Open Access Article
Open Access ArticleImproving the activity of horseradish peroxidase in betaine-based natural deep eutectic systems†
Liane
Meneses
 a,
Nicolás F.
Gajardo-Parra
a,
Nicolás F.
Gajardo-Parra
 b,
Esteban
Cea-Klapp
b,
Esteban
Cea-Klapp
 c,
José Matías
Garrido
c,
José Matías
Garrido
 c,
Christoph
Held
c,
Christoph
Held
 b,
Ana Rita
Duarte
a and
Alexandre
Paiva
*a
b,
Ana Rita
Duarte
a and
Alexandre
Paiva
*a
aDepartamento de Química, Faculdade de Ciências e Tecnologia, LAQV/REQUIMTE, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal. E-mail: alexandre.paiva@fct.unl.pt
bDepartment of Biochemical and Chemical Engineering, Laboratory of Thermodynamics, TU Dortmund, Emil-Figge-Str. 70, 44227 Dortmund, Germany
cDepartamento de Ingeniería Química, Universidad de Concepción, 4070386 Concepción, Chile
First published on 11th April 2023
Abstract
In this work, the activity of horseradish peroxidase (HRP) in betaine-based natural deep eutectic systems (NADESs) was measured and studied by molecular simulations. Focus was laid on enzymatic activity in the NADESs under thermal stress as well as under the influence of water. Furthermore, the structure of HRP under these different conditions was measured by circular dichroism (CD). As a result, HRP remains enzymatically active in all NADESs upon incubation for 24 h at 37 °C and 60 °C and after 4 h at 80 °C, especially when incubated in a NADES composed of betaine, trehalose, glycerol, and water, in a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. The CD studies have shown that high activity is obtained in the systems that promoted higher α-helix contents. The molecular simulations showed that using a NADES instead of buffer solvent reduces HRP flexibility, and we found that enzymatic activity correlates with Gibbs energy of solvation of HRP. Finally, hydrophobic hydration interactions govern the stabilization mechanism of the HRP folded state as shown by a drastic enzymatic activity drop upon 5 wt% water addition using a betaine: glycerol NADES as solvent.
5. The CD studies have shown that high activity is obtained in the systems that promoted higher α-helix contents. The molecular simulations showed that using a NADES instead of buffer solvent reduces HRP flexibility, and we found that enzymatic activity correlates with Gibbs energy of solvation of HRP. Finally, hydrophobic hydration interactions govern the stabilization mechanism of the HRP folded state as shown by a drastic enzymatic activity drop upon 5 wt% water addition using a betaine: glycerol NADES as solvent.
Sustainability spotlightEnzymes are widely used in industry as biocatalysts; however, their use is limited by their stability. Strategies to improve enzymatic stability are constantly sought. This work shows the use of natural deep eutectic systems (NADESs) for the stabilization of horseradish peroxidase (HRP) under thermal stress. Besides using green solvents, that comply with several sustainable and green chemistry principles, we have also used predictive methods that allow the time, reagents and consumables used during trial-and-error experiments to be decreased. By stabilizing the activity of enzymes, and thus improving their performance, we are narrowing the gap for implementing this technology in industry. This work is in alignment with SDGs 9 (industry) and 13 (climate action). |
1. Introduction
Enzymes are widely used in industrial processes since they are regarded as highly efficient biocatalysts. Horseradish peroxidase (HRP) is an enzyme widely used in several biomedical applications, being applied for cancer therapy or in biosensors.1 This enzyme is also widely used as a cross-linking agent.2 The enzyme has been thoroughly studied and has a well characterized catalytic centre,3 crystal structure4 and amino acid sequence.5 The structure of HRP is mostly α-helical with small β-sheet regions. The heme-group is located in the centre of HRP, between the distal and proximal domains.3 HRP can carry out oxidation reactions on a series of organic and inorganic compounds using hydrogen peroxide as the substrate, which makes it a great model protein to be used in stability studies.3,6However, before the application of enzymes at an industrial scale, there are numerous conditions that must be taken into consideration due to a lack of stability regarding temperature, pH or solvent. Hence, the stabilization of enzymes becomes crucial for their application in industrial processes such as treatment of wastewaters, tissue engineering, biosensors, or drug delivery systems.7,8 One of the strategies used to stabilize enzymes is immobilization. For several industrial processes, immobilization has become a routine process and made several industrial processes feasible and greener.9
Since the early 90s, when green chemistry was first introduced, chemists have combined efforts to change their laboratory practices towards compliance with the 12 principles of green chemistry. The use of alternative solvents to replace traditional organic solvents was one of the implemented changes, and ionic liquids (ILs) gained attention as green alternatives to traditional solvents, mostly due to their low vapor pressure and low flammability.10 From the first time ILs and enzymes were combined and reported in the literature, they have been used in biocatalysis for several purposes such as co-solvent, additives to the reaction or agents to stabilize or immobilize enzymes.10 There are examples of the use of ILs as solvents with several enzymes, such as lipases, oxidoreductases, peroxidases, etc., and whole microorganisms as well.10,11 However, and due to the fact that the “greenness” of ILs started to raise questions, deep eutectic systems (DESs) have appeared as a great alternative to ILs.10 DESs were first described by Abbott et al. as a mixture of two components with a deep melting point depression compared to the original components.12 Later, Choi et al. disclosed natural deep eutectic systems (NADESs), prepared from the combinations of only naturally occurring compounds such as sugars, polyols, organic acids, or amino acids.13 DESs have been successfully used to activate, crystallize,14 stabilize or act as reaction media for several types of enzymes, such as lipases oxidoreductases,15 hydrolases,15 and laccases,16 among others.
The stabilization of HRP has already been studied by some researchers using mostly choline-based DESs. Wu et al. studied the activity of HRP in a series of choline chloride (ChCl) and choline acetate (ChAc) based DESs.6 Under the conditions studied, HRP's enzymatic activity was slightly higher in solutions containing a ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol ratio (2
glycerol ratio (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) compared with control buffer and slightly worse when using ChAc. Furthermore, structural studies have shown that the systems that allow HRP to have a higher α-helix content and stabilization of the tertiary structure helped increase the enzymatic activity of HRP.6 In another study, the influence of several ChCl and ethyl ammonium chloride (EAC) based DESs on the activity and structure of two heme-containing enzymes – HRP and cytochrome C (cyt C) was studied.17 The work has shown that while cyt C's activity is improved in the presence of DESs, HRP's activity decreased for all the systems and concentrations tested.
1) compared with control buffer and slightly worse when using ChAc. Furthermore, structural studies have shown that the systems that allow HRP to have a higher α-helix content and stabilization of the tertiary structure helped increase the enzymatic activity of HRP.6 In another study, the influence of several ChCl and ethyl ammonium chloride (EAC) based DESs on the activity and structure of two heme-containing enzymes – HRP and cytochrome C (cyt C) was studied.17 The work has shown that while cyt C's activity is improved in the presence of DESs, HRP's activity decreased for all the systems and concentrations tested.
The diverse nature of NADESs can cause various effects on enzymes, so obtaining information about process thermodynamics or kinetics effects requires extensive experimental trial and error procedures.18 Predictive methods have emerged as a tool in process design to get physicochemical properties that could relate to observed enzyme properties.19 Equations of state and thermodynamic methods have been used in the literature to demonstrate the effect of different cosolvents on enzymatic properties, including kinetics20,21 and thermal stability.22 Diverse molecular dynamics methods, including classical molecular dynamics (MD)23,24 and quantum mechanics/molecular mechanics (QM/MM)25 have been used to describe HRP and different mutations.26 However, the effect of deep eutectic solvent has not been studied yet as it has been on other proteins such as lipases,27,28 lysosome29 or alcohol dehydrogenase.30 Nevertheless, MD methods are necessary for a detailed study of solvation to observe the protein surface hydration dynamics when interacting with a cosolvent. As proteins are complex molecular systems, techniques directly consider both the shape and the density of water/co-solvent around the enzyme.31 Minimum-distance distribution functions and Kirkwood–Buff integrals have recently been used to directly describe protein solvation using sorbitol urea mixtures, osmolytes, and ionic liquids.32,33 Finding valuable properties by applying these methods allows researchers to have “tailor-made” new green solvents (i.e. DESs or ILs), thinking about the final application, which significantly reduces the dependence on a great experimental effort.
In a previous study by this group,34 several betaine-based NADESs were fully characterized in terms of water content and activity, viscosity and density. Moreover, the influence of these NADESs on the activity and structure of HRP was also accessed. It was observed that several systems were able to improve enzyme's activity and that they were able to change HRP's structure to more stable conformations in which the enzyme had higher unfolding and denaturation temperatures. Hence, in this work, we went further to study the activity and stability of HRP after exposure to higher temperatures (60 and 80 °C) for longer periods of time. Betaine was used in these systems as a substitute for ChCl. The study of the utilization of such systems is important since the use of choline derivatives is restricted for some applications, mostly concerning health. It has also been recently published that ChCl based DESs and NADESs have high environmental impact.35
2. Materials and methods
2.1. Materials
Lyophilized powder of peroxidase from horseradish (HRP, type I, 89.63 U per mg solid, CAS 9003-99-0) was purchased from Sigma-Aldrich (St. Louis, Missouri, USA) and used without further purification. D-(+)-Xylose (≥99%, CAS 58-86-6), glycerol (≥99.5% CAS 56-81-5), D-sorbitol (≥98%, CAS 50-70-4), DL-proline (99%, CAS 609-36-9), phenol-4-sulfonic acid sodium salt dihydrate (PSA, 98%, CAS 10580-19-5), 4-aminoantipyrine (4-AAP, ≥99%, CAS 83-07-8) and hydrogen peroxide 30% solution (CAS 7722-84-1) were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Trehalose dihydrate (CAS 6138-23-4) was kindly provided by Hayashibara Co. (Okayama, Japan). Betaine anhydrous (>97%, CAS 107-43-7) was obtained from TCI (Tokyo, Japan) and sucrose (CAS 57-50-1) was purchased from Cmd Chemicals (Funchal, Portugal).2.2. NADES preparation and characterization
All systems were prepared by using the heating and stirring method36 and are listed in Table 1. The NADESs used have been fully characterized elsewhere.34 Using the system BGly, previously prepared as described in Table 1, water was added to prepare four formulations, with different water contents, namely, 5, 10, 15 and 20 wt%. The water content (wt%) and water activity (aw) of the BGly formulations were determined as reported elsewhere.34| Code | Component A | Component B | Component C | Component D | Molar ratio |
|---|---|---|---|---|---|
| BXylW | Betaine anhydrous | D-(+)-Xylose | Water | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6 |
| BTrehGlyW | Betaine anhydrous | Trehalose dihydrate | Glycerol | Water | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
| BSorbW | Betaine anhydrous | D-Sorbitol | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| BSucProW | Betaine anhydrous | Sucrose | DL-Proline | Water | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
| BGly | Betaine anhydrous | Glycerol | — | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
2.3. Enzymatic activity assay
Throughout this work, the enzymatic activity of HRP was assessed as described elsewhere.342.4. Evaluation of HRP activity and thermostability in NADESs
Enzyme activity of HRP was tested upon incubation in NADESs for 6 h and 24 h at 37 °C. To assess HRP activity under thermal stress (that will be further addressed as “thermostability”), two temperatures were tested, 60 and 80 °C. For the assays at 60 °C, the enzyme was incubated for 4 h and 24 h. For the assays at 80 °C, the tested incubation times were 2 h and 4 h. For all temperatures tested, after each incubation time, 3 mL of PBS were added to each mixture and these solutions were used to assess the activity of HRP, using the enzymatic activity assay described before. PBS (100 mM, pH 7) was used as a control medium for all the conditions tested. All incubations were performed in triplicate. Control experiments, using NADES solutions without additional incubation time (considered as 0 h), were performed for all the systems, as well as for PBS.2.5. Impact of water activity on enzymatic activity
Water is usually used to reduce the viscosity of a NADES; however, it can play a key role in protein hydration shell dynamics, modifying the enzyme activity. To evaluate the impact of the water activity of BGly on HRP activity, the enzyme was suspended in the BGly formulations with different water contents (5–20 wt%) and incubated at 37 °C, for 6 h and for 24 h. As described before, after each incubation time, PBS (100 mM, pH 7) was added to the incubated mixtures and the enzyme's activity was assessed using the enzymatic activity assay described before.2.6. Circular dichroism
Circular dichroism (CD) spectra were obtained between 190 and 250 nm using a Chirascan qCD spectrometer, from Applied Photophysics, provided with a Quantum Northwest TC125 temperature controller. After incubation under the conditions described before, HRP and the NADES were dissolved in PBS (100 mM, pH 7) to achieve a final concentration of 5 μM and 5 wt%, respectively. Spectra were obtained at 25 °C, using a 0.1 mm path length. The secondary structure contents were calculated using the Provencher & Glockner method, with reference data set SP175,37 in the DICROWEB web server (https://dichroweb.cryst.bbk.ac.uk).2.7. Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out to gain insights into the conformational and solvation effects of NADESs on HRP. The simulation was based on the X-ray structure of HRP C1A (PDB code: 1HCH), and the protonation states of the histidine residues were assigned using the PDB2PQR service.38 HRP was described with the CHARMM36m force field for proteins in conjunction with a modified TIP3P water model.39 Betaine was modelled with the force field developed by Ganguly et al., where the protein–betaine van der Waals interactions are scaled by 0.75.40 All the sugars and polyols were modelled with the ADD force field,41 which improved the description of interactions with proteins using the KBP force field.42Initial configurations were prepared using PACKMOL in a cubic box.43 One HRP structure was placed in the center and was solvated with an appropriate number of solvent molecules for each system (see Table S1†). For a fair comparison, the number of water molecules was kept the same in all box simulations (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules). Additional Cl− ions were used as counter ions to neutralize the net charge of HRP, which was 4 in all simulations. For each box, energy was minimized using the steepest descent algorithm for 50
000 molecules). Additional Cl− ions were used as counter ions to neutralize the net charge of HRP, which was 4 in all simulations. For each box, energy was minimized using the steepest descent algorithm for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps and equilibration in the canonical NVT ensemble, with all the non-hydrogen atoms of HRP fixed. After this, an equilibration using the isothermal–isobaric NPT ensemble ran for 6 ns, where the last 1 ns ran without positions being restrained. Finally, the production in the NPT ensemble ran for 200 ns. Three replicates were run for each system using the previous procedure.
000 steps and equilibration in the canonical NVT ensemble, with all the non-hydrogen atoms of HRP fixed. After this, an equilibration using the isothermal–isobaric NPT ensemble ran for 6 ns, where the last 1 ns ran without positions being restrained. Finally, the production in the NPT ensemble ran for 200 ns. Three replicates were run for each system using the previous procedure.
All MD simulations were carried out using the GROMACS software package (version 2021.2).44,45 The system temperature was kept constant at 310 K using the velocity rescaling thermostat46 with a time constant of 0.2 ps. The pressure was maintained at 1.0 bar using the Parrinello–Rahman barostat47,48 with a time constant of 2 ps. van der Waals interactions were described with a cut-off radius of 1.2 nm with a smooth force switch to zero between 1.0 and 1.2 nm. Furthermore, electrostatic interactions were handled using the particle mesh Ewald (PME) method49 with an interpolation order of 4 and a cut-off of 1.2 nm. The protein–solvent minimum-distance distribution functions (MDDFs) and the Kirkwood–Buff integral (KBI) were computed using the open-source software “ComplexMixtures.jl”.31 The root-mean-square deviation of atomic positions (RMSDCα), the radius of gyration (Rg), solvent-accessible surface areas (SASAs), and the number of hydrogen bonds were obtained directly from GROMACS.
3. Results and discussion
3.1. HRP activity after incubation at 37 °C
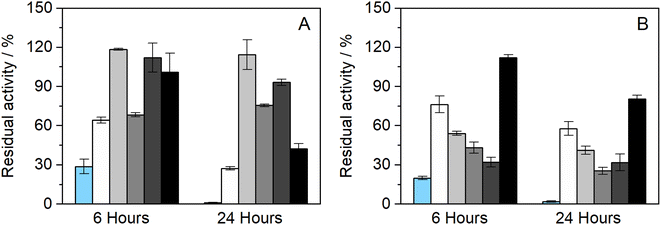
The influence of the presence of betaine-NADESs on the enzyme activity (i.e., the initial reaction rate) was assessed prior to the stability studies as shown in Fig. 1(A). For this purpose, HRP was suspended in NADESs and immediately dissolved in PBS (100 mM, pH 7). It was observed that, with the exception of the system BXylW, the presence of all the systems tested increased the reaction rate, indicating an increase in enzymatic activity. The increase in enzymatic activity, compared to PBS (100 mM, pH 7), was more pronounced in the presence of BSorbW, with enzymatic activity increasing by 2-fold, as represented in Fig. 1(A).34 These results, obtained from a previous study, were also used to set the standard for all the assays performed.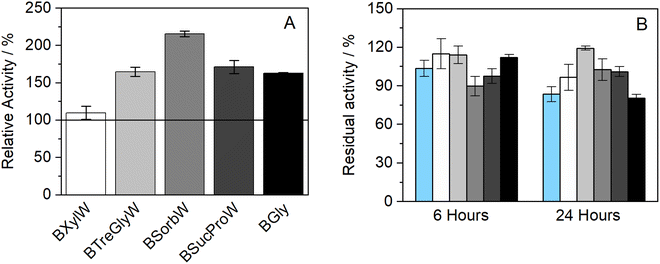 | ||
| Fig. 1 (A) Relative enzymatic activity of HRP at t = 0 h (n = 3, retrieved from ref. 34); (B) residual enzymatic activity of HRP after incubation for 6 and 24 h at 37 °C in BTrehGlyW (light gray), BSucProW (dark gray), BSorbW (gray), BGly (black), BXylW (white) and PBS (light blue) (100 mM, pH 7), (n = 3). | ||
The impact of the presence of NADESs on enzymatic activity was also evaluated upon incubation in a pure NADES and PBS, which was used as the control. Two incubation-time points were tested for a temperature of 37 °C, namely 6 h and 24 h. The activity under these conditions was determined using the same activity protocol as described before and then reported as “residual activity”, which was normalized to the value obtained at the start of the measurements (t = 0). The results obtained are illustrated in Fig. 1(B). Overall, all the systems tested, including PBS, kept HRP active throughout the 24 h of incubation. From Fig. 1(B) it is also possible to observe that PBS and BGly caused a decrease of about 20% compared to initial activity, while BSorbW, BSucProW, and BXylW maintained it close to the initial 100%, and BTrehGlyW increased it by 20% of the HRP's activity, after 6 h of incubation, which was maintained until 24 h. The reasons leading to the maintenance/increase in HRP's activity will be further discussed in combination with the other results.
3.2. Thermostability of HRP in the presence of NADESs
Further studies were performed to understand if the chosen NADES could also impact the enzyme's thermostability over longer periods. Therefore, to assess the thermostability under thermal stress, HRP was incubated in the pure NADES at two different temperatures, 60 and 80 °C.The results obtained from the incubation at 60 °C (for 4 and 24 h) can be observed in Fig. 2(A). When exposed to this temperature, the differences in enzymatic activity are more noticeable than at 37 °C (see Fig. 1(B)). It was possible to observe that two of the tested NADESs maintained high enzymatic activity, namely, BTrehGlyW and BSucProW. In particular, in the presence of BSorbW there was a decrease in activity of ca. 25%, compared to the initial one, after 24 h of incubation in those systems. When incubated in the systems BGly and BXylW, the enzymatic activity was reduced to 42% and 27%, respectively. After incubation for 24 h at 60 °C in control media, enzymatic activity was reduced to residual values. Although HRP's activity was reduced in the presence of some NADESs, it was always higher than that in the presence of PBS alone.
Furthermore, the thermostability of HRP at 80 °C was also evaluated. In this case, the incubation time points studied were 2 and 4 h. As shown in Fig. 2(B), the enzymatic activity decreased with the incubation in all the media tested; however, the impact of incubation temperature on enzyme activity was not the same for all NADESs. Since the system BXylW had the lowest performance in the assays at 60 °C, it was discarded for the tests at 80 °C. The system with the best thermal stability on HRP was BTrehGlyW, in which the enzyme maintained ca. 60% of its initial activity after 4 h of incubation at 80 °C. When incubated in BSorbW for 4 h, the enzyme maintained 41% of its initial activity. BSucProW and BGly showed thermal stability decreasing to 30% of the initial value. The complete loss of enzyme activity was observed only in the PBS systems, in 24 h for systems incubated at 60 °C and in 4 h for systems incubated at 80 °C.
Betaine-based systems had already proven their thermal protective behaviour at high temperatures (80 and 90 °C) in other enzymes, such as laccase.50 Nonetheless, it was observed that among betaine-based DESs the results have wide variability and are dependent on their composition. Some researchers have studied the effect of sugars on the activity of HRP and observed that in systems with equal viscosity, sugars such as trehalose, glucose, and sorbitol had less impact on the enzymatic activity of HRP.51,52 In a recent study from our group, it was observed that the presence of betaine-based NADESs caused structural changes to HRP that were linked to its activity and denaturation profiles.34 It was also observed that the presence of betaine-based NADESs increased the unfolding temperature of HRP, with the highest increase observed for BSorbW. Moreover, protein aggregation was also observed in the presence of NADESs, which did not happen in the control buffer, showing the thermal protective effect of these NADESs towards HRP.34
3.3. Structural studies of HRP incubated in NADESs
As it has been discussed, the incubation of HRP in NADESs affects not only the enzyme activity, but also its thermostability. In order to understand the effect of NADESs on the enzyme structure, circular dichroism (CD) analysis was performed to determine alterations in the secondary structure of HRP. The CD analysis was performed at the final incubation time point, for each temperature, 24 h at 37 and 60 °C and 4 h at 80 °C. It had been previously determined that native HRP, dissolved in PBS (100 mM, pH 7) had the following secondary structure content: 31% α-helix, 9% β-sheet, 16% turns and 44% random coils.34Fig. 3 shows the results obtained for the secondary structure content after incubation in PBS and the five NADESs studied, at different temperatures. Overall, the results correlate with the enzyme activity. In general, in all the systems tested, including the control in PBS, the random coils (∼43%) and turns' (∼16%) content did not have significant changes, with some exceptions. The major changes in the secondary structure were observed between the α-helix and β-sheet contents. It is known that the catalytic activity of HRP is closely related to and promoted by higher contents of the α-helix structure.6 Solvents that promote this structure over the β-sheet have proven to be the ones in which the enzyme has higher activity rates, since they allow HRP to have a more relaxed tertiary structure.6,34
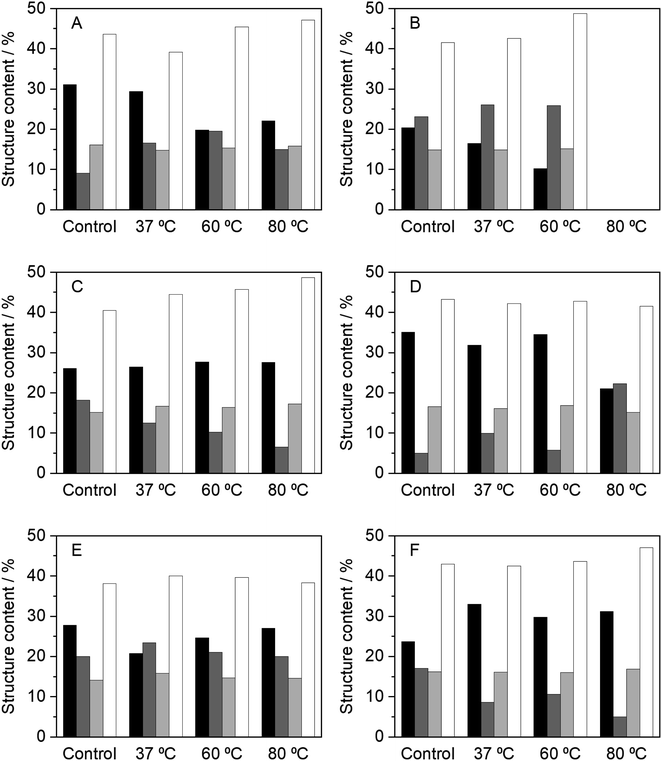 | ||
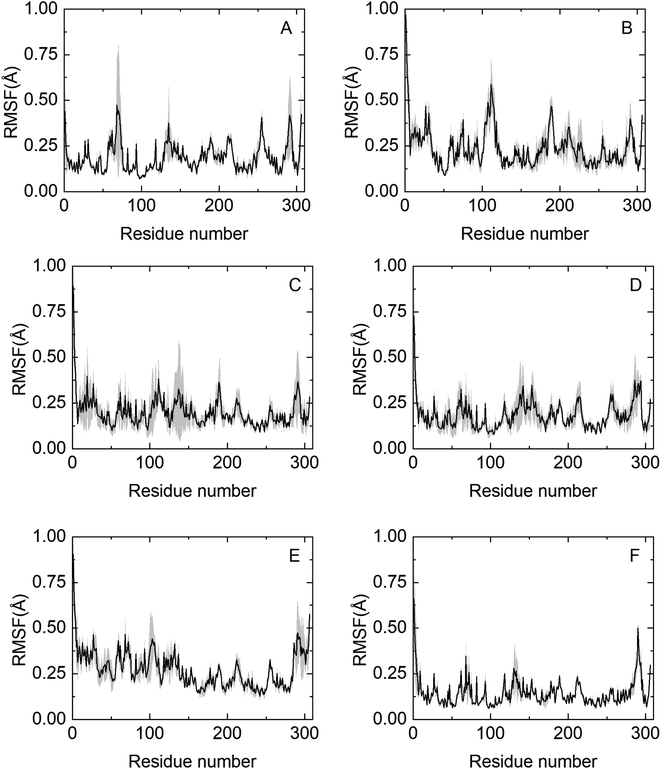
| Fig. 3 Relative content of the secondary structures of HRP, determined by CD, after 24 h of incubation, at 37 and 60 °C, and 4 h at 80 °C, in the different media tested, namely, (A) PBS (100 mM, pH 7), (B) BXylW, (C) BTrehGlyW, (D) BSorbW, (E) BSucProW and (F) BGly. α-Helix (black), β-sheet (dark gray), turns (light gray) and random coils (white). The control in each system represents the secondary structure content of HRP in the respective media, at initial time, without any incubation. These values are reported in Table S2.† | ||
Incubation in PBS at high temperatures resulted in a complete loss of enzymatic activity. It was expected that this loss represented a total loss of the secondary structure of the enzyme; however, the CD results (Fig. 3(A)) show that there was a decrease in the α-helix content at 60 and 80 °C, concomitant with an increase of the β-sheet, which correlates to the loss in activity. The same behaviour was observed for BXylW (Fig. 3(B)), which was the NADES with a less thermal protective effect on HRP.
In contrast, BTrehGlyW was the system in which HRP presented the highest thermostability. After incubation in this system (Fig. 3(C)), the secondary structure content shows that the α-helix content was stable at all the incubation temperatures tested, which correlates with higher activity. In BSorbW, the initial secondary structure content was different from the native enzyme and promoted a faster reaction rate, as discussed before.34 Incubation in this NADES highly promoted the presence of α-helix (Fig. 3(D)); however, this conformation did not provide increased protection at a higher temperature. After incubation in BSucProW (Fig. 3(E)), the presence of a β-sheet was favoured without causing a major decrease in the α-helix content; however, this was also reflected in the activity of the enzyme, which decreased. Finally, the results were very surprising when the enzyme was incubated in BGly. Fig. 3(F) shows that upon incubation at the three temperatures tested, there was an increase in α-helix with a corresponding decrease in the β-sheet content. However, in this system this did not correlate with an increase in activity, and this might be due to the fact that besides the structure, the hydration level of the environment i.e. the hydrogen bond network between protein–water and NADES–water influences enzyme activity not monotonically.14,53 In this case the system BGly presented a very low water content and water activity, which may be related to the lower activity in this system.
3.4. Influence of aw on HRP's activity
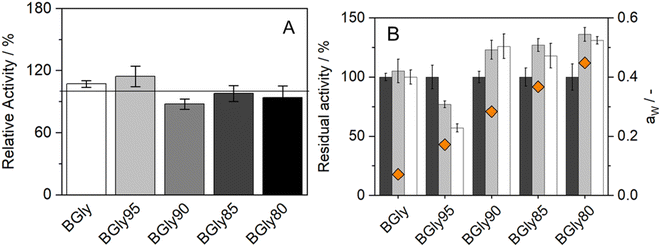
The impact of water activity (aw) on the enzymatic activity of HRP was also studied since this is one of the factors that highly influence enzymatic activity.51 For this purpose, the system BGly was used since it had the lowest water content (1.7 wt%, from residual water in the initial components). Table 2 shows the exact water content (wt%) and water activity (aw) of the BGly dilutions, prepared by adding water to the original NADES, finally yielding 4 different mixtures with different water activities. We observed that each 5 wt% increase in the water content caused an increase of ca. 0.1 in the water activity. Increasing the water content of BGly to 21.1 wt% allowed an aw of 0.448 to be achieved, which is close to the aw of the other systems used in this study,34 making the NADES comparable under nearly isoactivity conditions of water.| Formulation | Water added (wt%) | Water content (wt%) | a w, 37 °C |
|---|---|---|---|
| BGly | 0% | 1.7 ± 0.0 | 0.071 ± 0.004 |
| BGly95 | 5% | 5.8 ± 0.1 | 0.172 ± 0.001 |
| BGly90 | 10% | 11.5 ± 0.1 | 0.283 ± 0.003 |
| BGly85 | 15% | 16.9 ± 0.3 | 0.367 ± 0.001 |
| BGly80 | 20% | 21.1 ± 0.2 | 0.448 ± 0.000 |
The influence of aw on enzymatic activity was evaluated after incubation in each of the BGly formulations for 6 and 24 h, at 37 °C. The same procedure was used without any incubation time to determine the influence of the systems on the enzymatic activity shown in Fig. 4(A). It is important to mention that the relative activity obtained from BGly is different than the one presented in Fig. 1(A), since in that work a different enzyme batch was used.34 Based on our experience with enzymatic reactions in NADESs, we expected that increasing aw would increase the enzymatic activity; however, the results obtained were quite different. The influence of NADESs on the reaction rate is presented in Fig. 4(A), and it is noticeable that without incubation, i.e., time for the enzyme to adapt to the environment, the activity increases for BGly with 5 wt% of water, being lower for the remaining 3 water contents. However, Fig. 4(B) shows the results of activity after incubation in BGly, and it is visible that HRP's activity was promoted at 10, 15, and 20 wt% water, and higher water contents indeed yielded higher enzymatic activities, as expected. Nevertheless, the incubation with BGly95 caused a decrease in enzymatic activity down to 60% from the control value. Hence, it is possible to observe that while the presence of NADESs has impact on the reaction rate, the incubation time allows the enzyme to achieve a conformation that is more favourable for its catalytic activity. Possible explanations to this (hydrogen bond network disruption/reorganization induced by the water addition into dry BGly) are given later by MD simulations.
3.5. Protein flexibility upon NADES addition
To obtain molecular insights into NADES-induced enzyme stabilization, MD simulations were performed, primarily focused on the interactions between solvents and the surface of HRP at the nanosecond timescale. An extensive summary of relevant properties, calculated from the MD simulations, is shown in Table S3.† The effect of co-solvent on the dynamics of HRP was monitored by calculating the root mean square fluctuation (RMSF) of the Cα atoms of all 306 residues.The RMSF profile is overall similar for the NADESs used in this work, except for three highly flexible regions: the C-terminus and the N-terminus, where the RMSF values are noticeably high, reflecting the flexible nature of HRP in these regions. Secondly, the helix α5 region (residues 77–90) shows a remarkable decrease in the RMSF values as compared to native folds, as shown in Fig. 5. Helix α5 is close to the active site (residues 38–43), and its stabilization could contribute to maintaining a closed HRP conformation. A special flexibility suppression of Helix α5 is observed upon the addition of catalytic booster NADESs such as BSorb and BSucPro (Fig. 5(C) and (F)). Unlike the other NADESs, BXyl (Fig. 5(E)) shows a clear increment in the region of the residues 1–100 compared with the PBS system.
HRP remains folded in all the NADESs used in this work, as depicted by the root mean square deviation (RMSDCα), which indicates that Cα atoms coordinated with the equilibrated crystal structure as a reference, shown in Fig. S1.† A small RMSDCα (as for BSorbW) value implies that HRP stability of the folded state is promoted, reducing the conformational stability as shown in Fig. S1.†54 In an analogous fashion, the distance between the centers of mass of the active pocket residues, ARG-38 and HIS-42, is reduced in NADESs that are more catalytically active (i.e. BSorbW). Additionally, Fig. S1† shows the radius of gyration (Rg), which hints at the protein compactness in the NADES environment. The number of hydrogen bonds between HRP and water was computed for all systems as shown in Table S3.† The number of HRP–water hydrogen bonds is reduced in all NADESs, which might promote the experimentally observed increase in the enzymatic activity compared to PBS. One exception is BXylW, which promotes a higher density of water around the HRP surface due to a stronger protein volume-exclusion effect. Strong hydrogen bond donors such as sorbitol, sucrose, or glycerol compete with betaine and water for HRP surface association sites, displacing HRP hydrogen bonding sites. The direct interaction of NADES donor groups with the surface of HRP excludes water from the second shell solvation, thereby decreasing HRP–water hydrogen bonds. This type of stabilization, by the exclusion of water, is characteristic of the solvation of uncharged polar residues on the protein surface.54
3.6. Surface area and Gibbs energy of solvation of HRP
To explore the HRP–solvent (water or a NADES or water + NADES) dynamics, the solvent-accessible surface area (SASA) was studied. It is known that improved protein folded state stability of some enzymes appears to be related to reduced surface area, increased hydrophobicity and protein core-packed density.55Fig. 6(A) shows the total SASA and Fig. 6(B) shows Gibbs solvation energy calculated with the Eisenberg & McLachlan method of HRP in the NADES used in this work.56 A reduced SASA could be caused by a decrease in the volume of the substrate pocket, thus lowering protein flexibility and promoting catalytic activity. It is generally accepted that protein folded state stability then improves because a small surface area reduces the unfavourable surface energy and increases the attractive interior packing interactions. A low SASA (i.e. BSucProW) value suggests NADES component accumulations over the HRP surface, promoting thermostability and an increase in the enzymatic activity; meanwhile the SASA value is the highest in BXylW, the system with lower activity and thermostability as observed experimentally, suggesting an accumulation of water on the surface of HRP. This is in agreement with the results on hydrogen bonding in the previous section. Ultimately, the Gibbs solvation energy calculated from the SASA strongly correlates to the enzymatic activity discussed in this paper and previously reported work on the discussed NADESs.33 That is, low Gibbs energy of solvation values (i.e. BXylW and PBS) correlates with the lowest enzymatic activity obtained experimentally.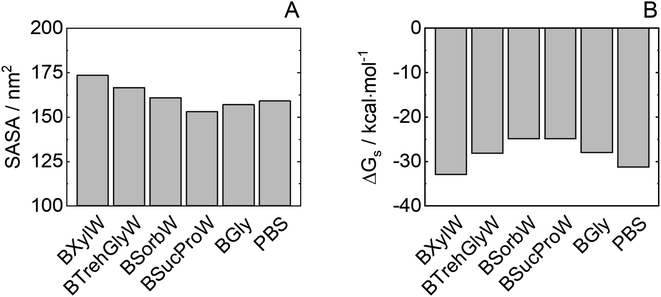 | ||
| Fig. 6 Protein (A) SASA and (B) Gibbs energy of solvation of HRP in NADESs from molecular dynamics. Average values from the last 50 ns of simulations and standard errors computed from 3 independent runs are shown in Table S3.† | ||
Although the accumulation of water molecules on the protein surface does not necessarily mean a decrease in enzymatic activity, it can cause the reorientation of the hydrophobic and hydrophilic side chains. Thus, the kind of interaction of NADESs with specific residues on the protein surface is responsible to promote or decrease the non-monotonic dynamics of water molecules in the protein active site. In the case of sorbitol, hydrophobic interactions around basic HRP residues (e.g., arginine) are encouraged; this could improve folded state protein stability due to the highly hydrophobic reaction pocket of wild-type HRP.57 Furthermore, an important correlation between HRP activity and Gibbs energy of solvation of hydrophobic residues can be observed for NADESs as shown in Fig. S2.† In contrast, xylitol interacts with the protein around non-polar aromatic residues, increasing water density on the HRP surface and increasing solvent-accessible surface area.
3.7. Protein solvation – MDDF and KBI
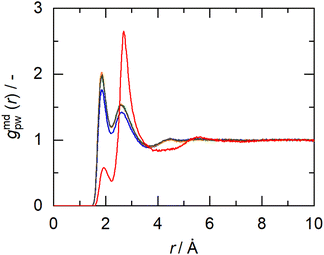
In this work, the minimum-distance distribution functions (MDDFs) were used as a practical way to obtain insights into the interaction between a solute with complex geometry (e.g., a protein surface) and a solvent (water or NADES components). Fig. 7 shows the MDDFs for HRP–water, reaffirming what was previously stated, showing a water accumulation in the first solvation shell due to the volume-exclusion effect, except for BSucProW where the water is repelled from the surface into larger distances. Fig. S3† displays the variation in the local density of water around the active pocket of HRP in the NADES. An accumulation of water can be observed in the protein solvation shell (at ∼1.8–2.5 Å), especially in the NADESs that allowed high enzymatic activity (i.e., BSorbW and BSucProW, cf.Fig. 1). This increase in water density occurs at hydrogen-bonding distances and at the second water layer (at ∼2.5–3.5 Å), most notably around residues ARG38, and HIS42. Interestingly, no significant changes were observed for BXylW compared to the PBS control, although BXylW displaces the most water to the surface (Fig. S3†). The MDDFs for all the systems are shown in Fig. S4 and S5.†To reflect the volume-exclusion effect associated with protein–solvent interactions at short distances, the KBI profiles were computed for the systems in this work according to eqn (1),
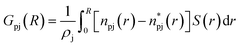 | (1) |
 is the number of minimum distances that would be observed at r if there were no solute–solvent interactions, and ρj is the molar density of the solvent j in the bulk. The KBI profiles determine whether there is excess or exclusion of each NADES constituent around the protein. At short distances, r < 1.5 Å, the KBIs drop to very negative values, which is caused by the excluded volume of protein and the solvent molecules; this could be compensated at long distances by favourable HRP–solvent interactions. As shown in Fig. S7,† the accumulation of water and betaine in the solvation shells offsets only a part of the exclusion volume. For most of the NADES' constituents, the KBI increases for r > 1.5 Å, but converges into negative values at long distances. A lower value of KBI indicates that the NADES constituents accumulate more effectively on the protein second solvation shell, promoting water molecules into the first solvation shell. In contrast, DL-proline accumulates in the first solvation shell, solvating the protein surface, as depicted by a prominent peak of proline at short distances in Fig. S5(E),† at r ∼ 3.0 Å.
is the number of minimum distances that would be observed at r if there were no solute–solvent interactions, and ρj is the molar density of the solvent j in the bulk. The KBI profiles determine whether there is excess or exclusion of each NADES constituent around the protein. At short distances, r < 1.5 Å, the KBIs drop to very negative values, which is caused by the excluded volume of protein and the solvent molecules; this could be compensated at long distances by favourable HRP–solvent interactions. As shown in Fig. S7,† the accumulation of water and betaine in the solvation shells offsets only a part of the exclusion volume. For most of the NADES' constituents, the KBI increases for r > 1.5 Å, but converges into negative values at long distances. A lower value of KBI indicates that the NADES constituents accumulate more effectively on the protein second solvation shell, promoting water molecules into the first solvation shell. In contrast, DL-proline accumulates in the first solvation shell, solvating the protein surface, as depicted by a prominent peak of proline at short distances in Fig. S5(E),† at r ∼ 3.0 Å.
Once the Gpc values have been computed, they were used to calculate the preferential hydration parameters according to eqn (2), usually used to correlate protein stability under the effect of a solvent.
| Γpc(R) ≈ ρc[Gpc(R) − Gpw(R)] | (2) |
3.8. Gibbs energy of solvation of HRP in NADES + water addition
It was shown that adding 5 wt% water to BGly (i.e., BGly95) causes a dramatic loss in enzymatic activity of HRP. The conditions of water content in BGly according to Table 2 were inherited also in MD simulations. As shown in Fig. S8,† 5 wt% water addition to BGly increases RMSDCα, thus promoting an increment in the HRP flexibility, and this correlates well with the drop that was experimentally found in enzymatic activity for BGly95 compared to BGly. Interestingly, an increase in the amount of protein–water hydrogen bonding (cf. Table S4†) and protein flexibility correlates with a decrease in enzymatic activity after incubation. This could be explained as a rearrangement of the hydrogen bond network due to the inclusion of water in the BGly system. After this disruption (>10 wt% water), probably caused by the competition between NADESs and HRP for the hydrogen bonds of the water, the BGly + water system seems to be stabilized in a re-organized structure, recovering the ability to prevent protein flexibility (decreasing values for RMSDCα and Rg). The water organization between the solvation shell and the bulk might directly affect the stabilization of the transition state.37 The same trend is observed for Gibbs energy of solvation of HRP in Fig. 8, whereupon water addition, the reorganization of the hydrogen bond network causes an increase in the SASA (Fig. S9†) and, consequently, a decrease in Gibbs energy of solvation. In a further step, we distinguished between hydrophilic and hydrophobic Gibbs energies of solvation based on the nature of the residues involved in the calculation. Particularly interesting is the prominent reduction in hydrophobic solvation energy shown in Fig. 8(B), which may be responsible for the inability of BGly95 to maintain enzyme stability, which is compensated for at higher water contents with a decrease in hydrophilic solvation energy. As shown in Fig. S6 and S8,† no significant changes in MDDFs are found in KBIs due to the similarity of the systems.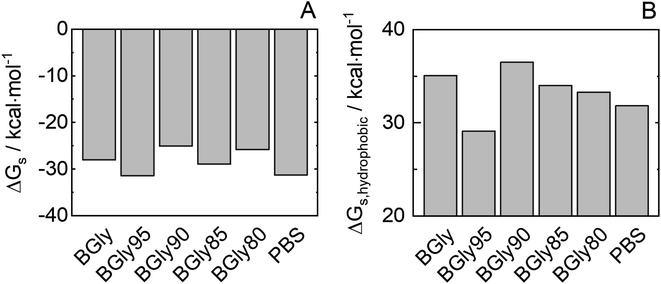 | ||
| Fig. 8 (A) Estimated Gibb energy of solvation of total HRP (ΔGs) and (B) of only the hydrophobic HRP groups for BGly + water used in this work. | ||
4. Conclusion
This work was developed as a proof-of-concept for enzyme stability without using choline-based compounds, which were the most used in the studies with DESs and enzymes in the past. The disadvantageous effect of choline-based DESs on HRP activity was overcome by using NADESs, in order to increase the scarce data and partly contradictory data on HRP. In this contribution, the folded-state HRP stability in buffer and in NADESs at moderate to high temperatures was evaluated. Our results show that it was possible to improve the activity of HRP in the presence of NADESs, especially BSorBW and BTrehGlyW. Moreover, HRP's activity was not impaired after 24 h incubation in BTrehGlyW at 60 °C, contrary to the results obtained for PBS and for all other NADESs under study. The system BTrehGlyW has also shown the best performance in protecting the enzyme during incubation at 80 °C. The structural studies conducted by CD analysis have shown that NADESs that improve enzyme activity the most facilitate higher contents of the α-helix structure in HRP. Besides thermostability, the effect of water activity on enzymatic activity was studied using the system BGly. The overall results have shown that increasing the water activity leads to an increase in enzymatic activity of HRP, but this advantage drops at a certain critical amount of water in BGly, namely at about 5 wt% water.Molecular dynamics simulations accompanied the experimental results. It was shown that the NADESs reduced the flexibility of HRP, leading to a greater stability of the active site. This is promoted by the water exclusion effect that is caused due to the interactions between NADESs and the surface of HRP. A correlation between enzyme activity and Gibbs energy of solvation was found, especially if the calculation breaks down into hydrophobic residues. Thus, predicting solvation energies allows estimating the beneficial effect of NADESs on HRP enzymatic activity. Furthermore, protein–solvent interactions were analysed using minimum distance functions and Kirkwood–Buff integrals, demonstrating that the interaction between the NADESs and the protein surface generates an increase in water on the protein surface, which can help stabilize the active site. Extrapolating these findings to more relevant and sensitive molecules will open new possibilities for biotechnological industries in the future.
Author contributions
L. M. – conceptualization, data curation, investigation, methodology, writing – original draft; N. F. G. P. – data curation, formal analysis, investigation, writing – original draft; E. C. K. – data curation; J. M. G. – supervision; C. H. – supervision, funding acquisition, writing – review and editing; A. R. C. D. – supervision, funding acquisition, writing – review and editing; A. P. – supervision, funding acquisition, writing – review and editing.Conflicts of interest
The authors declare no conflicts of interests.Acknowledgements
The authors are thankful for the funding from the European Union Horizon 2020 Programme under grant agreement number ERC-2016-CoG 725034 (ERC Consolidator Grant Des.solve). The authors also thank Laboratory for Green Chemistry – LAQV (UIDB/50006/2020), project CryoDES (PTDC/EQU-EQU/29851/2017) and the grant SFRH/BD/148510/2019 financed by national funds from FCT/MCTES. The authors thank the funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2033 – 390677874 – RESOLV. Nicolás Gajardo's work was supported by the German Academic Exchange Service (DAAD) under the Graduate School Scholarship Programme, 2020 (57516591). E. C.-K. acknowledges the scholarship from ANID, Chile. The authors wish to thank Elisabete Ferreira from the BioLab, supported by the Applied Molecular Biosciences Research Unit – UCIBIO and the Associated Laboratory for the Green Chemistry Research Unit – LAQV (UIDP/04378/2020, UIDB/04378/2020, UIDB/50006/2020, and UIDP/50006/2020, respectively). This research was partially supported by the supercomputing infrastructure of the Southern GPU Cluster – Fondequip EQM150134.References
- F. W. Krainer and A. Glieder, Appl. Microbiol. Biotechnol., 2015, 99, 1611–1625 CrossRef CAS PubMed.
- S. Saghati, A. B. Khoshfetrat, H. Tayefi Nasrabadi, L. Roshangar and R. Rahbarghazi, BMC Res. Notes, 2021, 14, 1–7 CrossRef PubMed.
- N. C. Veitch, Phytochemistry, 2004, 65, 249–259 CrossRef CAS PubMed.
- M. Gajhede, D. J. Schuller, A. Henriksen, A. T. Smith and T. L. Poulos, Nat. Struct. Biol., 1997, 4, 1032–1038 CrossRef CAS PubMed.
- K. G. Welinder, FEBS Lett., 1976, 72, 19–23 CrossRef CAS PubMed.
- B. P. Wu, Q. Wen, H. Xu and Z. Yang, J. Mol. Catal. B: Enzym., 2014, 101, 101–107 CrossRef CAS.
- B. W. Park, K. A. Ko, D. Y. Yoon and D. S. Kim, Enzyme Microb. Technol., 2012, 51, 81–85 CrossRef CAS PubMed.
- J. Chapman, A. E. Ismail and C. Z. Dinu, Catalysts, 2018, 8, 20–29 CrossRef.
- A. Basso and S. Serban, Mol. Catal., 2019, 479, 110607 CrossRef CAS.
- P. Domínguez de María and Z. Maugeri, Curr. Opin. Chem. Biol., 2011, 15, 220–225 CrossRef PubMed.
- P. Domínguez De María, Angew. Chem., Int. Ed., 2008, 47, 6960–6968 CrossRef PubMed.
- A. P. Abbott, S. S. M. Alabdullah, A. Y. M. Al-Murshedi and K. S. Ryder, Faraday Discuss., 2018, 206, 365–377 RSC.
- Y. H. Choi, J. van Spronsen, Y. Dai, M. Verberne, F. Hollmann, I. W. C. E. Arends, G.-J. Witkamp and R. Verpoorte, Plant Physiol., 2011, 156, 1701–1705 CrossRef CAS PubMed.
- B. D. Belviso, F. M. Perna, B. Carrozzini, M. Trotta, V. Capriati and R. Caliandro, ACS Sustainable Chem. Eng., 2021, 9, 8435–8449 CrossRef CAS.
- V. Gotor-Fernández and C. E. Paul, J. Biotechnol., 2019, 293, 24–35 CrossRef PubMed.
- M. L. Toledo, M. M. Pereira, M. G. Freire, J. P. A. Silva, J. A. P. Coutinho and A. P. M. Tavares, ACS Sustainable Chem. Eng., 2019, 7, 11806–11814 CrossRef CAS.
- A. A. Papadopoulou, E. Efstathiadou, M. Patila, A. C. Polydera and H. Stamatis, Ind. Eng. Chem. Res., 2016, 55, 5145–5151 CrossRef CAS.
- G. M. Kontogeorgis, R. Dohrn, I. G. Economou, J. C. De Hemptinne, A. Kate, S. Kuitunen, M. Mooijer, L. F. Zilnik and V. Vesovic, Ind. Eng. Chem. Res., 2021, 60, 4987–5013 CrossRef CAS PubMed.
- P. Braiuca, C. Ebert, A. Basso, P. Linda and L. Gardossi, Trends Biotechnol., 2006, 24, 419–425 CrossRef CAS PubMed.
- N. F. Gajardo-Parra, H. Akrofi-Mantey, M. Ascani, E. Cea-Klapp, J. M. Garrido, G. Sadowski and C. Held, Phys. Chem. Chem. Phys., 2022, 24, 27930–27939 RSC.
- M. Knierbein, A. Wangler, T. Q. Luong, R. Winter, C. Held and G. Sadowski, Phys. Chem. Chem. Phys., 2019, 21, 22224–22229 RSC.
- X. Tadeo, B. López-Méndez, D. Castaño, T. Trigueros and O. Millet, Biophys. J., 2009, 97, 2595–2603 CrossRef CAS PubMed.
- L. Vrbka, P. Jungwirth, P. Bauduin, D. Touraud and W. Kunz, J. Phys. Chem. B, 2006, 110, 7036–7043 CrossRef CAS PubMed.
- M. Filizola and G. H. Loew, J. Am. Chem. Soc., 2000, 122, 18–25 CrossRef CAS.
- E. Derat, S. Shaik, C. Rovira, P. Vidossich and M. Alfonso-Prieto, J. Am. Chem. Soc., 2007, 129, 6346–6347 CrossRef CAS PubMed.
- H. A. Heering, A. T. Smith and G. Smulevich, Biochem. J., 2002, 363, 571–579 CrossRef CAS PubMed.
- B. Nian, C. Cao and Y. Liu, J. Chem. Technol. Biotechnol., 2020, 95, 86–93 CrossRef CAS.
- M. Shehata, A. Unlu, U. Sezerman and E. Timucin, J. Phys. Chem. B, 2020, 124, 8801–8810 CrossRef CAS PubMed.
- P. Kumari, M. Kumari and H. K. Kashyap, J. Phys. Chem. B, 2020, 124, 11919–11927 CrossRef CAS PubMed.
- J. P. Bittner, N. Zhang, L. Huang, P. Domínguez De María, S. Jakobtorweihen and S. Kara, Green Chem., 2022, 24, 1120–1131 RSC.
- L. Martínez, J. Mol. Liq., 2022, 347, 12–17 CrossRef.
- V. Piccoli and L. Martínez, J. Mol. Liq., 2022, 363, 119953 CrossRef CAS.
- I. P. De Oliveira and L. Martínez, Phys. Chem. Chem. Phys., 2019, 22, 354–367 RSC.
- N. F. Gajardo-Parra, L. Meneses, A. R. C. Duarte, A. Paiva and C. Held, ACS Sustainable Chem. Eng., 2022, 10, 12873–12881 CrossRef CAS PubMed.
- Q. Zaib, M. J. Eckelman, Y. Yang and D. Kyung, Green Chem., 2022, 24, 7924–7930 RSC.
- L. Meneses, F. Santos, A. R. Gameiro, A. Paiva and A. R. C. Duarte, J. Visualized Exp., 2019, 2019, 1–5 Search PubMed.
- J. G. Lees, A. J. Miles, F. Wien and B. A. Wallace, Bioinformatics, 2006, 22, 1955–1962 CrossRef CAS PubMed.
- T. J. Dolinsky, J. E. Nielsen, J. A. McCammon and N. A. Baker, Nucleic Acids Res., 2004, 32, 665–667 CrossRef PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. de Groot, H. Grubmüller and A. D. MacKerell Jr, Nat. Methods, 2017, 14, 71–73 CrossRef CAS PubMed.
- P. Ganguly, D. Bubák, J. Polák, P. Fagan, M. Dračínský, N. F. A. Van Der Vegt, J. Heyda and J. E. Shea, J. Phys. Chem. Lett., 2022, 13, 7980–7986 CrossRef CAS PubMed.
- A. Arsiccio, P. Ganguly, L. La Cortiglia, J. E. Shea and R. Pisano, J. Phys. Chem. B, 2020, 124, 7779–7790 CrossRef CAS PubMed.
- T. Cloutier, C. Sudrik, H. A. Sathish and B. L. Trout, J. Phys. Chem. B, 2018, 122, 9350–9360 CrossRef CAS PubMed.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30(13), 2157–2164 CrossRef PubMed.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindah, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- S. Nosé and M. L. Klein, Mol. Phys., 1983, 59, 1055–1076 CrossRef.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- S. Khodaverdian, B. Dabirmanesh, A. Heydari, E. Dashtban-moghadam, K. Khajeh and F. Ghazi, Int. J. Biol. Macromol., 2018, 107, 2574–2579 CrossRef CAS PubMed.
- L. Neri, P. Pittia, G. Bertolo, D. Torreggiani and G. Sacchetti, J. Food Eng., 2010, 101, 289–295 CrossRef CAS.
- L. Neri, P. Pittia, C. D. Di Mattia, G. Bertolo, D. Mastrocola and G. Sacchetti, Food Biophys., 2014, 9, 260–266 CrossRef.
- A. Sanchez-Fernandez, M. Basic, J. Xiang, S. Prevost, A. J. Jackson and C. Dicko, J. Am. Chem. Soc., 2022, 144, 23657–23667 CrossRef CAS PubMed.
- Q. Qiao, J. Shi and Q. Shao, Phys. Chem. Chem. Phys., 2021, 23, 23372–23379 RSC.
- F. H. Arnold, Curr. Opin. Biotechnol., 1993, 4, 450–455 CrossRef CAS PubMed.
- D. Eisenberg and A. D. McLachlan, Nature, 1986, 319, 199–203 CrossRef CAS PubMed.
- S. Tatoli, C. Zazza, N. Sanna, A. Palma and M. Aschi, Biophys. Chem., 2009, 141, 87–93 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2su00127f |
| This journal is © The Royal Society of Chemistry 2023 |