Open Access Article
Open Access ArticleEngineering thermostable affinity proteins for use in high-throughput immunoassay formats†
Huan
Jia‡
 a,
Nazirulmubin Abdul
Moomen‡
a,
Jeanette
Leong‡
a,
Patthara
Kongsuphol‡
a,
Zhi Feng Sherman
Lim
a,
Nazirulmubin Abdul
Moomen‡
a,
Jeanette
Leong‡
a,
Patthara
Kongsuphol‡
a,
Zhi Feng Sherman
Lim
 a,
Carmen Sze Min
Pui
a,
Yuxuan
Tan
a,
Ki-Joo
Sung
b and
Hadley D.
Sikes
a,
Carmen Sze Min
Pui
a,
Yuxuan
Tan
a,
Ki-Joo
Sung
b and
Hadley D.
Sikes
 *ab
*ab
aSingapore-MIT Alliance for Research and Technology Centre, Antimicrobial Resistance, 1 CREATE Way #03-10/11 Innovation Wing, #03-12/13/14 Enterprise Wing, Singapore. E-mail: sikes@mit.edu
bMassachusetts Institute of Technology, Chemical Engineering, Cambridge, Massachusetts, USA
First published on 25th April 2023
Abstract
ELISA utilizes polystyrene materials for capture reagents. Natural absorption randomly orients the capture reagent, decreasing immobilization and target biomarker accessibility. A thermostable affinity protein rcSso7d can replace antibodies as a capture reagent in cellulose-based diagnostic tests. ‘RAFIASRRIRRP’ at the N-terminus of two repeating units of rcSso7d improved polystyrene immobilization for high-throughput ELISA.
1. Introduction
Immunoassays are bioanalytical techniques that quantify a target analyte on the basis of its interaction with affinity agents such as antibodies, aptamers, and small proteins.1–5 As detection reagents in the sandwich-based enzyme-linked immunosorbent assay (ELISA), match-paired antibodies that bind to complementary epitopes of the target biomarker are typically used. In recent years, as immunoassays and diagnostic tests have advanced, substitutions of antibodies have been used extensively in immunoassays to overcome drawbacks of antibodies, such as off-target binding, unpredictable thermostability, lengthy production times, high batch-to-batch variation, and complexity of modifications.6,7An engineered rcSso7d scaffold protein is a candidate as a detection reagent for the development of diagnostic tests. It has been shown to have great thermostability with a melting temperature of up to 98 °C and can be easily manufactured through bacterial production.8–10 Moreover, the RAPIDS (rapid affinity pair identification by directed selection) method can efficiently screen rcSso7d scaffold proteins as matched pairs for target biomarkers.11
For diagnostic test applications, an anchoring substrate is required for trapping the sandwich complex and differentiating it from the surrounding fluid. Polystyrene multi-well plates, nitrocellulose paper, and cellulose paper are common substrates.12 With tailored rcSso7d affinity agents with an additional cellulose binding domain (CBD), we have investigated the possibility of employing cellulose paper instead of nitrocellulose paper for point-of-care diagnostics.5,13
In contrast, 96-well polystyrene plates are commonly used in diagnostic tests, biochemical research, and pharmaceutical research in laboratory settings.14 The capability to conduct high-throughput experiments is an advantage of 96-well plates. Random orientation of the physiosorbed capture reagent on the polystyrene substrate has been recognized as a convenience that comes with limitations, as it significantly reduces the accessibility of the binding site to the target ligand. In addition, a small-sized capture reagent may lose functionality when immobilized on a polystyrene surface.15,16
Here, we investigate and compare methods for expanding the utilization of rcSso7d proteins to polystyrene 96-well immunoassay plates. Biomolecular modification strategies were employed to create modified rcSso7d proteins in an effort to increase immobilisation efficiency. The ELISA detection capability of modified rcSso7d proteins was also investigated.
2. Results
As a study model, a pair of engineered rcSso7d binders for interleukin-6 (IL-6) were utilized, namely Sso.E1 and Sso.E2.11,17 Sso.E1 was utilized as a capture binder for the modifications. Sso.E2 was fused to a maltose binding protein (MBP) and biotin acceptor (BA) to generate a reporter reagent BA-MBP-Sso.E2. In this investigation, we compared the immobilisation efficiency and ELISA performance of eight Sso.E1 constructs, as shown in Fig. 1A. Using SDS-PAGE, the isolated proteins encoded by these eight Sso.E1 genetic constructions and BA-MBP-Sso.E2 are depicted in Fig. 1B.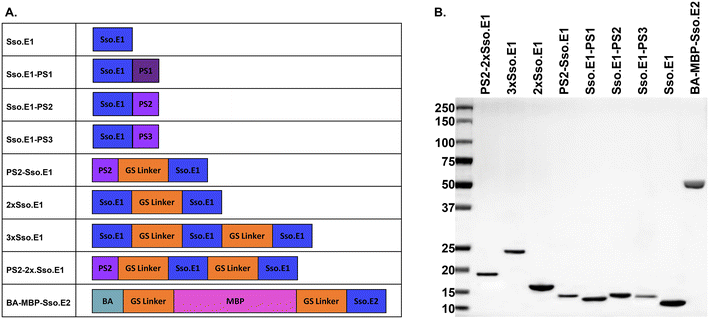 | ||
| Fig. 1 Information on protein constructs used in this study. A. A list of modified Sso.E1 constructs and BA-MBP-Sso.E2 construct, and B. purified proteins shown on SDS-PAGE. | ||
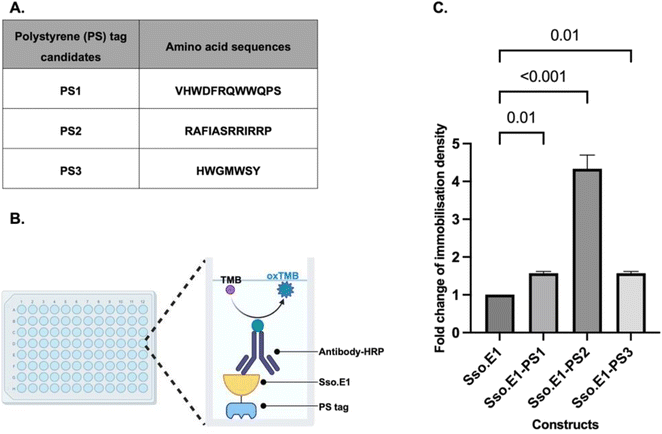
2.1 Small peptide polystyrene tag PS2 shows the greatest improvement in immobilisation of Sso.E1 onto polystyrene plates
A cellulose binding domain (CBD) was fused to the modified rcSso7d binders to improve their capture ability towards cellulose paper, hence enabling applications in cellulose-based point-of-care diagnostics.5,13,17–20 Using this biomolecular modification strategy as a guide, we explored the literature for fusion peptide sequences with the property of efficient capture towards polystyrene substrates. Three candidates listed in Fig. 2A have been identified and labelled as PS1, PS2 and PS3, which were isolated from a phage display library. These peptides exhibited consistent adhesion to polystyrene plates and were further characterised showing improved interaction with polystyrene substrates.21–23At the C terminus of Sso.E1, nucleotide sequences encoding PS1, PS2, and PS3 were inserted. All constructs were expressed and purified. Equimolar amounts of the Sso.E1-PS1, Sso.E1-PS2, Sso.E1-PS3 and Sso.E1 proteins were added to 96-well polystyrene plates, and each sample was tested with three replicates. Plates were incubated at room temperature for 10 to 120 minutes, and immobilisation densities were compared using an anti-His antibody linked to horseradish peroxidase (HRP) to mark the histidine tag at the N terminus of the proteins (Fig. 2B). At a brief immobilisation duration of 10 minutes, all PS tagged constructs exhibited improved immobilisation density in comparison to the original Sso.E1. The PS2 tag, with the amino acid sequence ‘RAFIASRRIRRP’, demonstrated the largest improvement of more than 3 fold (Fig. 2C). In addition, the immobilisation density of Sso.E1-PS2 improved with increasing incubation time in comparison to that of Sso.E1. At 60 minutes of incubation, the maximum fold change of immobilisation density between Sso.E1-PS2 and Sso.E1 was 20-fold (Fig. S1†).
2.2 The PS2-2xSso.E1 construct has optimal ELISA performance in detecting IL-6
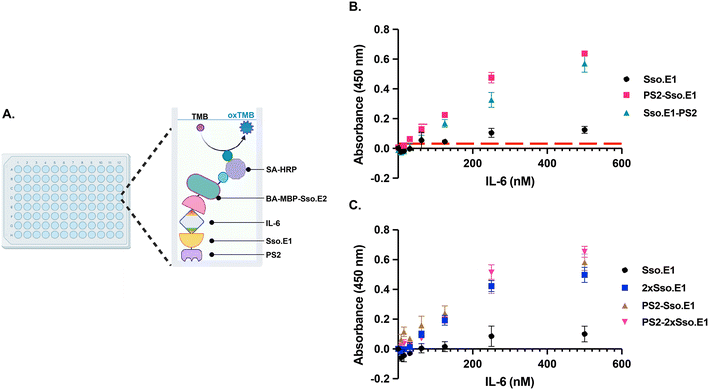
To further optimize the performance of Sso.E1 in ELISA, we examined the position of the PS2 tag at the N- or C-terminus, namely PS2-Sso.E1 and Sso.E1-PS2 (Fig. 1A). To conduct ELISA, Sso.E1 constructs were immobilised for 10 minutes in a 96-well polystyrene plate, followed by IL-6 antigen incubation at a range of doses. The IL-6 protein was labelled with BA-MBP-Sso.E2, and the signal was detected with HRP conjugated streptavidin (SA-HRP) and TMB (3,3′,5,5′-tetramethylbenzidine) (Fig. 3A). As depicted in Fig. 3B, PS2-Sso.E1 provides the greatest signal response for identifying the IL-6 antigen, compared to Sso.E1-PS2 and Sso.E1. The limit of detection (LoD) for the IL-6 antigen improved to 31.25 nM for PS2-Sso.E1 as compared to an LoD of 62.5 nM for Sso.E1-PS2 and LoD of 250 nM for Sso.E1.In a recent work, it was demonstrated that repeating units of an rcSso7d binder sequence targeting streptavidin (SA) as a model analyte can increase the density of binding sites, hence enhancing the detection capability of the SA analyte.24 The performance of repeating Sso.SA in capturing SA-HRP as a model analyte was evaluated, revealing a significant enhancement in the signal with 2xSso.SA, but a similar performance with 3xSso.SA, 4xSso.SA, and 5xSso.SA (Fig. S2A†). Thus, we designed 2xSso.E1 and 3xSso.E1 in order to test their sensitivity in identifying the IL-6 antigen in ELISA. As shown in Fig. S2B,† 2xSso.E1 and 3xSso.E1 exhibit comparable detection performance; hence 2xSso.E1 modification was chosen.
Next, a combination of the PS2 tag and repetitive units of Sso.E1 was designed to obtain PS2-2xSso.E1. The performance of PS2-2xSso.E1 in comparison to that of Sso.E1, PS2-Sso.E1, and 2xSso.E1 was evaluated by ELISA. Compared to the Sso.E1 construct, the PS2-Sso.E1 and 2xSso.E1 constructs resulted in a significant improvement in the detection signal, while the PS2-2xSso.E1 construct resulted in an even greater gain in performance (Fig. 3C).
3. Discussion and conclusions
Building upon our prior use of protein engineering technologies to evolve rcSso7d for protein detection assays,5,17,18 we demonstrated here the methods to broaden the use of the rcSso7d protein onto polystyrene 96 well-plates, a consumable often used for lab-based high-throughput ELISA tests.The method of protein engineering at the genetic level, such as the inclusion of the PS2 peptide sequence and the repeating units of the rcSso7d region, allows for the rapid and consistent modification of proteins. It eliminates post-translational modifications, which have a negative impact on site-selectivity, conjugation efficiency, and reproducibility.25–27 The enhanced performance of rcSso7d in detecting the IL-6 protein on a polystyrene substrate suggests the possibility of establishing a high-throughput plate-based assay with the rcSso7d protein. Though the limit of detection for the IL-6 antigen is yet to be comparable with those of commercially available antibody-based ELISAs,28,29 the reagent immobilisation time has been reduced significantly to 10 minutes. To achieve comparable sensitivity with antibody-based ELISAs, it will be important to replace the small reporter binder bearing one biotin coupled to at most one streptavidin-HRP with a system that provides comparable amplification with primary antibodies followed by labelled secondary antibodies. The focus of this study is immobilization of the capture reagent and the comparison of several approaches. We anticipate that thermostable rcSso7d proteins fused to polystyrene binding tags can be exploited in many fields of application, including polystyrene-based bioprocesses.
Author contributions
Huan Jia: supervision, conceptualization, data analysis, and writing and reviewing of manuscript. Nazirulmubin Abdul Moomen: writing – original draft, validation, and conceptualization. Jeanette Leong: investigation, validation, conceptualization, and vetting of original draft. Patthara Kongsuphol: resources, writing – review & editing, supervision, project administration, conceptualization, and methodology. Zhi Feng Sherman Lim: investigation. Carmen Sze Min Pui: investigation. Yuxuan Tan: investigation. Ki-Joo Sung: resources. Hadley D Sikes: conceptualization, supervision, resources, writing – review & editing, and funding acquisition.Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
This research is supported by the SMART Innovation Centre, Grant #ING-000770 BIO, and the Singapore National Research Foundation (NRF) via the Antimicrobial Resistance Interdisciplinary Research Group (AMR-IRG) within the Singapore-MIT Alliance in Research and Technology (SMART).References
- I. A. Darwish, International journal of biomedical science: IJBS, 2006, 2, 217 CAS.
- A. Chen and S. Yang, Biosens. Bioelectron., 2015, 71, 230–242 CrossRef CAS PubMed.
- R. Nezlin, Mol. Immunol., 2016, 70, 149–154 CrossRef CAS PubMed.
- P. Kongsuphol, H. Jia, H. L. Cheng, Y. Gu, B. D. O. Shunmuganathan, M. W. Chen, S. M. Lim, S. Y. Ng, P. A. Tambyah and H. Nasir, Communications Medicine, 2021, 1, 46 CrossRef PubMed.
- H. Jia, E. A. Miller, C. C. Chan, S. Y. Ng, M. Prabakaran, M. Tao, I. S.-Y. Cheong, S. M. Lim, M. W. Chen and X. Gao, Lab Chip, 2022, 22, 1321–1332 RSC.
- V. Marx, Nat. Methods, 2013, 10, 703–707 CrossRef CAS PubMed.
- J. Bordeaux, A. W. Welsh, S. Agarwal, E. Killiam, M. T. Baquero, J. A. Hanna, V. K. Anagnostou and D. L. Rimm, BioTechniques, 2010, 48, 197–209 CrossRef CAS PubMed.
- H. Baumann, S. Knapp, T. Lundbäck, R. Ladenstein and T. Härd, Nat. Struct. Biol., 1994, 1, 808–819 CrossRef CAS PubMed.
- F. Catanzano, G. Graziano, P. Fusi, P. Tortora and G. Barone, Biochemistry, 1998, 37, 10493–10498 CrossRef CAS PubMed.
- N. Gera, M. Hussain, R. C. Wright and B. M. Rao, J. Mol. Biol., 2011, 409, 601–616 CrossRef CAS PubMed.
- E. A. Miller, K.-J. Sung, P. Kongsuphol, S. Baniya, H. Q. Aw-Yong, V. Tay, Y. Tan, F. M. Kabir, K. Pang-Yeo and I. G. Kaspriskie, ACS Comb. Sci., 2019, 22, 49–60 CrossRef PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Frontiers in Bioengineering and Biotechnology, 2021, 8, 602659 CrossRef PubMed.
- S. Kim, E. Yee, E. A. Miller, Y. Hao, D. M. Tay, K.-J. Sung, H. Jia, J. M. Johnson, M. Saeed and C. R. Mace, ACS Appl. Mater. Interfaces, 2021, 13, 38990–39002 CrossRef CAS PubMed.
- J. W. Findlay, W. Smith, J. Lee, G. Nordblom, I. Das, B. DeSilva, M. Khan and R. Bowsher, J. Pharm. Biomed. Anal., 2000, 21, 1249–1273 CrossRef CAS PubMed.
- S. Sharma, B. Berne and S. K. Kumar, Biophys. J., 2010, 99, 1157–1165 CrossRef CAS PubMed.
- Y. Kumada, C. Zhao, R. Ishimura, H. Imanaka, K. Imamura and K. Nakanishi, J. Biotechnol., 2007, 128, 354–361 CrossRef CAS PubMed.
- K.-J. Sung, E. A. Miller and H. D. Sikes, Molecular Systems Design & Engineering, 2018, 3, 877–882 Search PubMed.
- E. A. Miller, M. W. Traxlmayr, J. Shen and H. D. Sikes, Molecular systems design & engineering, 2016, 1, 377–381 Search PubMed.
- S. Kim, Y. Hao, E. A. Miller, D. M. Tay, E. Yee, P. Kongsuphol, H. Jia, M. McBee, P. R. Preiser and H. D. Sikes, ACS sensors, 2021, 6, 1891–1898 CrossRef CAS PubMed.
- E. A. Miller, S. Baniya, D. Osorio, Y. J. Al Maalouf and H. D. Sikes, Biosens. Bioelectron., 2018, 102, 456–463 CrossRef CAS PubMed.
- J. M. Kogot, D. A. Sarkes, I. Val-Addo, P. M. Pellegrino and D. N. Stratis-Cullum, BioTechniques, 2012, 52, 95–102 CrossRef CAS PubMed.
- M. Vodnik, B. Štrukelj and M. Lunder, Anal. Biochem., 2012, 424, 83–86 CrossRef CAS PubMed.
- X. Qiang, K. Sun, L. Xing, Y. Xu, H. Wang, Z. Zhou, J. Zhang, F. Zhang, B. Caliskan and M. Wang, Sci. Rep., 2017, 7, 1–11 CrossRef CAS PubMed.
- J. M. Paloni, E. A. Miller, H. D. Sikes and B. D. Olsen, Biomacromolecules, 2018, 19, 3814–3824 CrossRef CAS PubMed.
- C. D. Spicer and B. G. Davis, Nat. Commun., 2014, 5, 1–14 Search PubMed.
- E. M. Sletten and C. R. Bertozzi, Angewandte Chemie International Edition, 2009, 48, 6974–6998 CrossRef CAS PubMed.
- W. H. So, Y. Zhang, W. Kang, C. T. Wong, H. Sun and J. Xia, Curr. Opin. Biotechnol., 2017, 48, 220–227 CrossRef CAS PubMed.
- S. Aldrich, Human IL-6 ELISA Kit, https://www.sigmaaldrich.com/US/en/product/sigma/rab0306?gclid=Cj0KCQjwz6ShBhCMARIsAH9A0qWap5tQZXO6BNjZ3k3EUv6Xv8aQBrZHRZezeTwbDp9vnDlhs4xl5C0aAoZCEALw_wcB, (accessed 2-April-2023) Search PubMed.
- K. B. Company, Human Interleukin 6 (IL-6) ELISA, https://www.kamiyabiomedical.com/pdf/KU-384.pdf, (accessed 3-April-2023) Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00041a |
| ‡ HJ, NAM, JL and PK contributed equally as co-first authors. |
| This journal is © The Royal Society of Chemistry 2023 |