 Open Access Article
Open Access ArticlePoint-of-care testing of infectious diseases: recent advances
Meiyun
Shang
a,
Jiuchuan
Guo
 a and
Jinhong
Guo
a and
Jinhong
Guo
 *ab
*ab
aThe M.O.E. Key Laboratory of Laboratory Medical Diagnostics, The College of Laboratory Medicine, Chongqing Medical University, #1 Yixueyuan Road, Yuzhong District, Chongqing, 400016, China
bSchool of Sensing Science and Engineering, Shanghai Jiaotong University, Shanghai, China. E-mail: guojinhong@sjtu.edu.cn
First published on 6th July 2023
Abstract
Infectious diseases have seriously threatened human health and caused enormous losses to the global economy. Rapid and accurate diagnosis of pathogens is crucial to the timely treatment of patients, improvement of their prognosis, and containment of disease transmission. The conventional methods for detecting pathogens are usually performed in well-equipped clinical laboratories that rely on sophisticated equipment and well-trained personnel. In addition, a series of pre-analytical procedures, such as long transport time, may bias test results and delay turnaround times (TAT), which are particularly detrimental to infectious disease control in resource-constrained areas. Advances in multidisciplinary technologies, shifts in health management models, and increased awareness of disease prevention have considerably driven the development of the point-of-care testing (POCT) market. Many portable, low-cost, and rapid POCT devices have been designed to promote health management, control disease spread, and improve patients' prognosis. This review focuses on a comprehensive summary of recently developed POCT methods for infectious diseases such as acquired immunodeficiency syndrome (AIDS), Zika virus disease, Coronavirus disease 2019 (COVID-19), Ebola virus disease (EVD), and malaria, highlights the utilization of different POCT devices in these diseases and reflects on the potential value of the internet of medical things (IoMT), big data, and artificial intelligence (AI) in the next-generation smart POCT. Finally, future perspectives, discussion and conclusions on detecting infectious diseases with POCT devices are listed.
1. Introduction
Infection is the process that involves pathogens breaking through the body's defense barriers, parasitizing and multiplying in the body, and causing infectious diseases. Diseases that can spread in the population are called communicable diseases. Infections can occur in many systems, such as cardiovascular, neurological, and urinary systems, and cause endocarditis,1 meningitis,2 urinary tract infection,3 and other diseases that can seriously threaten human health and even lead to death. In addition, an increasing number of reports have emphasized that some chronic diseases are closely related to infections. For example, Helicobacter pylori are closely related to chronic gastritis, gastric ulcers, and gastric carcinoma.4 Also, emerging and re-emerging infectious diseases, particularly communicable diseases that cause pandemics, continue to threaten global health. As such, the research on infectious diseases still deserves attention today, despite the changing disease spectrum.Timely diagnosis of pathogens avoids blind medication, curbs disease spread, and improves patients' prognosis. The standard procedures for pathogen identification in clinical laboratories are characterized by low positivity rates and long testing periods. Other methods used in the laboratory mainly include nucleic acid tests, immunological assays, and sequencing. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) and sequencing can provide direct evidence for pathogen diagnosis but require expensive instruments, skilled technicians, long hours, and high costs. Commonly used immunological assays, such as enzyme-linked immunosorbent assays (ELISA), are laborious and time-consuming. Accordingly, samples are usually transported to clinical laboratories for centralized testing. The prolonged transport process may lead to sample failure and affect the final results. Overall, the above methods are unsuitable for the immediate detection of pathogens, especially in low-resource areas.
In underdeveloped areas, the lack of diagnostic tools prevents infected individuals from realizing their conditions as early as possible, thereby delaying treatment and increasing the risk of disease transmission in the community. The development of point-of-care testing (POCT) has recently provided solutions to improve health surveillance and management. POCT is tests performed outside of large-scale laboratories and designed to shorten turnaround times (TAT), change the clinical management of patients, and improve patient prognosis.5 Compared to traditional methods, POCT is an ideal candidate for infectious disease diagnosis in underdeveloped areas due to user-friendliness, rapidity, high sensitivity, and specificity. Various POCT platforms, such as paper-based diagnostic platforms,6 lateral flow immunoassays (LFIA),7 plasmonic-based platforms,8 and smartphone-based biosensors,9 have been used for rapid diagnosis of infectious diseases to facilitate medical advances in remote areas.
Given the wide variety of pathogens that cause infectious diseases, presenting them all is difficult. Pathogens threatening the health of people in resource-poor areas are more often focused on our attention because people in these areas are vulnerable to these diseases due to poor sanitation, lagging medical technology, and special natural conditions. This review discussed POCT advancements for diagnosing infectious diseases over the past few years, using acquired immunodeficiency syndrome (AIDS), Zika virus disease, Coronavirus disease 2019 (COVID-19), Ebola virus disease (EVD), and malaria as examples. POCT-related methods were briefly described to facilitate the transition to specific POCT studies. Research advancements for these infectious diseases were then summarized, including microfluidic chip-based diagnostic platforms, isothermal amplification-based wearable devices, and LFIA. The last part is the future perspectives, discussion and conclusions of POCT in detecting infectious diseases.
2. A brief overview of detection methods
Pathogen detection methods mainly include isolation, culture and identification, morphological examination, gene sequencing, nucleic acid testing, and immunological testing. Differences in pathogen size, culture characteristics, and infectivity make the first two methods only suitable for high-level laboratories. Portable gene sequencers still present challenges in integrating library preparation, bioinformatics analysis, and accuracy.10 As such, the latter two methods are more suitable for POCT scenarios. Furthermore, intact virus particles can also be targeted for virus detection, and the methods mentioned in this paper for detecting intact viruses are described here.2.1. Nucleic acid testing
Nucleic acid amplification methods are widely used and divided into variable temperature amplification and isothermal amplification. Variable temperature amplification refers to the polymerase chain reaction (PCR), and exponential amplification is achieved by several “denaturation–annealing–extension”. Isothermal amplification is roughly subdivided into enzyme-free amplification reactions and enzyme-involved amplification reactions. The former is mainly based on the hybridization process represented by catalyzed hairpin assembly (CHA) and hybridization chain reaction (HCR). Among the enzyme-involved amplification reactions, loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are the more widely used methods. The unique advantage of LAMP is that the amplification products can be detected in various ways, including colorimetric assay, real-time fluorescence assay, and turbidimetric assay. Among them, the colorimetric method is performed by adding dyes such as calcein, pH-sensitive dyes, hydroxynaphthol blue (HNB), and SYBR Green I to the LAMP reaction system, and the results are interpreted by the color change before and after the reaction.11 RPA, another popular isothermal amplification technique, is characterized by low temperature, fast reaction kinetics, simple primer design, high sensitivity, and specificity and is more suitable for nucleic acid detection in POCT scenarios. To date, few reports of RPA based on color changes are available, except for those using SYBR Green I to indicate RPA-positive amplification products.12,13 Recently, Priti et al. used HNB to establish a colorimetric method for RPA, where HNB was added before amplification to indicate positive amplification products, and the color of the solution containing positive products changed from dark blue to sky blue.14 The RPA reaction system involves various proteins and has a strong buffering capacity, making exploring pH-sensitive dye-based RPA reactions difficult. A recent study by Tomar et al. initially investigated the mechanism of pH changes in RPA assays and provided a valuable guide for the in-depth study of RPA.15 Additionally, clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) systems consisting of guide RNA and Cas protein have become increasingly widespread for in vitro diagnostics. Since the sensitivity provided by CRISPR/Cas systems does not meet the detection needs for specific targets, such as the infectious diseases discussed here, the combination of CRISPR/Cas systems and nucleic acid amplification has become an important trend to improve analytical performance further, and has been applied to various detection platforms like microfluidics and biosensors. More introduction to LAMP, RPA, and CRISPR/Cas systems and learning about other isothermal amplification techniques can be accessed from the relevant literature.16–182.2. Immunological testing
Immunological testing mainly includes traditional serological assays (sedimentation assays, agglutination assays, and others) and labeled immunoassays (fluorescent-labeled immunoassay, enzyme-labeled immunoassay, and others). Traditional label-free immunoassays are difficult to meet the demand for detecting low-abundance targets. By introducing advanced materials into the sensors, label-free immunoassays have become feasible. For instance, the mutual benefit of each constituent in the nanocomposite formed by Zr-trimesic acid MOF (MOF-808) and carbon nanotube (CNT) produced enhanced electrochemical properties. By modifying MOF-808/CNT on the surface of the glassy carbon electrode and using streptavidin to increase the antibody binding sites, a label-free electrochemical sensor could be constructed to detect carbohydrate antigen 125 down to 0.5 pg ml−1.19 Another example is the label-free electrochemical aptasensor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection.20 Copper hydroxide nanorods (Cu(OH)2 NRs) were grown on the surface of screen-printed carbon electrodes, where the Cu(OH)2 NRs served both as a substrate for immobilizing the aptamers and as an electrochemical probe for the sensor. The complex formed by the binding of SARS-CoV-2 spike (S) protein to the aptamer blocked the electron transfer of Cu(OH)2 NRs on the electrode surface, which in turn caused a decrease in the peak current. This sensor showed good reproducibility and a wide dynamic range with detection limit of 0.03 ± 0.01 fg mL−1 for SARS-CoV-2 S protein. In recent years, the development of various novel labels, the advances in microfabrication technology, and the integration of different technologies have also contributed to advances in immunological assays, such as faster detection time, better detection performance, and easier methods for reading results, as well as making portable detection platforms based on LFIA, sensors and microfluidic chips accessible.21–26 The antigen–antibody reaction is performed on the surface of the chip channel, at the sensing interface of the sensor, or on the paper-based material, and then the signal readout method is selected according to the properties of the labels. Immunological methods based on different labels share the most procedures of classical ELISA. Thus, only ELISA is briefly described here. The basic principle of ELISA is coating a solid substrate with antigens or antibodies to capture the antibodies or antigens in the sample. Subsequently, enzyme-labeled antigens or antibodies are added to form sandwich complexes, and unbound material and impurities are removed by washing. Finally, the corresponding enzyme substrate is added for development reaction, and qualitative or semi-quantitative analysis is performed according to the color intensity. More introductions and experience with ELISA can be found in Aydin's review.272.3. Intact virus particle detection
The detection of intact virus particles covered in this article is molecular imprinting technology (MIT). MIT mimics the process of specific binding of enzymes to substrates or antigens to antibodies and involves three elements: templates, functional monomers, and cross-linkers. The core of MIT is imprinting a template in the synthetic polymer to form a cavity with the identical structure of the template. MIT simply means customizing a lock for the template, and when the template presents as a key, the specific binding between the lock and the key will trigger signal generation. These signals can be optical, electrical, thermal, or colorimetric, and therefore MIT can be integrated into different sensing platforms.28,29 MIT is now used in a wide range of fields, and research in pathogen detection is proliferating.29–33 Further details on the three elements and molecularly imprinted polymers (MIP) preparation methods can be obtained in the related review.28 Recently, the concept of green MIT has been proposed considering the problem of pollution to the environment and the hazards to the operators, and a detailed discussion is presented in the review by Arabi et al.34 Another conception to detect intact virus particles is based on the interaction between angiotensin-converting enzyme 2 (ACE2) and S protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). ACE2 serves as the receptor of SARS-CoV-2 and plays an essential role in the entry of SARS-CoV-2 into cells.35,36 Moreover, studies show that the affinity of ACE2 for the S protein of SARS-CoV-2 increases with viral mutation,37–40 and therefore ACE2 has the potential to be used as a recognition element in different platforms such as electrochemical sensors for the detection of SARS-CoV-2 and mutant strains of SARS-CoV-2.41–463. Research advances in the diagnosis of infectious diseases
3.1. Progress of POCT in AIDS diagnosis
AIDS is a chronic communicable disease caused by the human immunodeficiency virus (HIV). According to World Health Organization (WHO) statistics for 2021, approximately 38.4 million people worldwide were still living with HIV, and 0.65 million people died from HIV-related causes.47 Initial infection may be asymptomatic or have flu-like symptoms, including fever, headache, rash, and sore throat, followed by more pronounced symptoms such as swollen lymph nodes, weight loss, fever, diarrhea, and cough as the disease progresses. HIV gradually destroys the function of immune cells and eventually often leads to complications of various infections and tumors.48 However, AIDS-infected individuals may be reluctant to receive testing because of stigma or have no access to get tested. Although scientific endeavors have been numerous, the treatment of AIDS remains a significant challenge worldwide. Therefore, early diagnosis and timely treatment of AIDS are of great importance in managing AIDS and curbing the spread of the disease, and early detection of AIDS is the key to initiating antiretroviral therapy. Nucleic acid quantification can be used for the early diagnosis of HIV infection, and molecular-based virus load testing for HIV-positive individuals is considered a cost-effective and sustainable method to assess the effectiveness of antiviral treatment.49 Different isothermal amplification strategies have been adopted to eliminate the limitations of thermal cycling instruments. For example, RPA is a multienzyme-assisted isothermal amplification technique that can amplify nucleic acids at human body temperature. Kong et al. developed a wearable device for HIV-1 DNA detection using dorsal wrist temperature as the heat source for the RPA reaction.50 The entire procedure involved four steps: device assembly, RPA reaction, fluorescent detection, and image analysis (Fig. 1A). The smartphone-based fluorescence detection system further promoted the application of this rapid, sensitive, and easy-to-use wearable RPA device in low-resource AIDS-affected areas.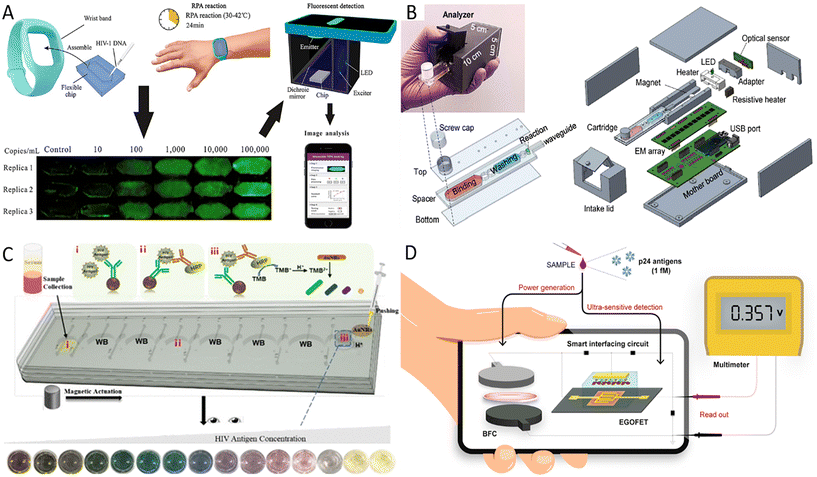 | ||
| Fig. 1 Advances in HIV POCT diagnosis. (A) The operational procedure of RPA-based wearable device for HIV-1 detection.50 (B) Overall design of the analyzer and microfluidic cartridge and their exposure views.51 (C) The principle of microfluidic integrated multicolor immunosensor for detecting HIV-24 antigen.54 i–iii are three reservoirs used to complete the immunoassay. (D) Schematic illustration of a self-powered smart sensing platform mainly consisting of EGOFET sensors and BFCs.56 | ||
Recently, Liu et al. developed a reverse transcription LAMP (RT-LAMP)-based streamlined HIV self-testing device consisting of a disposable reagent cartridge and a palm-sized analyzer (Fig. 1B).51 The analyzer with the USB interface integrated optical, thermal, and mechanical modules controlled by a microcontroller unit to perform sample preparation, purification, and real-time RT-LAMP reactions in a disposable microfluidic cartridge. Thus, this HIV testing platform removed the barrier of unavailable equipment for homes and poor laboratories. This method is highly promising and suitable for the general public because fingerpick whole blood is easy to collect, and the method is cost-effective and sensitive. In addition, the design of the disposable closed microfluidic cartridge effectively avoids cross-contamination. Secondly, the detection process is automated with minimal user intervention, and finally, the results are accessible to users with the help of a customized graphical user interface. Further improvements, such as lyophilization and pre-embedding of RT-LAMP reagents, are needed to lower cold chain transport and low-temperature storage requirements. This HIV device was designed for private self-testing in high-risk populations but can also be used for patient viral load monitoring. Therefore, it is worth exploring the iteration of the current ultra-compact analyzer into an internet of medical things (IoMT)-assisted device, as it would benefit medical centers in tracking HIV prevalence and clinicians in monitoring results online and adjusting treatment regimens as needed.
As well as nucleic acid amplification techniques, extensive studies have been conducted to detect the HIV P24 antigen, as it is considered an early virological biomarker of HIV infection and can be detected by sensitive immunological methods as early as day 14 after transmission.52 However, the cumbersome operation and the need for equipment of immunological methods may impede their use in resource-limited areas. The microfluidic chip is flexible enough to combine the functions of different partitions of the laboratory and enables the assembly of laboratory operations like sample preparation, reaction, and detection on a small platform with low reagent and sample consumption, high throughput, and fast response.53 Further, many portable microfluidic immunoassay chips have been developed to improve detection capabilities in under-resourced areas because microfluidic chips are compatible with various detection methods. For example, Liu et al. integrated cumbersome procedures of ELISA into a microfluidic chip and applied the optical properties of gold nanorods (AuNRs) to construct a multicolor immunosensor for p24 antigen detection.54 The mixture of sample and p24 capture antibodies-modified magnetic beads, horseradish peroxidase (HRP) labeled detection antibodies, and 3,3′,5,5′-tetramethylbenzidine (TMB) solutions were added to reservoirs i–iii, respectively. By moving the magnet, the liquid was moved from reservoir i to reservoir iii. When HRP oxidized TMB, AuNRs were etched to exhibit different colors after adding AuNRs and H+ solution (Fig. 1C). Sensitive, specific, and accurate results can be observed visually without interference from plasma proteins. This method's operation was extremely simplified, and the whole process could be completed within 1 hour. Besides, centrifugal microfluidics has an obvious application in massive field testing because it dramatically simplifies the experimental process by setting different parameters. In Li's research, an integrated and colorimetric-based platform consisting of a centrifugal device, a reusable 4 × 8 zigzag microchannel chip, and a smartphone was designed.55 The multi-step solution loading was required for this microchannel chip, but its design and fabrication are less challenging than other centrifugal chips. Application of the double antibody sandwich ELISA and biotin–(streptavidin–polyHRP) system allowed a minimum detection of 0.11 ng ml−1 p24 antigen in human plasma. Compared to traditional ELISA methods, this method consumes fewer reagents, costs less, and can analyze 32 samples simultaneously.
POCT devices that do not require external power sources are more portable and also reduce the need for resources, which is ideal for diagnosing diseases in remote areas, especially in areas where electricity is scarce. To this end, Sailapu et al. construct a self-powered smart sensing platform for P24 antigen detection.56 The platform used electrolyte-gated field-effect transistors (EGOFETs) as sensors and paper-based biofuel cells (BFCs) to obtain energy from the analyzed samples, with the smart interfacing circuit connecting the EGOFETs with the BFCs (Fig. 1D). Due to the low voltage required by the EGOFT, the 3.5 μL measured sample was sufficient for the BFC to provide a stable output voltage for this sensor. Based on effective capacitive coupling between the gate electrode and conducting electrolyte as well as between conducting electrolyte and semiconductor interface, the POCT platform can detect up to 1 fM P24 antigen after biorecognition between capture antibody immobilized on the gate electrode and P24 antigen, and no additional power supply was required. Another alternative strategy is the paper-based colorimetric detection system based on the enzymatic or color-change chemical reaction. The capillary action provided by the paper drives the liquid flow, which avoids the need for a pump in traditional microfluidic devices, and the colorimetric detection permits the results to be observed with the naked eye, so paper-based colorimetric detection systems can be free of resource limitations such as instruments. Unlike LFIA, where fluid flow is limited to one dimension and multi-step fluid manipulation is difficult, the microfluidic paper-based analytical devices based on the origami principle integrate sequential manipulation steps and enable fluid control by folding and pre-processing the paper, such as adjusting the fluid flow rate and controlling the fluid flow direction.57,58 For example, Chen et al. designed a sensor for infectious disease diagnosis that consists of a colorimetric ELISA-based 3D paper-based microfluidic analysis device (3D-tPADs), a 3D-printed holder, and a color chart diagram.59 Among them, the main components of 3D-tPADs included conjugate and detection wells, respectively encapsulated with detection and capture antibodies, and three absorbent pads pretreated with sucrose and pullulan. The multi-step operation of ELISA was performed by folding the 3D-tPADs. Notice that a ‘timer’ was integrated into the 3D-tPADs to indicate the appropriate time for the washing and color development steps by controlling the viscosity of the buffer flowing through the absorbent pads and the volume of the fluid path in the absorbent pads. By comparing the color on 3D-tPADs with the color chart diagram, this electricity- and instruments-independent sensor detected as low as 0.03 ng ml−1 p24 protein in the plasma matrix, further validating the feasibility of this device.
CD4+ cell counts are also an important indicator for clinical monitoring and often used to assess the patient's immune status. Flow cytometry (FCM) for counting CD4+ cells is usually used in well-equipped laboratories where professional operators are available to perform the project and manage the instrument. In addition, flow cytometers are expensive, which limits the further availability of the technology at the grassroots level. Therefore, simpler and more affordable platforms for counting CD4+ cells need to be developed to meet diagnostic needs. Sher et al. developed a magnet-driven method to quantify CD4+ cells from 30 μL whole blood based on cell lysate electrical impedance spectroscopy.60 With magnetic drive, CD4+ cells were first effectively captured with antibody-coated magnetic beads to form complexes, and then the complexes were sequentially washed and lysed. CD4+ cells were quantified based on the changes in solution conductivity at a specific frequency scan. The method requires no sample preparation procedure and allows quantitative CD4+ cell enumeration in 5 minutes with a detection limit of 25 cells per μL. In another study, Xiao et al. designed a fluorescent immunochromatographic system for indirect counting of CD4+ cells based on the blocking principle.61 Immune complexes formed by fluorescent anti-CD4+ cell antibodies and CD4+ cells were blocked at the filter pads, while free fluorescent antibodies that were not bound by CD4+ cells could bind to capture antibodies at the detection line. Thus, the fluorescence intensity at the detection line is inversely correlated with the number of CD4+ cells in the sample. The method reliably quantified CD4+ cells in whole blood samples without lysing red blood cells and had a high coincidence rate with FCM analysis, with potential application in resource-poor areas.
3.2. Progress of POCT in Zika virus disease diagnosis
Zika virus (ZIKV) is a positive-sense, single-stranded RNA virus. It was first isolated from Uganda in 1947, and WHO declared a global public health emergency in 2016.62 Eighty-nine countries and territories have reported evidence of mosquito-borne ZIKV infection, with a global spread trend emerging.63 ZIKV is spread primarily by infected Aedes mosquitoes. It can also be transmitted from mother to fetus during pregnancy or through sexual contact, blood or blood product transfusions, and organ transplants. Most individuals infected with ZIKV have no clinical symptoms or only mild or nonspecific clinical symptoms such as fever, rash, conjunctivitis, and headache that usually last 2–7 days.63 However, ZIKV also causes a range of severe complications like Guillain–Barré syndrome, microcephaly, and congenital Zika syndrome, which especially pose a significant threat to the health of newborns in impoverished areas.63 The massive ZIKV outbreak in 2015 and the severe complications associated with ZIKV infection alert us that portable and cost-effective diagnostic tools still need further development. Towards this aim, Mendes Duarte and co-workers developed a visual RT-LAMP method for directly detecting ZIKV from plasma samples using the Bst 3.0 enzyme and optimized conditions.64 This reaction amplified ZIKV nucleic acid in only 10 minutes at 72 °C with good specificity and sensitivity up to 20 zM with real plasma samples. Besides, clinical samples tested by RT-LAMP show 100% agreement with RT-qPCR. The highly sensitive and SYBR Green I-based RT-LAMP method facilitates early diagnosis of ZIKV in developing areas.Based on the RT-LAMP reaction, Jankelow et al. developed a more integrated detection system for ZIKV detection from the unprocessed blood sample.65 This system integrated sample processing, isothermal amplification, and result display into two separated microfluidic cartridges, and a detection device powered by batteries was used to provide heat and LED light sources. Nucleic acids were first released by chemical lysis and then mixed with RT-LAMP buffer before entering six parallel microfluidic channels for amplification. Finally, the results were real-time monitored by a smartphone clipped on the detection instrument, and the total time took less than 32 minutes with 103 virus particles detectable from the blood sample. Another strategy to detect ZIKV from whole blood is the POCT platform developed by Yang's team that includes a vial immunosensor for ZIKV (VISZIKV), platinum/gold core–shell nanoparticles coated with antibodies (AbZIKV-Pt@Au NPs), and a smartphone.66 By embedding a glass chip coated with capture antibodies in the VISZIKV lid, the assay was performed by simply dropping blood into a VISZIKV containing AbZIKV-Pt@Au NPs, shaking, transferring the lid and recording signals (Fig. 2A). The application of smartphones not only allows automatic access to the results by the customized application but also enables data storage, transmission and real-time geographical tagging. This high sensitivity, high specificity, robust, and portable detection platform is valuable.
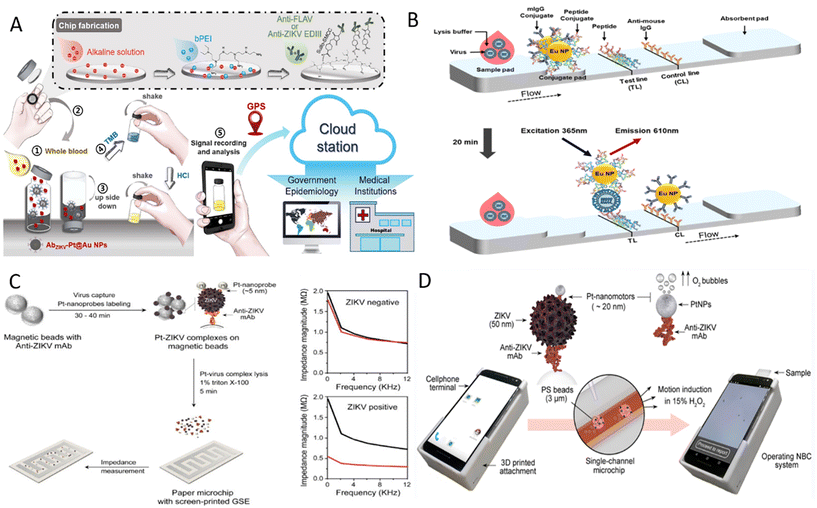 | ||
| Fig. 2 Detection of ZIKV on POCT devices. (A) Schematic diagram of the detection process for ZIKV antigen on an instrument-free POCT platform.66 (B) Illustration of peptide-based FICT.72 (C) Workflow for ZIKV detection based on the electrochemical paper-based chip and typical diagram of impedance magnitude with and without ZIKV samples.73 (D) Illustration of the NBC system for detecting ZIKV antigen.75 | ||
The sensitivity of traditional LFIA with colloidal gold as the labels is limited and cannot meet the detection needs. Nanoparticles with unique electrical, optical, magnetic, and catalytic properties have been widely used as signal amplifiers to improve detection performance further.67 LFIA based on magnetic nanoparticles, up-conversion particles, and other labels has been developed continuously to ensure that LFIA is more sensitive and specific.68–70 For example, the fluorescent carbon dots (CD)-based silica (FCS) colloids consisting of dendritic silica colloids and appropriate silanized CD were prepared by Xu et al.71 FCS-based test strips were then developed to detect NS1 in a sensitive, specific, and robust manner. The detection limit of FCS-based test strips was 100-fold lower than that of Au NP-based lateral flow (LF) strips. These developed FCS strips based on the sandwich method also showed good reproducibility and stability, and the results were obtained under UV light. To overcome the disadvantages of cross-reactivity, long preparation cycles, and the high costs of antibodies, a peptide pair-based flow immunochromatographic test strip (FICT) assay targeting the ZIKV E protein were established.72 Here, peptide aptamer B2.33 was directly coated to the detection line of the nitrocellulose (NC) membrane as the capture peptide and europium nanoparticle-linked peptide aptamer P6.1 in the conjugate pad as the FICT detector (Fig. 2B). The performance of the peptide pair B2.33-P6.1-linked FICT assay was confirmed using human urine and plasma samples, showing a comparable detection limit. This sensitive and specific method can detect ZIKV in 20 minutes and has the potential as POCT for screening ZIKV from urine and plasma samples.
Among the various nanoparticles, electroactive nanoparticles such as metal nanoparticles are often used in electrical or electrochemical methods to improve detection performance. One scheme is that metal nanoparticles are used to increase the electrical conductivity of virus lysates to improve detection sensitivity. Specifically, an interdigitated finger electrode printed paper chip using Pt nanoparticle (Pt NPs)-enhanced virus lysate electrical sensing test was developed by Shafiee's team.73 After magnetic beads capturing ZIKV combined with Pt NPs linked to anti-ZIKV antibodies and formed sandwich complexes, the charged substances of virus particles and Pt NPs were released by detergent. The change in the conductivity of the lysed solution can be measured by the impedance magnitude of the lysate loaded onto the chip, which is inversely proportional to the detected virus concentration (Fig. 2C). This method was proven affordable, highly sensitive, and without cross-reactivity with other yellow fever viruses, such as dengue virus. Even for complex biological sample matrices such as plasma, semen, and urine samples, detection limits of 100 particles per μl can be reached using this method. Moreover, the catalytic property of Pt NPs has been explored to develop POCT devices. For instance, Pt NPs catalyze hydrogen peroxide (H2O2) to produce electrochemically detectable molecules to generate currents or to produce gases to squeeze flexible photosensitive pressure sensors to produce the piezoelectric potential.67,74 Another conception is based on the catalytic properties of Pt NPs to convert chemical energy into mechanical energy.75 Here, the authors designed a Pt nanomotor-based bead-motion cellphone (NBC) system and determined whether the samples contained ZIKV by motional changes of sandwich complexes (Fig. 2D). When ZIKV presents in the sample, Pt nanomotors leveraged their catalytic properties to induce bead motion in a single-channel chip containing H2O2 solution, and results were reported in ∼2 minutes. The average velocity of beads in samples spiked with ZIKV was about 5.2 times higher than in virus-free samples. This NBC system is highly sensitive, specific, operationally simple, and applicable for the early detection of ZIKV in resource-limited areas.
MIT provides a viral sensing method with high recognition specificity by replicating the template cavity on the polymer, enabling highly sensitive detection of the target in combination with electrical methods. One example is the chip-based potentiometric sensor developed by Ricotta and his colleagues, and this sensor can detect ZIKV in 20 minutes.76 It was achieved by preparing thiol self-assembled monolayers (SAMs) with ZIKV-specific size and conformational cavities by MIT on gold chips and using the chips as working electrodes to establish three-electrode reaction systems. Similar to the process of enzyme–substrate binding, the binding of ZIKV to the imprinted cavity for single virion will cause a stable and significant potentiometric change. This method can detect as low as 10−1 plaque forming units (PFU) mL−1 ZIKV in buffer and 10 PFU mL−1 ZIKV in saliva, which the detection limit is comparable to that of molecular diagnostics. This simple and feasible protocol for ZIKV field testing is highly promising after further miniaturizing the detection system.
3.3. Progress of POCT in COVID-19 diagnosis
SARS-CoV-2, which was discovered at the end of 2019, is still circulating at a low level around the world, and the ongoing epidemic has brought a heavy burden to the global economy and public medical systems. After SARS-CoV-2 infection, the most common symptoms are fever, cough, tiredness, and loss of taste or smell. Some people may suffer from a range of symptoms, such as sore throat, headache, and diarrhea, with severe cases progressing to respiratory distress and multi-organ injury.77,78 Currently, COVID-19 treatment is still based on supportive therapy and lacks specific drugs. The keys to effective containment of the COVID-19 pandemic are wearing masks, frequent ventilation and hand washing, reducing crowd gathering, early diagnosis, and timely quarantine. Significant efforts have been made to develop a range of rapid and portable diagnostic methods that avoid sample transport, enable immediate testing in resource-poor areas, and facilitate outbreak management by providing rapid results, for example, in hospital clinics and customs. Moreover, the SARS-CoV-2 home self-testing, which is particularly suitable for the current setting, is also an effective way to know whether one is infected with SARS-CoV-2 and avoid unnecessary cross-infection during blind medical consultations. Therefore, POCT tools for SARS-CoV-2 are necessary to respond to outbreaks and to meet the needs of the residents for self-monitoring. New diagnostic platforms based on CRISPR/Cas system, exemplified by “Specific High-sensitivity Enzymatic Reporter unlocking” (SHERLOCK) and “DNA Endonuclease-Targeted CRISPR Trans Reporter” (DETECTR), have been widely studied for SARS-CoV-2 detection.79–81 For example, Broughton et al. developed a SARS-CoV-2 DETECTR platform targeting E (envelope) and N (nucleoprotein) genes based on RT-LAMP and CRISPR/Cas12 system (Fig. 3A).79 The extracted nucleic acids were pre-amplified using RT-LAMP, and subsequently, the predefined sequences were identified by the CRISPR/Cas system. Reporter molecules were then cleaved by the collateral cleavage activity of the Cas12 protein to obtain visual results identified by LF strips. Here, RNase P gene was applied as an internal reference gene to monitor the validity of sample collection and nucleic acid extraction process for further ensuring the quality of the SARS-CoV-2 assay. The DETECTR assay was proven to have comparable accuracy to RT-qPCR and can be completed in less than 40 minutes.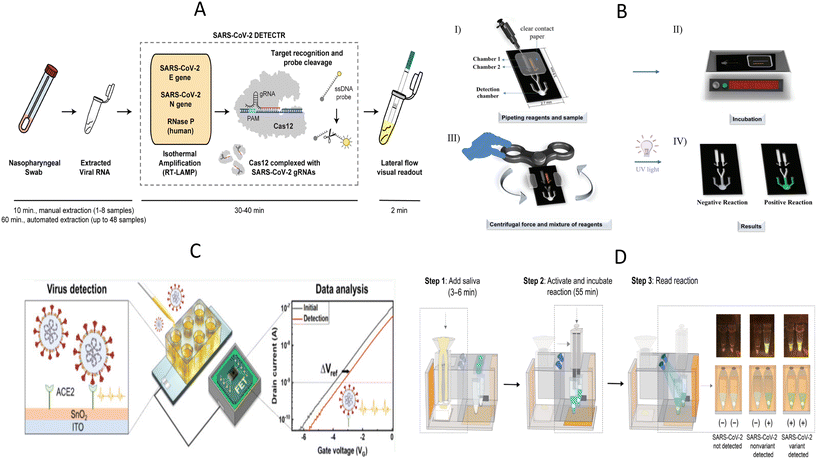 | ||
| Fig. 3 Illustration of POCT devices to detect SARS-CoV-2. (A) The workflow diagram of SARS-CoV-2 DETECTR.79 (B) Schematic diagram for the operation of the PS-T microdevice for visual detection of SARS-CoV-2.83 I) Addition of reagents and samples; II) incubation; III) centrifugation by fidget spinner and IV) visual detection by UV radiation. (C) Experimental setup of portable ACE2-based SARS-CoV-2 electrical sensor.44 (D) The procedure of “sample in-result out” miSHERLOCK platform for detection of SARS-CoV-2 and mutant strains.95 | ||
However, the SARS-CoV-2 DETECTR assay described above is likely to cause aerosol contamination due to the open-cap operation and multiple manual handling, which is often easily overlooked by novices and leads to misinterpretation of subsequent results. To reduce the risk of cross-contamination by lay users, Hu et al. designed a tube-in-tube vessel and then constructed a one-pot CRISPR Cas13a-based biosensor named ECS-CRISPR (easy to operate, contamination-free, and stable CRISPR).82 The tube-in-tube vessel consists of an inner tube and an outer tube. The inner tube was used for isothermal amplification, and then the amplification product was transferred to the outer tube for CRISPR detection through hydrophobic holes at the bottom of the inner tube. The integration of ECS-CRISPR and a portable device with incubation and centrifugation functions further overcame contamination, automation, and multiplexing limitations. This method detected SARS-CoV-2 nucleic acid down to 5 × 103 copies per mL in 25 minutes and had the potential to be applied in low-income areas. In another study, a polystyrene-toner (PS-T) centrifugal microfluidic device containing one RT-LAMP chamber, SYBR Green I chamber, and detection chamber was designed for SARS-CoV-2 detection.83 The fidget spinner, a handheld toy that spins freely by applying an external force, provides centrifugal force to automatically mix the amplicon and SYBR Green I in the chip, which enables the user to skip the pipetting step and visualize the results directly on the chip with the help of a UV light source, and thus avoid contamination (Fig. 3B). Rather quickly and affordably, this method costs less than $5 for the device and enables nucleic acid amplification in 10 minutes at 72 °C. Critically, the method provides a significantly lower detection limit than other LAMP-based microdevices for SARS-Cov-2 detection and has comparable performance to SARS-CoV-2 RT-qPCR.
As the epidemic progresses, there is an unmet need for the detection of SARS-CoV-2 variants because existing methods tend to miss SARS-CoV-2 variants and lead to false negative results. The fact that the genome and proteome of SARS-CoV-2 continue to mutate has posed new public health challenges for the diagnosis, treatment, and control of SARS-CoV-2.84 To respond these challenges, Lin et al. developed a single-tube genotyping assay by integrating recombinase-aided amplification (RAA) and the CRISPR–Cas12-mediated assay.85 Identification and detection of SARS-CoV-2 mutant strains, including alpha, beta, delta, and omicron sublineages BA.1 and BA.2 were achieved by designing a set of specific CRISPR RNAs (crRNAs) capable of recognizing single nucleotide mutations in the target sequences. The results can be read by fluorescence meters or naked eyes within 1.5–2 hours. It is true that CRISPR/Cas system is competent enough to detect SARS-CoV-2 variants, but another elaborate design of crRNAs is necessary for the new variants. In addition, protospacer adjacent motif (PAM) sequences need to be introduced into the primers when the mutant site does not contain PAM sequences.85–87 ACE2, as the receptor for SARS-CoV-2, has received increasing attention from scholars, and ACE2-based biosensors have been developed to detect SARS-CoV-2 and its variants.41,44,46 Without detailed sequence design, such sensors identify intact virus particles based on the interaction between SARS-CoV-2 and the ACE2 receptor. For example, de Lima et al. described a Low-cost Electrochemical Advanced Diagnostic assay (LEAD) consisting of a transducer made of graphite leads modified with ACE2 receptors and a plastic vial.41 LEAD allowed SARS-CoV-2 detection to be more affordable ($1.50 per unit), and faster (6.5 min) and the desired performance of the ACE2-based sandwich electrochemical assay was confirmed by testing different clinical sample types. The result of current change in SARS-CoV-2 UK variant 1.1.7.B was more pronounced than the wild strain is consistent with the finding that mutations in the S protein receptor binding region increase the binding affinity to ACE2 and further demonstrates the potential of ACE2-based biosensors for detecting SARS-CoV-2 variants.88 However, the application of traditional probe stations may partly make the test less portable. Park et al. developed a highly sensitive, portable, and virus-receptor-based SARS-CoV-2 biosensor, and dual-gate field-effect transistors were employed to enhance sensitivity (Fig. 3C).44 This portable electrical biosensor completed the assay in 20 minutes, and detection sensitivity was comparable to molecular diagnostic assays, even in portable form. Synthetic viruses with delta plus and kappa variants exhibited higher voltage shifts relative to wild-type SARS-CoV-2, demonstrating the feasibility of ACE2 as recognition elements to screen SARS-CoV-2 variants. Further clinical trials are needed before deployment to underserved populations.
Although less sensitive than nucleic acid testing in general, antigen detection with LFIA is still an essential supplement to RT-qPCR,89 especially for primary healthcare facilities, quarantined personnel, and residents with a need for self-testing. WHO has also indicated that paired serum samples from the acute and recovery phases can support the diagnosis of negative nucleic acid test results but with a strong epidemiological link to COVID-19.90 Therefore, immunology-based assays need to be urgently developed to better tailor pandemic control policies and provide timely epidemiological data to the government. To improve the sensitivity of traditional LFIA, Guo et al. used up-converting nanoparticles with mesoporous silica encapsulated nano-shell structure (UCNPs@mSiO2) labeled LFIA to quantitatively detect both nucleocapsid (N) protein and S protein of SARS-CoV-2 by a fluorescent sensor supporting 5G.69 The use of UCNPs@mSiO2 probes allows sensitive detection of S and N proteins down to 1.6 ng ml−1 and 2.2 ng ml−1, respectively. Furthermore, This IoMT-enabled fluorescence sensor can potentially upload relevant medical data to 5G cloud servers for big data analysis and storage to help people better understand diseases and establish diagnostic solutions.
Numerous studies have focused on developing electrochemical-based immunosensors for detecting proteins due to the characteristics of electrochemical sensing technology, such as high sensitivity, fast response time, economic sensing elements, and easy miniaturization.91 Currently, the modification of nanomaterials on the electrode surface has become an important way to improve the analytical performance of electrochemical immunosensors because this not only significantly enhances the ability of the electrode to transport electrons but also effectively reduces the detection time.92 For instance, Zeng et al. developed an electrochemical immunoassay of gold nanoparticle-modified screen-printed carbon electrodes for the rapid, low-cost, and sensitive detection of SARS-CoV-2 N protein.93 Antibodies coated on the surface of the working electrode recognize N protein and form immune complexes that decrease the redox efficiency of electroactive species due to the steric hindrance effect, which leads to the decrease in the peak current signal. This method exhibited a detection limit of 2.6 pg ml−1, provided a rapid response within 5 minutes, and was compatible with saliva samples. To further enhance the portability of electrochemical assays, a handheld potentiostat was used for SARS-CoV-2 N protein quantification. The measured current signal can be wirelessly transmitted to an adjunctive smartphone application and then converted into N protein concentration by its built-in algorithm. Impressively, the current intensities detected from the handheld device and the traditional electrochemical workstation were comparable. Thus, the fabricated handheld device has the potential to screen for SARS-CoV-2 in rural hard-to-reach areas, assuming it is validated in large-scale clinical trials.
In another study, a sample-in-answer-out microfluidic diagnostic platform integrating multiple electrochemical sensors for simultaneous detection of SARS-CoV-2 RNA and antibodies was developed to improve the overall accuracy of SARS-CoV-2 detection and to understand disease progression better.94 For nucleic acid detection, saliva samples and proteinase K were added to the sample preparation reservoir for incubation, and then pumped into the polyethersulfone (PES) membrane to capture RNA. LAMP solution for RNA amplification and the CRISPR mixture for amplicon recognition were sequentially pumped into the reaction reservoir and the results were finally obtained by an electrochemical sensing chip. Saliva for antibody detection was pumped onto multiplexed serology electrochemical sensor chips conjugated with S1, S1-RBD (receptor-binding domain), and N proteins to assess the patient's immune status based on the principle of indirect ELISA. Here, electrochemical signals for nucleic acid and antibody detection were further amplified by introducing the polystreptavidin-HRP/TMB-based reaction. However, further integration of electronics, peristaltic pumps and signal readout methods is necessary for better application in POCT scenarios. Besides that, de Puig et al. reported a minimally instrumented SHERLOCK (miSHERLOCK) platform that also enables sample-in-result-out detection for SARS-CoV-2 and its variants.95 This platform integrated nucleic acid extraction, in situ nucleic acid amplification, and Cas detection. After loading lysed saliva samples, SARS-CoV-2 nucleic acids were enriched onto the PES membrane and used for subsequent nucleic acids detection. As shown in the Fig. 3D, the operator only needs to complete three steps, namely loading the saliva sample containing the lysis buffer (nucleic acid extraction and enrichment), transferring column containing the PES membrane, and pushing the plunger to initiate the reaction (one-pot lyophilized SHERLOCK reactions) and reading transilluminated outputs by smartphone. The pushed-down plunger also acted as a cover for the one-pot reaction, which effectively ensures a safe testing environment. This affordable and all-in-one device simplified the experimental process and achieved the goal of “from sample input to result output”, and showed great promise for its application in field testing.
3.4. Progress of POCT in EVD diagnosis
EVD caused by Ebola virus (EBOV) is a zoonotic disease with high pathogenicity, infectiousness, and lethality.96 Since it was first reported in South Sudan and Congo in 1976,97 EBOV has caused several epidemics, especially in Africa.96 Infected individuals show non-specific clinical manifestations in the early stages, such as fever, fatigue, and muscle pain, which can be difficult to distinguish from other diseases based on symptoms alone. Even worse, if left untreated and uncontrolled, patients can easily develop severe clinical symptoms, including bleeding and impaired liver and kidney function.97 Based on these facts, timely diagnosis of EBOV may effectively curb the further progression of the disease. Laboratory-based diagnostic methods are difficult to meet the demands of EBOV clinical management because EBOV outbreaks often occur in resource-poor areas that lack sophisticated instrumentation and skilled personnel. EBOV is a relatively rare virus, but considering the high lethality of this virus and the need for EBOV detection in less developed regions, the development of POCT tools for EBOV early detection is still necessary to reduce large-scale spread and deaths from sudden EBOV outbreaks. To this end, Na et al. amplified nucleic acid samples by microbead-based rolling circle amplification (RCA), which allows highly sensitive detection of multiple viruses, including EBOV. Microbead-based RCA reactions are performed in a microfluidic system that can be simplified to a designed syringe and a microfluidic chip with multiple microchannels containing many sepharose microbeads (Fig. 4A).98 After the RCA reaction was initiated, a large number of DNA hydrogels (sufficiently amplified DNA products are entangled together) that fill the gaps between the sepharose beads were formed. Thus, the colored liquid could not flow to the other side of the channel after being injected into one side. Moreover, this method features rapid response (15 min), high sensitivity, and multi-target detection without electrical devices, rendering it suitable for the on-site screening of diseases.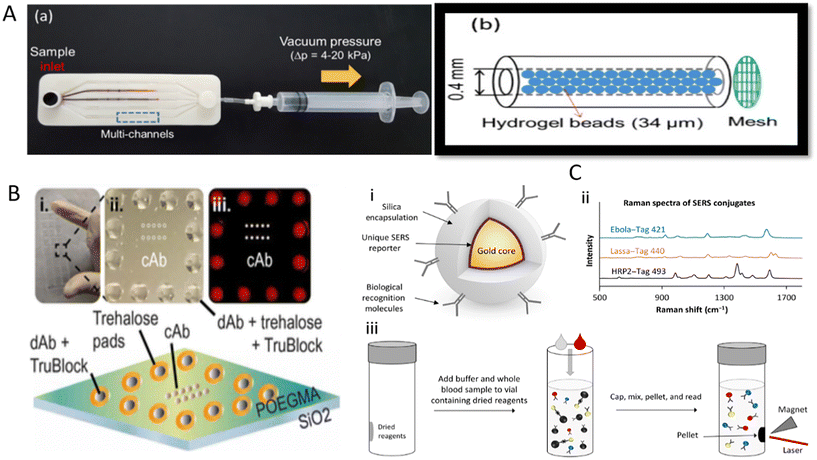 | ||
| Fig. 4 Application of various POCT devices for detecting EBOV. (A) The composition of a multiplex microfluidic device for RCA amplification-based detection of multiple pathogens involving EBOV. (a) A photograph of the experimental apparatus, (b) schematic illustration of the bead-packed microchannel. (B) Diagram of EBOV D4 chip.104 (i) Photograph of a glass slide containing 24 EBOV D4 assay chips. (ii) Photograph of a single EBOV D4 assay. (iii) Fluorescence image of incubation with sample containing sGP. (C) A schematic diagram of the SERS-based method for detecting EBOV, malaria, and LASV.107 (i) Schematic of BD’s SERS nanotag technology. (ii) Representative spectra from three SERS nanotags used to detect Ebola, Lassa, and malaria. (iii) Schematic of the homogeneous no-wash immunoassay using the SERS nanotags. | ||
Multimodal detection integrates multiple signal reading modalities to satisfy the need for reading results in different scenarios and further increases the practicality of POCT. Multimodal LFIA has been reported and demonstrated improved analytical performance.99–101 For example, a multimodal LFIA using colorimetric–fluorescent–magnetic nanoparticles as labels improved the sensitivity and the anti-interference ability of the assays due to the introduction of magnetic materials.99 This strategy also benefits EBOV early detection. Hu et al. developed a dual-signal readable RNs@Au nanosphere and used it as the reporter of bimodal LFIA.102 These nanospheres containing quantum dots and gold nanoparticles have stronger fluorescence and colorimetric signals than quantum dots or gold nanoparticles, and detection results can be read by colorimetric and fluorescence modes. Importantly, this method has been successfully used in field assays for detecting EBOV in spiked urine, plasma, and tap water samples. A series of data showed that this dual-mode LFIA is highly sensitive, specific, and robust, allowing for EBOV detection in less than 20 minutes.
EBOV is highly contagious and can be widely transmitted with the slightest mismanagement. In addition to early diagnosis of EBOV, researchers are also focusing on methodological studies for the co-detection of diseases with similar symptoms or the detection of different subtypes of EBOV genus so that a single test can more easily assist in clinical triage and patient management, as well as to further improve detection efficiency by providing results of EBOV subtypes or symptom-like diseases in a single test, which is particularly valuable in endemic areas where multiple disease co-infections often occur. An example is the optical microring resonators immobilized with different capture antibodies developed by Qavi et al. for the detection of EBOV subtypes.103 These sensors sensitively and specifically detect soluble glycoprotein (sGP) of EBOV and SDUV (Sudan virus) from serum using sandwich immunoassays, and the time span was under 40 min. Here, biotinylated detection antibodies were recognized by streptavidin HRP, and subsequently, the insoluble products were generated by HRP catalyzing the corresponding substrate, thus leading to the amplification step. These deposits onto the ring surface caused changes in the local refractive index and consequent shifts in the resonant wavelength within the microcavity. The final net shift was positively related to sGP concentration bound to the sensor surface. Regrettably, although this method is promising, the accompanying optical reading system is expensive and not portable, limiting the method to the field laboratory. Another platform developed to detect EBOV and clinically relevant EBOV species is the D4 assay, a low-cost antibody microarray chip.104 Different Ab pairs can be printed on a single D4 assay chip, allowing the detection of distinct antigens. Each unit consists of 10 capture antibody (cAb) dots surrounded by a circle of 12 fluorescently labeled dAb (FL-dAb) dots (Fig. 4B). When the sample is added, the trehalose pads will be dissolved. The FL-dAb printed on these pads binds to the analytes and forms complexes, which flow through the middle of the D4 chip and bind cAb to form sandwich complexes. The fluorescence signal at capture spots can be read with an affordable handheld detector. Although the experiment was performed in 1 hour, EBOV D4 displayed a sensitivity potentially better than that of RT-qPCR in a standard nonhuman primate model.
To distinguish Ebola from other febrile diseases in the epidemic area, Barnes et al. designed two CRISPR–Cas13a-based (SHERLOCK) protocols targeting Ebola and Lassa virus (LASV) and successfully tested clinical samples from Sierra Leone and Nigeria regions with this method.105 Moreover, the authors combined the SHERLOCK assays with a technique (HUDSON) that inactivates pathogens and releases nucleic acids by heat and chemical denaturation to simplify the procedure. After verifying the efficiency of the HUDSON protocol to inactivate viruses by safety testing, three sample types were used to further test the compatibility between the HUDSON method and the SHERLOCK assays, and saliva samples showed comparable results to GeneXpert. Despite a potential application of this developed method in infected areas, test strips-based readout is often prone to cause contamination of amplification products and a sealed external packaging is needed.
The ability of surface-enhanced Raman scattering (SERS) tags to encode different target molecules allows SERS to perform multiplexed assays. SERS tags usually consist of plasmonic nanoparticles, Raman reporters, protective shells, and ligands that recognize the target molecule.106 Target molecules in the sample are selectively captured by ligand molecules attached to the SERS tag. When SERS tags are excited, different target molecules can be identified according to the characteristic Raman spectra of different SERS tags. This strategy was adopted by Sebba et al. to develop an immunological method for detecting three pathogens simultaneously, i.e., EBOV, malaria, and LASV.107 In this POCT, different gap-enhanced Raman tags (GERTs) and magnetic nanoparticles were both coupled with antibodies against the three pathogens and then lyophilized together into a centrifuge tube. After whole blood sample and buffer were added to rehydrate the lyophilized reagents and mix homogeneously, the formed sandwich complex was fixed to the side of the tube wall by a magnet, and three different optical fingerprints were acquired by a standalone SERS reader (Fig. 4C). Most importantly, the method avoids repetitive washing and sample preparation and demonstrates high sensitivity and specificity with assays in clinical and non-human primate samples.
3.5. Progress of POCT in malaria diagnosis
Malaria is an acute febrile disease mainly transmitted by the bite of female Anopheles mosquitoes.108 Five Plasmodium parasites can cause malaria in humans, with Plasmodium falciparum (P. falciparum) and Plasmodium vivax (P. vivax) being the most harmful.109 Malaria remains a curable and non-negligible tropical disease. Although a series of antimalarial interventions have reduced global malaria morbidity and mortality, the resurgence of malaria deaths in the last two years may be attributed to lacking timely diagnosis and treatment during the COVID-19 pandemic.110 In clinical laboratories, microscopy and PCR are regarded as important methods for malaria diagnosis. However, microscopy is labor-intensive because it relies on experienced technicians to identify Plasmodium species and life cycle stages. Inadequate equipment further hinders the deployment of PCR methods in resource-constrained areas. In addition, magnetic beads are typically incubated and washed sequentially in traditional magnetic-based immunological assays, making this method challenging to integrate into low-cost diagnostic devices suitable for field testing. In view of the above, Ruiz-Vega et al. designed a microfluidic paper, double-sided, screen-printed carbon electrode (MP-dsSPCE) to simplify the operation of chip-based magnetic immunoassays and used this method to study six actual blood samples (Fig. 5A).111P. falciparum lactate dehydrogenase (PfLDH) was quantitatively detected from whole blood samples using a simple electrochemical cartridge in combination with a magnet, a whole blood filtration unit, and the absorption pads. Lysed blood samples, magnetic beads coupled with capture antibody, and biotin-modified detection antibody/streptavidin poly-HRP immunoconjugates were incubated for 5 min and added directly to the filtration unit, and the current was measured after the addition of wash buffer and substrate solution. This electrochemical POCT device quantifies PfLDH within 20 minutes with minimal human interference, and the lowest detectable concentration is 200 ng ml−1.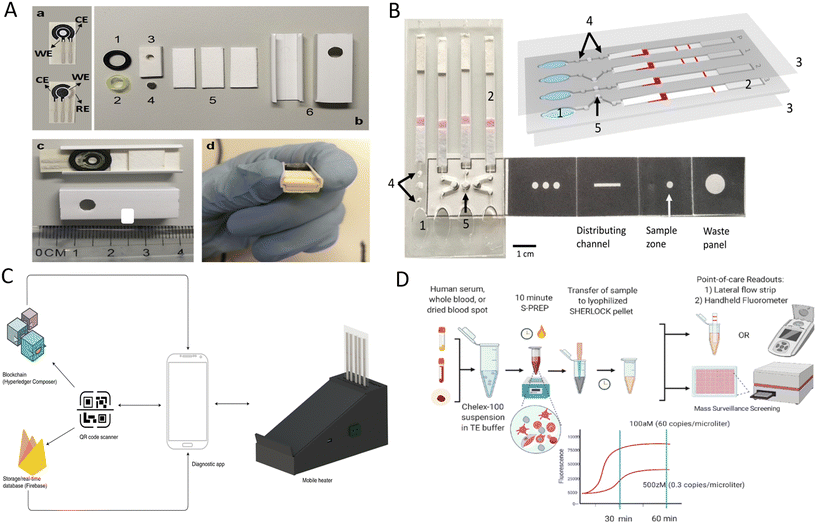 | ||
| Fig. 5 POCT devices for detecting Plasmodium parasites. (A) Components and assembly of disposable paper electrode microfluidic devices.111 a) Top and bottom sides of the MP-dsSPCE. b) Device components: (1) o-ring, (2) segment of a pipette tip, (3) silicon sheet with a perforation for the magnet (4), (5) absorbent pads, and (6) square cable trunking section. c and d) Assembled MP-dsSPCE. (B) Schematic representation of a LAMP-based paper-based microfluidic device for species-specific diagnosis of malaria.113 1, buffer chambers; 2, lateral flow DNA detection strip; 3, acetate films; 4, filter paper-based valves; and 5, filter paper for the LAMP reaction. (C) Malaria diagnostic system consisting of a heater, a smartphone, a paper-based microfluidic chip and the corresponding backend engine.114 (D) Schematic diagram of the SHERLOCK process for Plasmodium species detection.116 | ||
Different LFIA strategies have been applied to diagnose malaria and identify malaria species to increase detection capacity in poor malaria-affected areas. For multi-target detection, LFIA has to design detection lines that match the number of targets, and this requirement limits the application of LFIA for multiplexed detections. To overcome the limitations of traditional LFIA, Kim et al. developed a two-color multiplexed immunoassay based on the color-coding concept that allowed the identification of malaria species at single test line.112 The color and color intensity of test line determinates malaria species and antigen concentration levels, respectively. Moreover, to avoid the subjectivity of human eye interpretation, ‘Color Profiler’ plug-in command was used to obtain the image's RGB profile, and color discrimination protocol was also developed to distinguish malaria species. LFIA also serves to display the results of nucleic acid amplification by modifying primers and probes based on the labeled antibodies used in the test strips, thus reducing the need for nucleic acid readout instruments and ensuring better access in rural areas. For instance, Cooper's group developed a paper-based microfluidic device for malaria species-specific diagnosis.113 The device incorporated a paper-based vertical flow origami sample processing unit, LAMP, and strip-based results readou (Fig. 5B). The mixtures containing fingertip samples, lysis buffer containing magnetic beads, and binding buffer were added to the sample area of the origami device. After a series of simple folding and liquid loading operations, the prepared nucleic acids were transferred to LAMP reaction chambers for amplification. The amplicons were then transferred to the strips by finger squeezing and capillary action, and results were obtained within 1 minute. Further clinical studies in Uganda showed that this method was sensitive and specific and detected malaria in 98% of infected individuals within 50 minutes. Although this device has promising applicability, result readout is still subjective, and results also cannot be fed back to the central laboratory in time. Later, Cooper's team further developed a new malaria diagnosis platform consisting of a mobile heater, a smartphone, a paper-based microfluidic chip, and the backend engine (Fig. 5C).114 Deep learning algorithms that provide decision support and the edge computing capabilities of smartphones played a vital role in addressing the lack of skilled personnel to interpret results and varying levels of network perfection in under-resourced areas. Here, smartphones can power, geotag, control assay conditions, collect results, and serve as a tool to interact with cloud servers. Cooper's team also incorporated blockchain technology to ensure secure data transfer from the point-of-care facility to the central laboratory and revalidated the applicability of this diagnostic platform in Uganda.
Currently, species-specific detection of malaria is based on the differentiation between P. falciparum and nonfalciparum, while species-specific diagnosis of nonfalciparum is still a gap in malaria diagnosis. To this end, Lai et al. developed a multiplex LAMP reaction system, and combined with a homemade multiplex lateral flow strip for the simultaneous detection of P. falciparum, P. vivax, Plasmodium malariae (P. malariae), Plasmodium ovale (P. ovale) and Plasmodium knowlesi (P. knowlesi).115 The DNA extraction protocol without purification was applied to shorten the time and better suit resource-limited areas. With microscopy as the reference method, this established method tested 86 clinical samples with 100% sensitivity and 97.8% specificity. Besides, Lee et al. developed an ultra-sensitive, field-ready one-pot SHERLOCK platform for four Plasmodium species detection (P. falciparum, P. vivax, P. ovale, and P. malariae).116 A simplified sample preparation protocol was explored and used for rapid extraction of parasites, and results were read by fluorometric and test strip methods after species-specific detection by SHERLOCK reaction (Fig. 5D). The accuracy of the SHERLOCK platform was validated with different sample types (simulated whole blood, serum, and dried blood spot) and clinical samples of P. falciparum and P. vivax. Although four separate SHERLOCK assays were required to identify the four Plasmodium species, the results of this developed method showed good compatibility with these unextracted sample types, and the clinical samples demonstrated 100% sensitivity and specificity, which is highly applicable.
However, the above reports did not address the detection of antimalarial drug resistance. Artemisinin is the core component of the best drugs currently available to treat malaria, and artemisinin-based combination therapies have been recognized as the most effective options for treating malaria. The WHO release and studies from recent years have shown that the parasite has developed new resistance to artemisinin, which hinders malaria control.117,118 In the updated global technical strategy for malaria 2016–2030, WHO also underscores the importance of monitoring the efficacy of first- and second-line malaria therapies every two years.119 Antimalarial drug resistance surveillance is undoubtedly an urgent and non-negligible task at present. In view of this, Malpartida-Cardenas et al. successfully detected two single nucleotide polymorphisms (SNP) associated with artemisinin-resistant malaria: C580Y and Y493 using the USS–sbLAMP method.120 This method consists of SNP-based LAMP (sbLAMP) primers targeting the mutant allele and unmodified self-stabilizing (USS) competitive primers targeting the wild-type allele. USS competitive primers robustly delay or prevent non-specific sbLAMP amplification, which allows C580Y and Y493H to be identified via differences in time-to-positive values caused by delayed amplification of non-specific targets. The method showed high sensitivity, high efficiency, and high specificity, enabling the assay to be performed within 35 minutes. In a follow-up study, this group integrated the USS–sbLAMP method into a complementary metal-oxide-semiconductor based (CMOS-based) ion-sensitive field-effect transistor electrochemical sensor for P. falciparum diagnosis and artemisinin resistance detection to better serve in resource-constrained areas.121
Another noteworthy work on malaria detection is a second-scale, reagent-free, and non-invasive method for monitoring P. falciparum and P. vivax patients infected with varying parasitemia based on the near-infrared spectroscopy technique and machine learning algorithms.122 A miniature near-infrared spectrometer was used to collect characteristic absorption peaks of malaria from the ears, which can be used to distinguish infected from uninfected individuals and differentiate P. falciparum from P. vivax initially. The application of machine learning models further helps to predict infected and uninfected individuals with 100% sensitivity, even for very low parasitemia. Hence, the method allows for large-scale painless screening of populations to identify asymptomatic patients. Moreover, the spectrometer used in this study enables real-time cloud-based malaria monitoring, which allows physicians to make timely decisions and allocate medical resources more rationally. Because the sample size of this study was limited, a massive clinical trial is needed to optimize the predictive model further to improve the diagnostic efficacy. Whether the other three human-infecting Plasmodium species, including P. malariae, P. ovale, and P. knowlesi, can be differentiated and whether the effectiveness of malaria treatment can be real-time monitored by this method also deserves further exploration.
4. Future perspectives and hot spots
This paper focuses on five infectious diseases that seriously threaten the lives of people in poor areas and demonstrates the feasibility and necessity of POCT studies on infectious diseases. Timely and effective treatment regimens that control disease progression and significantly improve patient prognosis are especially critical for viruses with high lethality, such as EBOV, and diagnosis is a prerequisite for treatment and disease management. In developed areas, advanced instruments and skilled personnel ensure excellent medical resources and people's health. Instead, under-resourced areas even suffer from difficulties in securing water and electricity supplies, and the lack of testing capacity leads to disease spread in short periods. Affordable, sensitive, and specific POCT devices are an important way to increase access to testing in underserved populations. The current research on POCT is divided into two categories: diagnostic devices for outpatient, wards, grass-roots or other scenarios, and home self-testing. Etiological diagnosis often lags in hospital settings and delays patient treatment and increases the risk of nosocomial infection. The design of POCT devices should prioritize accuracy and time efficiency and improve analytical performance by introducing novel materials. For home self-testing, POCT devices should be designed to fully consider patients' compliance, such as selecting non-invasive sample collection methods to make them more acceptable to patients. Despite the significant progress of POCT in the diagnosis of infectious diseases, the majority of the studies mentioned above could not meet the “ASSURED” criteria (affordable, sensitive, specific, user-friendly, rapid/robust, equipment-free, and deliverable to end users) proposed by WHO.123 Besides meeting the basic requirements of high sensitivity and specificity to increase screening of asymptomatic individuals, an ideal pathogen POCT device should also be a simplified, integrated and smart device with the ability of quality control and multiplexed detection.4.1. Simplification
Pathogen POCT devices should consider the portability and friendliness of the devices to ensure that they can be used in a variety of situations, whether in the laboratory or the field and for experienced technicians or beginners. POCT diagnostic platforms are toward minimal or no equipment to approach the “equipment-free” standard. One trend is molecular diagnostics based on isothermal amplification technology,124 which thus lowers the threshold for POCT users and eliminates the need for professional staff to manage the device. Furthermore, immunology-based methods are increasingly integrated into different sensors or microfluidic chips to avoid tedious handling procedures. The development of wash-free homogeneous immunoassays is extremely attractive, and one example is Abionic's patented nanofluidic immunoassay, which avoids the washing process and obtains the results of pancreatic stone protein in 5 minutes due to its extremely high surface-to-volume ratio at the nanoscale.1254.2. Integration
The major difference between the POCT of pathogenic microorganisms and other physicochemical indicators such as troponin and uric acid is that there are certain biosafety requirements for detecting pathogenic microorganisms. Infectious samples have more or less infection risk and potentially harm the operator through aerosols, direct contact, and other routes. Even for testing physical and chemical indicators, there are cases where the patient suffers from infectious disease. As such, every unknown sample in medical testing should be treated as hazardous. In addition, if a nucleic acid amplification-based platform is used, a large number of amplicons are likely to interfere with the results of subsequent assays. Integrating “sample extraction, target detection, and result readout” into a completely sealed device minimizes human error, better protects the operator, and effectively avoids contamination. For highly infectious pathogens such as SARS-CoV-2 and EBOV, additional consideration should be given to the compatibility of the virus inactivation method with the detection method. Moreover, the integration of nucleic acid extraction steps in an automated platform is challenging due to the multiple operational steps that need to be introduced in the device. Excessive integration, such as the introduction of various valves and pump structures, complicates the equipment and compromises portability. Currently, nucleic acid extraction methods such as chemical lysis combined with thermal lysis and magnetic bead methods have been integrated into POCT platforms.126,127 However, these platforms require elaborate designs, which are demanding for mass production. The recently developed magnetically controlled droplet manipulation platform offers the ability to control the splitting, transport, and mixing of droplets by adjusting the magnetic field intensities, and enables the detection of analytes in whole blood and urine samples with a simple and cost-effective microfluidic chip.51,128 For immunological assays using plasma samples, fidget spinners have been shown to provide sufficient centrifugal force for plasma separation with extremely low matrix interference and high recoveries, making it a field-amenable sample preparation method.129 While realizing the integrated detection of “sample input to result out”, pathogen POCT should search for a balance between the complexity and automation of POCT devices.4.3. Quality control
Most POCT operators do not have professional knowledge, and POCT users tested at home may not even have medical knowledge. Even if POCT is performed in other departments outside the hospital laboratory, the frequent flow of clinicians can also increase the risk of errors. These operators lacking expertise in Laboratory Medicine have little awareness of quality control prior to testing and have no ability to analyze problems after testing. In addition to focusing on regular training, POCT devices should transform manual operations into standardized operations, such as embedding batch-specific calibration data into a chip for readout by POCT devices to reduce calibration operations. POCT devices also should have an individual quality control plan (IQCP).130 The device's internal quality control monitors its components and data processing module, and regular liquid quality control monitors the whole testing process. Due to biological differences within individuals, procedures for acceptable result intervals and unacceptable results should be embedded in POCT devices. When a life-threatening critical value occurs, the alarm system should be triggered. The locking system should also be embedded in POCT devices to achieve the purpose of quality control first and then testing.4.4. Multiplexed detection
The differential diagnosis of diseases with similar symptoms and multi-target joint detection of diseases has driven the development of POCT multiplexing technology. The similarity of the early symptoms of the disease makes the diagnosis based on clinical symptoms alone extremely challenging because multiple infectious diseases are often rampant in resource-constrained areas simultaneously. Multiplexed detection for pathogens provides clinicians with more valid information to adopt more tailored treatment strategies and better serve their patients. On the other hand, joint detection of different targets can improve the accuracy of disease diagnosis and obtain more precise diagnostic information, such as assisting physicians in understanding whether a patient is in the acute or recovery phase. Multiplexed detection can be performed in a single tube or parallel on platforms like microarrays. Single-tube multi-target detection often involves complex optimization of conditions because the amplification of different targets competes for common compositions. Hence, this approach is cumbersome and has limited multiplex detection capabilities. However, the latter can complete multiple reactions via an operating platform designed with multiple reaction units, permitting multiple assays in a parallel manner. Furthermore, POCT can reduce the financial burden on patients from the long-term perspective, but it is not less costly than laboratory testing from the manufacturing perspective as existing POCT is often designed to be disposable to obtain accurate results. For this reason, multiplexing is also an important way to reduce costs. Assuming that the RPA method is used to detect pathogens, commercial RPA reaction is approximately $7 per reaction. If 10 μL mixture is dispersed from the RPA mixture for the reaction using a centrifugal microfluidic chip, this would effectively reduce the cost to about $1.75. As such, the microfluidic-based POCT device can be used for multiplexed detection of target genes or even high-throughput detection at a more reasonable price by rationally designing the microchip structure.131,132 Some specific challenges also remain, such as the development of a detection platform that integrates drug resistance monitoring with routine disease surveillance for people infected AIDS and malaria and the multiplexed detection platform for the identification of five Plasmodium species needs to be further developed, especially for P. malariae, P. ovale, and P. knowlesi, which are relatively unavailable.4.5. Smart
Advances in IoMT, AI, cloud computing, and microfabrication technologies are driving medical testing toward intelligence, i.e., miniaturization, immediacy, portability, automation, and cloud-based approaches to provide timely medical services and reduce overall healthcare costs. Over the years, with the development of technologies such as electrochemical detection, microfluidics, and isothermal amplification, POCT has demonstrated great progress in terms of immediacy, miniaturization, and portability, such as detection speeds of minutes or even seconds and simplified readout methods based on handheld devices or smartphones.93 A primary goal in the development of POCT is to shorten the TAT and inform clinicians of the test results as soon as possible so that appropriate measures can be taken promptly to improve the patient's prognosis.133 As such, the current POCT model is very beneficial for hospital scenarios, but this model provides limited benefits to the home testing group because these people lack timely assessment and supervision by professionals. In reality, due to busy schedules or inconvenient transportation, people who self-test at home without feeling discomfort or pain often misinterpret the low median value of quantitative tests or weak positive results of qualitative tests and eventually miss the opportunity for early intervention. Therefore, the authors believe that the development of next-generation intelligent POCT should emphasize “automatic” and “communicative”. “automatic” is the automation of the entire detection process from sample processing to result reading, and allows POCT users to obtain results and suggestions directly from a supporting application based on AI algorithms, thus avoiding the interpretation of results by non-expert users. “Communicative” enables tridirectionally communication of information and opinions between POCT users, clinicians, and laboratory technicians to facilitate accurate detection, timely treatment and effective prevention. In addition, wearable devices are also an important trend towards developing smart sensors, especially for patients who need real-time monitoring and prevention of pathogenic infections, such as diabetic foot ulcers and burn patients. Based on a bacterial-responsive DNA hydrogel, Xiong et al. have designed a wireless and battery-free sensor that can be worn on wounds. Animal experiments demonstrated that the sensor can detect bacteria even before obvious manifestations of infection.134Combining IoMT, AI, and sensors has become an effective form of developing smart POCT devices. By training the data, AI plays an important role in the interpretation of test results, and the advent of IoMT enabled the connection between different medical devices and also between medical devices and health professionals.135 Through wireless networks, radio frequency identification, Bluetooth, and other peripheral support technologies, POCT results from various locations can be successively transmitted to the edge layer, fog layer, and cloud server for calculation and big data analysis. In diagnosing infectious diseases, smart POCT devices can potentially be used in self-monitoring scenarios, which is especially important for patients who require frequent medical visits. In the case of AIDS patients, they need to visit their doctors regularly to monitor the viral load in their bodies. Smart POCT devices can motivate AIDS patients to actively participate in treatment and reduce the discrimination they may encounter during offline medical visits because advances in IoMT allow doctors to monitor the condition of AIDS patients and adjust treatment plans promptly, as well as make remote consultation possible. Thanks to the location and transmission capabilities, IoMT-enabled devices are also applicable to areas lacking information management systems, so that governments and hospitals can receive real-time data to identify the location of infectious diseases immediately and take emergency measures to control disease outbreaks and allocate local medical resources to provide medical advice and assistance to infected individuals. However, widespread data communication may easily cause data leakage and raise security concerns. Some technological innovations or integrations that facilitate protecting patient privacy, such as federated learning and blockchain technology, should be fully considered.114,136
5. Discussion of POCT commercialization
The development pathway for POCT products involves a series of procedures such as clinical needs assessment and market analysis, technology development and economic analysis, supplemented sample testing, prototype production, and clinical trials, which leads researchers to spend years and invest sustained finances.137 Despite enormous research exploring the development of POCT detection platforms, the number of commercially available and widely used POCT products is still limited. We explored the reasons behind this and briefly gave some suggestions for readers' reference.5.1. Inadequate clinical evaluation
Many studies focus on method or device innovation itself, but not enough follow-up clinical trials were performed. During the study, the developed method was first initially evaluated by simulated samples, then the compatibility of this method with sample matrix was tested, and finally, a series of evaluation parameters were obtained by testing clinical samples with both the clinical method and the developed method. During this process, the number of clinical samples is generally small, and some biases exist in the process of collecting samples, such as lacking samples from the early stages of infection due to untimely patient visits, and the samples are often from a single center, so the obtained conclusions are often limited. As such, large-scale clinical trials with samples from multiple medical centers in different regions are needed to more objectively and comprehensively verify the practicality and reliability of the method.138 For studies designed to screen asymptomatic infected individuals, investigators should also actively collaborate with the community, clinical departments, customs, and Centers for Disease Control and Prevention to obtain low-concentration clinical samples promptly. Although this process is time-consuming and costly, the detection of clinical low-concentration samples can help investigators to optimize the experimental design and improve the detection performance of the methodology as early as possible, and to a certain extent, avoid the phenomenon that detection performance in the commercialization stage is inferior to that in the development stage.Peeling et al. noted several common problems associated with diagnostic evaluations, including inappropriate purpose and inappropriate reference standard test, which remain in the current study even though this article is older.139 In addition, existing studies lack more detailed and more comprehensive anti-interference assays. The components of clinical samples are complex, and various potentially non-targeted substances may affect the accuracy of test results. Beyond optimizing the experimental design, researchers should also collect specimens from patients in different health states and reasonably design anti-interference assays according to the detection principles to better label the instructions. Only after a systematic, comprehensive, and well-designed clinical evaluation can we ensure that the newly developed POCT provides stable test results and thus has the prospect of commercialization and user acceptance. Undeniably, clinical samples like EBOV are precious and indeed difficult to collect, which poses a significant obstacle to research.
5.2. Difficulties in mass production
The raw materials (sensing elements, such as nanocomposites) and devices (microfluidic chips, portable instruments, and others) of POCT determine the detection performance. The quantities of raw materials and devices needed for laboratory studies are small, but large-scale preparation is required at the commercialization stage to meet market demand. Mass production necessitates strict control of intra- and inter-batch variation of raw materials and devices, so researchers have to explore and carefully optimize the manufacturing process to ensure the stability of inter- and intra-batch assays. In addition to clinical departments, POCT also benefits resource-constrained areas and families. For this reason, developers should focus on the stability of the assay reagents and further explore method to allow the reagents to be stored at room temperature, thereby reducing the need for cold chain transportation and preservation.Tests with a shelf life of more than 18 months are recommended for use in poor areas.139 Furthermore, developers need to control the cost of mass production to make the cost of testing acceptable to most people. While new sensing elements are being explored in current research to achieve the desired detection performance, the improvement in detection performance may be accompanied by an increase in detection cost. Researchers need to find solutions to avoid situations where the cost of POCT makes obtaining regulatory approval difficult, even if the detection performance is desired.
5.3. The ultimate environment and users are ignored
The development of POCT is often performed in the laboratory and lacks performance testing under harsh conditions. Developers should test the prototype product following the original purposes. For example, for POCT products used in resource-limited areas, the tolerance of the product to the local environment needs to be tested in the field for better use in poor areas. In these underdeveloped areas, the high incidence of the studied diseases and the presence of co-infection with multiple pathogens helped the investigators to fully validate the specificity and accuracy of POCT. Another reason for conducting field testing is that the local testing environment is quite different from the laboratory environment. In less developed areas, even laboratories also suffer from varying degrees of physical constraints, such as limited clean water, unreliable power supplies, and large variations in temperature and humidity. These differences in the testing environment undoubtedly affect the accuracy and stability of test results.140 Secondly, researchers tend to sacrifice user-friendliness to further improve the ability to identify cases (e.g., using portable readout devices). Patient satisfaction is an essential factor in determining the success of a technology. Therefore, the POCT platform should strike a balance between detection ability and user-friendliness and be optimized based on users' feedback. In addition, at the technical level, more expectations can be focus on the development of a highly-sensitive POCT platform that delivers visual results, as well as the development of a smart and portable device that simplifies the operation and maintenance process and provides users with intuitive and easy-to-understand results, then user interest can be stimulated.6. Conclusions
Timely and effective diagnosis of infectious diseases is the key to anti-infection treatment and has become an urgent need today. The inaccessibility of testing prevents early detection of diseases, and erroneous or empirical treatment protocols undoubtedly delay the optimal stage of treatments and even cause the widespread of this disease. POCT is a crucial way to address the rapid diagnosis of infectious diseases, improve healthcare environment and effectively manage outbreaks in developing regions. Therefore, this review focuses on five infectious diseases that are often outbreaks in impoverished areas lacking medical equipment, summarizes recently developed POCT methods for diagnosing infectious diseases, and demonstrates how innovations in POCT devices can be used to diagnose these diseases. Emerging technologies such as AI, cloud computing, and IoMT are revolutionizing the POCT field in terms of results interpretation, data storage, and disease management. Increasingly popular smartphones also be used as optical sensors and corresponding applications can provide patients with more tailored management. The clinical scenarios of pathogen POCT devices are not widely used as well as some challenges in regulatory documents and technology still exist, but the potential of POCT as an accurate and effective diagnostic tool is unquestionable. Meanwhile, some other challenges also need to be addressed, like accurately handling data backup and recovery in the event of network outages. We firmly believe that these issues will be solved one by one as technology develops. This review will inspire researchers interested in developing POCT devices to establish the next-generation smart POCT platforms to serve human beings better.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China [grant numbers 62122017, 2017].References
- R. Rezar, et al., Infective endocarditis - A review of current therapy and future challenges, Hell. J. Cardiol., 2021, 62(3), 190–200 CrossRef PubMed.
- K. Putz, K. Hayani and F. A. Zar, Meningitis, Prim. Care, 2013, 40(3), 707–726 CrossRef PubMed.
- B. Foxman, Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden, Infect. Dis. Clin. North Am., 2014, 28(1), 1–13 CrossRef PubMed.
- W. Fischbach and P. Malfertheiner, Helicobacter Pylori Infection, Dtsch. Arztebl. Int., 2018, 115(25), 429–436 Search PubMed.
- J. H. Nichols, et al., Executive summary. The National Academy of Clinical Biochemistry Laboratory Medicine Practice Guideline: evidence-based practice for point-of-care testing, Clin. Chim. Acta, 2007, 379(1–2), 14–28 CrossRef CAS PubMed , discussion 29–30.
- S. Yadav, N. N. Sharma and J. Akhtar, Nucleic acid analysis on paper substrates (NAAPs): an innovative tool for Point of Care (POC) infectious disease diagnosis, Analyst, 2021, 146(11), 3422–3439 RSC.
- H. Kim, D. R. Chung and M. Kang, A new point-of-care test for the diagnosis of infectious diseases based on multiplex lateral flow immunoassays, Analyst, 2019, 144(8), 2460–2466 RSC.
- Z. Li, et al., Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care, Biotechnol. Adv., 2019, 37(8), 107440 CrossRef CAS PubMed.
- D. S. Y. Ong and M. Poljak, Smartphones as mobile microbiological laboratories, Clin. Microbiol. Infect., 2020, 26(4), 421–424 CrossRef CAS PubMed.
- L. R. Runtuwene, et al., On-Site MinION Sequencing, Adv. Exp. Med. Biol., 2019, 1129, 143–150 CrossRef CAS PubMed.
- Y. P. Wong, et al., Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms, J. Appl. Microbiol., 2018, 124(3), 626–643 CrossRef CAS PubMed.
- N. Singpanomchai, et al., Rapid detection of multidrug-resistant tuberculosis based on allele-specific recombinase polymerase amplification and colorimetric detection, PLoS One, 2021, 16(6), e0253235 CrossRef CAS PubMed.
- S. Zhang, et al., Development of a Directly Visualized Recombinase Polymerase Amplification-SYBR Green I Method for the Rapid Detection of African Swine Fever Virus, Front. Microbiol., 2020, 11, 602709 CrossRef PubMed.
- Priti, et al., A rapid field-based assay using recombinase polymerase amplification for identification of Thrips palmi, a vector of tospoviruses, J. Pest Sci., 2021, 94(2), 219–229 CrossRef CAS PubMed.
- S. Tomar, B. Lavickova and C. Guiducci, Recombinase polymerase amplification in minimally buffered conditions, Biosens. Bioelectron., 2022, 198, 113802 CrossRef CAS PubMed.
- J. S. Gootenberg, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356(6336), 438–442 CrossRef CAS PubMed.
- L. Li, et al., CRISPR-Cas-mediated diagnostics, Trends Biotechnol., 2022, 40(11), 1326–1345 CrossRef CAS PubMed.
- Y. Zhao, et al., Isothermal Amplification of Nucleic Acids, Chem. Rev., 2015, 115(22), 12491–12545 CrossRef CAS PubMed.
- S. Biswas, et al., Label-Free Electrochemical Immunosensor for Ultrasensitive Detection of Carbohydrate Antigen 125 Based on Antibody-Immobilized Biocompatible MOF-808/CNT, ACS Appl. Mater. Interfaces, 2021, 13(2), 3295–3302 CrossRef CAS PubMed.
- Z. Rahmati, et al., Label-free electrochemical aptasensor for rapid detection of SARS-CoV-2 spike glycoprotein based on the composite of Cu(OH)(2) nanorods arrays as a high-performance surface substrate, Bioelectrochemistry, 2022, 146, 108106 CrossRef CAS PubMed.
- L. Cohen, et al., Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay, ACS Nano, 2020, 14(8), 9491–9501 CrossRef CAS PubMed.
- B. Dahiya and P. K. Mehta, Detection of potential biomarkers associated with outrageous diseases and environmental pollutants by nanoparticle-based immuno-PCR assays, Anal. Biochem., 2019, 587, 113444 CrossRef CAS PubMed.
- J. Guo, et al., Nanomaterial Labels in Lateral Flow Immunoassays for Point-of-Care-Testing, J. Mater. Sci. Technol., 2021, 60, 90–104 CrossRef CAS.
- C. Hong, et al., A dual-signal electrochemiluminescence immunosensor for high-sensitivity detection of acute myocardial infarction biomarker, Biosens. Bioelectron., 2021, 194, 113591 CrossRef CAS PubMed.
- D. Liu, et al., Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen, Biosens. Bioelectron., 2020, 173, 112817 CrossRef PubMed.
- S. M. Yang, et al., Highly accurate multiprotein detection on a digital ELISA platform, Lab Chip, 2022, 22(16), 3015–3024 RSC.
- S. Aydin, A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA, Peptides, 2015, 72, 4–15 CrossRef CAS PubMed.
- L. Chen, et al., Molecular imprinting: perspectives and applications, Chem. Soc. Rev., 2016, 45(8), 2137–2211 RSC.
- J. Zhang, Y. Wang and X. Lu, Molecular imprinting technology for sensing foodborne pathogenic bacteria, Anal. Bioanal. Chem., 2021, 413(18), 4581–4598 CrossRef CAS PubMed.
- G. Guan, J. H. Pan and Z. Li, Innovative utilization of molecular imprinting technology for selective adsorption and (photo)catalytic eradication of organic pollutants, Chemosphere, 2021, 265, 129077 CrossRef CAS PubMed.
- G. Jamalipour Soufi, S. Iravani and R. S. Varma, Molecularly imprinted polymers for the detection of viruses: challenges and opportunities, Analyst, 2021, 146(10), 3087–3100 RSC.
- R. Tian, et al., Recent development in the design of artificial enzymes through molecular imprinting technology, J. Mater. Chem. B, 2022, 10(35), 6590–6606 RSC.
- W. Xu, et al., Fabrication of magnetic polymers based on deep eutectic solvent for separation of bovine hemoglobin via molecular imprinting technology, Anal. Chim. Acta, 2019, 1048, 1–11 CrossRef CAS PubMed.
- M. Arabi, et al., Molecular Imprinting: Green Perspectives and Strategies, Adv. Mater., 2021, 33(30), e2100543 CrossRef PubMed.
- M. Hoffmann, et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, 2020, 181(2), 271–280 CrossRef CAS PubMed , e8.
- A. C. Walls, et al., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell, 2020, 181(2), 281–292 CrossRef CAS PubMed , e6.
- S. M. Hirabara, et al., SARS-COV-2 Variants: Differences and Potential of Immune Evasion, Front. Cell. Infect. Microbiol., 2021, 11, 781429 CrossRef CAS PubMed.
- C. Laffeber, et al., Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants, J. Mol. Biol., 2021, 433(15), 167058 CrossRef CAS PubMed.
- C. S. Lupala, et al., Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor, Biochem. Biophys. Res. Commun., 2022, 590, 34–41 CrossRef CAS PubMed.
- J. Ou, et al., V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity, J. Virol., 2021, 95(16), e0061721 CrossRef PubMed.
- L. F. de Lima, et al., Minute-scale detection of SARS-CoV-2 using a low-cost biosensor composed of pencil graphite electrodes, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(30), e2106724118 CrossRef CAS PubMed.
- J. Han, et al., SARS-CoV-2 spike protein detection using slightly tapered no-core fiber-based optical transducer, Microchim. Acta, 2022, 189(9), 321 CrossRef CAS PubMed.
- J. H. Lee, et al., A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2), Biosens. Bioelectron., 2021, 171, 112715 CrossRef CAS PubMed.
- S. Park, et al., SARS-CoV-2 Variant Screening Using a Virus-Receptor-Based Electrical Biosensor, Nano Lett., 2022, 22(1), 50–57 CrossRef CAS PubMed.
- R. L. Pinals, et al., Rapid SARS-CoV-2 Spike Protein Detection by Carbon Nanotube-Based Near-Infrared Nanosensors, Nano Lett., 2021, 21(5), 2272–2280 CrossRef CAS PubMed.
- H. Wei, et al., Research progress of biosensors for detection of SARS-CoV-2 variants based on ACE2, Talanta, 2022, 251, 123813 CrossRef PubMed.
- World Health Organization, Key facts and latest estimates on the global HIV epidemic - 2021. 1 August 2022; Available from: https://cdn.who.int/media/docs/default-source/hq-hiv-hepatitis-and-stis-library/key-facts-hiv-2021-26july2022.pdf?sfvrsn=8f4e7c93_5.
- World Health Organization, Fact sheets of HIV. 1 August 2022; Available from: https://www.who.int/news-room/fact-sheets/detail/hiv-aids.
- S. L. Lecher, et al., HIV Viral Load Monitoring Among Patients Receiving Antiretroviral Therapy - Eight Sub-Saharan Africa Countries, 2013-2018, Morb. Mortal. Wkly. Rep., 2021, 70(21), 775–778 CrossRef CAS PubMed.
- M. Kong, et al., A wearable microfluidic device for rapid detection of HIV-1 DNA using recombinase polymerase amplification, Talanta, 2019, 205, 120155 CrossRef CAS PubMed.
- T. Liu, et al., Fingerpick Blood-Based Nucleic Acid Testing on A USB Interfaced Device towards HIV self-testing, Biosens. Bioelectron., 2022, 209, 114255 CrossRef CAS PubMed.
- E. R. Gray, et al., p24 revisited: a landscape review of antigen detection for early HIV diagnosis, Aids, 2018, 32(15), 2089–2102 CrossRef CAS PubMed.
- K. Wu, et al., Recent progress of microfluidic chips in immunoassay, Front. Bioeng. Biotechnol., 2022, 10, 1112327 CrossRef PubMed.
- D. Liu, et al., Microfluidic-Integrated Multicolor Immunosensor for Visual Detection of HIV-1 p24 Antigen with the Naked Eye, Anal. Chem., 2020, 92(17), 11826–11833 CrossRef CAS PubMed.
- F. Li, et al., Smartphone assisted immunodetection of HIV p24 antigen using reusable, centrifugal microchannel array chip, Talanta, 2019, 203, 83–89 CrossRef CAS PubMed.
- S. K. Sailapu, et al., Standalone operation of an EGOFET for ultra-sensitive detection of HIV, Biosens. Bioelectron., 2020, 156, 112103 CrossRef CAS PubMed.
- T. Akyazi, L. Basabe-Desmonts and F. Benito-Lopez, Review on microfluidic paper-based analytical devices towards commercialisation, Anal. Chim. Acta, 2018, 1001, 1–17 CrossRef CAS PubMed.
- E. Noviana, et al., Microfluidic Paper-Based Analytical Devices: From Design to Applications, Chem. Rev., 2021, 121(19), 11835–11885 CrossRef CAS PubMed.
- C. A. Chen, et al., An electricity- and instrument-free infectious disease sensor based on a 3D origami paper-based analytical device, Lab Chip, 2021, 21(10), 1908–1915 RSC.
- M. Sher and W. Asghar, Development of a multiplex fully automated assay for rapid quantification of CD4(+) T cells from whole blood, Biosens. Bioelectron., 2019, 142, 111490 CrossRef CAS PubMed.
- W. Xiao, et al., A Rapid, Simple, and Low-Cost CD4 Cell Count Sensor Based on Blocking Immunochromatographic Strip System, ACS Sens., 2019, 4(6), 1508–1514 CrossRef CAS PubMed.
- World Health Organization, The history of Zika virus. 20 July 2022; Available from: https://www.who.int/news-room/feature-stories/detail/the-history-of-zika-virus.
- World Health Organization, Zika virus. 21 July 2022; Available from: https://www.who.int/en/news-room/fact-sheets/detail/zika-virus.
- P. F. N. Estrela, et al., Ten-minute direct detection of Zika virus in serum samples by RT-LAMP, J. Virol. Methods, 2019, 271, 113675 CrossRef CAS PubMed.
- A. M. Jankelow, et al., Smartphone clip-on instrument and microfluidic processor for rapid sample-to-answer detection of Zika virus in whole blood using spatial RT-LAMP, Analyst, 2022, 147(17), 3838–3853 RSC.
- Y. P. Hsu, et al., A serological point-of-care test for Zika virus detection and infection surveillance using an enzyme-free vial immunosensor with a smartphone, Biosens. Bioelectron., 2020, 151, 111960 CrossRef CAS PubMed.
- N. J. Wittenberg and C. L. Haynes, Using nanoparticles to push the limits of detection, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1(2), 237–254 CAS.
- Q. Bayin, et al., Anti-SARS-CoV-2 IgG and IgM detection with a GMR based LFIA system, Talanta, 2021, 227, 122207 CrossRef CAS PubMed.
- J. Guo, et al., 5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19, Biosens. Bioelectron., 2021, 181, 113160 CrossRef CAS PubMed.
- Q. Liu, et al., Near-Infrared Lanthanide-Doped Nanoparticles for a Low Interference Lateral Flow Immunoassay Test, ACS Appl. Mater. Interfaces, 2020, 12(4), 4358–4365 CrossRef CAS PubMed.
- L. D. Xu, et al., Luminous silica colloids with carbon dot incorporation for sensitive immunochromatographic assay of Zika virus, Analyst, 2021, 146(2), 706–713 RSC.
- A. T. V. Nguyen, et al., Development of a peptide aptamer pair-linked rapid fluorescent diagnostic system for Zika virus detection, Biosens. Bioelectron., 2022, 197, 113768 CrossRef CAS PubMed.
- M. S. Draz, et al., Nanoparticle-enhanced electrical detection of Zika virus on paper microchips, Nanoscale, 2018, 10(25), 11841–11849 RSC.
- L. Zhu, et al., Digital multimeter-based point-of-care immunoassay of prostate-specific antigen coupling with a flexible photosensitive pressure sensor, Sens. Actuators, B, 2021, 343, 130121 CrossRef CAS.
- M. S. Draz, et al., Motion-Based Immunological Detection of Zika Virus Using Pt-Nanomotors and a Cellphone, ACS Nano, 2018, 12(6), 5709–5718 CrossRef CAS PubMed.
- V. Ricotta, et al., A chip-based potentiometric sensor for a Zika virus diagnostic using 3D surface molecular imprinting, Analyst, 2019, 144(14), 4266–4280 RSC.
- C. Huang, et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, 2020, 395(10223), 497–506 CrossRef CAS PubMed.
- World Health Organization, Coronavirus disease (COVID-19). 18 September 2022; Available from: https://www.who.int/health-topics/coronavirus#tab=tab_3.
- J. P. Broughton, et al., CRISPR-Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed.
- B. Casati, et al., Rapid, adaptable and sensitive Cas13-based COVID-19 diagnostics using ADESSO, Nat. Commun., 2022, 13(1), 3308 CrossRef CAS PubMed.
- J. Joung, et al., Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing, N. Engl. J. Med., 2020, 383(15), 1492–1494 CrossRef CAS PubMed.
- F. Hu, et al., A one-pot CRISPR/Cas13a-based contamination-free biosensor for low-cost and rapid nucleic acid diagnostics, Biosens. Bioelectron., 2022, 202, 113994 CrossRef CAS PubMed.
- K. G. de Oliveira, et al., Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection, Analyst, 2021, 146(4), 1178–1187 RSC.
- C. Badua, K. A. T. Baldo and P. M. B. Medina, Genomic and proteomic mutation landscapes of SARS-CoV-2, J. Med. Virol., 2021, 93(3), 1702–1721 CrossRef CAS PubMed.
- H. Lin, et al., Combination of Isothermal Recombinase-Aided Amplification and CRISPR-Cas12a-Mediated Assay for Rapid Detection of Major Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern, Front. Microbiol., 2022, 13, 945133 CrossRef PubMed.
- S. Y. Li, et al., CRISPR-Cas12a-assisted nucleic acid detection, Cell Discovery, 2018, 4, 20 CrossRef PubMed.
- X. Zhu, et al., Rapid, Ultrasensitive, and Highly Specific Diagnosis of COVID-19 by CRISPR-Based Detection, ACS Sens., 2021, 6(3), 881–888 CrossRef CAS PubMed.
- B. O. Villoutreix, et al., In Silico Investigation of the New UK (B.1.1.7) and South African (501Y.V2) SARS-CoV-2 Variants with a Focus at the ACE2-Spike RBD Interface, Int. J. Mol. Sci., 2021, 22(4), 1695 CrossRef CAS PubMed.
- S. Pickering, et al., Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study, Lancet Microbe, 2021, 2(9), e461–e471 CrossRef CAS PubMed.
- World Health Organization, Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. 31, October, 2022; Available from: https://www.who.int/publications/i/item/10665-331501.
- Y. Dai and C. C. Liu, Recent Advances on Electrochemical Biosensing Strategies toward Universal Point-of-Care Systems, Angew. Chem., Int. Ed., 2019, 58(36), 12355–12368 CrossRef CAS PubMed.
- Z. Zhang, et al., Nanomaterials-based Electrochemical Immunosensors, Micromachines, 2019, 10(6), 397 CrossRef PubMed.
- R. Zeng, et al., Smartphone-Based Electrochemical Immunoassay for Point-of-Care Detection of SARS-CoV-2 Nucleocapsid Protein, Anal. Chem., 2022, 94(43), 15155–15161 CrossRef CAS PubMed.
- D. Najjar, et al., A lab-on-a-chip for the concurrent electrochemical detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva and plasma, Nat. Biomed. Eng., 2022, 6(8), 968–978 CrossRef CAS PubMed.
- H. de Puig, et al., Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants, Sci. Adv., 2021, 7(32), eabh2944 CrossRef PubMed.
- S. T. Jacob, et al., Ebola virus disease, Nat. Rev. Dis. Primers, 2020, 6(1), 13 CrossRef PubMed.
- World Health Organization, Ebola virus disease. 23 September 2022; Available from: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease.
- W. Na, et al., Rapid molecular diagnosis of infectious viruses in microfluidics using DNA hydrogel formation, Biosens. Bioelectron., 2018, 108, 9–13 CrossRef CAS PubMed.
- J. Hu, et al., Colorimetric-Fluorescent-Magnetic Nanosphere-Based Multimodal Assay Platform for Salmonella Detection, Anal. Chem., 2019, 91(1), 1178–1184 CrossRef CAS PubMed.
- X. Liu, et al., Multifunctional Nano-Sunflowers with Color-Magnetic-Raman Properties for Multimodal Lateral Flow Immunoassay, Anal. Chem., 2021, 93(7), 3626–3634 CrossRef CAS PubMed.
- P. Y. You, et al., Colorimetric and Fluorescent Dual-Mode Immunoassay Based on Plasmon-Enhanced Fluorescence of Polymer Dots for Detection of PSA in Whole Blood, ACS Appl. Mater. Interfaces, 2019, 11(10), 9841–9849 CrossRef CAS PubMed.
- J. Hu, et al., Dual-Signal Readout Nanospheres for Rapid Point-of-Care Detection of Ebola Virus Glycoprotein, Anal. Chem., 2017, 89(24), 13105–13111 CrossRef CAS PubMed.
- A. J. Qavi, et al., Rapid detection of an Ebola biomarker with optical microring resonators, Cells Rep. Methods, 2022, 2(6), 100234 CrossRef CAS PubMed.
- C. M. Fontes, et al., Ultrasensitive point-of-care immunoassay for secreted glycoprotein detects Ebola infection earlier than PCR, Sci. Transl. Med., 2021, 13(588), eabd9696 CrossRef CAS PubMed.
- K. G. Barnes, et al., Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time, Nat. Commun., 2020, 11(1), 4131 CrossRef CAS PubMed.
- H. Liu, et al., SERS Tags for Biomedical Detection and Bioimaging, Theranostics, 2022, 12(4), 1870–1903 CrossRef CAS PubMed.
- D. Sebba, et al., A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases, Sci. Transl. Med., 2018, 10(471), eaat0944 CrossRef CAS PubMed.
- A. Rajagopalan and C. M. Ilboudo, Malaria, Pediatr. Rev., 2019, 40(3), 151–153 CrossRef PubMed.
- World Health Organization, Malaria. 26 October 2022; Available from: https://www.who.int/news-room/fact-sheets/detail/malaria.
- World Health Organization, World malaria report 2021. 28 October 2022; Available from: https://apps.who.int/iris/handle/10665/350147.
- G. Ruiz-Vega, et al., Electrochemical POC device for fast malaria quantitative diagnosis in whole blood by using magnetic beads, Poly-HRP and microfluidic paper electrodes, Biosens. Bioelectron., 2020, 150, 111925 CrossRef CAS PubMed.
- J. Kim, et al., A two-colour multiplexed lateral flow immunoassay system to differentially detect human malaria species on a single test line, Malar. J., 2019, 18(1), 313 CrossRef PubMed.
- J. Reboud, et al., Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(11), 4834–4842 CrossRef CAS PubMed.
- X. Guo, et al., Smartphone-based DNA diagnostics for malaria detection using deep learning for local decision support and blockchain technology for security, Nat. Electron., 2021, 4(8), 615–624 CrossRef CAS.
- M. Y. Lai, et al., Point-of-Care Diagnosis of Malaria Using a Simple, Purification-Free DNA Extraction Method Coupled with Loop-Mediated Isothermal Amplification-Lateral Flow, Tropical Medicine and Infectious Disease, 2023, 8(4), 199 CrossRef PubMed.
- R. A. Lee, et al., Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(41), 25722–25731 CrossRef CAS PubMed.
- World Health Organization, Tackling emerging antimalarial drug resistance in Africa. 21 March 2023; Available from: https://www.who.int/news/item/18-11-2022-tackling-emerging-antimalarial-drug-resistance-in-africa.
- J. Naß and T. Efferth, Development of artemisinin resistance in malaria therapy, Pharmacol. Res., 2019, 146, 104275 CrossRef PubMed.
- World Health Organization, Global technical strategy for malaria 2016-2030, 2021 update. 23 March 2023; Available from: https://www.who.int/publications/i/item/9789240031357.
- K. Malpartida-Cardenas, et al., Allele-Specific Isothermal Amplification Method Using Unmodified Self-Stabilizing Competitive Primers, Anal. Chem., 2018, 90(20), 11972–11980 CrossRef CAS PubMed.
- K. Malpartida-Cardenas, et al., Quantitative and rapid Plasmodium falciparum malaria diagnosis and artemisinin-resistance detection using a CMOS Lab-on-Chip platform, Biosens. Bioelectron., 2019, 145, 111678 CrossRef CAS PubMed.
- G. A. Garcia, et al., Malaria absorption peaks acquired through the skin of patients with infrared light can detect patients with varying parasitemia, PNAS Nexus, 2022, 1(5), pgac272 CrossRef PubMed.
- D. Mabey, et al., Diagnostics for the developing world, Nat. Rev. Microbiol., 2004, 2(3), 231–240 CrossRef CAS PubMed.
- K. T. L. Trinh and N. Y. Lee, Fabrication of Wearable PDMS Device for Rapid Detection of Nucleic Acids via Recombinase Polymerase Amplification Operated by Human Body Heat, Biosensors, 2022, 12(2), 72 CrossRef CAS PubMed.
- Abionic PSP PRODUCT LEAFLET, 15 October 2022; Available from: https://www.abionic.com/sites/default/files/pdf/D4haab_221108_PSP_Brochure_V2.1_JGA.pdf.
- N. Li, et al., A self-contained and fully integrated fluidic cassette system for multiplex nucleic acid detection of bacteriuria, Lab Chip, 2020, 20(2), 384–393 RSC.
- X. Zhu, et al., A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection, Micromachines, 2020, 11(2), 186 CrossRef PubMed.
- B. Shu, et al., A pocket-sized device automates multiplexed point-of-care RNA testing for rapid screening of infectious pathogens, Biosens. Bioelectron., 2021, 181, 113145 CrossRef CAS PubMed.
- C. H. Liu, et al., Blood Plasma Separation Using a Fidget-Spinner, Anal. Chem., 2019, 91(2), 1247–1253 CrossRef CAS PubMed.
- A. I. Khan, Best Laboratory Practices Regarding POCT in Different Settings (Hospital and Outside the Hospital), eJIFCC, 2021, 32(2), 124–130 Search PubMed.
- P. Chen, et al., Integrated and finger-actuated microfluidic chip for point-of-care testing of multiple pathogens, Talanta, 2021, 224, 121844 CrossRef CAS PubMed.
- J. Yin, et al., A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens, Lab Chip, 2020, 20(5), 979–986 RSC.
- P. Mitra and P. Sharma, POCT in Developing Countries, eJIFCC, 2021, 32(2), 195–199 Search PubMed.
- Z. Xiong, et al., A wireless and battery-free wound infection sensor based on DNA hydrogel, Sci. Adv., 2021, 7(47), eabj1617 CrossRef CAS PubMed.
- B. Pradhan, S. Bhattacharyya and K. Pal, IoT-Based Applications in Healthcare Devices, J. Healthc. Eng., 2021, 2021, 6632599 Search PubMed.
- Y. Chang, C. Fang and W. Sun, A Blockchain-Based Federated Learning Method for Smart Healthcare, Comput. Intell. Neurosci., 2021, 2021, 4376418 Search PubMed.
- P. Wang and L. J. Kricka, Current and Emerging Trends in Point-of-Care Technology and Strategies for Clinical Validation and Implementation, Clin. Chem., 2018, 64(10), 1439–1452 CrossRef CAS PubMed.
- J. Wang, et al., Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA, Clin. Microbiol. Infect., 2020, 26(8), 1076–1081 CrossRef CAS PubMed.
- R. W. Peeling, P. G. Smith and P. M. Bossuyt, A guide for diagnostic evaluations, Nat Rev Microbiol, 2006, 4(9 Suppl), S2–S6 CrossRef PubMed.
- J. Hu, et al., Portable microfluidic and smartphone-based devices for monitoring of cardiovascular diseases at the point of care, Biotechnol. Adv., 2016, 34(3), 305–320 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
