Open Access Article
Open Access ArticleTuning atomic-scale sites in metal–organic framework-based nanozymes for sensitive biosensing
Yating
Wen
a,
Weiqing
Xu
a,
Liuyong
Hu
b,
Miao
Xu
a,
Wenling
Gu
*a,
Hongcheng
Sun
 c and
Chengzhou
Zhu
c and
Chengzhou
Zhu
 *a
*a
aNational Key Laboratory of Green Pesticide, International Joint Research Center for Intelligent Biosensing Technology and Health, College of Chemistry, Central China Normal University, Wuhan, 430079, P.R. China. E-mail: wlgu@ccnu.edu.cn; czzhu@ccnu.edu.cn
bHubei Key Laboratory of Plasma Chemistry and Advanced Materials, Hubei Engineering Technology Research Center of Optoelectronic and New Energy Materials, Wuhan Institute of Technology, Wuhan 430205, P.R. China
cCollege of Material Chemistry and Chemical Engineering, Key Laboratory of Organosilicon Chemistry and Material Technology, Ministry of Education, Hangzhou Normal University, Hangzhou 311121, P. R. China
First published on 29th September 2023
Abstract
Nanozymes have gained significant attention within the scientific community due to their high enzyme-like activity, low cost, high stability, and ease of preparation. Among the various nanomaterials, metal–organic frameworks (MOFs) are a type of nanozyme with a unique composition and adjustable structures. By fine-tuning at the atomic level, the catalytic activity and selectivity of MOFs can be improved, thereby expanding their application range in the field of biosensing. Herein, we summarize the recent advances on the atomic-level design of MOFs and their derivatives for sensitive biosensing. First, tuning metal ions/cluster activities and organic ligands, constructing defects and introducing axial ligands are focused on optimizing the electronic and geometrical structure of atomic-scale sites. Specifically, the underlying enzyme-like catalytic mechanisms at the atomic scale are demonstrated. Then, MOF-based nanozymes with tuned atomic-scale sites for biosensing of small biomolecules, biomacromolecules and toxic pollutants are summarized. Finally, we provide a summary and outlook on the further refinement of MOFs and their expansion in the field of biosensing applications.
1. Introduction
Enzymes are efficient and selective biocatalysts that are commonly used in biosensing systems. However, most natural enzymes are isolated from living organisms, which is a costly, deactivation-prone, and difficult process.1–3 To address these limitations, researchers have utilized nanomaterials with enzyme-like activity,4 such as metal nanoparticles,5–8 quantum dots,9 graphene,10 nitride,11 carbon nanotubes12 and metal–organic frameworks (MOFs),13–15 to replace natural enzymes and develop nanozyme-based biosensing systems. Compared with natural enzymes, nanozymes have advantages such as low cost, simple preparation, good tunability, and stability,16–18 but they often exhibit unsatisfactory activity and selectivity in sensing applications.19,20 Nevertheless, precise atomic-level control of nanomaterials provides great opportunities to tune the microenvironment of the catalytic center, greatly enhancing the catalytic activity and selectivity of nanozymes and thus improving the performance of nanozyme-based biosensing.21–23MOFs are crystalline materials formed through the self-assembly of metal ions/clusters and organic ligands. The interaction between metal ions/clusters and organic ligands results in the unique framework structure of MOFs, providing them with a high pore volume and surface area.24–28 These materials have abundant metal sites, ordered structural frameworks, and high surface areas, making them potential substitutes for natural enzymes.29,30 Metal ions/clusters offer potential catalytic sites, while organic ligands have rich chemical functional groups or different configurations that can modulate the microenvironment of the catalytic center or anchor new metal active sites.31 Therefore, various strategies such as atomic-level control of metal ions/clusters, ligand modification, structural defect construction, and multi-component introduction have been reported for improving the catalytic activity and selectivity of MOFs, thereby constructing high-performance biosensors.20,32 Furthermore, MOF-derived atomically dispersed catalysts are a class of high-performance nanozymes with high atom-utilization efficiency and well-defined catalytic structures.33 Fine-tuning these catalysts at the atomic level by changing the central metal element, doping with heteroatoms, introducing multiple metal sites, constructing defects, and introducing axial ligands can provide infinite possibilities for constructing high-performance biosensors.34
In this review, we systematically introduce the latest advances on the atomic-level control of MOFs and their derivatives, and the enzyme-like catalytic mechanisms are highlighted. Then, sensitive detection of small biomolecules, biomolecules, and toxic pollutants are presented using MOF nanozymes and their derivatives. Finally, we discuss the application prospects of MOF nanozymes and their derivatives in biosensing.
2. Regulation of atomic-scale sites
2.1. Pristine MOFs
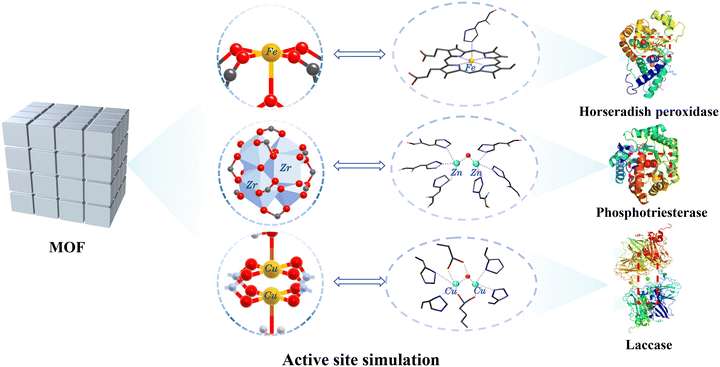
To enhance the catalytic activity and expand the application scope of MOFs, precise regulation of the metal nodes and organic ligands is necessary to achieve a customized MOF design.35 The regulation of metal nodes directly impacts the catalytic activity of MOFs, while the control and modification of ligands affect the activity and selectivity by influencing the microenvironment of catalytic centers, catalyst structure, and other factors.30 This regulatory approach of MOFs has found extensive applications in various fields, including catalysis, gas storage and separation, sensors, and drug delivery.36–39 This chapter will focus on the specific regulation of metal nodes and organic ligands.2.1.1.1. Monometallic MOFs. Metal ions/clusters serve as the catalytic activity center in MOFs, and the catalytic activity and selectivity of MOFs can be regulated by adjusting their type (Fig. 1). For example, Fe-based MOFs have received significant attention owing to their abundant iron sites, which resemble those of natural peroxidases. Numerous iron-based MOFs, such as MIL-100(Fe),42 NH2-MIL-101(Fe),43 NH2-MIL-88B(Fe),44 and PCN-600(Fe),45 have exhibited excellent peroxidase-like (POD-like) activities. In addition, the simulation of oxidase-like (OXD-like) performance by MOFs has also been extensively studied. In 2022, Wang et al. synthesized a Cu-MOF (HT-STAM-17-OEt) functionalized with histidine and tryptophan.46 The atomically dispersed Cu center in MOFs, combined with the ligand amino acids, mimics the catalytic center structure of ascorbate oxidase, imparting excellent OXD-like activity (Fig. 2a). Similarly, by doping with varying levels of cysteine, Co-based MOFs with different degrees of defects in the Co-MOF were reported.47 It was found that the balance between Co and N in the MOF played a critical role in regulating the OXD-like activity of the Co-based MOF with Co as the catalytic center. Moreover, Ce4+ and Zr2+ are frequently used to synthesize MOFs with hydrolase-like activities due to their strong Lewis acidity. For example, after incorporating imidazole and its derivatives into the pores of Zr-based-MOF-808 (Im@MOF-808), the resultant composites exhibit a similar active site and corresponding linked histidine residues of phosphotriesterase.48 The as-prepared Im@MOF-808 can hydrolyze nerve toxin analogues in pure water and a liquid-free humid environment (Fig. 2b). Likewise, Lewis acid catalysts based on cerium MOFs (Ce-FMA) with fumaric acid as the ligand also demonstrate efficient hydrolytic performance. The shorter chain length of the ligands increases the density of Lewis acid active sites in Ce-FMA, enabling the Ce-FMA to hydrolyze phosphate, amide, and glycosidic bonds simultaneously, even complex biological membranes, showing broad application prospects.49
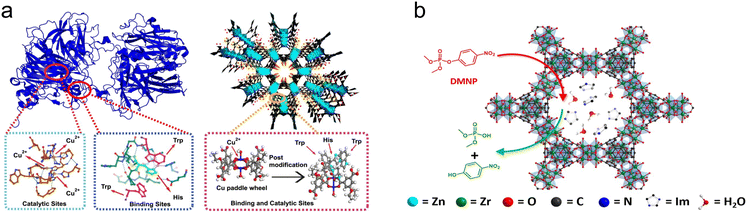 | ||
| Fig. 2 (a) Schematic illustration of the structure of natural ascorbate oxidase and HT-STAM-17-OEt.46 Copyright (2023) Springer Nature. (b) Schematic illustration of the hydrolysis process of nerve toxin analogues by Im@MOF-808.48 Copyright (2021) American Chemical Society. | ||
2.1.1.2. Heterometallic MOFs. Heterometallic MOFs primarily refer to a class of MOFs where the metal nodes that constitute the MOFs contain different metals. The synthesis of heterometallic MOFs can be achieved by incorporating different metal precursor species during the initial synthesis through a one-pot method, or by introducing heterometals into pre-synthesized monometallic MOFs. The introduction of heterometals into the metal nodes can enhance the catalytic activity and selectivity of MOFs, thereby broadening the scope of their enzyme-mimicking applications.42 Recently, Qu's group synthesized a multi-metal MOF (MOF-Au) by mixing Au and Fe precursors along with ligands using a one-pot hydrothermal method.50 Compared to mononuclear Fe-based MOFs, the multi-nuclear MOF-Au exhibited synergistic catalytic activity, resulting in superior POD-like performance. And then, a Ce complex with DNA hydrolytic capability was integrated with MOF-Au, endowing MOF-Au-Ce with double enzyme-like activity. The resultant MOF-Au-Ce demonstrated excellent bactericidal properties and prevented further proliferation, thereby expanding the application scope of MOFs as nanozymes. Similarly, inspired by the Cu/Zn bimetallic active site in the natural superoxide dismutase (SOD), MOF-818 with Cu/Zr bimetallic centers was reported51 (Fig. 3a). Compared to monometallic MOF mimics, MOF-818 with bimetallic active centers demonstrated higher SOD-like activity. It should be noted that the synergistic catalysis of the Cu/Zr bimetallic centers in MOF-818 is of great significance, where Zr-modulated Cu catalytic active sites reduce the energy barrier for the conversion between Cu(II) and Cu(I) and enhance the overall catalytic performance, similar to the catalytic mechanism of natural SOD.
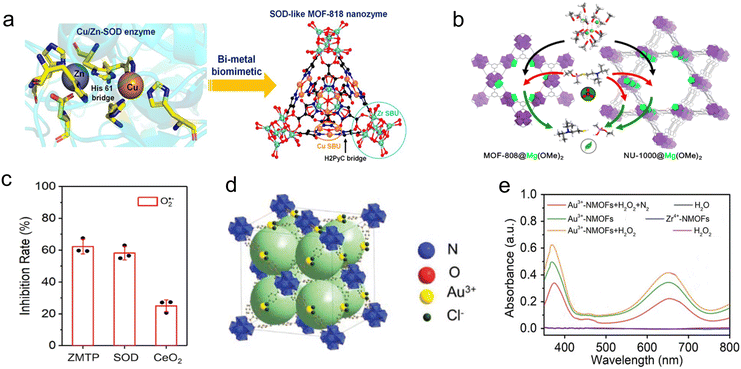 | ||
| Fig. 3 (a) Schematic illustration of the structure of SOD and MOF-818.51 Copyright (2022) American Chemical Society. (b) Schematic illustration of the detoxification of a nerve agent in MOF-808@Mg(OMe)2 and NU-1000@Mg(OMe)2.52 Copyright (2019) American Chemical Society. (c) Evaluation of SOD-like activity.55 Copyright (2022) Springer Nature. (d) Schematic illustration of the structure and (e) OXD and POD-like activity of Au3+-NMOFs.56 Copyright (2022) Wiley. | ||
Moreover, introducing heterogeneous metals into monometallic MOFs can also generate heterometallic MOFs. Zr-MOFs are commonly encountered MOFs known for their hydrolytic capabilities. To further enhance their hydrolytic performance and investigate the hydrolysis mechanism, Navarro's group employed a mixed-exchange approach by reacting a bulky reagent [Mg–(OMe)2(MeOH)2]4 with Zr-MOF. The resultant Zr-MOF@Mg(OMe)2 significantly enhanced the hydrolytic performance of the MOFs52 (Fig. 3b). Further investigations have revealed that the main factor contributing to this enhancement is the effective rise in material alkalinity brought about by the formation of secondary building units of MgZr5O2(OH)6. Meanwhile, the charge gradient generated by the heterometal doping facilitated the polarization of P–X bonds during the hydrolysis process. The combined effect of these two phenomena significantly enhanced the ability of Zr-MOFs to hydrolyze P–X (X = F, OR, SR).
2.1.2.1. Heterometallic MOFs. Organic ligands play a crucial role in MOFs as they not only act as connecting agents in the framework structure but can also incorporate metal sites with catalytic activity. Dispersed metal sites on the ligands enable MOFs to exhibit efficient catalytic performance. Typically, the one-pot method and the post-synthetic modification method are two common approaches to introduce metal active sites into organic ligands to obtain heterometallic MOFs. For instance, Han's group employed the one-pot method to incorporate Ni into the Zn-based zeolitic imidazole framework (Ni-ZIF-8).54 Due to the unique electronic configuration of Ni, it is exclusively located on the surface ligands of the MOFs. The structure of Ni coordinated with surface ligands serves as a vivid simulation of the active structure of efficient homogeneous ethylene dimerization catalysts, thus exhibiting excellent catalytic performance. Similarly, Shi's group utilized the one-pot method to synthesize bimetallic two-dimensional nanosheets by combining Zn2+ with the ligand MnTCPP (ZMTP).55 The geometric coordination structure of the Mn metal in the ligand MnTCPP is similar to the active center of natural superoxide dismutase (Mn-SOD), endowing ZMTP with SOD-like performance (Fig. 3c). Furthermore, the incorporation of Zn2+ with MnTCPP effectively regulates the redox potential on MnTCPP+, further enhancing the SOD-like activity of ZMTP. The resultant ZMTP exhibited great potential in the field of anti-oxidation. In addition to introducing metal active sites on ligands through the one-pot approach, the other common method involves modifying metal active sites onto ligands after the synthesis of MOFs. For instance, UiO-67 was synthesized by combining Au3+ ions and 2,2′-bipyridine-5,5′-dicarboxylic acid, and then Au3+ was modified onto the ligand to create Au3+-NMOF56 (Fig. 3d). Au3+ introduced as catalytic active sites on the ligands of MOFs endows Au3+-NMOF with excellent OXD and POD-like activity (Fig. 3e).
2.1.2.2. Defective MOFs. Modulators, a type of ligand with similar functionality to linker ligands, can coordinate with the metal nodes in MOFs and compete with linker ligands. The addition of modulator ligands during the synthesis of MOFs can induce structural defects, expose more active sites, and modulate the electronic structure of metal active sites, thereby tuning the catalytic activity of MOFs.57 For instance, trifluoroacetic acid as a partial ligand substitute was introduced into UiO-66 during the synthetic process.58 Upon thermal activation, the removal of trifluoroacetic acid created additional Lewis acid sites on UiO-66, exposing a large number of active sites and enhancing the catalytic activity. In addition, amino acids with a single carboxylic acid group are also a commonly used class of modulators. By introducing cysteine (Cys) as a modulator, which exhibits a strong affinity for Co2+, it coordinates with the metal nodes and partially replaces the linker ligands, thereby resulting in structural defects in Co-MOF.47 The exposure of metal active sites greatly enhances the POD-like performance of Co-MOF. At the same time, the introduction of Cys causes sulfur doping and disrupts the balance between Co and N in MOFs, leading to lattice distortion. As a consequence, Co-MOF facilitated the adsorption of oxygen and achieved the enhancement of POD-like activity. Similarly, by adding another amino acid, histidine (His), as a modulator during the synthesis of MIL-101, our group synthesized His-MIL-101 with defect structures.59 Notably, the introduction of His not only exposes active sites by inducing structural defects but also vividly mimics the catalytic pocket of natural peroxidases through the coordination between His and the Fe active site. The optimized electronic structure of the Fe active site further enhances the POD-like activity of His-MIL-101.
2.1.2.3. Functionalized ligands. Compared to pristine MOFs, introducing electron-withdrawing/electron-donating functional groups or incorporating chiral ligands with different configurations into MOFs can significantly improve their activity and selectivity. Specifically, the introduction of electron-withdrawing/electron-donating functional groups on the ligands of MOFs can influence the electronic states of metal sites within the MOFs, thereby impacting the catalytic activity of MOF-based nanozymes. Taking inspiration from naturally occurring metalloenzymes with well-defined coordination structures, Wei's group explored the relationship between the introduction of functional groups in MOF ligands and the activity of the resulting MOFs60 (Fig. 4a). Both experimental results and theoretical calculations indicated that the incorporation of electron-withdrawing functional groups is favorable for enhancing OXD-like activity. Modulation of orbital energies of the atomic-scale Fe catalytic center was realized through π-conjugation with the electron-withdrawing groups. This modulation lowers the frontier molecular orbital energies, facilitating catalytic reactions and thus enhancing the OXD-like activity of MOFs. Simultaneously, by introducing an electron-withdrawing group –NO2 and an electron-donating group –NH2 onto the ligands of MIL-101(Fe), our group obtained MOFs with different catalytic activities. We further investigated the effect of functional groups in ligands on catalytic activity.61 Theoretical calculations demonstrated that the incorporation of an electron-withdrawing group –NO2 can enhance the binding strength of intermediate products. At the same time, this modification optimizes the electronic structure of the catalytic center Fe (Fig. 4b), adjusts the geometric configuration of adsorbed intermediates in catalytic reactions, lowers the energy barrier for the reaction (Fig. 4c), and ultimately enhances the POD-like activity of MOFs (Fig. 4d). Furthermore, we applied nitro-functionalized MIL-101(Fe) for the practical evaluation of acetylcholinesterase activity and detection of organophosphates, demonstrating the potential of high-performance MOF nanozymes in biosensing.
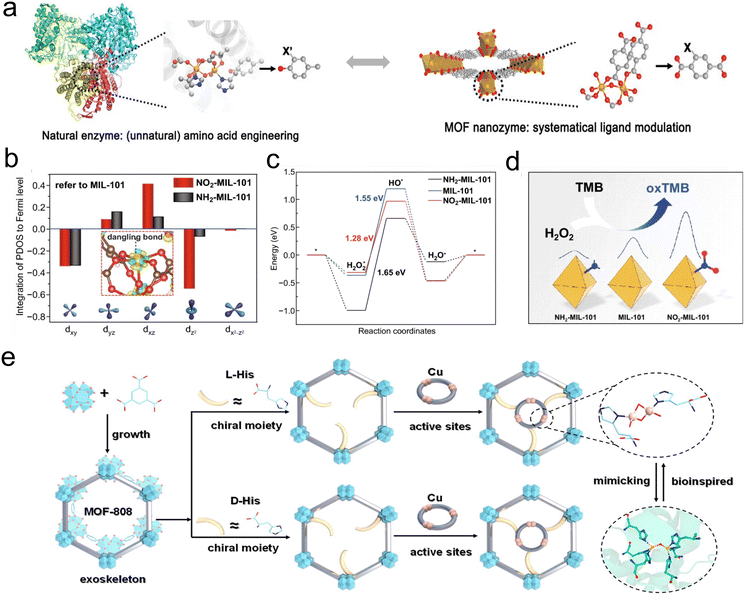 | ||
| Fig. 4 (a) Protein-engineering-inspired MOF nanozyme regulation.60 Copyright (2020) Wiley-VCH. (b) The projected electronic density of states (PDOS) and Fermi level of the Fe 3d orbit on different nanozymes. (c) Energy change illustration of the reaction on different nanozymes. (d) Schematic illustration of the MOF-based nanozymes to mimic peroxidase.61 Copyright (2020) Springer Nature. (e) Schematic illustration of the synthesis of MOF-L(D)-His-Cu.65 Copyright (2023) American Chemical Society. | ||
In addition to modulating the activity of MOFs by introducing electron-withdrawing/electron-donating functional groups on ligands, it is also possible to control the selectivity of MOFs by introducing functional groups with specific preferences. For instance, abnormal concentrations of homocysteine (Hcy) in the blood are risk signals for Alzheimer's disease and cardiovascular diseases. To achieve specific recognition of Hcy, Wang et al. introduced aldehyde groups with selective recognition capabilities for Hcy into the MOF ligands as specific recognition units.62 The introduction of functional groups enables the MOF to possess excellent specificity for Hcy recognition, thus offering tremendous potential for clinical applications. Notably, for simultaneous detection and removal of Hg2+ in water, Wang's group employed a ligand exchange strategy to introduce –SH groups into the original MOFs (NU66), resulting in the synthesis of functionalized MOFs (NUS66) enriched with –SH.63 Partial defects generated during the ligand exchange process enhanced the adsorption capacity of NUS66 for Hg2+, thereby improving the sensitivity of Hg2+ detection. Additionally, the introduction of –SH, which has a strong affinity for Hg2+, significantly enhanced the selectivity of MOFs for Hg2+. The incorporation of –SH achieved synchronous enhancement of both the activity and selectivity of MOFs, demonstrating significant application value in the field of simultaneous detection and removal of Hg2+ in water. Apart from introducing functional groups into MOF ligands to modulate selectivity, the introduction of chiral ligands with different configurations in MOFs can also enhance their selectivity. By introducing chiral L-proline into MIL-53(Al) and DUT-5(Al) ligands, Nießing's group transform the MOFs from non-selective catalysts to catalysts with enantiomeric excess (ee) values of 16% and 19%, respectively.64 The introduction of chiral ligands endowed the MOFs with unique chiral selectivity. Furthermore, our group also incorporated the dicopper metal center into the chiral L/D-histidine (L/D-His) ligand coordinated by the MOF-808 framework (Fig. 4e).65 The structure of the dicopper metal center simulates the catalytic center of natural catechol oxidase. The introduction of chiral ligands makes the catalytic reaction have good stereo-selectivity for chiral substrate L/D-DOPA. Theoretical calculations show that the spatial effect between the amino group in the substrate and the active site of the material is an important factor in the stereo-selectivity, highlighting the importance of chiral ligands. This work provides a promising path for designing chiral catalysts and deepens our understanding of the catalytic mechanism.
2.2. MOF-derivatives
MOFs, with their tunable composition, porous structure, and unique crystalline shape, have become excellent precursors and templates for preparing nanomaterials via high-temperature pyrolysis. Upon thermal transformation, MOFs can be converted into metal oxides, carbon-based materials, metal carbides, metal sulfides, and metal phosphides, displaying vast potential applications in sensing, gas storage and separation, energy storage and conversion, and catalysis.34,66 Transition metal and nitrogen co-doped carbon materials with atomically dispersed metal sites (M–N–C ADMs, where M = Fe, Co, Mg, etc.) derived from MOFs have become a hotspot in the field of catalysis due to their high metal utilization efficiency, well-defined structure and good catalytic performance.67–70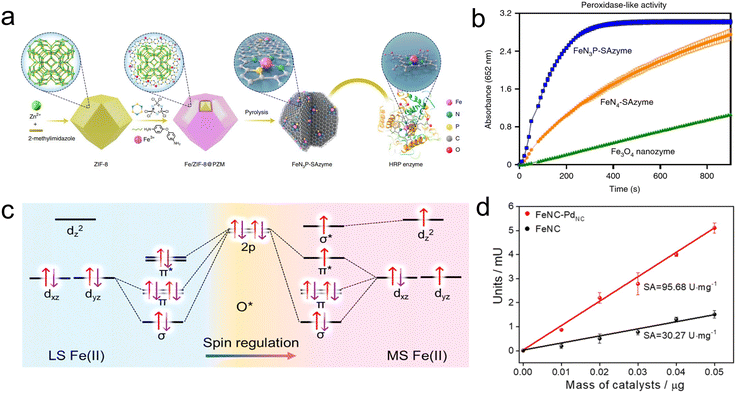 | ||
| Fig. 5 (a) Scheme illustration of the synthesis process of FeN3P-SAzyme. (b) Evaluation of the POD-like activity.74 Copyright (2021) Springer Nature. (c) Orbital interaction between low spin Fe(II) atoms, medium spin Fe(II) atoms and *O adsorbates. (d) Evaluation of the specific activity of FeNC-PdNC and FeNC.75 Copyright (2022) Royal Society of Chemistry. | ||
In most studies, the utilization efficiency of active sites in M–N–C SACs is far below the theoretical value because most of the active sites are buried in the carbon matrix and not fully exposed, which limits the catalytic activity. Therefore, defect engineering is essential to further improve the catalytic activity of M–N–C SACs. Fan's group designed an Fe-based SAC through edge-site engineering, which can form defect-exposed Fe–N4 single-atom sites in the layered mesoporous structure through reasonable design, thereby improving the catalase-like performance.77 Theoretical calculations indicated that the formation of defects leads to the transfer of electrons from Fe to the carbon-based material, enhancing the activity of Fe sites and increasing their interaction with H2O2. This promotes the cleavage of O–O bonds and enhances the catalytic activity of the catalyst. In addition, inspired by the highly efficient natural enzyme catalysts, the spatial configuration of the catalytic center can also be optimized by introducing axial ligands onto the existing planar catalytic site of M–N–C SACs, breaking the activity limitations and applications of traditional M–N–C SACs. For example, Dong's group synthesized Fe–N5 SACs with axial ligands coordinated to the catalytic site, and the synergistic effect and electron-pushing mechanism between axial ligands and the catalytic center made the Fe–N5 SACs highly efficient as oxidase mimics, breaking the limitations of traditional M–N–C SACs.78
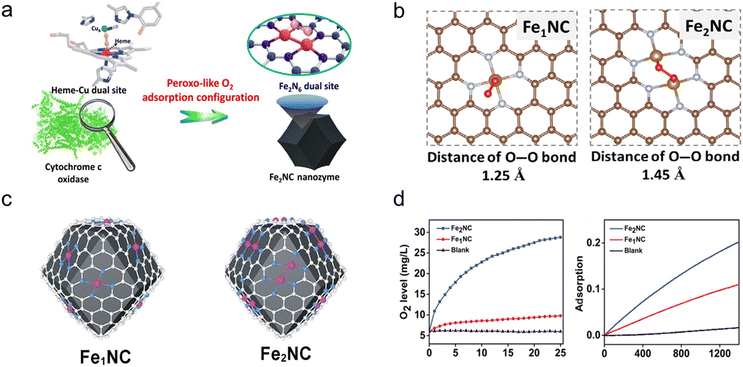 | ||
| Fig. 6 (a) Schematic illustration of the peroxo-like O2 adsorption configuration of the Fe2NC nanozyme. (b) Adsorption structure of O2 at different nanozymes.80 Copyright (2021) Springer Nature. (c) Scheme illustration of the structure of Fe1NC and Fe2NC. (d) Evaluation of the activities of oxidase-like and catalase-like enzymes to nanozymes.81 Copyright (2022) Wiley-VCH. | ||
To further broaden the activity and application of bimetallic catalysts, Liu's group used a similar method as mentioned above to synthesize Fe2–N–C and then embedded the material in the Se-MOF shell to form Fe2–N–C@Se with multi-enzyme cascading properties.81 Compared with single-atom Fe–N–C materials, the synergistic effect between Fe–Fe sites endows Fe2–N–C with higher catalytic activity for catalase, superoxide dismutase, and oxidase-like activity (Fig. 6c and d). The introduction of the Se shell endowed the material with the activity of glutathione peroxidase and also improved the stability of Fe2–N–C. The obtained Fe2–N–C@Se with multi-enzyme activity can excellently treat oxidative damage and inhibit neuronal apoptosis.
3. Applications in biosensing
The growing demand for healthcare has prompted researchers to focus on developing advanced biosensing technology that can detect disease biomarkers and harmful substances rapidly and economically.82 MOF-based nanozymes have gained wide interest in biosensing due to their unique structural adjustability and good enzyme-like activity.83 The atomic-scale control of MOFs can enhance their catalytic activity, enabling specific catalysis, and thereby facilitating selective and sensitive detection of small biomolecules (such as dopamine, hydrogen peroxide, uric acid, and glucose), biomacromolecules (such as bacterial lipopolysaccharides, proteins, and nucleic acids), and toxic pollutants (such as organophosphorus pesticides, aflatoxin and toxic ions).3.1. Small biomolecules
Dopamine is an important neurotransmitter that plays a crucial role in normal physiological activities in the human body. Abnormal levels of dopamine can significantly affect human emotional cognition, and may even lead to mental disorders such as Alzheimer's disease, Parkinson's disease, and schizophrenia.84 Therefore, the timely detection of dopamine is of great importance in clinical diagnosis and treatment. By utilizing MOF-based nanozymes as catalysts to catalyze the reaction of dopamine, a color change is generated to achieve signal output, enabling sensitive detection of dopamine. Recently, an amorphous MOF (I-Cu) with trinuclear copper centers was synthesized by water-induced Cu2+ and imidazole precipitation. I-Cu exhibits excellent tyrosinase and catechol OXD-like activity, which can generate color changes in dopamine.85 Based on I-Cu nanozymes, a dopamine colorimetric and smartphone detection method was designed with a detection limit of 0.412 μM. In living organisms, the chirality of molecules is of great significance to life activities. Generally, molecules with chirality only have one chiral molecule that is beneficial to the organism, while the other cannot be utilized by the organism and may even be toxic, making chiral identification of molecules very important. Our group introduced chiral histidine and Cu into the MOF-808 framework to generate MOF-His (L/D)-Cu with catechol oxidase-like activity.65 Based on MOF-His (L/D)-Cu, a sensor for the sensitive detection of dopamine was designed, with detection limits of 0.59 μM and 0.51 μM for L-DOPA and D-DOPA, respectively. Besides, due to steric hindrance effects, chiral substrates can be recognized by the catalyst. The MOF-His (L/D)-Cu sensor not only facilitates the highly sensitive detection of DOPA but also exhibits the ability to discriminate the chirality of DOPA.H2O2 produced by cell metabolism is a common reactive oxygen species in the body. Excessive H2O2 can affect cell homeostasis, damage tissues, cells, or organs, and induce cardiovascular or mental disorders.86 In addition, H2O2 can establish connections with most biological macromolecules in the form of intermediates or oxidation products. Therefore, the significance of detecting H2O2 as a biomarker is extraordinary. In general, we use materials with POD-like activity to catalyze the oxidation of other substrates (such as 3,3′,5,5′-tetramethylbenzidine (TMB), luminol, etc.) by H2O2 to achieve signal output (colorimetric, chemiluminescence, etc.), enabling sensitive detection of H2O2 concentrations. A bimetal-doped Fe3Ni-MOF by directly mixing Ni2+, Fe3+ and the ligand was obtained using a one-pot method. The introduction of Ni enhanced the POD-like activity of Fe3Ni-MOF by promoting the generation of hydroxyl radicals (˙OH) (Fig. 7a). The Fe3Ni-MOF with excellent performance enables ultra-sensitive detection of H2O2, with a linear range of 0.02–15 μM and a detection limit of 11 nM.87 Similarly, Fe3+ was introduced into NH2-UiO-66 through a post-modification method. The introduction of Fe3+ enhanced the POD-like performance of NH2-UiO-66. This modification enables sensitive detection of H2O2, with a detection limit of 1.0 μM.88 Additionally, the detection of glucose can be achieved through the indirect detection of the oxidized product H2O2. Cu2+ chelated onto the UiO-type MOFs generates Cu2+-NMOF, which produces chemiluminescence in the presence of luminol and H2O2. By detecting the H2O2 produced from glucose under the action of glucose oxidase, sensitive detection of glucose is accomplished.53
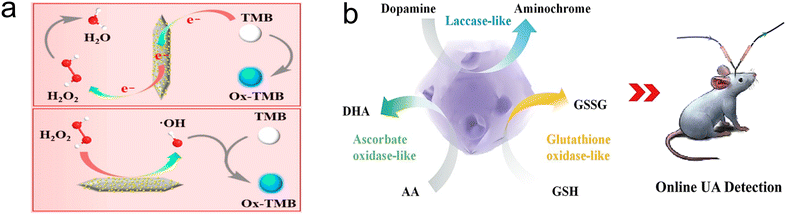 | ||
| Fig. 7 (a) Schematic illustration of the POD-like activity of Fe3Ni-MOF.87 Copyright (2022) American Chemical Society. (b) Schematic illustration of analysis of uric acid with ZIF-L-Co.47 Copyright (2021) American Chemical Society. | ||
In addition to dopamine and H2O2, uric acid, adrenaline, and glutathione have also received widespread attention from researchers due to their unique biological roles. For example, Lin's group introduced defect structures into a Co-containing imidazole framework (ZIF-L-Co) through cysteine doping, and as the degree of defects increased, the activities of ascorbate oxidase, glutathione oxidase and laccase-like activities were greatly improved.47 The obtained ZIF-L-Co was applied to the upstream anti-interference of the electrochemical detection of uric acid microreactors, achieving successful removal of interference from ascorbate, dopamine, and DOPA in the sample (Fig. 7b). Moreover, a bimetallic MOF-919 (Fe–Cu), which exhibited OXD-like activity due to the introduction of Cu metal and POD-like activity due to the introduction of Fe metal.89 This dual-enzyme functional material can oxidize adrenaline, resulting in a color change even in the absence of H2O2, with the color deepening gradually as the concentration of adrenaline increases. Based on this, a “signal on” colorimetric biosensor for adrenaline detection was developed. Glutathione, as a reducing agent, can partially inhibit the occurrence of oxidation reactions. PCN-224-Mn, which exhibits excellent OXD-like activity, can oxidize TMB without the presence of H2O2, leading to signal output.90 By adding glutathione as a reducing agent to the detection system, the oxidation reaction of TMB can be effectively suppressed. Based on this, a “signal off” colorimetric detection method for glutathione has been established, with a linear detection range of 0.5–60 μM and a detection limit of 0.233 μM.
3.2. Biomacromolecules
Recently, MOF nanozymes have also been applied to detect large biomolecules, such as bacterial lipopolysaccharides, alkaline phosphatase, phosphorylated proteins, nucleic acids, and so on. For instance, a Cu2+-modified MOF (Cu2+-NMOF) for the electrochemical detection of bacterial lipopolysaccharides (LPS) was designed.91 LPS immobilized on the electrode can capture Cu2+-NMOF with POD-like activity, which further oxidizes dopamine to form an intense electrochemical oxidation signal. The linear detection range of LPS content using this sensor can reach 0.0015–750 ng mL−1, and the detection limit is 6.1 × 10−4 ng mL−1. Li's group utilized a similar material, Zr-MOF-Cu, to detect the activity of alkaline phosphatase (ALP) through its POD-like activity.91 ALP catalyzes the L-ascorbic acid-2-phosphate to ascorbic acid, which is further oxidized to dehydroascorbic acid in the presence of Zr-MOF-Cu. Dehydroascorbic acid reacts with o-phenylenediamine to generate a fluorescent probe, resulting in an 8-fold signal amplification. Moreover, the introduction of other proteins into the detection system does not interfere with the detection process. The Zr-MOF-Cu-based sensing platform exhibits good specificity, enabling efficient signal output for alkaline phosphatase (ALP) activity detection. Similarly, for detecting ALP activity, an inhibitory colorimetric biosensor was also developed.92 They utilized Fe–Zn MOF as a precursor and synthesized Fe–N–C SACs with excellent POD-like activity. The ascorbic acid generated by ALP catalyzing L-ascorbic acid-2-phosphate inhibits the coloration reaction of the Fe–N–C/TMB system. Based on this, an inhibitory colorimetric biosensor for detecting ALP activity was designed.Phosphorylated proteins play a crucial role in cellular signal transduction, and their timely detection is of great importance in clinical medicine. A hierarchically mesoporous cerium metal–organic framework (Ce-HMMOF), having a mesopore size of up to 9.18 nm, exhibits outstanding alkaline phosphatase-like activity.93 Ce-HMMOF directly reacts with phosphorylated proteins, resulting in a color change and completing the signal output. Based on this, an enhanced colorimetric biosensing platform for detecting phosphorylated proteins has been designed. Moreover, Zhou's group developed a bimetallic organic framework, MOF-818, for detecting phosphorylated proteins. The bimetallic synergistic catalysis within the MOF effectively mimics the natural Cu/Zn SOD, exhibiting good SOD-like activity.51 However, when the phosphate group in phosphorylated proteins interacts with the Zr site in the MOF, the SOD-like activity of MOF-818 is inhibited.
Based on this, an inhibitory sensing detection platform for phosphorylated proteins has been designed. In recent years, due to the spread of the COVID-19 epidemic all over the world, it is of great significance to develop simple, rapid, and sensitive methods to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for human health. Recently, a bimetallic MOF, MIL-101(CuFe), with excellent POD-like activity was developed.94 They chemically coupled the universal receptor CD147 of SARS-CoV-2 with MIL-101(CuFe) to establish the MIL-101(CuFe)-CD147 biosensor. After SARS-CoV-2 binds to the universal receptor CD147, it blocks the active site on the surface of MIL-101(CuFe), resulting in inhibition of the POD-like activity of MIL-101(CuFe) and reducing the amount of TMB oxidation by MIL-101(CuFe) in the presence of H2O2, thereby reducing the color change (Fig. 8). The sensor has an extremely low detection limit of only 3 PFU mL−1, demonstrating significant application value.
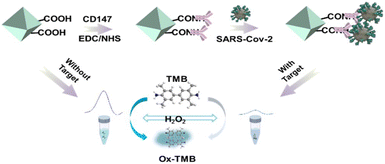 | ||
| Fig. 8 Schematic of the preparation of MIL-101(CuFe)-CD147 and application for the colorimetric detection of SARS-CoV-2.94 Copyright (2023) American Chemical Society. | ||
3.3. Toxic pollutants
With the increasing use of pesticides, pesticide residues have become a major concern, and the development of sensitive, rapid, and low-cost detection methods has become urgent.95,96 Researchers have shown interest in using MOF nanozymes for colorimetric detection. Pesticides can inhibit the enzyme-like activity of MOF nanozymes with OXD-like or POD-like activities, resulting in the generation of inhibitory signal output. Also, they can be hydrolyzed by MOF nanozymes with hydrolase-like activities, further leading to direct signal output. For example, as shown in Fig. 9, a simple and fast pesticide detection method was established by synthesizing bimetallic Mn/Fe-MIL(53).97 With the introduction of Mn into Fe-MIL(53), the OXD-like activity was improved. At the same time, the introduction of Mn made the material susceptible to choline damage and loss of OXD-like activity. Acetylcholinesterase catalyzes the decomposition of acetylcholine to generate choline, thereby inhibiting the OXD-like activity of Mn/Fe-MIL(53). In the presence of organophosphorus pesticides, the activity of acetylcholinesterase is suppressed, resulting in a decrease in the production of choline and a reduced inhibition of Mn/Fe-MIL's OXD-like performance. Based on this, a colorimetric biosensor for organophosphate pesticides based on Mn/Fe-MIL(53) can achieve sensitive detection of common pesticides such as methyl parathion and chlorpyrifos. Moreover, by using a bimetallic MOF Zn-ZrMOF, a semi-quantitative colorimetric analysis method was developed for detecting profenofos, which can be visually analyzed.98 Due to the excellent hydrolase-like activity of Zn-ZrMOF, it can hydrolyze profenofos to produce 4-bromo-2-chlorophenol, which can react with the color reagent 4-amino antipyrine in the presence of the oxidant K3[Fe(CN)6] to generate a pink substance for colorimetric detection.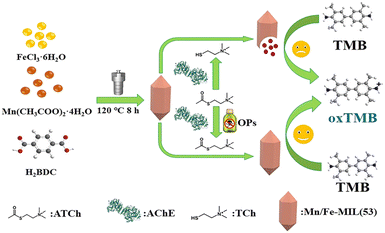 | ||
| Fig. 9 Schematic illustration of the determination of organophosphorus pesticides using the MnFe-MIL(53)-based biosensor.97 Copyright (2023) Elsevier. | ||
Fungal toxins are also common substances that harm human life and health in daily life. They exist in the agricultural products that we consume daily and have strong carcinogenicity. As shown in Fig. 10, to achieve sensitive detection of toxins, Zhang's group designed a dual-modal fluorescence/colorimetric detection method for detecting aflatoxin B1 (AFB1) by combining DNA probes with MOFs.99 First, a heme-like ligand FeTCPP was integrated into UiO-66 to generate FeTCPP⊂UiO-66, which endowed the material with POD-like activity and enhanced its fluorescence quenching ability. After AFB1 is captured by the aptamer, blocking DNA will be released from magnetic beads (MBs), and then MBs are removed by magnetic adsorption, only blocking DNA is left. Then, the blocking DNA interacts with FeTCPP⊂UiO-66, which inhibits the peroxidase activity of the FeTCPP⊂UiO-66/H2O2/ABTS system, leading to a weaker colorimetric signal output. Meanwhile, the addition of blocking DNA triggers a hybrid chain reaction between H1 and H2, resulting in a fluorescent signal output. Based on this, the dual-modal detection method has a colorimetric channel detection limit of 0.97 nM and a fluorescent channel detection limit of 0.068 nM for AFB1. Furthermore, the persistent accumulation of toxic ions (such as S2− and Hg2+) from wastewater in the human body poses significant health risks, thus it is critical for sensitive toxic ion detection. Because these toxic ions can affect the activity of MOF-based nanozymes, sensitive detection of these toxic ions can be achieved. For instance, a colorimetric sensing platform for S2− based on a bimetallic NiFe two-dimensional MOF was developed.100 NiFe MOF has good POD-like activity, which can oxidize TMB in the TMB/H2O2/NiFeMOF system, and the addition of S2− will compete with TMB, leading to a decrease in signal output. In real water sample testing, some common anions and cations do not have obvious interference with the TMB/H2O2/NiFeMOF system. The inhibition of the reaction by S2− exhibits specificity. Based on this principle, S2− can be detected with a colorimetric method, with a linear detection range of 0.5–60 μM and a detection limit of 28 nM. Similarly, to achieve highly sensitive detection of Hg2+, Kataria's group constructed a Cu-MOF-based sensing platform.101 Cu-MOF with POD-like activity can catalyze the oxidation of TMB by H2O2 to produce a colorimetric signal output. The introduction of Hg2+ further amplifies the intensity of signals. Furthermore, the addition of other ions to the system does not significantly enhance the signal, demonstrating excellent specificity.
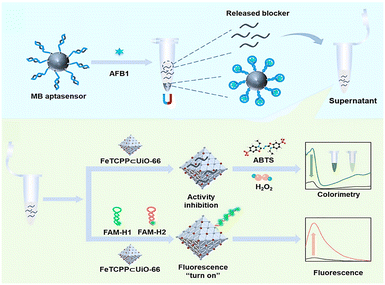 | ||
| Fig. 10 Schematic illustration of the dual-channel sensing strategy for AFB1 detection based on the MOF–DNA interaction.99 Copyright (2023) American Chemical Society. | ||
4. Conclusion and perspective
In conclusion, we summarize the methods of controlling MOFs at the atomic scale to obtain highly catalytic and selective catalysts and discuss the applications of the controlled MOFs in the field of biosensing. Although there has been some progress in the atomic-level control of MOFs and the application of high-performance MOFs has driven the development of biosensing, there are still many areas for improvement.Developing regulatory strategies for boosted enzyme-like activity
Despite the significant improvements achieved through atomic-level control, the catalytic performance of MOFs still needs to be further enhanced to match that of natural enzymes. To reach this goal, more precise and effective control methods are required to construct the optimized atomically dispersed active sites and spatial structure of MOFs with the same precision as natural enzymes. This will be a significant challenge, but one that we must face together to further improve the catalytic performance of MOFs.Understanding underlying catalytic mechanisms
Controlling MOFs leads to a more complex catalytic center environment and catalytic mechanism, making it challenging to understand the catalytic mechanism at the atomic scale. Advanced characterization techniques and accurate computational models are essential to assist our understanding and guide further research.Integrating recognition targets for good selectivity
Low selectivity is a major obstacle to the application of nanozyme-based biosensing. The broad range of substrates catalyzed by MOFs interferes with the sensing and detection process, leading to inaccurate output signals. One approach is improving catalytic specificity by adjusting the configuration of catalytic active sites, mimicking the catalytic pockets of natural enzymes. Additionally, integrating natural enzymes, DNA, antibodies, or other biomolecules with specific recognition sites into MOF nanozymes can also enhance the selectivity of the reactions. This represents a promising area for future research.Expanding biosensing applications
Despite the establishment of many biosensing systems based on MOF nanozymes, the range of detectable analytes remains limited. For instance, there is still significant scope for further development in nucleic acid analysis, protein detection, and immunodetection. Future research should focus on exploring new analytes for biosensing systems based on MOFs and integrating them with other advanced signal output modes to achieve more sensitive and accurate sensing.Currently, the atomic-level control of MOFs and their application in biosensing have received widespread attention from the scientific community. The use of more sophisticated technologies to address the catalytic activity and selectivity issues of MOFs is expected to expand the application of MOF-based nanozymes in the field of biosensing.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support by the Fundamental Research Funds for the Central Universities (no. CCNU22JC006), the Open Research Fund of the Key Laboratory of Ministry of Education, Hangzhou Normal University (no. KFJJ2023009) and the Program of Introducing Talents of Discipline to Universities of China (111 program, B17019).References
- S. J. Benkovic and S. Hammes-Schiffer, Science, 2003, 301, 1196–1202 CrossRef CAS PubMed.
- L. Gao and X. Yan, Sci. China: Life Sci., 2016, 59, 400–402 CrossRef PubMed.
- Q. Wang, H. Wei, Z. Zhang, E. Wang and S. Dong, TrAC, Trends Anal. Chem., 2018, 105, 218–224 CrossRef CAS.
- Y. Wu, W. Xu, L. Jiao, W. Gu, D. Du, L. Hu, Y. Lin and C. Zhu, Chem. Soc. Rev., 2022, 51, 6948–6964 RSC.
- I. Koh and L. Josephson, Sensors, 2009, 9, 8130–8145 CrossRef CAS PubMed.
- M. K. Masud, J. Na, M. Younus, M. S. A. Hossain, Y. Bando, M. J. A. Shiddiky and Y. Yamauchi, Chem. Soc. Rev., 2019, 48, 5717–5751 RSC.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- Y. Wang, J. Dostalek and W. Knoll, Anal. Chem., 2011, 83, 6202–6207 CrossRef CAS PubMed.
- Y. Wang, R. Hu, G. Lin, I. Roy and K. T. Yong, ACS Appl. Mater. Interfaces, 2013, 5, 2786–2799 CrossRef CAS PubMed.
- M. Pumera, Mater. Today, 2011, 14, 308–315 CrossRef CAS.
- Y. Hu, X. J. Gao, Y. Zhu, F. Muhammad, S. Tan, W. Cao, S. Lin, Z. Jin, X. Gao and H. Wei, Chem. Mater., 2018, 30, 6431–6439 CrossRef CAS.
- C. M. Tilmaciu and M. C. Morris, Front. Chem., 2015, 3, 59 Search PubMed.
- T. Y. Huang, C. W. Kung, Y. T. Liao, S. Y. Kao, M. Cheng, T. H. Chang, J. Henzie, H. R. Alamri, Z. A. Alothman, Y. Yamauchi, K. C. Ho and K. C. Wu, Adv. Sci., 2017, 4, 1700261 CrossRef PubMed.
- W. Xu, L. Jiao, H. Yan, Y. Wu, L. Chen, W. Gu, D. Du, Y. Lin and C. Zhu, ACS Appl. Mater. Interfaces, 2019, 11, 22096–22101 CrossRef CAS PubMed.
- W. Xu, Y. Wu, L. Jiao, W. Gu, D. Du, Y. Lin and C. Zhu, Curr. Anal. Chem., 2022, 18, 739–752 CrossRef CAS.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- H. Sun, Y. Zhou, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2018, 57, 9224–9237 CrossRef CAS PubMed.
- X. Zhang, D. Wu, X. Zhou, Y. Yu, J. Liu, N. Hu, H. Wang, G. Li and Y. Wu, TrAC, Trends Anal. Chem., 2019, 121, 115668 CrossRef CAS.
- H. Jin, D. Ye, L. Shen, R. Fu, Y. Tang, J. C. Jung, H. Zhao and J. Zhang, Anal. Chem., 2022, 94, 1499–1509 CrossRef CAS PubMed.
- X. Niu, X. Li, Z. Lyu, J. Pan, S. Ding, X. Ruan, W. Zhu, D. Du and Y. Lin, Chem. Commun., 2020, 56, 11338–11353 RSC.
- L. Jiao, H. Yan, Y. Wu, W. Gu, C. Zhu, D. Du and Y. Lin, Angew. Chem., Int. Ed., 2020, 59, 2565–2576 CrossRef CAS PubMed.
- J. Mao, J. Li, J. Pei, Y. Liu, D. Wang and Y. Li, Nano Today, 2019, 26, 164–175 CrossRef CAS.
- W. Wu, L. Huang, E. Wang and S. Dong, Chem. Sci., 2020, 11, 9741–9756 RSC.
- G. Ferey, C. Mellot-Draznieks, C. Serre and F. Millange, Acc. Chem. Res., 2005, 38, 217–225 CrossRef CAS PubMed.
- R. J. Hill, D. L. Long, N. R. Champness, P. Hubberstey and M. Schroder, Acc. Chem. Res., 2005, 38, 335–348 CrossRef CAS PubMed.
- S. T. Meek, J. A. Greathouse and M. D. Allendorf, Adv. Mater., 2011, 23, 249–267 CrossRef CAS PubMed.
- S. Qiu and G. Zhu, Coord. Chem. Rev., 2009, 253, 2891–2911 CrossRef CAS.
- W. Xu, Y. Wu, L. Jiao, M. Sha, X. Cai, Y. Wen, Y. Chen, W. Gu and C. Zhu, Nano Res., 2022, 16, 3364–3371 CrossRef.
- L. Jiao, J. Wang and H.-L. Jiang, Acc. Mater. Res., 2021, 2, 327–339 CrossRef CAS.
- M. Sha, W. Xu, Q. Fang, Y. Wu, W. Gu, C. Zhu and S. Guo, Chem Catal., 2022, 2, 2552–2589 CrossRef CAS.
- J. Gascon, A. Corma, F. Kapteijn and F. X. Llabrés i Xamena, ACS Catal., 2013, 4, 361–378 CrossRef.
- Y. Shen, T. Pan, L. Wang, Z. Ren, W. Zhang and F. Huo, Adv. Mater., 2021, 33, e2007442 CrossRef PubMed.
- L. Jiao, W. Xu, Y. Wu, H. Yan, W. Gu, D. Du, Y. Lin and C. Zhu, Chem. Soc. Rev., 2021, 50, 750–765 RSC.
- X. Wei, X. Luo, N. Wu, W. Gu, Y. Lin and C. Zhu, Nano Energy, 2021, 84, 105817 CrossRef CAS.
- L. Jiao, Y. Wang, H. L. Jiang and Q. Xu, Adv. Mater., 2018, 30, e1703663 CrossRef PubMed.
- P. Horcajada, R. Gref, T. Baati, P. K. Allan, G. Maurin, P. Couvreur, G. Ferey, R. E. Morris and C. Serre, Chem. Rev., 2012, 112, 1232–1268 CrossRef CAS PubMed.
- Z. Hu, B. J. Deibert and J. Li, Chem. Soc. Rev., 2014, 43, 5815–5840 RSC.
- L. E. Kreno, K. Leong, O. K. Farha, M. Allendorf, R. P. Van Duyne and J. T. Hupp, Chem. Rev., 2012, 112, 1105–1125 CrossRef CAS PubMed.
- B. Li, H. M. Wen, Y. Cui, W. Zhou, G. Qian and B. Chen, Adv. Mater., 2016, 28, 8819–8860 CrossRef CAS PubMed.
- S. L. Griffin and N. R. Champness, Coord. Chem. Rev., 2020, 414, 213295 CrossRef CAS.
- L. Chen, H. F. Wang, C. Li and Q. Xu, Chem. Sci., 2020, 11, 5369–5403 RSC.
- G. Jiang, Z. Yang, K. Zhu, S. Zong, L. Wu, Z. Wang and Y. Cui, Nano Res., 2022, 16, 1141–1148 CrossRef.
- L. Wang, L. Wen, S. Zheng, F. Tao, J. Chao, F. Wang and C. Li, Sens. Actuators, B, 2022, 361, 131688 CrossRef CAS.
- B. Yang, L. Ding, H. Yao, Y. Chen and J. Shi, Adv. Mater., 2020, 32, e1907152 CrossRef PubMed.
- K. Wang, D. Feng, T. F. Liu, J. Su, S. Yuan, Y. P. Chen, M. Bosch, X. Zou and H. C. Zhou, J. Am. Chem. Soc., 2014, 136, 13983–13986 CrossRef CAS PubMed.
- Z. Wang, Y. Huang, K. Xu, Y. Zhong, C. He, L. Jiang, J. Sun, Z. Rao, J. Zhu, J. Huang, F. Xiao, H. Liu and B. Y. Xia, Nat. Commun., 2023, 14, 69 CrossRef CAS PubMed.
- G. Ren, F. Dong, Z. Zhao, K. Li and Y. Lin, ACS Appl. Mater. Interfaces, 2021, 13, 52987–52997 CrossRef CAS PubMed.
- H. B. Luo, A. J. Castro, M. C. Wasson, W. Flores, O. K. Farha and Y. Liu, ACS Catal., 2021, 11, 1424–1429 CrossRef CAS PubMed.
- S. Li, Z. Zhou, Z. Tie, B. Wang, M. Ye, L. Du, R. Cui, W. Liu, C. Wan, Q. Liu, S. Zhao, Q. Wang, Y. Zhang, S. Zhang, H. Zhang, Y. Du and H. Wei, Nat. Commun., 2022, 13, 827 CrossRef CAS PubMed.
- Z. Liu, F. Wang, J. Ren and X. Qu, Biomaterials, 2019, 208, 21–31 CrossRef CAS PubMed.
- T. Wu, S. Huang, H. Yang, N. Ye, L. Tong, G. Chen, Q. Zhou and G. Ouyang, ACS Mater Lett., 2022, 4, 751–757 CrossRef CAS.
- R. Gil-San-Millan, E. Lopez-Maya, A. E. Platero-Prats, V. Torres-Perez, P. Delgado, A. W. Augustyniak, M. K. Kim, H. W. Lee, S. G. Ryu and J. A. R. Navarro, J. Am. Chem. Soc., 2019, 141, 11801–11805 CrossRef CAS PubMed.
- W. H. Chen, M. Vazquez-Gonzalez, A. Kozell, A. Cecconello and I. Willner, Small, 2018, 14, 1703149 CrossRef PubMed.
- C. Chen, M. R. Alalouni, X. Dong, Z. Cao, Q. Cheng, L. Zheng, L. Meng, C. Guan, L. Liu, E. Abou-Hamad, J. Wang, Z. Shi, K. W. Huang, L. Cavallo and Y. Han, J. Am. Chem. Soc., 2021, 143, 7144–7153 CrossRef CAS PubMed.
- B. Yang, H. Yao, J. Yang, C. Chen and J. Shi, Nat. Commun., 2022, 13, 1988 CrossRef CAS PubMed.
- M. M. Pan, Y. Ouyang, Y. L. Song, L. Q. Si, M. Jiang, X. Yu, L. Xu and I. Willner, Small, 2022, 18, e2200548 CrossRef PubMed.
- S. Dissegna, K. Epp, W. R. Heinz, G. Kieslich and R. A. Fischer, Adv. Mater., 2018, 30, e1704501 CrossRef PubMed.
- F. Vermoortele, B. Bueken, G. Le Bars, B. Van de Voorde, M. Vandichel, K. Houthoofd, A. Vimont, M. Daturi, M. Waroquier, V. Van Speybroeck, C. Kirschhock and D. E. De Vos, J. Am. Chem. Soc., 2013, 135, 11465–11468 CrossRef CAS PubMed.
- M. Sha, W. Xu, Y. Wu, L. Jiao, Y. Chen, J. Huang, Y. Tang, W. Gu and C. Zhu, Sens. Actuators, B, 2022, 366, 131927 CrossRef CAS.
- J. Wu, Z. Wang, X. Jin, S. Zhang, T. Li, Y. Zhang, H. Xing, Y. Yu, H. Zhang, X. Gao and H. Wei, Adv. Mater., 2021, 33, e2005024 CrossRef PubMed.
- W. Xu, Y. Kang, L. Jiao, Y. Wu, H. Yan, J. Li, W. Gu, W. Song and C. Zhu, Nano-Micro Lett., 2020, 12, 184 CrossRef CAS PubMed.
- J. Wang, Y. Liu, M. Jiang, Y. Li, L. Xia and P. Wu, Chem. Commun., 2018, 54, 1004–1007 RSC.
- L. Zhang, J. Wang, H. Wang, W. Zhang, W. Zhu, T. Du, Y. Ni, X. Xie, J. Sun and J. Wang, Nano Res., 2020, 14, 1523–1532 CrossRef.
- S. Nießing and C. Janiak, Mol. Catal., 2019, 467, 70–77 CrossRef.
- M. Sha, L. Rao, W. Xu, Y. Qin, R. Su, Y. Wu, Q. Fang, H. Wang, X. Cui, L. Zheng, W. Gu and C. Zhu, Nano Lett., 2023, 23, 701–709 CrossRef CAS PubMed.
- X. Huang, Y. Ma and L. Zhi, Acta Phys.-Chim. Sin., 2022, 38(2), 2011050 Search PubMed.
- S. Fu, C. Zhu, D. Su, J. Song, S. Yao, S. Feng, M. H. Engelhard, D. Du and Y. Lin, Small, 2018, 14, 1703118 CrossRef PubMed.
- C. Lan, Y. Chu, S. Wang, C. Liu, J. Ge and W. Xing, Acta Phys.-Chim. Sin., 2023, 39(8), 2210036 Search PubMed.
- S. Dang, Q.-L. Zhu and Q. Xu, Nat. Rev. Mater., 2017, 3, 17075 CrossRef.
- R. Zhang, X. Yan and K. Fan, Acc. Mater. Res., 2021, 2, 534–547 CrossRef CAS.
- Z. Li, F. Liu, C. Chen, Y. Jiang, P. Ni, N. Song, Y. Hu, S. Xi, M. Liang and Y. Lu, Nano Lett., 2023, 23, 1505–1513 CrossRef CAS PubMed.
- Y. Hou, Y. Lu, X. Zhang and Y. Huang, Sens. Actuators, B, 2022, 370, 132409 CrossRef CAS.
- Y. Zhou, R. Lu, X. Tao, Z. Qiu, G. Chen, J. Yang, Y. Zhao, X. Feng and K. Mullen, J. Am. Chem. Soc., 2023, 145, 3647–3655 CrossRef CAS PubMed.
- S. Ji, B. Jiang, H. Hao, Y. Chen, J. Dong, Y. Mao, Z. Zhang, R. Gao, W. Chen, R. Zhang, Q. Liang, H. Li, S. Liu, Y. Wang, Q. Zhang, L. Gu, D. Duan, M. Liang, D. Wang, X. Yan and Y. Li, Nat. Catal., 2021, 4, 407–417 CrossRef CAS.
- X. Wei, S. Song, W. Song, Y. Wen, W. Xu, Y. Chen, Z. Wu, Y. Qin, L. Jiao, Y. Wu, M. Sha, J. Huang, X. Cai, L. Zheng, L. Hu, W. Gu, M. Eguchi, T. Asahi, Y. Yamauchi and C. Zhu, Chem. Sci., 2022, 13, 13574–13581 RSC.
- X. Wei, S. Song, W. Cai, X. Luo, L. Jiao, Q. Fang, X. Wang, N. Wu, Z. Luo, H. Wang, Z. Zhu, J. Li, L. Zheng, W. Gu, W. Song, S. Guo and C. Zhu, Chem, 2023, 9, 181–197 CAS.
- R. Zhang, B. Xue, Y. Tao, H. Zhao, Z. Zhang, X. Wang, X. Zhou, B. Jiang, Z. Yang, X. Yan and K. Fan, Adv. Mater., 2022, 34, e2205324 CrossRef PubMed.
- L. Huang, J. Chen, L. Gan, J. Wang and S. Dong, Sci. Adv., 2019, 5, eaav5490 CrossRef CAS PubMed.
- H. Shang and D. Liu, Nano Res., 2023, 16, 6477–6506 CrossRef CAS.
- L. Jiao, W. Ye, Y. Kang, Y. Zhang, W. Xu, Y. Wu, W. Gu, W. Song, Y. Xiong and C. Zhu, Nano Res., 2021, 15, 959–964 CrossRef.
- R. Tian, H. Ma, W. Ye, Y. Li, S. Wang, Z. Zhang, S. Liu, M. Zang, J. Hou, J. Xu, Q. Luo, H. Sun, F. Bai, Y. Yang and J. Liu, Adv. Funct. Mater., 2022, 32, 2204025 CrossRef CAS.
- Z. S. Nishat, T. Hossain, M. N. Islam, H. P. Phan, M. A. Wahab, M. A. Moni, C. Salomon, M. A. Amin, A. A. Sina, M. S. A. Hossain, Y. V. Kaneti, Y. Yamauchi and M. K. Masud, Small, 2022, 18, e2107571 CrossRef PubMed.
- H.-S. Wang, Coord. Chem. Rev., 2017, 349, 139–155 CrossRef CAS.
- D. Wen, W. Liu, A. K. Herrmann, D. Haubold, M. Holzschuh, F. Simon and A. Eychmuller, Small, 2016, 12, 2439–2442 CrossRef CAS PubMed.
- J. Wang, R. Huang, W. Qi, R. Su and Z. He, J. Hazard. Mater., 2022, 429, 128404 CrossRef CAS PubMed.
- R. Shen, P. Liu, Y. Zhang, Z. Yu, X. Chen, L. Zhou, B. Nie, A. Zaczek, J. Chen and J. Liu, Anal. Chem., 2018, 90, 4478–4484 CrossRef CAS PubMed.
- Z. Mu, S. Wu, J. Guo, M. Zhao and Y. Wang, ACS Sustainable Chem. Eng., 2022, 10, 2984–2993 CrossRef CAS.
- T. Li, Y. Bao, H. Qiu and W. Tong, Anal. Chim. Acta, 2021, 1152, 338299 CrossRef CAS PubMed.
- S. Kulandaivel, C. H. Lin and Y. C. Yeh, Chem. Commun., 2022, 58, 569–572 RSC.
- X. Lai, Y. Shen, S. Gao, Y. Chen, Y. Cui, D. Ning, X. Ji, Z. Liu and L. Wang, Biosens. Bioelectron., 2022, 213, 114446 CrossRef CAS PubMed.
- Z. Li, G. Dai, F. Luo, Y. Lu, J. Zhang, Z. Chu, P. He, F. Zhang and Q. Wang, Microchim. Acta, 2020, 187, 415 CrossRef CAS PubMed.
- Q. Chen, S. Li, Y. Liu, X. Zhang, Y. Tang, H. Chai and Y. Huang, Sens. Actuators, B, 2020, 305, 127511 CrossRef CAS.
- R. Gao, N. Ye, X. Kou, Y. Shen, H. Yang, T. Wu, S. Huang, G. Chen and G. Ouyang, Chem. Commun., 2022, 58, 12720–12723 RSC.
- X. Zhao, Z. Yang, R. Niu, Y. Tang, H. Wang, R. Gao, Y. Zhao, X. Jing, D. Wang, P. Lin, H. Guan and L. Meng, Anal. Chem., 2023, 95, 1731–1738 CAS.
- Y. Tang, Y. Wu, W. Xu, L. Jiao, W. Gu, C. Zhu, D. Du and Y. Lin, Adv. Agrochem., 2022, 1, 12–21 CrossRef.
- X. Luo, J. Zhao, M. Li, X. Zhao, X. Wei, Z. Luo, W. Gu, D. Du, Y. Lin and C. Zhu, Mater. Today, 2023, 64, 121–137 CrossRef CAS.
- L. Luo, Y. Ou, Y. Yang, G. Liu, Q. Liang, X. Ai, S. Yang, Y. Nian, L. Su and J. Wang, J. Hazard. Mater., 2022, 423, 127253 CrossRef CAS PubMed.
- L. Hou, X. Chen, X. Zhang, Y. Tang, Y. Yao, T. Lin and S. Zhao, Sens. Actuators, B, 2023, 375, 132902 CrossRef CAS.
- D. Xiong, J. Cheng, F. Ai, X. Wang, J. Xiao, F. Zhu, K. Zeng, K. Wang and Z. Zhang, Anal. Chem., 2023, 95, 5470–5478 CrossRef CAS PubMed.
- X. Zhang, Y. Li, Q. Chen and Y. Huang, Sens. Actuators, B, 2022, 369, 132365 CrossRef CAS.
- Kirandeep, J. Kaur, I. Sharma, E. Zangrando, K. Pal, S. K. Mehta and R. Kataria, J. Mater. Res. Technol., 2023, 22, 278–291 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2023 |