 500078, India
First published on 25th November 2022
Abstract
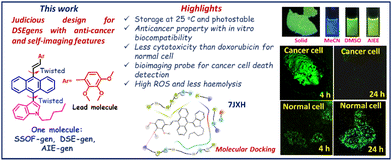
Dual-state emissive fluorogens (DSE-gens) are currently defining their importance as a transpiring tool in biological and biomedical applications. This work focuses on designing and synthesizing indole-anthracene-based solid-state emitting twisted π-conjugates using a metal-free protocol to achieve AIE-active DSE-gens, expanding their scope in biological applications. Special effort has been made to introduce proficient and photo/thermostable DSE-gens that inhibit cancer but not normal cells. Here, the lead DSE-gen initially detects cancer and normal cells by bioimaging; however, it could also confirm and distinguish cancer cells from normal cells by its abated fluorescence signal after killing cancer cells. In contrast, the fluorescence signals for a normal cell remain unscathed. Surprisingly, these molecules displayed decent anticancer properties against FaDu and 4T1 but not MCF-7 cell lines. From a series of newly designed indole-based molecules, we report one single 2,3,4-trimethoxybenzene-linked DSE-gen (the lead), exhibiting high ROS generation, less haemolysis, and less cytotoxicity than doxorubicin (DOX) for normal cells, crucial parameters for a biocompatible in vitro anticancer probe. Thus, we present a potentially applicable anticancer drug, offering a bioactive material with bioimaging efficacy and a way to detect dead cancer cells selectively. The primary mechanism behind the identified outcomes is deciphered with the support of experimental (steady-state and time-resolved fluorescence, biological assays, cellular uptake) and molecular docking studies.
1. Introduction
The dual-state emission (DSE) feature of aggregation-induced emission (AIE) active solid-state organic fluorogens (SSOF-gens) enables their effectiveness in biotechnology and biomedical applications due to the constant presence of emission in various states such as solutions, aggregates, and solids.1–4 Besides, simple administration into cells and flexibility in structural modification are the main benefits of organic fluorescent molecules for their versatile application in biomedicine avoiding the ACQ effects that limit their ultimate use in image-guided treatment.1,5 Hence, biologically non-toxic solid-state emitting AIE-active DSE-gens claim prominence in bioimaging, biosensing, and image-guided therapy systems (IGTs).1,6,7 Moreover, small but efficient π-conjugated organic fluorogens are primarily desired in comparison to various inorganic-organic hybrid materials due to their easy synthesis/modification, high thermo/photostability, and cost-effectiveness.8–13Recently, cancer cell detection by simple bio-imaging techniques using AIE-gens and DSE-gens has become a dependable, convenient, and cost-effective technique in cancer treatment.7b,14–16 Notably, molecular imaging techniques can detect cancer mass based on the photophysical properties enabling cell visualization in living organisms at the cellular and molecular level without any disorder. Some earlier identified molecular probes for in vitro and in vivo bio-imaging have inadequate effectivity due to their photobleaching tendency, acute toxicity, and low-fluorescence efficacy.17 Highly active commercial drugs are known to treat several types of cancers but with severe side effects.18–21 Nevertheless, many admirable anticancer drugs are available, but with no bioimaging ability,22,23 while numerous bioimaging agents are known without anticancer properties.24 Hence, the discovery of an anticancer agent with bioimaging ability is a looming challenge but a surging need of the time.25
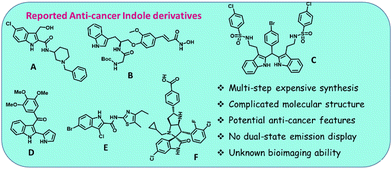
To address this necessity, we herein focus on designing an indole-based fluorophore by incorporating anthracene and indole as the primary scaffold. Anthracene and indole are individually realized as effective anticancer agents (vide infra). A few essential indole-based molecules A–F and their pros and cons are presented in Fig. 1.26–31
Our thoughtful design makes the typical ACQ-fluorophore anthracene emissive in the aggregate and solid states. An additional conjugation was needed to enhance the emission intensity. The newly designed indole-anthracene-based dyes could be easily accessed economically and mainly established as efficient AIE-active DSE-gens. Here, a few aggressive and obdurate cell lines were chosen: (i) FaDu (one of the most aggressive hypopharyngeal carcinoma cell lines),32–34 (ii) 4T1 (a highly destructive triple-negative mouse breast cancer cell line (TNBC), with a lack of treatment scope),35–39 (iii) MCF-7 (a breast cancer cell line, but not triple negative),40 and (iv) the non-cancerous human kidney embryonic cell line HEK-293 to investigate normal cell cytotoxicity and for comparison in bioimaging. Most approved marketed therapeutic drugs for breast cancer (sporadic for 4T1) are not emissive and have diverse side effects.37 Among various substituents, the highly electron-rich 2,3,4-trimethoxybenzene (TMB) linked molecule only exhibits promising activity against the FaDu and 4T1 cell lines but is surprisingly not responsive to the MCF-7 cell line. The non-toxic nature towards the normal cell line HEK-293 was also verified. The pyridyl-linked emitters are active against FaDu and 4T1 but are cytotoxic to normal cell lines. Apart from inhibiting, the TMB-linked emitter could image both FaDu and 4T1 cell lines with green emission before cell death but the fluorescence signal was diminished 24 h after cell death. In addition, commendable ROS generation and less haemolysis exhibited by the lead molecule (SB4) enhance the scope of utilization. Thus, this report (Fig. 2) reports the discovery of new AIE(E)-active DSE-gens as cancer-inhibiting molecules with bioimaging ability. Relevant steady-state and time-resolved photophysical studies, biological assays, and docking studies mechanistically support the observed outcomes.
2. Results and discussion
2.1. Molecular engineering
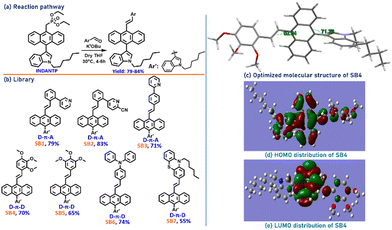
Considering the earlier identified design for organic solid-state emitters, specially DSEgens, we intended to obtain twisted organic molecules, having donor(D)–π–acceptor(A) and D–π–D combinations.41–43 Thus, the flat indole (an established motif with anticancer properties) with a moderately long alkyl chain was tagged onto the flat anthracene ring. This was made possible through an easy electrophilic aromatic substitution reaction44 using freshly prepared alkylated (n-hexyl) indole45 as a nucleophile. These two cores, indole and anthracene, prefer to occupy different planes. Such a twisted structure would prohibit π–π stacking in aggregates, resulting in relaxation through a radiative channel and strong emission.The designed D–π–A (SB1-SB3) and D–π–D combinations (SB4-SB7, Fig. 3) could be generated by Horner–Wadsworth–Emmons (HWE)-reactions of indole-anthracenyl phosphonates at room temperature through a metal/ligand-free protocol.46
The twisted structure and extended π-conjugation favorably compromise to emit with high efficiency in solution and solid states. Moreover, the indole and anthracene cores are individually potential anticancer agents.47–51 Molecules with extended conjugation with the core consisting of pyridine, TMB, phenothiazine, and triphenylamine were conveniently created because these are well-known motifs for bioimaging and anticancer applications.52–56 Unlike many previous reports, all synthesized molecules were intended to be non-charged, in order not to compromise with the fluorescence quantum yield,57,58 despite a possible reduction in cellular permeability.59 Nevertheless, the long alkyl chain and many hydrophobic units in our synthesized probes assist in achieving commendable cellular uptake capacities (vide infra). Notably, these probes are significantly stable and storable at room temperature, in contrast to many other complex organic molecules due to chiral/spiro centers or complex functionalization.60,61 Introduction of a conventional ‘spacer’ was also avoided to keep the fluorescence efficacy intact because it might disrupt the π-conjugation and introduce molecular motion, leading to quenched emission.62
Despite several attempts to generate a single crystal of these molecules, a suitable crystal for X-ray diffraction studies could not be generated. However, the viability of the molecular design and electronic effects on the lead molecule SB4 were validated by determining its optimized molecular structure using density functional theory (DFT) calculations at the B3LYP/6-31G(d,p) level. The calculated gas-phase HOMO, LUMO, and optimized molecular structure are presented in Fig. 3c–e. The molecular structure is vastly twisted with a dihedral angle of 71.3° (anthracene/indole) and ∼53° (anthracene-TMB). The HOMO is mainly distributed on the indole-anthracene part with a fine dispersion on the TMB ring, and the LUMO is mainly located at the core of anthracene. As the anthracene core contributes to both the HOMO and LUMO, generation of a charge transfer state is unexpected.
2.2. Photophysical studies in the solid state
We primarily focused on emission in solution, aggregates, and the solid-state for all these synthesized D–π–A (SB1-SB3) and D–π–D (SB4-SB7) molecules to make them suitable for biological applications. Notably, all these synthesized π-conjugates are SSOF-gens with green/greenish-yellow emission under 365 nm UV light except SB7, whose emission was orange (Fig. 4). The solid-state absorbance, emission, and other related parameters are tabulated elsewhere (see ESI† Fig. S1 and Table S1). In the solid-state UV-Vis studies, SB1 shows maximum λabs at 426 nm with a bent phenyl-pyridinyl core while SB2 has maximum λabs at 458 nm due to the suitably placed cyano group, improving π-conjugation in the ground state. In the emission profile, SB1 displays emission at 510 nm with higher brightness (absolute quantum yield φf = 7.04%), but SB2 is feebly emissive (φf = 1.29%) with λem at 505 nm. Another pyridinyl-conjugated dye SB3 performed as an intense solid-state emitter (φf = 26.64%) with λem at 517 nm. Out of the two TMB-linked isomeric D–π–D systems, SB4 with 2,3,4-TMB emits at λmax. = 534 nm (φf = 10.28%) with a 33 nm red-shift compared to its 3,4,5-TMB analog SB5 (λmax. = 501 nm, φf = 7.00%), possibly due to the presence of –OMe at both the ortho and para position. The bulky and conformationally twisted triphenylamine provided intense emission for SB6 (λmax. = 517 nm, φf = 20.11%). Still, the bowl-shaped phenothiazine moiety introduced a weak red emission in SB7 (λmax. = 589 nm, φf = 2.66%).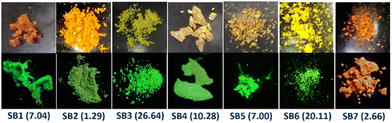 | ||
| Fig. 4 The solid-state images of the molecules taken under sunlight (top row) and a 365 nm UV lamp (bottom row). The φf% values of these molecules are stated in parentheses. | ||
2.3. Dual-state emission (DSE) feature studies
To examine the solution state emission property of the SSOF-gens, DMSO was chosen as a solvent due to its numerous applications in biology and medicine.63 However, MeCN was also used as another polar solvent. Moreover, both DMSO and MeCN are water-miscible and, thus, suitable for AIE studies. All the synthesized SSOF-gens (SB1-SB7) were identified as dual-state emitters because weak/decent emissions were observed even in such polar solutions (Fig. 5a, b and Fig. S2, Table S2, ESI†) and the solid state (Fig. 4 and Fig. S1, Table S1, ESI†). The DSE-gen behaviors were well-supported by TD-DFT calculations performed with the representative D–π–A molecule SB2 and D–π–D molecule SB4. The excited singlet state (S1) achieves more planarity than the ground singlet S0 state for SB2 and SB4 in both solvents, resulting in emissions in solutions (Fig. 5c, d and Fig. S3, S4, Table S3, ESI†). Compounds SB2 and SB4 both have a lower φf in DMSO than in MeCN, most possibly due to a relatively higher root mean square displacement (RMSD) (between S0 and S1) and weaker emission oscillator strength in DMSO.64 The HOMO–LUMO energy gap in such polar solvents would be smaller to afford a weakly emissive solution. However, this energy gap was slightly larger for SB4 than SB2 (Fig. S5 and Table S3, ESI†), ensuring the slightly better φf for SB4.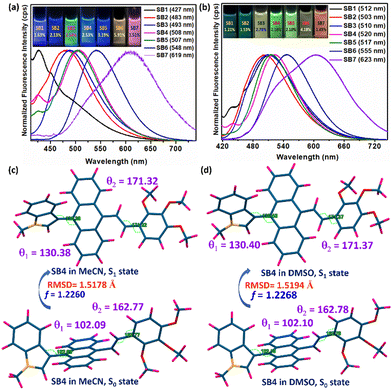 | ||
| Fig. 5 Solution-state fluorescence with images (under a 365 nm UV lamp) of SB1-SB7 in (a) MeCN and (b) DMSO with their emission λmax and respective relative φf (%), [error: (±) 5%] [reference: 10–5 M quinine sulfate solution]; (c) TD-DFT optimized structures of SB4 with selected torsion angles (θ1 and θ2). The DFT/TDDFT calculations were performed with N-methyl only as the long chain will not change its electronic properties (see ‘Computational details’ in the ESI†). | ||
2.4. Aggregation-induced emission (AIE) property of the DSE-gens
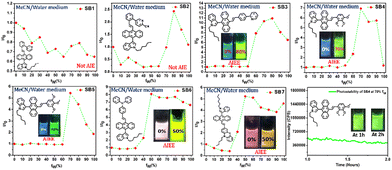
We aimed to verify the AIE activities of the synthesized DSE-gens and expected them to emit in the aggregated state as well. Notably, DSE-gens and SSOF-gens with AIE properties are more valuable in reporting biological information.4b,4g,65,66 Initially, the AIE properties of all the DSE-gens were tested in an acetonitrile/water mixture (fraction of water in acetonitrile fw: v/v%). All of the DSE-gens discussed here were not AIE-active. Especially, the admirable solid-state fluorophore SB1 undergoes ACQ (Fig. S6, ESI†) with a gradual increment of water fraction (fw), possibly due to the π-stacking of the phenyl/pyridine rings in the aggregated state.67,68 Introduction of the cyano group in SB2 might prevent π-stacking to a small extent, and the AIE-property of SB2 was somewhat better but not promising (Fig. S7, ESI†). The AIE feature of SB3 is more pleasing (Fig. S8, ESI†), with an 11 times fluorescence boost at fw = 80%. The aggregate formation was also verified from the DLS (dynamic light scattering) studies (Table 1). A notable AIE feature (7 times increment) was noticed at fw = 70% with SB4, containing three methoxy groups in an unsymmetrical fashion (Fig. 6a, b and Fig. S9a, ESI†). The indole with a long alkyl chain, twisted structure, and lack of symmetry may significantly restrict π-stacking and facilitate AIE. A simple viscofluorochromism study further supported this speculation. In MeOH, SB4 is weakly emissive, and upon consecutive addition of glycerol into MeOH, the overall viscosity of the medium is gradually increased, which would help restrict the molecular motion, resulting in enhanced emission. To our delight, the emission efficacy of SB4 was progressively enhanced with increasing viscosity up to a fG of 70% (Fig. 6c, d and Fig. S9b, ESI†), indicating viscofluorochromism in SB4.| SSOF-gen with AIE-property | λ max. abs. (nm) in agg. state | λ max. em. (nm) in agg. state | α-factor | Z-avg. (d nm) in agg. state | PDI | Life-time in agg. state (ps) |
|---|---|---|---|---|---|---|
| SB3 | 425 | 530 | 3.52 (8.94/2.54) | 194.5 | 0.296 | 64 |
| SB4 | 423 | 517 | 1.61 (4.07/2.53) | 186.1 | 0.178 | 66 |
| SB5 | 418 | 514 | 1.12 (3.57/3.19) | 190.4 | 0.197 | 48 |
| SB6 | 428 | 533 | 2.64 (10.52/3.91) | 219.9 | 0.295 | 51 |
| SB7 | 440 | 565 | 1.18 (2.25/1.91) | 190.7 | 0.185 | 49 |
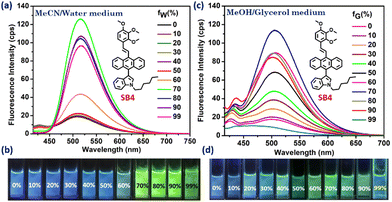 | ||
| Fig. 6 AIE-property (a and b) and viscofluorochromism (c and d) of SB4; fw is the fraction of water (v/v%) in MeCN and fG is the fraction of glycerol (v/v%) in MeOH. | ||
Another symmetrically different analog, SB5, was also identified as a good AIE-gen (Fig. S10, ESI†). The non-planar conformation of TPA in SB6 and bowl-shaped-phenothiazine in SB7 induce molecular twisting in the aggregate form, enabling destruction of the ACQ effect and the occurrence of AIE (Fig. S10, ESI†). An emission enhancement efficiency (I/I0) comparison (Fig. 7) for all the synthesized molecules was also performed to gain a clear view and the AIE-genic properties are tabulated for clarity (Table 1). The photostability of SB4 (Fig. 7) was investigated in its aggregated state (fw of 70%). SEM images of the AIE-gens in their respective aggregated states were captured with different morphologies (Fig. S11, ESI†).
We were interested in measuring the nanosecond scale excited state lifetime of one of the molecules, SB4 (as a lead; vide infra), in the solution, aggregate, and solid states. It was hard to measure the lifetime on a nanosecond scale due to its quick decay from the excited state. However, the decay profiles (ESI,† Fig. S12 and Table S4) were fitted better into bi or triexponential forms. The experiments were carefully repeated to confirm the lifetime in less than a nanosecond range. As observed in the emission intensity, the lifetime (ns) was enhanced in the aggregate/solid state, as previously reported for other relevant molecules.69 The radiative (kr) and nonradiative (knr) rate constants were measured (Table S4, ESI†) and the large nonradiative rate constant in solution explains the weak emission. The gradually improving kr in the aggregate and solid state explains the emission enhancement.
2.5. Biological applications
| GI50 (μM) for each cell-line | Doxorubicin | SB1 | SB2 | SB3 | SB4 | SB5 | SB6 | SB7 |
|---|---|---|---|---|---|---|---|---|
| FaDu | 0.24–0.3 | 39.27 | 37.53 | >100 | 24.94 | >100 | 27.21 | 69.27 |
| 4T1 | 0.85–2.2 | 49.75 | 32.99 | >100 | 20.76 | 82.11 | >100 | >100 |
| MCF-7 | 1.07–2.57 | 88.7 | >100 | >100 | >100 | >100 | >100 | >100 |
| HEK-293 | 3.3 | 35.63 | 33.72 | — | >100 | — | >100 | >100 |
From the cytotoxicity assay results using the cancer cell lines, only five active DSE-gens were selected to check their cytotoxicity against the non-cancerous human kidney embryonic cell line HEK-293. Notably, the DSE-gen SB4 shows the best anticancer activity against both FaDu and 4T1 and is less cytotoxic than DOX for HEK-293 normal cells. Surprisingly, it was inactive against the comparative cell line MCF-7. The table also indicates good responses from SB1 and SB2 against FaDu and 4T1, but these compounds are acutely toxic towards HEK-293 and, thus, inappropriate for further use. The TPA-analog SB6 also possesses favorable anti-cancer activity against FaDu but not 4T1.
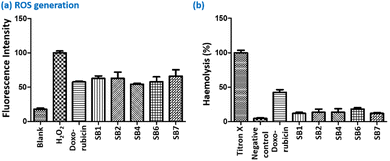
In addition, we treated red blood cells with these DSE-gens at their respective GI50 concentrations and then analyzed the treated cells by the haemolysis (rupture of red blood cells) assay. For the standard titron X, 100% haemolysis was considered, whereas DOX showed 40% haemolysis, a destructive impact. Interestingly, AIEgens SB1, SB2, SB4, SB6, and SB7 exhibited an inferior haemolyzing effect compared to DOX (Fig. 8b). It is noteworthy that SB1, SB2, SB4, SB6, and SB7 are less cytotoxic than DOX. Thus, the prime molecule SB4 offers a decent anticancer property against FaDu and 4T1 with excellent ROS generation and a weaker haemolyzing effect in comparison with DOX.
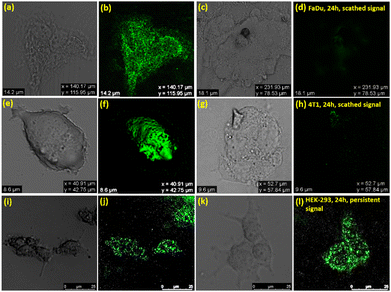
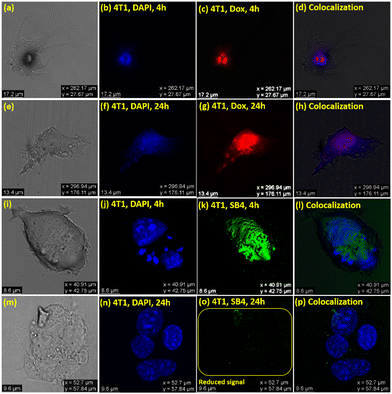
All the above results convincingly define SB4 as a bioimaging agent able to visualize the cancer cells FaDu, 4T1, and HEK-293 before and after inhibition. After four hours of SB4 treatment in both the FaDu and 4T1 cell lines at the GI50 concentration, the confocal experiment was performed, keeping the cells fixed, and the image was captured. Intense green fluorescence was noticed upon staining cells with SB4 at its GI50 concentration. As the cytotoxicity assay against cancer cell lines was conducted at a 24 h time-point, confocal images were again captured 24 h after treatment using the same cell lines. Regarding the anticancer properties, SB4 will inhibit the cells and will not provide any intense fluorescence signal to be observed by a confocal microscope at the 24 h time-point. But for HEK-293, a strong green fluorescence signal can be observed at 4 h and at 24 h because SB4 does not inhibit HEK-293 normal cells. Thus, SB4 can not only kill but also detect cancer cell death with its diminished fluorescence signal (Fig. 10).
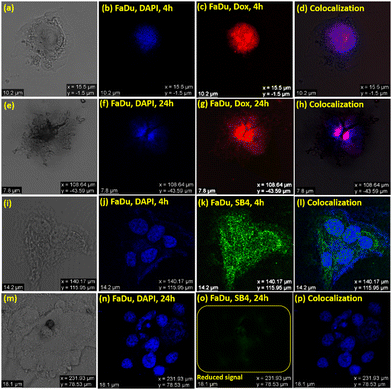
The utility of SB4 as a bioimaging probe for both cancer cell lines (Fadu and 4T1) was further investigated, and a comparative study was conducted with the standard drug DOX using DAPI’(4′,6-diamidino-2-phenylindole) as a typical stain.71 The experimental outcomes indicate that DAPI and DOX could stain FaDu and 4T1 at both the time points of 4 h and 24 h. But, as stated earlier, SB4 could stain cancer cells initially at the 4 h time-point, but a scathed fluorescence signal was noticed at the 24 h time-point for FaDu and 4T1 (Fig. 11 and 12). The used concentration of DAPI was 1 μg mL−1, and DOX was used at its GI50 concentration for bioimaging (0.3 μM for FaDu and 2.2 μM for 4T1). DAPI is a well-known bis-intercalator with DNA and a minor-groove binder.72 It can stain live and dead cells because the nucleus is not ruined instantly after the cell death. Interestingly, DOX also forms complexes by intercalation between DNA-base pairs,73 and it can stain live and dead cells like DAPI.74 These may be the reasons for recording fluorescence signals from both DAPI and DOX before and after the death of the cancer cells, respectively at 4 h and 24 h. However, the result is different for SB4 than the standard drug DOX. The colocalization studies showed that SB4 is not a nucleus-specific dye and stains the whole cell. Therefore, SB4- staining patterns remain intact for FaDu, 4T1, and HEK-293. As SB4 is nontoxic against HEK-293, the cell is not ruptured, and hence SB4 can possibly reside within the cell to provide a fluorescence signal even after 24 h and 4 h. Of note, aggregates are known to generate ROS.7c Likewise, the AIE-active DSE-gen SB4 generates ROS and exhibits more affinity towards the HER2 protein, ensuring cancer cell inhibition. Meanwhile, cell membranes might be ruptured in dead cells and permit the dye to leak out of the cell. Hence, the dye SB4 would tend to leak out of the cell, causing reduced fluorescence signals due to its lower concentration inside the cell. Nevertheless, the actual mechanism behind this observation needs further detailed investigation.
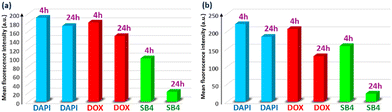
Further, different confocal images (Fig. S14, ESI†) utilizing DOX and SB4 were captured with colocalization comparison with DAPI to analyze and ultimately provide a quantitative contrast in a bar graph (Fig. 13) of mean fluorescence intensity using ‘ImageJ’ software.75 The Fig. 13 illustrates a comparative view to recognize the unique quenching of the mean fluorescent intensity of SB4 at a 24 h time-point for both Fadu and 4T1 cell lines.
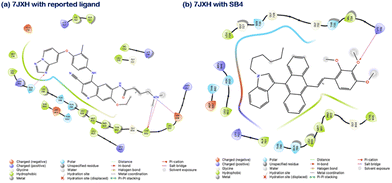
Here, the Protein Preparation Wizard module, LigPrep module and Glide module from Schrodinger suites 2022-1 were employed to prepare the crystal structures of protein obtained from the PDB (PDB IDs: 7JXH, 3RCD and 3PP0 for HER2 protein and 5X2C and 4I22 for EGFR protein) for ligand preparation and for performing molecular docking, respectively. The crystal structures were chosen based on the availability of inhibitor ligands at the tyrosine kinase binding site and their resolutions. For HER2/ErbB2 protein, the resolution of structures was: 3PP0 with 2.25 Å, 7JXH with 3.27 Å, and 3RCD with 3.21 Å. For EGFR protein, the resolution of structures was: 5X2C with 2.05 Å and 4I22 with 1.71 Å. Post protein preparation, the co-crystal ligands and inhibitor ligands were prepared using the LigPrep module. The crystal structures were analyzed to find the active site of the kinase domain where the inhibitor binds. For all the proteins, a 20 × 20 × 20 Å size grid was generated at the ligand binding site of the protein before docking, and the ligand was docked to the active site of all the proteins. Post-docking ligand protein interactions were analyzed. For the indole compound docked to the EGFR and HER2 proteins, the interacting amino acid residues are provided in Table 3. A comparison has been made with the interacting residues of the co-crystal ligands, suggesting the indole compound's affinity towards HER2 protein compared to EGFR protein (Fig. 14 and Fig. S15, ESI†). However, a further detailed investigation is highly needed for our future work.
| PDB ID | Crystal ligand interacting residues and Glide docking score | Indole compound interacting residues and Glide docking score |
|---|---|---|
| 3PP0 | Lys753, Leu796, Met801, Asp863, Phe864 | Arg811 |
| −14.784 | −5.931 | |
| 7JXH | Ser783, Cys805, Asp808 | Lys736 |
| −10.752 | −6.058 | |
| 3RCD | Lys753, Leu796, Met801, Asp863 | Phe1004 |
| −11.277 | −5.597 | |
| 5X2C | Met793 | NA |
| −9.854 | −6.502 | |
| 4I22 | Leu788, Met793 | NA |
| −10.065 | −7.519 |
3. Experimental
3.1. Materials and general conditions
All the syntheses were carried out in hot-air-oven-dried glassware. 1H-Indole, 1-bromohexane, 1-bromopentane, anthracene-9-carbaldehyde, diethyl phosphite, and piperazine were obtained from Alfa Aesar, Avra, and Merck and were utilized without further purification. Potassium hydroxide, potassium tert-butoxide, and methanesulfonic acid (MSA) were procured from Alfa Aesar and Sigma and were used as received. Dry DMSO, DCE, and THF were purchased from Finar. The rest of the chemicals were bought from Merck and Alfa Aesar and were used without any further purification. Spectroscopic grade solvents were used for UV-vis and fluorescence spectroscopy studies and procured from Sisco Research Laboratories (SRL). Column chromatography was performed by using silica gel (100–200 mesh), and the reactions were monitored via thin-layer chromatography on pre-coated silica gel 60 F254 plates (Merck & Co.) and observed using a 365 nm UV lamp. The NMR spectra were recorded at ambient temperature (ca. 20 °C) in CDCl3 solution. The signal of TMS was considered as the reference one with the deuterated chloroform solvent peak as an internal standard (7.26 ppm for 1H NMR, 77.0 ppm for 13C NMR). Chemical shifts are reported here in ppm denoting s (singlet), d (doublet), dd (doublet of doublet), t (triplet), q (quintet), and m (multiplet). All the experiments were performed at room temperature (298 ± 2K). IR spectra (4000–400 cm−1) were recorded using an FT/IR-4200 Jasco spectrometer (KBr pellets).3.2. Steady-state absorption and fluorescence measurements
Solid-state absorption spectra were recorded using a JASCO-500 spectrophotometer, and the solution-state absorption spectra were recorded using a UV-vis-NIR spectrophotometer (Hitachi F7000, Japan). Solution-state emission spectra were obtained using an FP-6300 spectrometer (JASCO), using a 10 mm path-length quartz cuvette, while the solid-state emission spectra were recorded with a fluorimeter (Fluorolog, HORIBA). The emission spectra were recorded at the corresponding absorption wavelengths.3.3. Absolute/relative quantum yield and lifetime decay measurement
For solid samples, the absolute quantum yield (φf) values were obtained by utilizing the calibrated integrating sphere method with a fluorimeter (Fluorolog, HORIBA). Absolute errors within ∼± 2% are included. For the relative quantum yield (relative φf) in the solution state, the error is ±5%.Time-resolved fluorescence measurements were performed using a time-correlated single-photon counting (TCSPC) unit (Horiba Deltaflex). The lasers used for all samples were 440 nm and 509 nm. All measurements were performed at room temperature. The decay fitting was completed, keeping the χ2 value close to unity.
3.4. Molecular docking
As mentioned earlier, Schrodinger suites 2022-1 were employed to prepare the crystal structures of protein obtained from the PDB, for ligand preparation and performing molecular docking, respectively.3.5. Cell-culture
The cell lines were purchased from NCCS, Pune, India and ATCC and maintained in DMEM supplemented with 10% FBS with 1% (v/v) penicillin-streptomycin. Cells were incubated at 37 °C in a humidified atmosphere comprising 05/95 CO2/air. All culture supplies were procured from Thermo Fisher Scientific.3.6. Cytotoxicity – MTT assay
All cells were seeded into 96 well plates at a density of 5 × 103 cells per well. After proper adhesion, cells were incubated with the compounds for 24 h. Cells treated with DMSO (less than 1%) were used as a control group. After 24 h, the supernatant from the plate was discarded and the cells were rinsed with 100 μL of sterile PBS. Freshly prepared 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (MTT) (100 μL at 5 mg mL−1 in sterile PBS) was added. The 96 well plate was incubated at 37 °C for 4 h. The supernatant was removed after 4 h, and 100 μL per well DMSO was added to the 96 well plate. A multi-plate reader was used to read absorbance at a wavelength of 570 nm. The anticancer activities of the synthesized indole-anthracene-based AIE-gens were evaluated against three cell lines: FaDu, which is a robust cell line, 4T1, a triple-negative breast cancer cell line, and MCF-7, which is a non-TNBC cell line, and one normal cell line: HEK 293, which is a human embryonic cell line using MTT assay. DOX is a clinical anticancer drug that was used as a positive control.3.7. Cellular uptake (FACS)
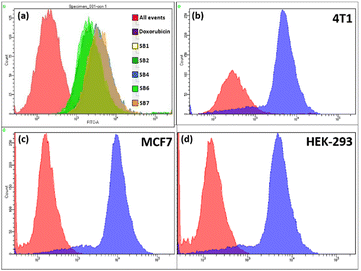
Cells were used for quantitative analysis of the uptake of SB4 using flow cytometry. Cell lines were seeded in a 12 well cell culture plate (1 × 106 cells per well) in DMEM (Dulbecco's Modified Eagle's Medium) containing 10% FBS with 1% antibiotics. After complete adhesion, cells were treated with different AIE-gens using their GI50 for 24 h. On the next day, cells were washed with sterile PBS followed by trypsinization of cells. Cells were collected in a 1.5 mL tube and centrifuged at 4 °C at 1000 rpm for 5 minutes. Cells were resuspended in 250 μL of sterile PBS (pH 7.4). A flow cytometer (BD FACS Aria™ III) was used for histogram generation of compound uptake.3.8. Reactive oxygen species (ROS) assay
FaDu cells were seeded in a 96 well plate using DMEM for 24 h. Later, cells were treated with the AIE-gens for 24 h. Cells were washed with PBS and incubated with a 10 μM 2′, 7′-Dichlorofluorescin diacetate (DCF-DA) solution for 30 min. The cells were washed 3 times with PBS and fluorescence was recorded using a Spectramax instrument at an excitation wavelength of 485 nm and an emission wavelength of 530 nm.3.9. Haemolysis assay
Rat blood was collected and centrifuged at 2500 rpm for 10 minutes. Red blood cells (RBCs) were carefully separated from plasma and collected RBCs were washed with PBS and centrifuged at 3000 rpm for 10 minutes. RBCs were resuspended into PBS to obtain a working 5% v/v solution. Triton-X 100 (1%) was used as a positive control and the PBS treated sample was used as a negative control. Compounds were incubated at their GI50 concentration for 1 h and at 37 °C. Later, the RBC solution was centrifuged at 8000 rpm for 1 minute and the supernatant was collected for absorbance at 576 nm using a Spectramax™ multiplate reader (Molecular Devices, US). The formula used to calculate haemolysis (%) = (As − A0)/(A100 − A0) × 100 where As is sample absorbance, A0 is the negative control absorbance treated with PBS and A100 is (Absorbance of titron X-100).3.10. Fixed cell confocal fluorescence imaging
Cell lines were seeded on a lysine coated sterile coverslip placed on a 12 well cell culture plate (1 × 105 cells per well) in DMEM containing 10% FBS with 1% antibiotics. Cells were allowed to adhere completely. After 24 h, cells were treated with different formulations using their GI50 for 4 h and 24 h. Later, cells were washed with sterile PBS and mounted on a glass slide. Cell imaging was done using a confocal microscope.3.11. Synthetic routes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The maroon color organic layer was separated and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (30% EtOAc into hexane (v/v)), which yielded (50%, 0.38 g) a gummy cyan fluorescent compound. IR (KBr, cm−1): 3057, 2930, 2854, 2355, 1586, 1462, 1390, 1254, 1161, 1057, 1021, 964, 740. M.p.: 79–81 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.39 (d, J = 8.92 Hz, 2H), 7.92 (d, J = 8.76 Hz, 2H), 7.54–7.45 (m, 3H), 7.29–7.21 (m, 4H), 7.05–6.98 (m, 2H), 4.33–4.25 (m, 4H, PCH2 doublet signals are merged), 4.01–3.85 (m, 4H), 2.00–1.93 (m, 2H), 1.42–1.26 (m, 6H), 1.12 (t, J = 7.04 Hz, 6H), 0.88 (t, J = 6.48 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 136.1, 131.7, 130.8, 130.5, 129.8, 128.9, 128.3, 125.7, 125.1, 124.7, 123.3, 121.8, 120.5, 119.5, 111.9, 109.7, 62.3 (d, J= 6.84 Hz), 46.6, 31.5, 30.3, 27.3 (d, J = 140 Hz), 26.8, 22.6, 16.3 (d, J= 5.98 Hz), 14.0. 31P NMR (162 MHz, CDCl3, 25 °C, δ): 26.08. Mass spectrometry: HRMS (ESI) m/z: calcd for C33H38NO3P 527.2589, found: 528.2445 [M + H]+.
H2O. The maroon color organic layer was separated and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (30% EtOAc into hexane (v/v)), which yielded (50%, 0.38 g) a gummy cyan fluorescent compound. IR (KBr, cm−1): 3057, 2930, 2854, 2355, 1586, 1462, 1390, 1254, 1161, 1057, 1021, 964, 740. M.p.: 79–81 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.39 (d, J = 8.92 Hz, 2H), 7.92 (d, J = 8.76 Hz, 2H), 7.54–7.45 (m, 3H), 7.29–7.21 (m, 4H), 7.05–6.98 (m, 2H), 4.33–4.25 (m, 4H, PCH2 doublet signals are merged), 4.01–3.85 (m, 4H), 2.00–1.93 (m, 2H), 1.42–1.26 (m, 6H), 1.12 (t, J = 7.04 Hz, 6H), 0.88 (t, J = 6.48 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 136.1, 131.7, 130.8, 130.5, 129.8, 128.9, 128.3, 125.7, 125.1, 124.7, 123.3, 121.8, 120.5, 119.5, 111.9, 109.7, 62.3 (d, J= 6.84 Hz), 46.6, 31.5, 30.3, 27.3 (d, J = 140 Hz), 26.8, 22.6, 16.3 (d, J= 5.98 Hz), 14.0. 31P NMR (162 MHz, CDCl3, 25 °C, δ): 26.08. Mass spectrometry: HRMS (ESI) m/z: calcd for C33H38NO3P 527.2589, found: 528.2445 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, (v/v)) was poured into it and stirred for 5 min again. Next, Pd(PPh3)4 (0.09 mmol, 0.08 equiv., 0.09 g), K2CO3 (1.63 mmol, 1.5 equiv., 0.22 g), and Tetrabutylammonium bromide (0.09 mmol, 0.08 equiv., 0.03 g) were added into the reaction solution, stirred for 15 min and the N2 environment was maintained. Then, the reaction mixture was refluxed at 90 °C for 18 h. The progress of the reaction was monitored by eluting TLC into hexane/EtOAc (20% EtOAc in hexane (v/v)) medium to observe an intense product spot formed at Rf = 0.32. The reaction mixture was worked up with 30 mL of an EtOAc and brine water mixture of 1
1, (v/v)) was poured into it and stirred for 5 min again. Next, Pd(PPh3)4 (0.09 mmol, 0.08 equiv., 0.09 g), K2CO3 (1.63 mmol, 1.5 equiv., 0.22 g), and Tetrabutylammonium bromide (0.09 mmol, 0.08 equiv., 0.03 g) were added into the reaction solution, stirred for 15 min and the N2 environment was maintained. Then, the reaction mixture was refluxed at 90 °C for 18 h. The progress of the reaction was monitored by eluting TLC into hexane/EtOAc (20% EtOAc in hexane (v/v)) medium to observe an intense product spot formed at Rf = 0.32. The reaction mixture was worked up with 30 mL of an EtOAc and brine water mixture of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (81%, 0.16 g) a solid whitish compound. IR (KBr, cm−1): 3055, 2898, 2768, 1677, 1599, 1469, 1252, 1191, 836. M.p.: 60–62 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 9.98 (s, 1H), 8.72–8.67 (m, 2H), 8.09–8.05 (t, J = 8.33 Hz, 1H), 7.75–7.56 (m, 3H), 7.46–7.29 (m, 2H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 191.3, 150.1, 149.3, 141.8, 137.2, 133.9, 133.8, 133.7, 131.0, 128.7, 128.5, 123.2. Mass spectrometry: LCMS (ESI) m/z: calcd for C12H9NO 184.0684, found: 184.1500 [M]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (81%, 0.16 g) a solid whitish compound. IR (KBr, cm−1): 3055, 2898, 2768, 1677, 1599, 1469, 1252, 1191, 836. M.p.: 60–62 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 9.98 (s, 1H), 8.72–8.67 (m, 2H), 8.09–8.05 (t, J = 8.33 Hz, 1H), 7.75–7.56 (m, 3H), 7.46–7.29 (m, 2H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 191.3, 150.1, 149.3, 141.8, 137.2, 133.9, 133.8, 133.7, 131.0, 128.7, 128.5, 123.2. Mass spectrometry: LCMS (ESI) m/z: calcd for C12H9NO 184.0684, found: 184.1500 [M]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, (v/v)) was poured into it and stirred for 5 min again. Next, Pd(PPh3)4 (0.09 mmol, 0.08 equiv., 0.1 g), K2CO3 (1.64 mmol, 1.5 equiv., 0.23 g), and Tetrabutylammonium bromide (0.09 mmol, 0.08 equiv., 0.03 g) were added into the reaction solution, stirred for 15 min and the N2 environment was maintained. Then, the reaction mixture was refluxed at 90 °C for 18 h. The progress of the reaction was monitored by eluting TLC into hexane/EtOAc (20% EtOAc into hexane (v/v)) medium to observe an intense product spot formed at Rf = 0.36. The reaction mixture was worked up with 30 mL of an EtOAc and brine water mixture of 1
1, (v/v)) was poured into it and stirred for 5 min again. Next, Pd(PPh3)4 (0.09 mmol, 0.08 equiv., 0.1 g), K2CO3 (1.64 mmol, 1.5 equiv., 0.23 g), and Tetrabutylammonium bromide (0.09 mmol, 0.08 equiv., 0.03 g) were added into the reaction solution, stirred for 15 min and the N2 environment was maintained. Then, the reaction mixture was refluxed at 90 °C for 18 h. The progress of the reaction was monitored by eluting TLC into hexane/EtOAc (20% EtOAc into hexane (v/v)) medium to observe an intense product spot formed at Rf = 0.36. The reaction mixture was worked up with 30 mL of an EtOAc and brine water mixture of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (71%, 0.16 g) a solid whitish compound. IR (KBr, cm−1): 3040, 2925, 2782, 2232, 1688, 1595, 1462, 1260, 1197, 992. M.p.: 120–122 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 9.98 (s, 1H), 8.75–8.74 (m, 1H), 8.08–8.06 (d, J = 7.68 Hz, 1H), 7.87–7.65 (m, 4H), 7.42–7.39 (d, J = 7.58 Hz). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 190.6, 151.3, 139.6, 137.8, 134.1, 133.9, 133.7, 133.2, 131.1, 130.7, 129.8, 127.8, 117.0. Mass spectrometry: LCMS (ESI) m/z: calcd for C13H8N2O 208.0637, found: 209.2000.[M]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (71%, 0.16 g) a solid whitish compound. IR (KBr, cm−1): 3040, 2925, 2782, 2232, 1688, 1595, 1462, 1260, 1197, 992. M.p.: 120–122 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 9.98 (s, 1H), 8.75–8.74 (m, 1H), 8.08–8.06 (d, J = 7.68 Hz, 1H), 7.87–7.65 (m, 4H), 7.42–7.39 (d, J = 7.58 Hz). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 190.6, 151.3, 139.6, 137.8, 134.1, 133.9, 133.7, 133.2, 131.1, 130.7, 129.8, 127.8, 117.0. Mass spectrometry: LCMS (ESI) m/z: calcd for C13H8N2O 208.0637, found: 209.2000.[M]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (79%, 0.16 g) a maroon sticky solid compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3452, 3178, 2966, 2852, 1728, 1597, 1453, 1429, 1380, 1328, 1190, 1119, 1062, 1015. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.73 (s, 1H), 8.53–8.51 (dd, J = 1.6 Hz, 1.6 Hz 1H), 8.35 (d, J = 8.76 Hz, 2H), 8.15–8.13 (d, J = 7.56 Hz, 1H), 7.99–7.89 (m, 3H), 7.79–7.76 (m, 1H), 7.61–7.39 (m, 6H), 7.30–7.25 (m, 5H), 7.11–6.89 (m, 3H), 4.29 (t, J = 7.04 Hz, 2H), 2.02–1.95 (quint, J =7.36 Hz, 2H), 1.39–1.28 (m, 6H), 0.89 (t, J = 7.02 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 150.2, 148.3, 137.4, 137.1, 136.5, 136.1, 136.0, 135.4, 132.3, 131.5, 130.5, 130.3, 129.7, 129.5, 128.8, 128.6, 128.1, 127.9, 127.7, 126.5, 125.8, 125.2, 124.8, 123.0, 121.8, 120.6, 119.5, 111.8, 109.6, 46.6, 31.4, 29.7, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C41H36N2 556.2878, found: 557.2743 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (79%, 0.16 g) a maroon sticky solid compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3452, 3178, 2966, 2852, 1728, 1597, 1453, 1429, 1380, 1328, 1190, 1119, 1062, 1015. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.73 (s, 1H), 8.53–8.51 (dd, J = 1.6 Hz, 1.6 Hz 1H), 8.35 (d, J = 8.76 Hz, 2H), 8.15–8.13 (d, J = 7.56 Hz, 1H), 7.99–7.89 (m, 3H), 7.79–7.76 (m, 1H), 7.61–7.39 (m, 6H), 7.30–7.25 (m, 5H), 7.11–6.89 (m, 3H), 4.29 (t, J = 7.04 Hz, 2H), 2.02–1.95 (quint, J =7.36 Hz, 2H), 1.39–1.28 (m, 6H), 0.89 (t, J = 7.02 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 150.2, 148.3, 137.4, 137.1, 136.5, 136.1, 136.0, 135.4, 132.3, 131.5, 130.5, 130.3, 129.7, 129.5, 128.8, 128.6, 128.1, 127.9, 127.7, 126.5, 125.8, 125.2, 124.8, 123.0, 121.8, 120.6, 119.5, 111.8, 109.6, 46.6, 31.4, 29.7, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C41H36N2 556.2878, found: 557.2743 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (83%, 0.18 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3436, 3123, 2924, 2848, 1690, 1569, 1464, 1419, 1364, 1021, 963. M.p.: 168–170 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.67 (s, 1H), 8.33 (d, J = 8.76 Hz), 8.21–8.15 (m, 2H), 8.0–7.97 (m, 3H), 7.82–7.60 (m, 2H), 7.53–7.40 (m, 5H), 7.30–7.27 (m, 2H), 7.24 (s, 1H), 7.10–6.86 (m, 3H), 5.51 (s, 1H), 4.30 (t, J = 7.04 Hz, 2H), 1.98 (quint, J = 7.36 Hz, 2H), 1.45–1.26 (m, 6H), 0.89 (t, J = 7.1 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 148.9, 148.1, 139.5, 138.3, 136.5, 136.2, 136.1, 135.1, 131.9, 131.5, 130.5, 130.4, 129.7, 129.6, 129.5, 129.3, 129.1, 128.9, 128.3, 128.2, 127.9, 126.7, 125.7, 125.3, 124.8, 121.9, 121.8, 120.6, 119.5, 109.6, 46.6, 31.4, 30.2, 26.8, 22.6, 13.9. Mass spectrometry: HRMS (ESI) m/z: calcd for C42H35N3 581.2831, found: 582.2720 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (12% EtOAc into hexane (v/v)), which yielded (83%, 0.18 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3436, 3123, 2924, 2848, 1690, 1569, 1464, 1419, 1364, 1021, 963. M.p.: 168–170 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.67 (s, 1H), 8.33 (d, J = 8.76 Hz), 8.21–8.15 (m, 2H), 8.0–7.97 (m, 3H), 7.82–7.60 (m, 2H), 7.53–7.40 (m, 5H), 7.30–7.27 (m, 2H), 7.24 (s, 1H), 7.10–6.86 (m, 3H), 5.51 (s, 1H), 4.30 (t, J = 7.04 Hz, 2H), 1.98 (quint, J = 7.36 Hz, 2H), 1.45–1.26 (m, 6H), 0.89 (t, J = 7.1 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 148.9, 148.1, 139.5, 138.3, 136.5, 136.2, 136.1, 135.1, 131.9, 131.5, 130.5, 130.4, 129.7, 129.6, 129.5, 129.3, 129.1, 128.9, 128.3, 128.2, 127.9, 126.7, 125.7, 125.3, 124.8, 121.9, 121.8, 120.6, 119.5, 109.6, 46.6, 31.4, 30.2, 26.8, 22.6, 13.9. Mass spectrometry: HRMS (ESI) m/z: calcd for C42H35N3 581.2831, found: 582.2720 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (71%, 0.15 g) a solid yellowish compound under room light and a bright but slightly yellowish-green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3445, 3048, 2944, 2849, 1580, 1467, 1431, 1345, 1322, 1233, 1147, 1017, 958. M.p.: 154–156 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.74 (s, 1H), 8.46 (d, J = 8.76 Hz, 2H), 8.13–7.93 (m, 5H), 7.82–7.75 (m, 4H), 7.53–7.43 (m, 3H), 7.32–7.23 (m, 5H), 7.14–7.01 (m, 3H), 4.30 (t, J = 7.04 Hz, 2H), 1.99 (quint, J = 7.32 Hz, 2H), 1.46–1.23 (m, 6H), 0.90 (t, J = 7.04 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 156.9, 149.8, 138.8, 138.1, 136.9, 136.1, 132.4, 131.6, 130.2, 129.8, 129.7, 128.9, 127.9, 127.3, 127.2, 127.0, 126.2, 126.1, 125.3, 124.9, 122.2, 121.8, 120.6, 120.5, 119.5, 111.9, 109.6, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C41H36N2 556.2878, found: 557.2674 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (71%, 0.15 g) a solid yellowish compound under room light and a bright but slightly yellowish-green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3445, 3048, 2944, 2849, 1580, 1467, 1431, 1345, 1322, 1233, 1147, 1017, 958. M.p.: 154–156 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.74 (s, 1H), 8.46 (d, J = 8.76 Hz, 2H), 8.13–7.93 (m, 5H), 7.82–7.75 (m, 4H), 7.53–7.43 (m, 3H), 7.32–7.23 (m, 5H), 7.14–7.01 (m, 3H), 4.30 (t, J = 7.04 Hz, 2H), 1.99 (quint, J = 7.32 Hz, 2H), 1.46–1.23 (m, 6H), 0.90 (t, J = 7.04 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 156.9, 149.8, 138.8, 138.1, 136.9, 136.1, 132.4, 131.6, 130.2, 129.8, 129.7, 128.9, 127.9, 127.3, 127.2, 127.0, 126.2, 126.1, 125.3, 124.9, 122.2, 121.8, 120.6, 120.5, 119.5, 111.9, 109.6, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C41H36N2 556.2878, found: 557.2674 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (75%, 0.16 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3447, 3401, 2959, 2934, 2854, 1596, 1465, 1378, 1324, 1287, 1091. M.p.: 125–127 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.47 (d, J = 8.72 Hz, 2H), 7.94–7.90 (m, 3H), 7.62–7.42 (m, 4H), 7.31–7.00 (m, 7H), 6.84 (d, J = 8.76 Hz, 1H), 4.31 (t, J = 7.06 Hz, 2H), 3.92 (s, 6H), 3.88 (s, 3H), 1.99 (quint, J = 7.36 Hz, 2H), 1.45–1.28 (m, 6H), 0.90 (t, J = 7.34 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 153.6, 151.9, 142.7, 136.1, 133.3, 131.9, 131.6, 129.8, 129.7, 128.9, 127.8, 126.3, 125.0, 124.8, 124.7, 121.7, 121.1, 120.6, 119.4, 112.0, 109.6, 107.9, 61.4, 61.0, 56.2, 46.6, 31.5, 29.7, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C39H39NO3 569.2930, found: 570.2792 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (75%, 0.16 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3447, 3401, 2959, 2934, 2854, 1596, 1465, 1378, 1324, 1287, 1091. M.p.: 125–127 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.47 (d, J = 8.72 Hz, 2H), 7.94–7.90 (m, 3H), 7.62–7.42 (m, 4H), 7.31–7.00 (m, 7H), 6.84 (d, J = 8.76 Hz, 1H), 4.31 (t, J = 7.06 Hz, 2H), 3.92 (s, 6H), 3.88 (s, 3H), 1.99 (quint, J = 7.36 Hz, 2H), 1.45–1.28 (m, 6H), 0.90 (t, J = 7.34 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 153.6, 151.9, 142.7, 136.1, 133.3, 131.9, 131.6, 129.8, 129.7, 128.9, 127.8, 126.3, 125.0, 124.8, 124.7, 121.7, 121.1, 120.6, 119.4, 112.0, 109.6, 107.9, 61.4, 61.0, 56.2, 46.6, 31.5, 29.7, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C39H39NO3 569.2930, found: 570.2792 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (77%, 0.17 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3417, 3064, 2955, 2930, 2359, 1580, 1504, 1461, 1417, 1319, 1231, 1128, 1013. M.p.: 108–110 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.43 (d, J = 8.76 Hz, 2H), 7.9–7.89 (m, 3H), 7.54–7.43 (m, 3H), 7.32–7.28 (m, 4H), 7.13–6.91 (m, 5H), 4.32 (t, J = 7.04 Hz, 2H), 3.99 (s, 6H), 3.94 (s, 3H), 2.00 (quint, J = 7.38 Hz, 2H), 1.45–1.24 (m, 6H), 0.91 (t, J = 7.1 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 153.6, 138.3, 137.3, 136.1, 134.3, 133.2, 132.3, 131.6, 130.1, 129.8, 129.7, 128.9, 127.9, 126.1, 125.2, 124.8, 121.8, 120.6, 119.5, 111.9, 109.6, 103.7, 61.1, 56.3, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C39H39NO3 569.2930, found: 570.2744 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (10% EtOAc into hexane (v/v)), which yielded (77%, 0.17 g) a solid yellowish compound under room light but a green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3417, 3064, 2955, 2930, 2359, 1580, 1504, 1461, 1417, 1319, 1231, 1128, 1013. M.p.: 108–110 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.43 (d, J = 8.76 Hz, 2H), 7.9–7.89 (m, 3H), 7.54–7.43 (m, 3H), 7.32–7.28 (m, 4H), 7.13–6.91 (m, 5H), 4.32 (t, J = 7.04 Hz, 2H), 3.99 (s, 6H), 3.94 (s, 3H), 2.00 (quint, J = 7.38 Hz, 2H), 1.45–1.24 (m, 6H), 0.91 (t, J = 7.1 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 153.6, 138.3, 137.3, 136.1, 134.3, 133.2, 132.3, 131.6, 130.1, 129.8, 129.7, 128.9, 127.9, 126.1, 125.2, 124.8, 121.8, 120.6, 119.5, 111.9, 109.6, 103.7, 61.1, 56.3, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C39H39NO3 569.2930, found: 570.2744 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (5% EtOAc into hexane (v/v)), which yielded (84%, 0.20 g) a solid greenish compound under room light and a bright green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3029, 2928, 2860, 2351, 2323, 1588, 1505, 1494, 1326, 1280, 1173. M.p.: 190–192 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.46 (d, J = 8.72 Hz, 2H), 7.93–7.87 (m, 3H), 7.60–7.42 (m, 5H), 7.32–7.28 (m, 8H), 7.19–7.16 (m, 6H), 7.13–6.93 (m, 5H), 4.31 (t, J = 7.04 Hz, 2H), 1.99 (quint, J = 7.34 Hz, 2H), 1.49–1.26 (m, 6H), 0.90 (t, J = 7.12 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 147.7, 147.6, 139.9, 136.8, 136.1, 132.8, 131.7, 131.6, 129.8, 129.7, 129.6, 129.3, 128.9, 127.9, 127.5, 126.2, 125.0, 124.8, 124.5, 123.8, 123.6, 123.1, 121.7, 120.6, 119.5, 113.1, 113.0, 112.9, 111.9, 109.6, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C48H42N2 646.3348, found: 647.3156 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (5% EtOAc into hexane (v/v)), which yielded (84%, 0.20 g) a solid greenish compound under room light and a bright green fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3029, 2928, 2860, 2351, 2323, 1588, 1505, 1494, 1326, 1280, 1173. M.p.: 190–192 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.46 (d, J = 8.72 Hz, 2H), 7.93–7.87 (m, 3H), 7.60–7.42 (m, 5H), 7.32–7.28 (m, 8H), 7.19–7.16 (m, 6H), 7.13–6.93 (m, 5H), 4.31 (t, J = 7.04 Hz, 2H), 1.99 (quint, J = 7.34 Hz, 2H), 1.49–1.26 (m, 6H), 0.90 (t, J = 7.12 Hz, 3H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 147.7, 147.6, 139.9, 136.8, 136.1, 132.8, 131.7, 131.6, 129.8, 129.7, 129.6, 129.3, 128.9, 127.9, 127.5, 126.2, 125.0, 124.8, 124.5, 123.8, 123.6, 123.1, 121.7, 120.6, 119.5, 113.1, 113.0, 112.9, 111.9, 109.6, 46.6, 31.5, 30.3, 26.8, 22.6, 14.0. Mass spectrometry: HRMS (ESI) m/z: calcd for C48H42N2 646.3348, found: 647.3156 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc
2 EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (5% EtOAc into hexane (v/v)), which yielded (63%, 0.19 g) a solid orange compound (it becomes solid after 2–3 days though initially may remain as gummy) under room light but an orange-red fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3441, 3061, 2953, 2927, 2856, 1573, 1463, 1334, 1247. M.p.: 119–121 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.41 (d, J = 8.76 Hz, 2H), 7.93–7.83 (m, 3H), 7.64–7.37 (m, 5H), 7.31–7.23 (m, 4H), 7.17–7.00 (m, 4H), 6.96–6.85 (m, 4H), 4.29 (t, J = 7 Hz, 2H), 3.87 (quint, J = 6.28 Hz, 2H), 1.99 (quint, J = 7.12 Hz, 2H), 1.86 (quint, J = 7.14 Hz, 2H), 1.45–1.26 (m, 10H), 0.94–0.86 (m, 6H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 145.1, 144.9, 136.1, 132.7, 132.0, 131.6, 129.9, 129.8, 129.7, 128.9, 127.9, 127.5, 127.3, 126.2, 125.9, 125.4, 125.1, 124.8, 124.4, 123.6, 122.5, 121.8, 120.6, 119.5, 115.9, 115.5, 115.4, 114.8, 111.9, 109.6, 47.9, 47.6, 46.6, 31.5, 30.3, 29.2, 26.8, 26.7, 22.7, 22.5, 14.1. Mass spectrometry: HRMS (ESI) m/z: calcd for C47H46N2S 670.3382, found: 671.3252 [M + H]+.
H2O. The organic layer was separated, passed through a Na2SO4 layer, and made into a slurry with silica gel (60–120 mesh) to perform column chromatography (5% EtOAc into hexane (v/v)), which yielded (63%, 0.19 g) a solid orange compound (it becomes solid after 2–3 days though initially may remain as gummy) under room light but an orange-red fluorescent compound under a UV-365 nm bulb. IR (KBr, cm−1): 3441, 3061, 2953, 2927, 2856, 1573, 1463, 1334, 1247. M.p.: 119–121 °C. NMR spectroscopy: 1H NMR (400 MHz, CDCl3, 25 °C, δ): 8.41 (d, J = 8.76 Hz, 2H), 7.93–7.83 (m, 3H), 7.64–7.37 (m, 5H), 7.31–7.23 (m, 4H), 7.17–7.00 (m, 4H), 6.96–6.85 (m, 4H), 4.29 (t, J = 7 Hz, 2H), 3.87 (quint, J = 6.28 Hz, 2H), 1.99 (quint, J = 7.12 Hz, 2H), 1.86 (quint, J = 7.14 Hz, 2H), 1.45–1.26 (m, 10H), 0.94–0.86 (m, 6H). 13C NMR (100 MHz, CDCl3, 25 °C, δ): 145.1, 144.9, 136.1, 132.7, 132.0, 131.6, 129.9, 129.8, 129.7, 128.9, 127.9, 127.5, 127.3, 126.2, 125.9, 125.4, 125.1, 124.8, 124.4, 123.6, 122.5, 121.8, 120.6, 119.5, 115.9, 115.5, 115.4, 114.8, 111.9, 109.6, 47.9, 47.6, 46.6, 31.5, 30.3, 29.2, 26.8, 26.7, 22.7, 22.5, 14.1. Mass spectrometry: HRMS (ESI) m/z: calcd for C47H46N2S 670.3382, found: 671.3252 [M + H]+.
Conclusions
In conclusion, this work introduces a new class of N-alkyl indole-linked anthracenyl twisted π-conjugates as efficient DSE-gens. All these emitters were tested in anticancer studies, especially with the aggressive and robust FaDu and TNBC 4T1 cell lines. Among all these molecules, the 2,3,4-TMB substituted probe SB4 exhibited potential and was a convincing AIE-active DSE-gen that not only inhibited both FaDu and 4T1 (not MCF-7 and HEK-293) at 24 h but could also detect cell death by its diminished fluorescence signal. The HEK-293 normal cells and cancer cells could be easily distinguished by comparing the intensity of intact fluorescence signals. The noncytotoxic nature, high ROS generation, and less haemolysis make SB4 a commendable biocompatible drug with imaging capability, even in comparison to DOX. The intrinsically anticancer indole and anthracene play a crucial role in providing anticancer features with their affinity to HER2 protein, as suggested by the primary docking studies. The hydrophobicity of the lead probe may have assisted in penetrating the cell membrane (validated by FACS) to form a closer interaction and provide an image of the cells with DSE properties. Hence, a new class of easily accessible indole-anthracenyl π-conjugate compounds offers a promising solution to kill cancer cells selectively and detect the damaged cells through diminished fluorescence signals. This research showcases the prospects of such newly designed DSE-gens in challenging cancer research. Finding a detailed mechanistic path to decipher the selectivity of 4T1 and modification of the probe to improve the sensitivity and specificity are currently underway in our laboratory.Author contributions
SB: conceptualization of the whole work, planning, molecular structure designing, synthesis and characterization, steady-state and time-resolved photophysical studies, analysis of the TD-DFT optimized structures, assistance in confocal microscopy, confocal image quantification, collaborative discussions, writing the original manuscript; PS: cell culture, cytotoxicity, ROS, haemolysis, FACS, confocal microscopy and suggestions in manuscript editing; PC: molecular docking and analysis, manuscript editing and assistance in confocal image analysis; OK: time to time discussion in the biological part and manuscript revision; MC: overall supervision, scientific guidance, analysis through theoretical and experimental support, a detailed plan on manuscript presentation, revision, continuous discussions and monitoring of the work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank LSRB-389/FSH & ABB/2021, India, CSIR [02(289)/17], India, for their financial support. The DST-FIST facility is also partially accredited. The BITS-Pilani Hyderabad campus NMR facility is highly acknowledged. MC and SB earnestly thank P. Sivasakthi, and Dr Pralok Samanta for theoretical studies. SB thanks Mr. Dwaipayan Bhattacharya for his cordial assistance in HEK-293 cell preparation for imaging. SB also thanks BITS-Pilani Hyderabad for the ‘Institute Research Fellowship’ and the ‘Central Analytical Laboratory (CAL) facility; special thanks to technician Mr. G. Mallesh for tireless assistance in confocal imaging.Notes and references
- K. Li, T. B. Ren, S. Huan, L. Yuan and X. B. Zhang, Progress and Perspective of Solid-State Organic Fluorophores for Biomedical Applications, J. Am. Chem. Soc., 2021, 143, 21143–21160 CrossRef CAS PubMed.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Aggregation-Induced Emission: Together We Shine, United We Soar!, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- Y. Hong, J. W. Y. Lam and B. Z. Tang, Aggregation-induced emission, Chem. Soc. Rev., 2011, 40, 5361–5388 RSC.
- (a) J. L. B. Vazquez, Y. A. A. Sanchez, L. A. R. Cortes and B. R. Molina, Dual-State Emission (DSE) in Organic Fluorophores: Design and Applications, Chem. Mater., 2021, 33, 7160–7184 CrossRef; (b) F. Yu, H. Zhao, Y. Li, G. Xia and H. Wang, D–A-Type fluorophores with efficient dual-state emission for imaging at ultralow concentration, Mater. Chem. Front., 2022, 6, 155 RSC; (c) S. K. Behera, S. Y. Park and J. Gierschner, Dual Emission: Classes, Mechanisms, and Conditions, Angew. Chem., Int. Ed., 2021, 60, 22624–22638 CrossRef CAS PubMed; (d) C. Han, S. B. Sun, X. Ji and J. W. Wang, A novel fluorescent probe with ACQ-AIE conversion by alkyl chain engineering for simultaneous visualization of lipid droplets and lysosomes, Spectrochim. Acta-A: Mol. Biomol. Spectrosc., 2023, 285, 121884 CrossRef CAS PubMed; (e) H. V. Humeniuk, A. Rosspeintner, G. Licari, V. Kilin, L. Bonacina, E. Vauthey, N. Sakai and S. Matile, White-Fluorescent Dual-Emission Mechanosensitive Membrane Probes that Function by Bending Rather than Twisting, Angew. Chem., Int. Ed., 2018, 57, 10559–10563 CrossRef CAS PubMed; (f) J. Lao, J. Li and H. Zou, Palladium-Catalyzed Cascade Reaction in Water to Imidazo[1,2-a]pyridazines as Switchable DSEgens, AIEgens, and ACQgens, Chem. Eur. J, 2022, e202202179 CAS; (g) M. Tan, Y. Li, W. Guo, Y. Chen, M. Wang, Y. Wang, B. Chi, H. Wang, G. Xia and H. Wang, Accessing conjugated and twisted structures for efficient dual-state emission fluorophore and its sensitive lysosomal imaging, Dyes Pigm., 2022, 201, 110243 CrossRef CAS.
- X. Ren, S. Zhang, L. Liu, B. Xu and W. Tian, Recent advances in assembled AIEgens for image-guided anticancer therapy, Nanotechnology, 2021, 32, 502008 CrossRef CAS PubMed.
-
(a) W. Yu, H. Zhang, P. A. Yin, F. Zhou, Z. Wang, W. Wu, Q. Peng, H. Jiang and B. Z. Tang, Restriction of Conformation Transformation in Excited State: An Aggregation-Induced Emission Building Block Based on Stable Exocyclic C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N Group, iScience, 2020, 23, 101587 CrossRef CAS PubMed;
(b) Y. Zhang, Y. Qu, J. Wu, Y. Rui, Y. Gao and Y. Wu, Naphthalimide end-capping molecular rotors with different donor cores: Tuning emission in wide gamut and cell imaging, Dyes Pigm., 2020, 179, 108431 CrossRef CAS.
N Group, iScience, 2020, 23, 101587 CrossRef CAS PubMed;
(b) Y. Zhang, Y. Qu, J. Wu, Y. Rui, Y. Gao and Y. Wu, Naphthalimide end-capping molecular rotors with different donor cores: Tuning emission in wide gamut and cell imaging, Dyes Pigm., 2020, 179, 108431 CrossRef CAS. - (a) F. Yin, B. Gu, J. Li, N. Panwar, Y. Liu, Z. Li, K. T. Yong and B. Z. Tang, In vitro anticancer activity of AIEgens, Biomater. Sci., 2019, 7, 3855 RSC; (b) W. Dai, P. Liu, S. Guo, Z. Liu, M. Wang, J. Shi, B. Tong, T. Liu, Z. Cai and Y. Dong, Triphenylquinoline (TPQ)-Based Dual-State Emissive Probe for Cell Imaging in Multicellular Tumor Spheroids, ACS Appl. Bio Mater., 2019, 2, 3686–3692 CrossRef CAS PubMed; (c) Z. Huang, F. Tang, F. He, L. Kong, J. Huang, J. Yang and A. Ding, Pyrene and triphenylamine substituted cyanostyrene and cyanostilbene derivatives with dual-state emission for high-contrast mechanofluorochromism and cell imaging, Org. Chem. Front., 2022, 9, 5118 RSC; (d) H. Lv, L. Wei, S. Guo, X. Zhang, F. Chen, X. Qin, C. Wei, B. Jiang and Y. Gong, Ionic Rigid Organic Dual-State Emission Compound With Rod-Shaped and Conjugated Structure for Sensitive Al3+ Detection, Front. Chem., 2022, 10, 807088 CrossRef CAS PubMed; (e) Y. Liu, L. Teng, C. Xu, T. B. Ren, S. Xu, X. Lou, L. Yuan and X. B. Zhang, An Integration Strategy to Develop Dual-State Luminophores with Tunable Spectra, Large Stokes Shift, and Activable Fluorescence for High-Contrast Imaging, CCS Chem., 2022, 4, 2153–2164 CrossRef CAS.
- W. Liu, Y. M. Wang, Y. H. Li, S. J. Cai, X. B. Yin, X. W. He and Y. K. Zhang, Fluorescent Imaging-Guided Chemotherapy-and-Photodynamic Dual Therapy with Nanoscale Porphyrin Metal–Organic Framework, Small, 2017, 13(17), 1603459 CrossRef PubMed.
- M. Dulski, K. Malarz, M. Kuczak, K. Dudek, K. Matus, S. Sulowicz, A. M. Wilczkiewicz and A. Nowak, An Organic-Inorganic Hybrid Nanocomposite as a Potential New Biological Agent, Nanomaterials, 2020, 10, 2551 CrossRef CAS PubMed.
- E. I. Ayisigi and O. Y. Celiktas, Silica-based organic-inorganic hybrid nanoparticles and nanoconjugates for improved anticancer drug delivery, Eng. Life Sci., 2018, 18, 882–892 CrossRef PubMed.
- F. Duan, M. Hu, C. Guo, Y. Song, M. Wang, L. He, Z. Zhang, R. Pettinari and L. Zhou, Chromium-based metal–organic framework embedded with cobalt phthalocyanine for the sensitivity impedimetric cytosensing of colorectal cancer (CT26) cells and cell imaging, Chem. Eng. J., 2020, 398, 125452 CrossRef CAS.
- J. W. M. Osterrieth and D. F. Jimenez, Metal–Organic Framework Composites for Theragnostics and Drug Delivery Applications, Biotechnol. J., 2020, 16(2), 2000005 CrossRef PubMed.
- W. A. Shakarchi, A. Alsuraifi, A. Curtis and C. Hoskins, Dual Acting Polymeric Nano-Aggregates for Liver Cancer Therapy, Pharmaceutics, 2018, 10, 63 CrossRef PubMed.
- D. M. J. Seema, B. Saifullah, M. Selvanayagam, S. Gothai, M. Z. Hussein, S. K. Subbiah, N. M. Esa and P. Aruselvan, Designing of the Anticancer Nanocomposite with Sustained Release Properties by Using Graphene Oxide Nanocarrier with Phenethyl Isothiocyanate as Anticancer Agent, Pharmaceutics, 2018, 10, 109 CrossRef CAS PubMed.
- R. S. Bhosale and V. Sing, Advances in Aggregation Induced Emission Materials in Biosensing and Imaging for Biomedical Applications- Part B, Progress in Molecular Biology and Translational Science, Book Series, 2021, vol. 185, pp. 1–234 Search PubMed.
- (a) C. Zhang, S. Jin, S. Li, X. Xue, J. Liu, Y. Huang, Y. Jiang, W. Q. Chen, G. Zou and X. J. Liang, Imaging Intracellular Anticancer Drug Delivery by Self-Assembly Micelles with Aggregation-Induced Emission (AIE Micelles), ACS Appl. Mater. Interfaces, 2014, 6, 5212–5220 CrossRef CAS PubMed; (b) B. Muthuraj, S. Mukherjee, C. R. Patra and P. K. Iyer, Amplified Fluorescence from Polyfluorene Nanoparticles with Dual State Emission and Aggregation Caused Red Shifted Emission for Live Cell Imaging and Cancer Theranostics, ACS Appl. Mater. Interfaces, 2016, 8(47), 32220–32229 CrossRef CAS PubMed.
- N. H. Ly and S. W. Joo, Recent advances in cancer bioimaging using a rationally designed Raman reporter in combination with plasmonic gold, J. Mater. Chem. B, 2020, 8, 186 RSC.
- S. Thangudu, P. Kalluru and R. Vankayala, Preparation, Cytotoxicity, and In Vitro Bioimaging of Water Soluble and Highly Fluorescent Palladium Nanoclusters, Bioengineering, 2020, 7, 20 CrossRef CAS PubMed.
- J. Dhuguru and R. Skouta, Role of Indole Scaffolds as Pharmacophores in the Development of Anti-Lung Cancer Agents, Molecules, 2020, 25, 1615 CrossRef CAS PubMed.
- R. Oun, Y. Moussa and N. Wheate, The side effects of platinum-based chemotherapy drugs: a review for chemists, Dalton Trans., 2018, 47, 6645–6653 RSC.
- C. Carvalho, R. X. Santos, S. Cardosa, S. Correia, P. J. Oliveira, M. S. Santos and P. I. Moreira, Doxorubicin: The Good, the Bad and the Ugly Effect, Curr. Med. Chem., 2009, 16, 3267–3285 CrossRef CAS PubMed.
- Y. Qin, T. Guo, Z. Wang and Y. Zhao, The role of iron in doxorubicin-induced cardiotoxicity: recent advances and implication for drug delivery, J. Mater. Chem. B, 2021, 9, 4793–4803 RSC.
- H. Cong, X. Zhao, B. T. Castle, E. J. Pomeroy, B. Zhou, J. Lee, Y. Wang, T. Bian, Z. Miao, W. Zhang, Y. Y. Sham, D. J. Odde, C. E. Eckfeldt, C. Xing and C. Zhuang, An Indole-Chalcone Inhibits Multidrug-Resistant Cancer Cell Growth by Targeting Microtubules, Mol. Pharmaceutics, 2018, 15, 3892–3900 CrossRef CAS PubMed.
- (a) H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, New Strategies for Fluorescent Probe Design in Medical Diagnostic Imaging, Chem. Rev., 2010, 110(5), 2620–2640 CrossRef CAS PubMed; (b) H. C. Hao, G. Zhang, Y. N. Wang, R. Sun, Y. J. Xu and J. F. Ge, Distinguishing cancer cells from normal cells with an organelle-targeted fluorescent marker, J. Mater. Chem. B, 2022, 10, 5796 RSC.
- S. Liu, W. Song, X. Gao, Y. Su, E. Gao and Q. Gao, Discovery of Nonpeptide, Reversible HER1/HER2 Dual-Targeting Small-Molecule Inhibitors as Near-Infrared Fluorescent Probes for Efficient Tumor Detection, Diagnostic Imaging, and Drug Screening, Anal. Chem., 2019, 91, 1507–1515 CrossRef CAS PubMed.
- X. Ma, C. Zhang, L. Feng, S. H. Liu, Y. Tan and J. Yin, Construction and bioimaging application of novel indole heptamethine cyanines containing functionalized tetrahydropyridine rings, J. Mater. Chem. B, 2020, 8, 9906–9912 RSC.
- F. A. M. Mohamed, H. A. M. Gomma, O. M. Hendawy, A. T. Ali, H. S. Farghaly, A. M. Gouda, A. H. Abdelazeem, M. H. Abdelrahman, L. Trembleau and B. G. M. Youssif, Design, synthesis, and biological evaluation of novel EGFR inhibitors containing 5-chloro-3-hydroxymethyl-indole-2- carboxamide scaffold with apoptotic antiproliferative activity, Bioorg. Chem., 2021, 112, 104960 CrossRef CAS PubMed.
- Y. Zhang, P. Yang, C. J. Chou, C. Liu, X. Wang and W. Xu, Development of N-Hydroxycinnamamide-Based Histone Deacetylase Inhibitors with an Indole-Containing Cap Group, ACS Med. Chem. Lett., 2013, 4, 235–238 CrossRef CAS PubMed.
- A. O’Dea, C. Sondergard, P. Sweeney and C. K. Arnatt, A Series of Indole-Thiazole Derivatives Act as GPER Agonists and Inhibit Breast Cancer Cell Growth, ACS Med. Chem. Lett., 2018, 9, 901–906 CrossRef PubMed.
- R. Pingaew, P. Mandi, V. Prachayasittikul, A. Thongnum, S. Prachayasittikul, S. Ruchirawat and V. Prachayasittikul, Investigations on Anticancer and Antimalarial Activities of Indole-Sulfonamide Derivatives and In Silico Studies, ACS Omega, 2021, 6, 31854–31868 CrossRef CAS PubMed.
- G. L. Regina, R. Bai, W. M. Rensen, E. D. Cesara, A. Coluccia, F. Piscitelli, V. Famiglini, A. Reggio, M. Nalli, S. Pelliccia, E. D. Pozzo, B. Costa, I. Granata, A. Porta, B. Maresca, A. Soriani, M. L. Iannitto, A. Santoni, J. Li, M. M. Cona, F. Chen, Y. Ni, A. Brancale, G. Dondio, S. Vultaggio, M. Varasi, C. Mercurio, C. Martini, E. Hamel, P. Lavia, E. Novellino and R. Silvestri, Toward Highly Potent Cancer Agents by Modulating the C-2 Group of the Arylthioindole Class of Tubulin Polymerization Inhibiltors, J. Med. Chem., 2013, 56, 123–149 CrossRef PubMed.
- S. R. S. Rangan, A New Human Cell Line (FaDu) From A Hypopharyngeal Carcinoma, Cancer, 1972, 29, 117–121 CrossRef CAS PubMed.
- J. Ma, S. Lu, L. Yu, J. Tian, J. Li, H. Wang and W. Xu, FaDu cell characteristics induced by multidrug resistance, Oncol. Rep., 2011, 26, 1189–1195 CAS.
- A. S. M. Noman, R. R. Parag, M. I. Rashid, S. Islam, M. Z. Rahman, A. A. Chowdhury, A. Sultana, C. Jerin, A. Siddiqua, L. Rahman, J. Nayeem, S. Akther, S. Baidya, R. K. Shil, M. Rahman, A. Shirin, R. Mahmud, S. M. I. Hossain, S. A. Sumi, A. Chowdhury, S. B. Basher, A. Hasan, S. Bithy, J. Aklima, N. Chowdhury, M. N. Hasan, T. Banu, S. Chowdhury, M. M. Hossain, H. Yeger, W. A. Farhat and S. S. Islam, Chemotherapeutic resistance of head and neck squamous cell carcinoma is mediated by EpCAM induction driven by IL-6/p62 associated Nrf2-antioxidant pathway activation, Cell Death Dis., 2020, 11, 663 CrossRef CAS PubMed.
- H. Wang, H. Bai, J. Wang, X. Zhou, H. Chen, L. Wang, H. Ren, Z. Liu, W. Zhuo, Z. Zhou, J. Tang, Z. Li, J. Wang, Y. Shen, T. Zhou and X. Liu, Nanoprodrug ratiometrically integrating autophagy inhibitor and genotoxic agent for treatment of triple-negative breast cancer, Biomaterials, 2022, 283, 121458 CrossRef CAS PubMed.
- B. Schrors, S. Boegel, C. Albrecht, T. Bukur, V. Bukur, C. Holtstrater, C. Ritzel, K. Manninen, A. D. Tadmor, M. Vormehr, U. Sahin and M. Lower, Multi-Omics Characterization of the 4T1 Murine Mammary Gland Tumor Model, Front. Oncol., 2020, 10, 1195 CrossRef PubMed.
- R. Acharya, S. Chacko, P. Bose, A. Lapenna and P. Pattanayak, Structure Based Multitargeted Molecular Docking Analysis of Selected Furanocoumarins against Breast Cancer, Sci. Rep., 2019, 9, 15743 CrossRef PubMed.
- S. Nagini, Breast Cancer: Current Molecular Therapeutic Targets and New Players, Anti-Cancer Agents Med. Chem., 2017, 17, 152–163 CrossRef CAS PubMed.
- Q. Li, M. Li, K. Zheng, S. Tang and S. Ma, Expression pattern analysis and drug differential sensitivity of cancer-associated fibroblasts in triple-negative breast cancer, Trans. Oncol., 2021, 14, 100891 CrossRef CAS PubMed.
- S. Comsa, A. M. Cimpean and M. Raicia, The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research, Anticancer Res., 2015, 35, 3147–3154 CAS.
- M. Ito, E. Ito, M. Hirai and S. Yamaguchi, S. Donor–π–Acceptor Type Unsymmetrical Triarylborane-Based Fluorophores: Synthesis, Fluorescence Properties, and Photostability, J. Org. Chem., 2018, 83, 8449–8456 CrossRef CAS PubMed.
- Z. M. Essam, G. E. Ozmen, D. Setiawan, R. R. Hamid, R. M. A. El-Aal, R. Aneja, D. Hamelberg and M. Henary, Donor acceptor fluorophores: synthesis, optical properties, TD-DFT and cytotoxicity studies, Org. Biomol. Chem., 2021, 19, 1835–1846 RSC.
- V. Hrobarikova, P. Hrobarik, P. Gajdos, I. Fitilis, M. Fakis, P. Persephonis and P. Zahradnik, Benzothaizole-Based Fluorophores of Donor–π–Acceptor–π–Donor Type Displaying High Two-Photon Absorption, J. Org. Chem., 2010, 75, 3053–3068 CrossRef CAS PubMed.
- M. Z. K. Baig, G. Pallikonda, P. Trivedi, R. N. P. Tulichala, B. G. Ghosh and M. Chakravarty, M. Metal-Free Arylation to Access Distinct Anthracenylphophonates and Anticancer Activities for These and Allied Phosphonates, ChemistrySelect, 2016, 1, 4332–4339 CrossRef CAS.
- E. Grenet and J. Waser, Iridium- and Rhodium-Catalyzed Directed C–H Heteroarylation of Benzaldehydes with Benziodoxolone Hypervalent Iodine Reagents, Org. Lett., 2018, 20, 1473–1476 CrossRef CAS PubMed.
- M. Z. K. Baig, B. Prusti, D. Roy, P. K. Sahu, M. Sarkar, A. Sharma and M. Chakravarty, Weak Donor-/Strong Acceptor-Linked Anthracenyl π-Conjugates as Solvato(fluoro)chromophore and AEEgens: Contrast between Nitro and Cyano Functionality, ACS Omega, 2018, 3, 9114–9125 CrossRef CAS PubMed.
- D. H. Hua, K. Lou, J. Havens, E. M. Perchellet, Y. Wang, J. P. Perchellet and T. Iwamoto, Synthesis and in vitro antitumor activity of substituted anthracene-1,4-diones, Tetrahedron, 2004, 60, 10155–10163 CrossRef CAS.
- R. B. Weiss, The anthracyclines: will we ever find a better doxorubicin?, Semin. Oncol., 1992, 19, 670–686 CAS.
- D. W. Fry, Biochemical pharmacology of anthracenediones and anthrapyrazoles, Pharmacol. Ther., 1991, 52, 109–125 CrossRef CAS PubMed.
- J. Yan, J. Chen, S. Zhang, J. Hu, L. Huang and X. Li, Synthesis, Evaluation, and Mechanism Study of Novel Indole-Chalcone Derivatives Exerting Effective Antitumor Activity Through Microtubule Destabilization in vitro and in vivo, J. Med. Chem., 2016, 59, 5264–5283 CrossRef CAS PubMed.
- A. Andreani, S. Burnelli, M. Granaiola, A. Leoni, A. Locatelli, R. Morigi, M. Rambaldi, L. Varoli, L. Landi, C. Prata, M. V. Berridge, C. Grasso, H. H. Fiebig, G. Kelter, A. M. Burger and M. W. Kunkel, Antitumor Activity of Bis-indole Derivatives, J. Med. Chem., 2008, 51, 4563–4570 CrossRef CAS PubMed.
- M. El-Naggar, H. Almahli, H. S. Ibrahim, W. M. Eldehna and H. A. Abdel-Aziz, Pyridine-Ureas as Potential Anticancer Agents: Synthesis and In Vitro Biological Evaluation, Molecules, 2018, 23, 1459 CrossRef PubMed.
- M. Yousuf, S. Jinka, S. S. Adhikari and R. Banerjee, Methoxy-enriched cationic stilbenes as anticancer therapeutics, Bioorg. Chem., 2020, 98, 103719 CrossRef CAS PubMed.
- S. I. Omoruyi, O. E. Ekpo, D. M. Semenya, A. Jardine and S. Prince, Exploitation of a novel phenothiazine derivative for its anti-cancer activities in malignant glioblastoma, Apoptosis, 2020, 25, 261–274 CrossRef CAS PubMed.
- M. Arulkumar, K. Yang, N. Wang, S. Penislusshiyan, T. Palvannan, K. Ramalingam, F. Chen, S. H. Luo, Y. J. Zhou and Z. Y. Wang, Synthesis of benzimidazole/triphenylamine-based compounds, evaluation of their bioactivities and an in silico study with receptor tyrosine kinases, J. Chem., 2022, 46, 675–685 CAS.
- J. Qian and B. Z. Tang, AIE Luminogens for Bioimaging and Theranostics: From Organelles to Animals, Chem, 2017, 3, 56–91 CAS.
- S. Bhuin, P. K. Samanta and M. Chakravarty, Efficient and reversible acidofluorochromic features on a solid platform for reusable security writing: A structure-property relationship study on anthracenyl π-conjugates, Dye. Pigm., 2022, 197, 109944 CrossRef CAS.
- S. A. Tucker and W. E. Acree Jr., Polycyclic Aromatic Nitrogen Heterocycles. Part VI. Fluorescence Emission and Quenching Behavior of Select Phenyl- and Alkyl-derivatives Dissolved in Nonelectrolyte Solvents, Polycyclic Aromat. Compd., 1993, 3, 221–229 CrossRef CAS.
- J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. V. Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed.
- W. H. Brooks, W. C. Guida and K. G. Daniel, The Significance of Chirality in Drug Design and Development, Curr. Top. Med. Chem., 2011, 11, 760–770 CrossRef CAS PubMed.
- Y. J. Zheng and C. M. Tice, The utilization of spirocyclic scaffolds in novel drug discovery, Expert Opin. Drug Delivery, 2016, 11(9), 831–834 CrossRef PubMed.
- Y. Yamaguchi, Y. Matsubara, T. Ochi, T. Wakamiya and Z. Yoshida, How the π Conjugation Length Affects the Fluorescence Emission Efficiency, J. Am. Chem. Soc., 2008, 130, 16442 CrossRef CAS.
- B. Gironi, Z. Kahveci, B. McGiill, B. D. Lechner, S. Pagliara, J. Metz, A. Morresi, F. Palombo, P. Sassi and P. G. Petrov, Effect of DMSO on the Mechanical and Structural Properties of Model and Biological Membranes, Biophys. J., 2020, 119, 274–286 CrossRef CAS PubMed.
- L. Zou, S. Guo, H. Lv, F. Chen, L. Wei, Y. Gong, Y. Liu and C. Wei, Molecular design for organic luminogens with efficient emission in solution and solid state, Dyes Pigm., 2022, 198, 109958 CrossRef CAS.
- X. C. Cai and B. Liu, Aggregation-Induced Emission: Recent Advances in Materials and Biomedical Applications, Angew. Chem., 2020, 59, 9868–9886 CrossRef CAS PubMed.
- Y. Huang, J. Xing, Q. Gong, L. C. Chen, G. Liu, C. Yao, Z. Wang, H. L. Zhang, Z. Chen and Q. Zhang, Reducing aggregation caused quenching effect through co-assembly of PAH chromophores and molecular barriers, Nat. Commun., 2019, 10, 169 CrossRef PubMed.
- (a) C. Wang and Z. Li, Molecular conformation and packing: their critical roles in the emission performance of mechanochromic fluorescence materials, Mater. Chem. Front., 2017, 1, 2174–2194 RSC; (b) Q. Li and Z. Li, The Strong Light-Emission Materials in the Aggregated State: What Happens from a Single Molecule to the Collective Group, Adv. Sci., 2017, 1600484 CrossRef PubMed.
- Y. Dai, H. Liu, T. Geng, F. Ke, S. Niu, K. Wang, Y. Qi, B. Zou, B. Yang, W. L. Mao and Y. Lin, Pressure-induced excimer formation and fluorescence enhancement of an anthracene derivative, J. Mater. Chem. C, 2021, 9, 934–938 RSC.
- M. Z. K. Baig, D. Majhi, R. N. P. Tulichala, M. Sarkar and M. Chakravarty, Easy access to new anthracenyl π-conjugates: generation of distinct AIE-active materials, J. Mater. Chem. C, 2017, 5, 2380 RSC.
- V. Aggarwal, H. S. Tuli, A. Varol, F. Thakral, M. B. Yerer, K. Sak, M. Varol, A. Jain, M. A. Khan and G. Sethi, Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements, Biomolecules, 2019, 9, 735 CrossRef CAS PubMed.
- (a) J. Phuchareon, A. van Zante, J. B. Overdevest, F. McCormick, D. W. Eisele and O. Tetsu, c-Kit Expression is Rate-Limiting for Stem Cell Factor-Mediated Disease Progression in Adenoid Cystic Carcinoma of the Salivary Glands, Transl. Oncol., 2014, 7, 537–545 CrossRef PubMed; (b) Y. Zhou, R. Sridhar, L. Shan, W. Sha, X. Gu and S. Sukumar, Loperamide, an FDA-Approved Antidiarrhea Drug, Effectively Reverses the Resistance of Multidrug Resistant MCF-7/MDR1 Human Breast Cancer Cells to Doxorubicin-Induced Cytotoxicity, Cancer Invest., 2012, 30, 119–125 CrossRef CAS PubMed; (c) J. G. Rosch, A. L. Brown, A. N. DuRoss, E. L. DuRoss, G. Sahay and C. Sun, Nanoalginates via Inverse-Micelle Synthesis: Doxorubicin-Encapsulation and Breast-Cancer Cytotoxicity, Nanoscale Res. Lett., 2018, 13, 350 CrossRef PubMed.
- F. A. Tanious, J. M. Veal, H. Buczak, L. S. Ratmeyer and W. D. Wilson, DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites, Biochemistry, 1992, 31, 3103–3112 CrossRef CAS PubMed.
- P. C. Lyon, V. Suomi, P. Jakeman, L. Campo, C. Coussios and R. Carlisle, Quantifying cell death induced by doxorubicin, hyperthermia or HIFU ablation with flow cytometry, Sci. Rep., 2021, 11, 4404 CrossRef CAS PubMed.
- O. Tacar, P. Sriamornsak and C. R. Dass, Doxorubicin: an update on anticancer molecular action,toxicity and novel drug delivery systems, J. Pharm. Pharmacol., 2012, 65, 157–170 CrossRef PubMed.
- V. Vangala, N. V. Nimmu, S. Khalid, M. Kuncha, R. Sistla, R. Banerjee and A. Chaudhuri, Combating Glioblastoma by Codelivering the Small-Molecule Inhibitor of STAT3 and STAT3siRNA with α5β1 Integrin Receptor Selective Liposomes, Mol. Pharmaceutics, 2020, 17, 1859–1874 CrossRef CAS PubMed.
- P. Chakraborty, S. B. Ahil, T. Jamma and P. Yogeeswari, Combining structure-based and 3D QSAR pharmacophore models to discover ligands against EGFR in oral cancer, Future Med. Chem., 2022, 14, 463–478 CrossRef CAS PubMed.
- R. Pola, E. Bohmova, M. Filipova, M. Pechar, J. Pankrac, D. Vetvicka, T. Olejar, M. Kabesova, P. Pouckova, L. Sefc, M. Zábrodský, O. Janoušková, J. Broucek and T. Etrych, Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors, Pharmaceutics, 2020, 12, 31 CrossRef CAS PubMed.
- N. Iqbal and N. Iqbal, Human Epidermal Growth Factor Receptor2 (HER2) in Cancers: Overexpression and Therapeutic Implications, Mol. Biol. Int., 2014, 852748 Search PubMed.
- K. Aertgeerts, R. Skene, J. Yano, B. C. Sang, H. Zou, G. Snell, A. Jennings, K. Iwamoto, N. Habuka, A. Hirokawa, T. Ishikawa, T. Tanaka, H. Miki, Y. Ohta and S. Sogabe, Structural Analysis of the Mechanism of Inhibition and Allosteric Activation of the kinase Domain of HER2 Protein, J. Biol. Chem., 2011, 286, 18756–18765 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb02078e |
| This journal is © The Royal Society of Chemistry 2023 |