Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bacterial-mediated selenium nanoparticles as highly selective antimicrobial agents with anticancer properties†
David
Medina-Cruz
a,
Linh B.
Truong
a,
Eduardo
Sotelo
 b,
Lidia
Martínez
b,
Lidia
Martínez
 c,
María Ujué
González
c,
María Ujué
González
 d,
Yves
Huttel
d,
Yves
Huttel
 c,
Thomas J.
Webster
c,
Thomas J.
Webster
 e,
José Miguel
García-Martín
e,
José Miguel
García-Martín
 d and
Jorge L.
Cholula-Díaz
d and
Jorge L.
Cholula-Díaz
 *b
*b
aDepartment of Chemical Engineering, Northeastern University, Boston, MA 02115, USA
bSchool of Engineering and Sciences, Tecnologico de Monterrey, Eugenio Garza Sada 2501, Monterrey 64849, NL, Mexico. E-mail: jorgeluis.cholula@tec.mx
cInstituto de Ciencia de Materiales de Madrid (ICMM), CSIC (CEI UAM + CSIC), Sor Juana Inés de la Cruz 3, Madrid 28049, Spain
dInstituto de Micro y Nanotecnología, IMN-CNM, CSIC (CEI UAM+CSIC), Isaac Newton 8, Tres Cantos 28760, Spain
eSchool of Health Sciences and Biomedical Engineering, Hebei University of Technology, Tianjin 300401, China
First published on 10th July 2023
Abstract
The green synthesis of nanomaterials offers advantages over traditional chemical methods, such as the production of biocompatible nanostructures in an environmentally friendly and cost-effective manner. Among all of the available green synthesis routes, the use of bacteria offers a high-throughput and versatile synthesis of nanoparticles that can be used in a wide range of biomedical approaches. In this article, we present a controllable and versatile synthesis of selenium nanoparticles (SeNPs) using bacterial isolates of both Gram negative and Gram positive bacteria. The SeNPs were characterized in terms of their physicochemical properties and tested in both antimicrobial and cytotoxicity assays, showing a selective dose-dependent antibacterial activity in a selected range of concentrations (especially when SeNPs synthesized by a particular bacterial isolate were exposed to that isolate) and a mild cytotoxicity when exposed to human dermal fibroblasts. Furthermore, the SeNPs were tested for their anticancer activity by exposure to melanoma cells (skin cancer) in in vitro models, showing a significant dose–response cytotoxic behavior that was associated to the production of reactive oxygen species. Therefore, this work presents a robust and versatile method to produce SeNPs using selected bacterial isolates for numerous biomedical applications.
Sustainability spotlightThe application of nanomaterials in different scientific and technological fields is spreading widely. However, one of the major drawbacks that has impeded their intensive application is the way they are produced, i.e., normally, their synthesis implies the use of toxic reactants and the generation of potentially dangerous by-products. Hence, green nanotechnology has recently emerged as a potential solution for the synthesis of nanomaterials. This report has the objective to produce Se nanoparticles using bacterial isolates as natural biofactories. Important to notice is that the SeNPs showed a selective and effective inhibition when the NPs were exposed to the same bacteria strain that was used for their synthesis. Our work emphasizes the importance of the following UN sustainable development goals: Responsible Consumption and Production (SDG 12). |
1 Introduction
Antimicrobial resistance (AMR) refers to the ability of microorganisms to change their genetic information or properties that help them evade the antimicrobial effects of biomedical agents. Resistant strains of pathogens can lead to more severe symptoms and higher mortality rates compared to common strains.1 Although the resistant mechanisms occur naturally, the AMR crisis has worsened significantly by the overuse and misuse of antibiotics in healthcare, agricultural and environmental settings.2,3 Unfortunately, the rate of the traditional discovery of new antibiotics cannot keep up with the acquired resistance in pathogens, leading to potentially dangerous infections with no suitable treatments. AMR is projected to kill more than 10 million people worldwide annually while causing up to $100 trillion in financial costs.4 With such a bleak future, the AMR crisis demands more urgent and innovative approaches.Presently, the treatment of bacterial infections still mainly involves a combination of traditional antibiotics and the limited use of a novel class of drugs in fear of further resistance. Very few antimicrobials have been developed over the past decades from traditional screening, and virtually no drugs have exhibited a new mechanism of action.5 Eventually, the use of every traditional form of antibiotics will incur resistance, adding to the problem at hand. Therefore, multiple alternatives to treating infections have been studied, with the most promising approaches using antimicrobial peptides (AMP) and phage therapy (PT).6,7 However, AMP discovery remains a time-consuming, expensive, and highly inefficient process, making the number of AMP drug candidates going through clinical settings low.8 PT demonstrates a reduction in resistance due to the uniqueness and importance of its target, but it is still poorly understood in terms of efficacy and safety profiles, leading to an uncertain clinical and regulatory framework.9,10 Lastly, the COVID-19 pandemic has brought more attention to AMR, as the overuse and misuse of antibiotics in SARS-CoV-2 patients has been linked to the development of drug-resistant bacteria (with the increased use of antibiotics to treat COVID-19), along with disruptions in healthcare systems and changes in prescribing practices which may exacerbate AMR problems.11 Additionally, the emergence of new variants of the virus that are resistant to existing treatments could lead to an increased demand for antibiotics.12 Overall, the COVID-19 pandemic has the potential to worsen the already serious problem of antimicrobial resistance.
Nanotechnology can present a novel solution in the fight against the formation of “superbugs”. Specifically, nanoparticles (NPs), mainly made of metals, are an optimal form of therapeutics due to their natural antimicrobial properties.13 Several hypotheses of the mechanism of action of metallic NPs in killing bacteria have been proposed, including the adhesion of NPs onto the surface of the cells; the penetration and destruction of NPs to intracellular organs; the induction of toxicity and oxidative stress; and the regulation of cellular signalling pathways.14–16 The most common metallic NPs used in antimicrobial therapy have been silver NPs (AgNPs).17,18 Nevertheless, while possessing strong antimicrobial properties, the use of AgNPs can lead to drawbacks such as the higher risk of mammalian cell toxicity, further bacterial mutations, and increased horizontal resistance of gene transfer.19,20
Furthermore, inorganic nanoparticles (including AgNPs) have emerged as a promising tool in cancer treatment due to their unique physical and chemical properties.21 They can be engineered to enhance specificity and drug delivery efficiency, minimizing side effects typically associated with traditional chemotherapy.22 NPs such as gold, silver, and iron oxide have been widely studied for their ability to carry therapeutic agents directly to tumor cells, enabling targeted therapy.23 Furthermore, they can also serve as agents in photothermal therapy, where they generate heat under specific light exposure to kill cancer cells.24 Additionally, due to their unique optical properties, they can be used as contrast agents in diagnostic imaging, thus contributing to the early detection of cancer.25
Other metal and non-metal elements should be explored to reduce the burden on AgNPs and overcome their biomedical limitations. One such element is selenium (Se), a chalcogen element that is essential for many organisms. Se nanoparticles (SeNPs) exhibit antimicrobial properties in their nanoscale form, increased biocompatibility, and bioavailability, while also adding antioxidant and anticancer promises.26,27 These advantages have encouraged more research to explore the potential of SeNPs as an effective treatment to both common and resistant strains of bacterial infections.28,29
While the physical and chemical synthesis of SeNPs are both possible, the green synthesis of these nanostructures presents a more effective, environmental-friendly, and biocompatible approach to obtain Se-based nanomaterials compared to traditional production. Moreover, biogenic SeNPs possess additional organic molecular signatures from the host organisms, potentially providing them with increased efficacy and bioavailability compared to traditionally synthesized NPs.30–32 Furthermore, it was suggested that pathogenic organisms could act as biofactories to produce SeNPs, and these products showed increased potency toward the pathogens they were made from, presenting an opportunity for a tuneable, selective treatment for every bacterial infection.33,34
In this study, the biomedical potential of SeNPs synthesized from four pathogenic bacterial strains, namely Escherichia coli referred as EC-SeNPs, Staphylococcus aureus referred as SA-SeNPs, multidrug-resistant Escherichia coli referred as MDR-EC-SeNPs, and methicillin-resistant Staphylococcus aureus referred as MRSA-SeNPs, was explored upon an improved version of a synthetic method developed by our team.35 These bacterial-mediated SeNPs show the following novel features: (a) a selective antimicrobial behavior, whose outcome was associated with the protein corona of the SeNPs; (b) no antibacterial resistance triggered compared to commercial Ag nanomaterials and antibiotics; and (c) a potent anticancer activity. Moreover, through the study of the mechanism of action, we showed a dependence of the reported biomedical activities with the production of reactive oxygen species (ROS). These findings allow us to present bacterial-mediated SeNPs as a suitable biomedical agent with strong potential to the topical treatment of bacterial infections. Furthermore, suitable cytocompatibility and promising anticancer effects of the produced SeNPs are also reported here.
2 Materials and methods
2.1 Bacterial-mediated synthesis of selenium nanoparticles (SeNPs)
Escherichia coli, referred as EC (K-12 HB101), multidrug-resistant Escherichia coli, referred as MDR-EC (ATCC BAA-2471; Manassas, VA), Staphylococcus aureus, referred as SA (ATCC-12600, Manassas, VA) and methicillin-resistant Staphylococcus aureus, referred as MRSA (ATCC-4330, Manassas, VA), were used as the bacterial organisms for the synthesis of the SeNPs, as well as for antibacterial tests. Alternatively, Pseudomonas aeruginosa, referred as PA (Schroeter, Migula, ATCC 15442, Manassas, VA) and Staphylococcus epidermidis, referred as SE (Winslow and Winslow, ATCC, Manassas, VA) were used to conduct additional antimicrobial tests. Each bacterial strain was briefly cultured in Luria–Bertani broth (LB) (Sigma-Aldrich, St. Louis, MO, US) on agar plates and maintained at 4 °C. Cultures were transferred from an agar plate and inoculated with 40 mL of LB broth in a 50 mL conical centrifuge tube. The stock bacterial solution was incubated at 37 °C at 200 rpm for 24 h. Afterwards, the stock solution was centrifuged at 6000 rpm for 10 min. Pellets were resuspended in 40 mL of media. The solution was inoculated with sodium selenite (Na2SeO3) (Sigma-Aldrich) to obtain a concentration of 2 mM and incubated in a shaking incubator at 37 °C, 200 rpm for 24 h. The samples were centrifuged at 7500 rpm for 10 min, with the pellets resuspended in Milli-Q water and sonicated for 2–3 min to disrupt the bacterial membrane. Another centrifugation at 7500 rpm and 2 min was performed, and the SeNPs-rich supernatant was collected. Final separation was performed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min (repeating the cycle twice more if a deeper removal of any organic material was needed). The pellet phase was collected and stored in a glass vial, resuspended in Milli-Q water, and then lyophilized into a fine powder using a freeze-dryer (Labconco Freezone 4.5) overnight. To quantitatively follow the progression of the synthesis of the SeNPs, a kinetic study coupled with ultraviolet-visible (UV-vis) spectroscopy was carried out, along with a reactive oxygen species (ROS) quantification to elucidate the involvement of these species in the process, whose methods have been described in the ESI (Sections S1–S3).†
000 rpm for 30 min (repeating the cycle twice more if a deeper removal of any organic material was needed). The pellet phase was collected and stored in a glass vial, resuspended in Milli-Q water, and then lyophilized into a fine powder using a freeze-dryer (Labconco Freezone 4.5) overnight. To quantitatively follow the progression of the synthesis of the SeNPs, a kinetic study coupled with ultraviolet-visible (UV-vis) spectroscopy was carried out, along with a reactive oxygen species (ROS) quantification to elucidate the involvement of these species in the process, whose methods have been described in the ESI (Sections S1–S3).†
2.2 Cell fixation for the synthesis process
A cell fixation protocol was used with Scanning Electron Microscopy (SEM, SU3800/SU3900, Hitachi) to observe the changes of the bacteria during the synthesis protocol. The experiment was performed over 6 h after the inoculation with 2 mM Na2SeO3 (see Sections S1 and S2† for details about the selection of the inoculation time and Se salt concentration). Bacteria were added to 4 mL of LB media and incubated overnight at 37 °C, and a spectrophotometer was used to analyze the optical density further to dilute the solution to 106 CFU mL−1. A concentration of 75 μg mL−1 of the four bacteria-solutions was obtained by mixing with LB media and transferred to a 6-well plate (Fisher Scientific), and a pre-treated coverslip was added to the bottom of the well (Sigma-Aldrich). After incubation for 6 h at 37 °C, coverslips were treated with a fixative solution of 2.5% glutaraldehyde (C5H8O2) (Sigma-Aldrich) and 0.1 M sodium cacodylate (C2H7AsO2) (Sigma-Aldrich) for 1 h. A buffer solution of 0.1 M C2H7AsO2 replaced the fixative solution, and then the coverslips were treated with a post-fixative solution of 1% osmium tetroxide (OsO4) (Sigma-Aldrich) for 1 h. The coverslips were washed with 30, 50, 70, 80, 95, and 100% ethanol and dehydrated. Samples were dried with liquid CO2–ethanol in a Samdri®-PVT-3D Critical Point Dryer and were mounted with adhesive tabs on SEM stubs. Lastly, the coverslips were individually treated with liquid graphite and sputter-coated with a thin layer of platinum (Pt) using a Sputter Coater (Polaron Range). Images of the cells were then obtained using a SEM with a 10 kV acceleration voltage and a 400 pA beam current.2.3 Morphological, chemical, and structural characterization
A thorough morphological characterization of the nanostructures was accomplished using transmission electron microscopy (TEM) (JEM-1010 TEM). To prepare the samples for imaging, a drop of the different NPs suspensions was dried on 300 mesh copper-coated carbon grids (Electron Microscopy Sciences, Hatfield, PA) and placed inside the scope for observation. Additionally, SEM, energy-dispersive X-ray (EDX), and X-ray photoemission spectroscopy (XPS) characterization were completed, with methods and results shown in the ESI section (Sections S4–S6),† along with a stability analysis (TEM and determination of the zeta potential of colloids) to quantify the physical stability of the SeNPs in both dried and colloidal forms (Section S8†).Additionally, to evaluate the crystallographic nature of the Se nanomaterials, a volume of 2 mL of each sample was dried on a sample holder for the Rigaku MiniFlex 600 XRD instrument. The operational conditions of the XRD included a voltage of 40 kV, a current of 15 mA, and Cu-Kα radiation (λ = 1.542 Å). The XRD patterns were recorded at room temperature with a step width of 0.05° (2θ, diffraction angle) and a scan speed of 0.25° min−1. To study the chemical composition of the SeNPs, Fourier-transformer infrared spectroscopy (FTIR) (PerkinElmer 400 FT-IR/FT-NIR) was used under the attenuated total reflectance (ATR) mode. The samples were prepared by drop-casting of the nanostructure colloids on a sample holder heated to 50 °C. The FTIR spectra were scanned in the range of 500 to 4000 cm−1. The individual spectra were normalized using Spectrum™ software.
2.4 In vitro therapeutic application of the SeNPs
An extensive study was conducted following a protocol that was divided into two main categories: (1) direct analysis, in which SeNPs made by X bacteria – for instance, EC-SeNPs were tested towards the X bacterial strain, both antibiotic-resistant and standard phenotypes, for example, EC and MDR-EC; and (2) indirect analysis, in which SeNPs made by X bacteria – for instance, EC-SeNPs were tested towards the Y bacterial strain, both antibiotic-resistant and standard phenotypes – for example, SA and MRSA. From these experiments, log reduction, minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values were obtained. Finally, a resistance study was done in two synthesized nanosystems: (a) EC-SeNPs and (b) SA-SeNPs through serial exposure cycles. Moreover, a comparison was made between the bacterial-mediated SeNPs with commercially available nanoparticles, i.e., AgNPs (particle size of 80 nm, Alfa Aesar) and SeNPs (particle size of 80 nm, Sigma), as well as vancomycin (Sigma), methicillin (Sigma) and penicillin (Sigma), as antibiotic controls.
Lastly, a cell fixation protocol was followed to observe the interaction between the SeNPs and bacterial cells, following a methodology described in the ESI section (Section S9).†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 of MTS
5 of MTS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) medium). After adding the solution, the 96-well plate was incubated for 4 h in the incubator to allow for a color change. Then, the absorbance was measured at 490 nm on an absorbance plate reader (SpectraMAX M3, Molecular Devices) for cell viability assays after exposure to the SeNPs at different concentrations. Cell viability was calculated by dividing the average absorbance obtained for each sample by the control sample and then multiplied by 100. Controls containing cells and media, and just media, were also included in the 96-well plate to identify the normal growth of cells without SeNPs and to determine the absorbance of the media itself. A cell fixation protocol was followed to observe the interaction between the nanoparticles and human cells. IC50 values were calculated by means of mathematical regression using a licensed software upon data collection (Prism, GraphPad 2021).
medium). After adding the solution, the 96-well plate was incubated for 4 h in the incubator to allow for a color change. Then, the absorbance was measured at 490 nm on an absorbance plate reader (SpectraMAX M3, Molecular Devices) for cell viability assays after exposure to the SeNPs at different concentrations. Cell viability was calculated by dividing the average absorbance obtained for each sample by the control sample and then multiplied by 100. Controls containing cells and media, and just media, were also included in the 96-well plate to identify the normal growth of cells without SeNPs and to determine the absorbance of the media itself. A cell fixation protocol was followed to observe the interaction between the nanoparticles and human cells. IC50 values were calculated by means of mathematical regression using a licensed software upon data collection (Prism, GraphPad 2021).
2.5 Statistical analysis
To verify the accuracy of the results, all tests were conducted three times (N = 3) unless otherwise stated. Student's t-tests were used to determine statistical significance; an alpha value of 0.05 or less was considered statistically significant. The mean and standard deviation are reported.3 Results and discussion
3.1 Synthesis of bacterial-mediated SeNPs
The synthesis of the bacterial-mediated SeNPs was completed by inoculating the bacterial isolates in suspension with different concentrations of Na2SeO3 (1, 2, 3, 5 and 10 mM), producing different bacterial growth and nanoparticle synthesis patterns (Fig. S1 in the ESI section†), which allowed us to find out the best concentration of raw material to maximize the production of SeNPs (Fig. S2†). After inoculation of the bacterial cultures with 2 mM Na2SeO3, a switch from yellowish to an orange color was visible after just a few minutes, with an increase in coloration intensity until the dispersion reached a dark red color after 24 h of reaction (Fig. S3A and B†). The change of color was attributed to the reduction of SeIV in SeO32− ions, presented in the media, into Se0 nanoparticles (SeNPs). The absorption spectra of both samples showed a wide band with a maximum at around 300 nm (Fig. S3C and D†). This optical behavior has been observed for SeNPs with particle sizes in the range of 70–100 nm.36,37 Moreover, this agrees with the particle sizes obtained by TEM analysis for our synthesized SeNPs (vide infra). It has been shown that for several bacterial strains, the reduction of SeIV in SeO32− may occur under the action of nitrate enzymes (as happened with EC) or nitrite reductases.38,39 Therefore, the reduction of SeO32− is usually linked to denitrification in bacteria. Furthermore, thiol functional groups (R–SH), like the well-known glutathione, or GSH, present inside the cytoplasm of bacterial cells,40 are involved in reducing SeIV in SeO32−. From these experiments, it was clear that the employed bacteria could transform the toxic SeO32− in the media to insoluble and non-toxic SeNPs, through a process of detoxification.41,42 Despite the complex system of enzymatic reactions involved in the production of SeNPs and the wide variety of reactions and physical processes that might be involved, the mechanism can be described in one single elementary chemical reaction: SeIVO32− + 4e− + 6H+ → Se0 + 3H2O.The mechanisms employed by different bacterial strains to trigger the reduction of SeIV are not fully understood due to the wide diversity of these entities and the active use of several metabolic pathways, enzymes, and proteins in the reduction process. Therefore, to reach a higher level of understanding, two studies were conducted in parallel: a kinetic control using UV-vis spectroscopy to quantify the rate and quantity of SeNPs production, and a ROS analysis to observe how the production of the SeNPs was related to the ability of bacteria to cope with such species. The results can be found in ESI (Sections S1–S3),† and they showed that the bacterial strains are exposed to an increased concentration of ROS once the Se salt precursor was added (Fig. S4A and B†), which led to a harsh environment. The subsequent production of SeNPs and the decrease in the levels of Se salts in the media were accompanied by a decline in ROS levels, which indicated that the bacterial strains used the production of SeNPs to cope with the toxicity in the media, hence restoring the ROS levels to intra-cellular standards and triggering normal growth.
3.2 Characterization of bacterial-mediated SeNPs
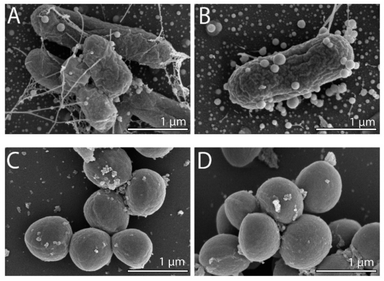
To observe any morphological changes around the bacteria over the synthesis process, SEM imaging was performed for fixed MDR-EC and MRSA. The obtained micrographs are shown in Fig. 1. The SEM images suggest that SeNPs growth can happen either inside of the cells (to then be exported outside of the cells) or in the extracellular side of the bacterial membranes.43 This process can occur simultaneously in the cells, with both intra- and extracellular synthesis happening, as it has been observed in bacteria and other microorganisms.44–46 From the observations, MDR-EC bacteria produced spheroidal and non-agglomerated SeNPs that were spread over the cell surfaces (Fig. 1A and B). The SeNPs were evenly dispersed with no disruptions to the cellular membranes. Conversely, MRSA formed smaller but agglomerated nanoparticles present over the surface of the membrane (Fig. 1C and D).It is important to mention that the produced SeNPs present some instability if left in culture in the presence of the cells and/or biomolecules coming from the cells, which can lead to the formation of rods or larger structures, as shown in the SEM data (see the ESI Section S4†). This instability can be explained in terms of nucleation and further coalescence of the particles due to the presence of active biomolecules with reducing properties in the media, that continue reducing the selenium salt and incorporate them in the form of clusters into the nanoparticles beyond the end of the synthesis protocol.47 For instance, when the nanoparticle core exceeds a critical size, morphological instability develops, which makes the core asymmetric and allows for extension, towards the nanoparticle surface, leading to morphologies that differ from the original spherical core.48,49 To avoid these problems, a purification process is needed, in which the SeNPs are completely separated from any reducing biomolecules and stored in an appropriate dispersion medium (such as water) for future experiments, as was done with the purified SeNPs used in the stability studies.
It is also important to mention that the SeNPs release may be related to vesiculation, a mechanism for the secretion of materials involving the outer membrane of the cells. This mechanism, which appears in response to stress, happens when a small portion of the outer membrane shrinks and forms a spherical vesicle. Once created, the vesicle gets separated from the cell enclosing periplasmatic materials.50 Nevertheless, no vesicles were found after observation enclosing the SeNPs, which might indicate that the synthesis did not happen exclusively in the periplasmatic space, once more indicating that there are several mechanisms involved in the synthesis of the SeNPs.51
TEM characterization revealed the presence of SeNPs around the cells for EC (Fig. 2A) and SA (Fig. 2B). It is well known that the growth of the NPs and their subsequent assembly into larger structures can happen both inside and outside the cells. Once the SeNPs were isolated employing the purification process, the presence of an organic layer was observed surrounding the spheres for both SeNPs produced by EC (Fig. 2C) and SA (Fig. 2D). As can be seen in the TEM images, smaller SeNPs can be found within the cytoplasm, while larger nanostructures were observed in the external membrane outer surface. However, we are not able to elucidate yet if the SeNPs within the external membrane were synthesized inside or outside the cells. Finally, the particle size of the different Se nanostructures extracted from the TEM measurements is reported in Table 1.
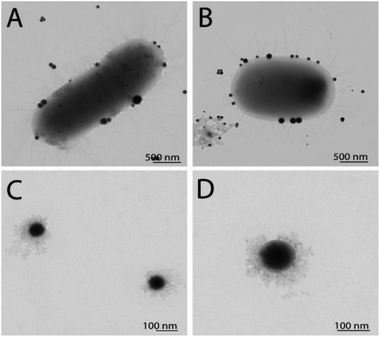 | ||
| Fig. 2 TEM characterization of SeNPs synthesized by EC before (A) and after purification (C), and by SA before (B) and after purification (D). | ||
| Nanostructure type | Particle sizes of SeNPs (nm) |
|---|---|
| MRSA-SeNPs | 62 ± 12 |
| SA-SeNPs | 72 ± 9 |
| MDR-EC-SeNPs | 90 ± 20 |
| EC-SeNPs | 100 ± 30 |
Besides, once the SeNPs were released from the cells, they remained relatively stable in the dispersion medium. This behavior was mainly attributed to an organic coating action surrounding the SeNPs, referred to as the protein corona. This corona could be represented as a double-layer coating wrapping the SeNPs, consisting of a first tightly bound monolayer, usually referred to as hard corona, composed of proteins with high affinity for the nanoparticle surface. The outer layer, termed soft corona, consists of a loose protein layer that reflects the abundance of serum proteins.39 In contrast with Se nanostructures synthesized chemically, microbiologically produced SeNPs contain proteins, among other biomolecules. These proteins are thought to have a stabilizing role of the SeNPs in the colloid.52,53 The stability tests performed on the samples, whose methods and results are shown in the ESI Material section (Section S8),† evidenced that for all the studied systems, once they are completely separated from any reducing biomolecules and stored in an appropriate dispersion medium are stable after 120 days, showing no significant aggregation and decrease in surface charge.
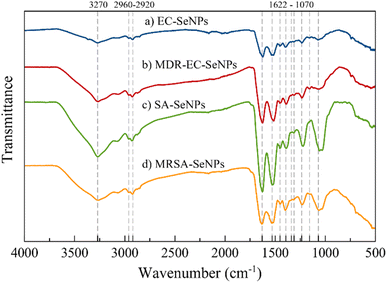
The FT-IR spectra of samples EC-, MDR-EC-, SA- and MRSA-SeNPs are shown in Fig. 3. In general, the samples showed a broad signal around 3270 cm−1 that is characteristic of the OH bond and a very weak asymmetrical stretching band centered at 2960 cm−1 that is representative of CH3. There is an asymmetrical vibration at the wavenumber 2925 cm−1 due to CH2 that is found in proteins. Around 1625, 1520, and 1234 cm−1, other protein vibrational stretching signals were found that represent amide I, II, and III bonds.54,55 Carboxylate (COO−) signals related to amino acids can be located at 1452 and around 1390 cm−1. These bands correspond to the bending and symmetrical vibrations of the COO− ion.55 The vibrational band localized at 1313 cm−1 may be related to the C–H deformation signal that normally occurs in proteins.56 All the samples, excluding MDR-EC-SeNPs, contain a signal at 1157 cm−1 common on proteins with a CH2 wagging vibration. Finally, all SeNPs present a band around 1070 cm−1 that is characteristic of stretching vibrations in CO bonds. According to the overall assignment of the vibrational signals found in the FT-IR spectra, there might be a correlation of threonine-related proteins being functionalized more frequently for the Se-based NPs.56
As mentioned above, FT-IR spectroscopic results are consistent with other characterization methods, in that they provided evidence that bacterial-mediated SeNPs may be covered by various surface-associated biomolecules, including proteins, polysaccharides, and lipids.57 Particularly for the SeNPs capping layer proteins, their secondary structure can differ from that of natural cellular proteins, as featured by the positions of various spectral components of the amide I band (typically within the region 1620–1680 cm−1).55 In any case, the microbial synthesis of SeNPs cannot be regarded as a purely chemical process (such as “Ostwald ripening”, which is characteristic of nanoparticles obtained by classical “wet” methods, or the Gibbs–Thomson law), since it involved the use of different biomolecules and biological entities (such as enzymes).58
3.3 Therapeutic relevance of the bacterial-mediated SeNPs
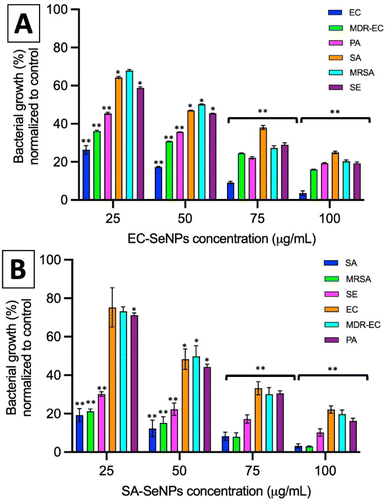
The impact of the SeNPs on the morphology of bacteria after exposure in antimicrobial tests was reported in the ESI section (Section S9 and Fig. S18)† and the in-direct analysis is presented in Section S10, Fig. S19 and S20.† The results showed that those bacteria exposed to SeNPs made by the same bacterial strain presented typical morphological disruption patterns as those seen with similar NPs,59,60 mainly swelling of the membranes and breaking of the structure were observed. This behaviour was not observed when different bacterial strains were exposed to such SeNPs. It is important to notice that most of the SeNPs do not penetrate the bacterial cells but adhere to the cell wall, which can be explained by the relatively big nanoparticle size compared with the pores in the cell wall (4–16 nm).61
For both systems, log reductions, MIC and MBC values were calculated. Log reduction express the relative number of living bacteria that were eliminated by the NPs. Alternatively, MIC is defined as the lowest concentration of NPs necessary to inhibit visible growth, while MBC is the minimum concentration of NPs that result in bacterial death. It is generally understood that the smaller the MIC and MBC values are, the more effective the antimicrobial effect when the antibacterial agent is exposed to bacteria. The results presented in Tables 2 and 3 demonstrate that the SeNPs studied here showed a superior competitive advantage over other microbiologically produced SeNPs and those made by traditional methods. For instance, Srivastava et al. showed that bacterial-mediated SeNPs produced by the R. eutropha biomass, testing against SA and EC, rendered a MIC value of 100 μg mL−1,62 while Hariharan et al. reported that microbially-synthesized SeNPs toward EC and SA produced a MIC value of 30 μg mL−1.63 On the other hand, SeNPs prepared via a reduction of Na2SO3 by ascorbic acid (vitamin C) and stabilized by polysorbate 20 showed MIC values of 82 μg mL−1 against SA,64 while SeNPs prepared by a chemical reduction method using ascorbic acid as a reductant and polyvinyl alcohol (PVA) as stabilizer rendered MIC values towards EC of 250 μg mL−1.26
| Gram staining | Bacteria | Log reduction (8 h) | MIC (μg mL−1) | MBC (μg mL−1) |
|---|---|---|---|---|
| Negative | EC | 6.2 | 5 | 10 |
| MDR-EC | 5.6 | 10 | 10 | |
| PA | 4.1 | 25 | 25 | |
| Positive | SA | 3.6 | 50 | 50 |
| MRSA | 3.4 | 50 | 50 | |
| SE | 3.1 | 25 | 50 |
| Gram staining | Bacteria | Log reduction (8 h) | MIC (μg mL−1) | MBC (μg mL−1) |
|---|---|---|---|---|
| Negative | SA | 6.2 | 3 | 5 |
| MRSA | 5.6 | 5 | 10 | |
| SE | 4.1 | 5 | 10 | |
| Positive | EC | 3.6 | 50 | 50 |
| MDR-EC | 3.4 | 50 | 50 | |
| PA | 3.1 | 40 | 50 |
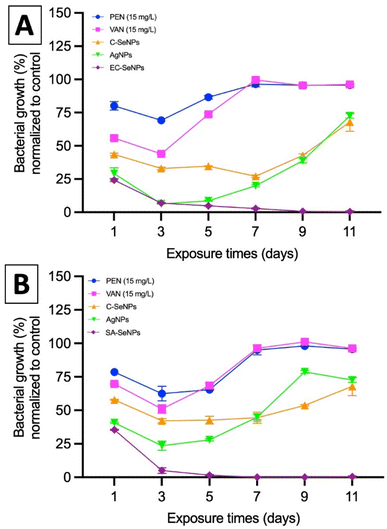
Lastly, the potential resistance that the bacteria showed towards the exposure of SeNPs was evaluated by means of MIC values compared to commercial silver nanoparticles (AgNPs, 80 nm, Alfa Aesar), commercially available SeNPs (Sigma), and the antibiotics penicillin, methicillin, and vancomycin. Briefly, both EC and SA were exposed to a fixed concentration of these agents (50 μg mL−1) over 11 days, and the results are presented in Fig. 5A and B for both EC- and SA-SeNPs, respectively. The results showed a clear increase in MIC values and the development of bacteria resistance for penicillin, methicillin, and vancomycin, as well as a delayed, yet obvious, increase in MIC values and development of resistance for AgNPs, and a significantly more delayed to C-SeNPs. On the other hand, a sustained and constant MIC record was found for EC-SeNPs (Fig. 5A) and SA-SeNPs (Fig. 5B), with no apparent development of resistance upon the final cycles of the experiment. Bacterial resistance to Ag (both in ionic and nanoparticle form) has been previously reported.65 For instance, it has been shown that two strains of EC and PA can develop resistance to AgNPs after repeated exposure due to the bacterial production of the adhesive flagellum protein flagellin, which triggers the aggregation of the nanoparticles and reduced their effect.66,67 Although it was not established before, we hypothesize that the bacteria resistance trigger of the NPs must be influenced by the mechanisms of induced cell death. In the specific case of AgNPs, four different mechanisms of cell death are observed: (a) associated to NP adhesion to the cell surface, hence damaging the membrane and altering the transport activity; (b) NP penetration inside the cells and interaction with cellular organelles and biomolecules; (c) significant increase in ROS inside the microbial cells leading to cell damage and; (d) modulation of cellular signal system leading to dysfunction within the cells.68–71 On the other hand, the antibacterial effect of SeNPs is widely associated to the production of ROS (vide infra).72 When comparing both Se- and AgNPs, previous studies have shown that it is important to understand in detail how ionic silver (Ag+) and AgNPs exert their toxicity through silver ions and to understand how bacteria acquire silver resistance through mechanisms triggered by the interaction of such ions with bacteria,73,74 while not much has been reported in terms of SeNPs and how they might or not induce resistance. Nevertheless, the present results indicated that whether this resistance will eventually happen after longer exposure cycles not studied here, the mechanisms of resistance are hindered and delayed in comparison to those found in AgNPs and commonly used antibiotics.
In summary, these results showed that the use of the SeNPs can exert a selective antimicrobial behavior when exposed to different bacteria isolated, following a simple rule that those SeNPs produced by a specific bacterium will better inhibit the growth of that specific bacterium. Additionally, as the log reduction and MIC/MBC values data showed, the SeNPs can also provide an antibacterial effect towards other strains and species, whose impact slightly decrease when moving far from the producer bacteria but keeping the same structural similarity (for instance, within the Gram-negative spectrum), and significantly decrease when attacking bacterial strains of different structural kind (for instance, Gram-negative vs. Gram-positive). Furthermore, it has been shown that there is not an apparent resistance arising from the interaction of the SeNPs with bacteria upon 7 cycles of continuous exposure, a trend that might be related to the mechanism of cell death exposed by SeNPs and its clear differences with those found in AgNPs and antibiotics.
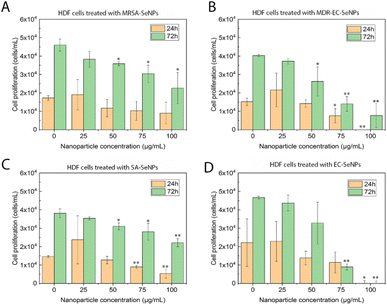
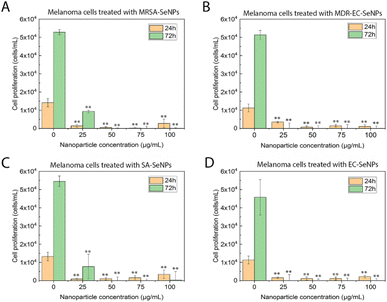
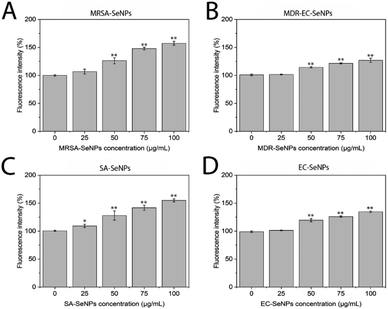
On the other hand, melanoma cells were used to evaluate the potential of the SeNPs as anticancer agents in the second set of experiments (Fig. 7). A dose-dependent cell proliferation decay was found when the bacterial-mediated SeNPs were cultured with melanoma cells for 24 and 72 h. MRSA-SeNPs (Fig. 7A) showed a dose-dependent inhibition of melanoma cells in the extended range of concentrations. With a similar behavior, MDR-EC- (Fig. 7B), SA- (Fig. 7C), and EC-SeNPs (Fig. 7D) presented a remarkable decrease in cell proliferation. Since concentrations of 25 μg mL−1 showed a significant decrease in cell proliferation for all the systems, it was confirmed that a low concentration of SeNPs could trigger a significant anti-tumoral cell efficacy while showing a relatively low cytotoxicity effect when cultured with healthy cells.
The analysis mentioned above showed that the studied bacterial-produced SeNPs could provide acceptable and selective antibacterial effects and excellent anticancer effects, showing an almost negligible cytotoxicity for healthy fibroblasts. It was further suggested that the protein corona surrounding the SeNPs significantly impacted the cytotoxic behavior and properties of the SeNPs when exposed to these biological systems. As such, it has been hypothesized that the coating would allow for a continuous and more targeted cell attachment, leading to external and internal damage.75,76 Indeed, some studies have reported an essential reduction of nanostructure toxicity when green-synthesized NPs are employed compared to traditional NPs.77,78 In the specific case of cancer cells, a deteriorated ROS protective mechanism would account for a large component of the causes of cell death but would fail to entirely explain the SeNPs efficiency inhibiting tumor cell proliferation (since there might be other mechanisms involved). On the other hand, deteriorated membranes in cancer cells would also have led to an increase of the enhanced permeability and retention effect, for which the NPs (especially those with smaller size) would have tended to accumulate in the environment and interior of cancer cells much more than they do in healthy cells.79,80
The half-maximal inhibitory concentration (IC50) was measured to show the potency of the SeNPs in inhibiting the proliferation of the cancerous cells. IC50 values were calculated to study further the cell response to the Se nanostructures (Table 4).
| Sample | 1 day IC50 (μg mL−1) | 3 day IC50 (μg mL−1) |
|---|---|---|
| MRSA-SeNPs | 20 | 15 |
| SA-SeNPs | 12 | 11 |
| MDR-EC-SeNPs | 8 | 9 |
| EC-SeNPs | 8 | 15 |
These values were compared with some found in similar articles. For example, Vekariya et al. investigated the anticancer effect of green synthesized SeNPs tested against an early-stage breast cancer cell line (MCF-7), with an IC50 value of 25 μg mL−1 after a 1 day treatment.81 Moreover, Chen et al. synthesized SeNPs and tested them against different cell lines (A375, CNE2, Hep G2 and MCF-7) with IC50 values ranging from 3.0 to 14.1 μM.82 It becomes worth mentioning that the cytotoxicity of the presented bacterial-mediated SeNPs towards human cells does not mean a problem in the current study and potential future experiments. It is important to mention that the toxicity limits of selenium ions (400 μg d−1) are completely different from those that could be achieved through nanoscale Se, in its elemental form, and would not easily react with biological components as the ionic form does.83,84
4 Conclusions
In this study, four bacterial strains were used to produce selenium nanoparticles (SeNPs), which underwent extensive characterization. These SeNPs demonstrated selective and potent antimicrobial properties, with the greatest effect observed when exposed to the bacterial strain used in their synthesis. A slightly decreased antibacterial effect was noted with similar Gram-category strains and significantly reduced activity with different Gram-category bacteria. Importantly, the SeNPs did not induce bacterial resistance unlike silver nanoparticles and commonly used antibiotics (e.g., penicillin, vancomycin, and methicillin). SeNPs also exhibited potent anticancer effects against melanoma cells, with minimal cytotoxicity to healthy fibroblasts. These anticancer effects were attributed to the generation of reactive oxygen species. Thus, bacterial-mediated SeNPs have shown significant therapeutic potential as antimicrobial and anticancer agents, providing a sustainable solution for combating bacterial infections in the post-antibiotic era.Author contributions
Conceptualization, D. M.-C., L. B. T.; software, D. M.-C., L. B. T.; methodology, D. M.-C.; validation, D. M.-C., L. B. T.; formal analysis, D. M.-C., L. M., Y. H., M. U. G.; investigation, D. M.-C., E. S., L. M., Y. H., M. U. G.; resources, L. M., Y. H., M. U. G., J. L. C.-D., J. M. G.-M., T. J. W.; data curation, D. M.-C., L. B. T.; writing—original draft preparation, D. M.-C., L. B. T., E. S.; writing—review and editing, D. M.-C., L. M., Y. H., M. U. G., J. L. C.-D., J. M. G.-M.; visualization, D. M.-C., E. S., L. M., Y. H., M. U. G., J. M. G.-M.; supervision, J. M. G.-M., T. J.-W., J. L. C.-D.; project administration, T. J. W.; funding acquisition, T. J. W., M. U. G., L. M., J. L.C.-D. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have a financial interest in the start-up company SynCell Biotechnology that has been created because of the research presented in this article. Specifically, D. M. C., L. B. and T. W. are founders and/or shareholders of the company. The authors also declare that they have followed all relevant ethical guidelines and have obtained appropriate institutional review board approval for the research described in this article. The authors believe that their financial interest in the start-up company does not compromise the integrity of the research, or the results presented in this article. However, in the interest of full transparency, the authors have disclosed their financial interest in the start-up company and any potential conflicts of interest.Acknowledgements
The authors from Tecnologico de Monterrey and CSIC thank the bilateral project BILTC22001 for funding. The authors from Tecnologico de Monterrey also thank the Department of Sciences – Chemistry and Nanotechnology at Tecnologico de Monterrey, campus Monterrey. TJW thanks Art Zafiropoulo for funding. The authors acknowledge the service from the MiNa Laboratory at IMN, funded from CM (project SpaceTec, S2018/NMT-4291 TEC2SPACE), MINECO (project CSIC13-4E-1794), and EU (FEDER, FSE).Notes and references
- M. A. Abushaheen, Muzaheed, A. J. Fatani, M. Alosaimi, W. Mansy, M. George, S. Acharya, S. Rathod, D. D. Divakar, C. Jhugroo, S. Vellappally, A. A. Khan, J. Shaik and P. Jhugroo, Disease-a-Month, 2020, 66, 100971 CrossRef.
- E. Christaki, M. Marcou and A. Tofarides, J. Mol. Evol., 2019, 88(1), 26–40 CrossRef PubMed.
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. R. Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. K. Hamadani, E. A. P. Kumaran, B. McManigal, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. de Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, N. Mturi, T. Munera-Huertas, P. Musicha, M. M. Mussi-Pinhata, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, A. Y. Peleg, C. Perrone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, K. E. Rudd, N. Russell, J. Schnall, J. A. G. Scott, M. Shivamallappa, J. Sifuentes-Osornio, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, H. Vu, T. Walsh, S. Waner, T. Wangrangsimakul, T. Wozniak, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- P. Dadgostar, Infect. Drug Resist., 2019, 12, 3903 CrossRef CAS PubMed.
- C. Nathan, Nat. Rev. Microbiol., 2020, 18(5), 259–260 CrossRef CAS PubMed.
- S. Kapil and V. Sharma, Can. J. Microbiol., 2021, 67, 119–137 CrossRef CAS PubMed.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute, A. L. Ferguson, A. Cherkasov, M. N. Seleem, C. Pinilla, C. de la Fuente-Nunez, T. Lazaridis, T. Dai, R. A. Houghten, R. E. W. Hancock and G. P. Tegos, Lancet Infect. Dis., 2020, 20, e216–e230 CrossRef CAS PubMed.
- K. Browne, S. Chakraborty, R. Chen, M. D. P. Willcox, D. S. Black, W. R. Walsh and N. Kumar, Int. J. Mol. Sci., 2020, 21, 7047 CrossRef CAS PubMed.
- D. Saha and R. Mukherjee, IUBMB Life, 2019, 71, 781–790 CrossRef CAS PubMed.
- S. Hesse and S. Adhya, Annu. Rev. Microbiol., 2019, 73, 155–174 CrossRef CAS PubMed.
- H. Getahun, I. Smith, K. Trivedi, S. Paulin and H. H. Balkhy, Bull. W. H. O., 2020, 98, 442 CrossRef PubMed.
- C. C. Lai, S. Y. Chen, W. C. Ko and P. R. Hsueh, Int. J. Antimicrob. Agents, 2021, 57, 106324 CrossRef CAS.
- A. S. Balderrama-González, H. A. Piñón-Castillo, C. A. Ramírez-Valdespino, L. L. Landeros-Martínez, E. Orrantia-Borunda and H. E. Esparza-Ponce, Int. J. Mol. Sci., 2021, 22, 12890 CrossRef PubMed.
- S. A. Bansal, V. Kumar, J. Karimi, A. P. Singh and S. Kumar, Nanoscale Adv., 2020, 2, 3764–3787 RSC.
- K. McNamara and S. A. M. Tofail, Adv. Phys. X, 2017, 2, 54–88 CAS.
- N. T. K. Thanh and L. A. W. Green, Nano Today, 2010, 5, 213–230 CrossRef CAS.
- F. Paladini and M. Pollini, Materials, 2019, 12, 2540 CrossRef CAS PubMed.
- J. S. Kim, E. Kuk, K. N. Yu, J. H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park, C. Y. Hwang, Y. K. Kim, Y. S. Lee, D. H. Jeong and M. H. Cho, Nanomedicine, 2007, 3, 95–101 CrossRef CAS PubMed.
- R. Vishwanath and B. Negi, Curr. Res. Green Sustainable Chem., 2021, 4, 100205 CrossRef CAS.
- P. G. Jamkhande, N. W. Ghule, A. H. Bamer and M. G. Kalaskar, J. Drug Deliv. Sci. Technol., 2019, 53, 101174 CrossRef CAS.
- E. Ilhan-Ayisigi and O. Yesil-Celiktas, Eng. Life Sci., 2018, 18, 882–892 CrossRef CAS PubMed.
- S.-J. Choi, G. E. Choi, J.-M. Oh, Y.-J. Oh, M.-C. Park and J.-H. Choy, J. Mater. Chem., 2010, 20, 9463–9469 RSC.
- Y. Chen, H. Chen and J. Shi, Mol. Pharm., 2014, 11, 2495–2510 CrossRef CAS PubMed.
- A. Gharatape, S. Davaran, R. Salehi and H. Hamishehkar, RSC Adv., 2016, 6, 111482–111516 RSC.
- D. Kim, J. Kim, Y. I. Park, N. Lee and T. Hyeon, ACS Cent. Sci., 2018, 4, 324–336 CrossRef CAS.
- T. Huang, J. A. Holden, D. E. Heath, N. M. O'Brien-Simpson and A. J. O'Connor, Nanoscale, 2019, 11, 14937–14951 RSC.
- X. Huang, X. Chen, Q. Chen, Q. Yu, D. Sun and J. Liu, Acta Biomater., 2016, 30, 397–407 CrossRef CAS.
- G. M. Khiralla and B. A. El-Deeb, LWT–Food Sci. Technol., 2015, 63, 1001–1007 CrossRef CAS.
- N. Filipović, D. Ušjak, M. T. Milenković, K. Zheng, L. Liverani, A. R. Boccaccini and M. M. Stevanović, Front. Bioeng. Biotechnol., 2021, 8, 1591 Search PubMed.
- M. C. Escobar-Ramírez, A. Castañeda-Ovando, E. Pérez-Escalante, G. M. Rodríguez-Serrano, E. Ramírez-Moreno, A. Quintero-Lira, E. Contreras-López, J. Añorve-Morga, J. Jaimez-Ordaz and L. G. González-Olivares, Fermentation, 2021, 7, 130 CrossRef.
- E. Cremonini, E. Zonaro, M. Donini, S. Lampis, M. Boaretti, S. Dusi, P. Melotti, M. M. Lleo and G. Vallini, Microb. Biotechnol., 2016, 9, 758–771 CrossRef CAS PubMed.
- S. A. Wadhwani, U. U. Shedbalkar, R. Singh and B. A. Chopade, Appl. Microbiol. Biotechnol., 2016, 100, 2555–2566 CrossRef CAS PubMed.
- H. Alam, N. Khatoon, M. A. Khan, S. A. Husain, M. Saravanan and M. Sardar, J. Clust. Sci., 2020, 31, 1003–1011 CrossRef CAS.
- H. Zhang, Z. Li, C. Dai, P. Wang, S. Fan, B. Yu and Y. Qu, Environ. Res., 2021, 194, 110630 CrossRef CAS PubMed.
- D. M. Cruz, G. Mi and T. J. Webster, J. Biomed. Mater. Res., Part A, 2018, 106, 1400–1412 CrossRef PubMed.
- A. Husen and K. S. Siddiqi, J. Nanobiotechnol., 2014, 12, 1–10 CrossRef PubMed.
- Z. H. Lin and C. R. C. Wang, Mater. Chem. Phys., 2005, 92, 591–594 CrossRef CAS.
- S. Shoeibi, P. Mozdziak and A. Golkar-Narenji, Top. Curr. Chem., 2017, 375, 1–21 CrossRef CAS.
- A. V. Tugarova and A. A. Kamnev, Talanta, 2017, 174, 539–547 CrossRef CAS PubMed.
- Y. Tan, R. Yao, R. Wang, D. Wang, G. Wang and S. Zheng, Microb. Cell Fact., 2016, 15, 157 CrossRef.
- M. Vogel, S. Fischer, A. Maffert, R. Hübner, A. C. Scheinost, C. Franzen and R. Steudtner, J. Hazard. Mater., 2018, 344, 749–757 CrossRef CAS.
- S. Lampis, E. Zonaro, C. Bertolini, D. Cecconi, F. Monti, M. Micaroni, R. J. Turner, C. S. Butler and G. Vallini, J. Hazard. Mater., 2017, 324, 3–14 CrossRef CAS PubMed.
- M. Shakibaie, M. R. Khorramizadeh, M. A. Faramarzi, O. Sabzevari and A. R. Shahverdi, Biotechnol. Appl. Biochem., 2010, 56, 7–15 CrossRef CAS.
- A. J. Kora and L. Rastogi, J. Environ. Manage., 2016, 181, 231–236 CrossRef CAS PubMed.
- S. Shoeibi and M. Mashreghi, J. Trace Elem. Med. Biol., 2017, 39, 135–139 CrossRef CAS.
- N. Srivastava and M. Mukhopadhyay, Powder Technol., 2013, 244, 26–29 CrossRef CAS.
- J. Dobias, E. I. Suvorova and R. Bernier-Latmani, Nanotechnology, 2011, 22, 195605 CrossRef CAS PubMed.
- M. Quintana, E. Haro-Poniatowski, J. Morales and N. Batina, Appl. Surf. Sci., 2002, 195, 175–186 CrossRef CAS.
- Z. H. Lin, F. C. Lin and C. R. C. Wang, J. Chin. Chem. Soc., 2004, 51, 239–242 CrossRef CAS.
- N. S. Khoei, S. Lampis, E. Zonaro, K. Yrjälä, P. Bernardi and G. Vallini, Nat. Biotechnol., 2017, 34, 1–11 CAS.
- C. M. Debieux, E. J. Dridge, C. M. Mueller, P. Splatt, K. Paszkiewicz, I. Knight, H. Florance, J. Love, R. W. Titball, R. J. Lewis, D. J. Richardson and C. S. Butler, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13480–13485 CrossRef CAS PubMed.
- S. Rajeshkumar, P. Veena and R. v. Santhiyaa, Nanotechnology in the Life Sciences, 2018, pp. 63–79 Search PubMed.
- S. Menon, H. Agarwal, S. V. Kumar and S. Rajeshkumar, Green Synthesis, Characterization and Applications of Nanoparticles, 2019, pp. 165–197 Search PubMed.
- A. J. Kora and L. Rastogi, J. Environ. Manage., 2016, 181, 231–236 CrossRef CAS.
- A. A. Kamnev, P. v. Mamchenkova, Y. A. Dyatlova and A. v. Tugarova, J. Mol. Struct., 2017, 1140, 106–112 CrossRef CAS.
- A. Barth, Biochim. Biophys. Acta, 2007, 1767, 1073–1101 CrossRef CAS PubMed.
- R. Jain, N. Jordan, S. Weiss, H. Foerstendorf, K. Heim, R. Kacker, R. Hübner, H. Kramer, E. D. van Hullebusch, F. Farges and P. N. L. Lens, Environ. Sci. Technol., 2015, 49, 1713–1720 CrossRef CAS PubMed.
- W. Zhang, Z. Chen, H. Liu, L. Zhang, P. Gao and D. Li, Colloids Surf., B, 2011, 88, 196–201 CrossRef CAS PubMed.
- J. Dorazilová, J. Muchová, K. Šmerková, S. Kočiová, P. Diviš, P. Kopel, R. Veselý, V. Pavliňáková, V. Adam and L. Vojtová, Nanomaterials, 2020, 10, 1971 CrossRef PubMed.
- L. D. Geoffrion, T. Hesabizadeh, D. Medina-Cruz, M. Kusper, P. Taylor, A. Vernet-Crua, J. Chen, A. Ajo, T. J. Webster and G. Guisbiers, ACS Omega, 2020, 5, 2660–2669 CrossRef CAS PubMed.
- T. Huang, J. A. Holden, E. C. Reynolds, D. E. Heath, N. M. O'Brien-Simpson and A. J. O'Connor, ACS Appl. Mater. Interfaces, 2020, 12, 55696–55709 CrossRef CAS PubMed.
- N. Srivastava and M. Mukhopadhyay, Bioprocess Biosyst. Eng., 2015, 38, 1723–1730 CrossRef CAS.
- H. Hariharan, A. Al-Dhabi, P. Karuppiah and S. K. Rajaram, Chalcogenide Lett., 2012, 9, 509–515 CAS.
- V. Bartůněk, J. Junková, J. Šuman, K. Kolářová, S. Rimpelová, P. Ulbrich and Z. Sofer, Mater. Lett., 2015, 152, 207–209 CrossRef.
- S. Silver, L. T. Phung and G. Silver, J. Ind. Microbiol. Biotechnol., 2006, 33, 627–634 CrossRef CAS PubMed.
- L. M. Stabryla, K. A. Johnston, N. A. Diemler, V. S. Cooper, J. E. Millstone, S. J. Haig and L. M. Gilbertson, Nat. Nanotechnol., 2021, 16(9), 996–1003 CrossRef CAS PubMed.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek, O. Tomanec and R. Zbořil, Nat. Nanotechnol., 2017, 13(1), 65–71 CrossRef PubMed.
- W. R. Li, X. B. Xie, Q. S. Shi, H. Y. Zeng, Y. S. Ou-Yang and Y. B. Chen, Appl. Microbiol. Biotechnol., 2010, 85, 1115–1122 CrossRef CAS PubMed.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, 1701503 CrossRef PubMed.
- X. Yan, B. He, L. Liu, G. Qu, J. Shi, L. Hu and G. Jiang, Metallomics, 2018, 10, 557–564 CrossRef CAS PubMed.
- C. You, C. Han, X. Wang, Y. Zheng, Q. Li, X. Hu and H. Sun, Mol. Biol. Rep., 2012, 39, 9193–9201 CrossRef CAS PubMed.
- F. Martínez-Esquivias, J. M. Guzmán-Flores, A. Pérez-Larios, N. González Silva and J. S. Becerra-Ruiz, J. Nanosci. Nanotechnol., 2021, 21, 5383–5398 CrossRef PubMed.
- S. L. Percival, P. G. Bowler and D. Russell, J. Hosp. Infect., 2005, 60, 1–7 CrossRef CAS PubMed.
- A. Hamad, K. S. Khashan and A. Hadi, J. Inorg. Organomet. Polym. Mater., 2020, 30, 4811–4828 CrossRef CAS.
- S. Menon, H. Agarwal, S. Rajeshkumar, P. J. Rosy and V. K. Shanmugam, Bionanoscience, 2020, 10, 122–135 CrossRef.
- C. Ferro, H. F. Florindo and H. A. Santos, Adv. Healthcare Mater., 2021, 10, 2100598 CrossRef CAS.
- M. Ikram, B. Javed, N. I. Raja and Z. U. R. Mashwani, Int. J. Nanomed., 2021, 16, 249 CrossRef.
- B. K. Ndwandwe, S. P. Malinga, E. Kayitesi and B. C. Dlamini, Int. J. Food Sci. Technol., 2021, 56, 2640–2650 CrossRef CAS.
- D. Kalyane, N. Raval, R. Maheshwari, V. Tambe, K. Kalia and R. K. Tekade, Mater. Sci. Eng., C, 2019, 98, 1252–1276 CrossRef CAS PubMed.
- A. Nel, E. Ruoslahti and H. Meng, ACS Nano, 2017, 11, 9567–9569 CrossRef CAS PubMed.
- K. K. Vekariya, J. Kaur and K. Tikoo, Nanomedicine, 2012, 8, 1125–1132 CrossRef CAS PubMed.
- T. Chen, Y. S. Wong, W. Zheng, Y. Bai and L. Huang, Colloids Surf., B, 2008, 67, 26–31 CrossRef CAS.
- Y. N. Larimi, M. H. Mallah, M. A. Moosavian and J. Safdari, J. Radioanal. Nucl. Chem., 2013, 298, 1511–1518 CrossRef CAS.
- B. Hosnedlova, M. Kepinska, S. Skalickova, C. Fernandez, B. Ruttkay-Nedecky, Q. Peng, M. Baron, M. Melcova, R. Opatrilova, J. Zidkova, G. Bjørklund, J. Sochor and R. Kizek, Int. J. Nanomed., 2018, 13, 2107 CrossRef CAS.
- G. Zhao, X. Wu, P. Chen, L. Zhang, C. S. Yang and J. Zhang, Free Radical Biol. Med., 2018, 126, 55–66 CrossRef CAS PubMed.
- M. Guo, Y. Li, Z. Lin, M. Zhao, M. Xiao, C. Wang, T. Xu, Y. Xia and B. Zhu, RSC Adv., 2017, 7, 52456–52464 RSC.
- Y. Huang, L. He, W. Liu, C. Fan, W. Zheng, Y. S. Wong and T. Chen, Biomaterials, 2013, 34, 7106–7116 CrossRef CAS PubMed.
- Y. Zhuang, L. Li, L. Feng, S. Wang, H. Su, H. Liu, H. Liu and Y. Wu, Nanoscale, 2020, 12, 1389–1396 RSC.
- S. Menon, K. S. Shrudhi Devi, R. Santhiya, S. Rajeshkumar and S. Venkat Kumar, Colloids Surf., B, 2018, 170, 280–292 CrossRef CAS PubMed.
- S. K. Torres, V. L. Campos, C. G. León, S. M. Rodríguez-Llamazares, S. M. Rojas, M. González, C. Smith and M. A. Mondaca, J. Nanopart. Res., 2012, 14, 1–9 Search PubMed.
- X. Wang, K. Sun, Y. Tan, S. Wu and J. Zhang, Free Radical Biol. Med., 2014, 72, 1–10 CrossRef CAS PubMed.
- F. Gao, J. Zhao, P. Liu, D. Ji, L. Zhang, M. Zhang, Y. Li and Y. Xiao, Int. J. Biol. Macromol., 2020, 142, 265–276 CrossRef CAS PubMed.
- B. Yu, Y. Zhang, W. Zheng, C. Fan and T. Chen, Inorg. Chem., 2012, 51, 8956–8963 CrossRef CAS.
- A. Guleria, S. Chakraborty, S. Neogy, D. K. Maurya and S. Adhikari, Chem. Commun., 2018, 54, 8753–8756 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Spectroscopic kinetics analysis. Toleration time study. SEM, EDX, XPS, XRD and stability over time characterization of the bacterial-mediated SeNPs. See DOI: https://doi.org/10.1039/d3su00145h |
| This journal is © The Royal Society of Chemistry 2023 |