Open Access Article
Open Access ArticleThe development of matrix-metalloproteinase responsive sensors for the machine-independent detection of oral inflammation†
Björn
ter Mors
a,
Marc D.
Driessen
ab,
Axel
Seher
c,
Imme R.
Haubitz
de,
Martina
Raschig
a,
Magdalena
Nowak
a,
Yvonne
Jockel-Schneider
de,
Christian
Linz
c and
Lorenz
Meinel
 *ade
*ade
aInstitute for Pharmacy and Food Chemistry, University of Wuerzburg, Am Hubland, DE-97074 Wuerzburg, Germany. E-mail: lorenz.meinel@uni-wuerzburg.de
bInstitute for Molecular Medicine I, Heinrich-Heine-University Duesseldorf, Building 22.07.U1, DE-40225 Duesseldorf, Germany
cDepartment of Oral and Maxillofacial Plastic Surgery, University Hospital Wuerzburg, D-97070 Wuerzburg, Germany
dDepartment of Periodontology, University Hospital Wuerzburg, Pleicherwall 2, Wuerzburg, D-97070, Germany
eHelmholtz Institute for RNA-based Infection Biology (HIRI), Josef-Schneider-Straße 2/D15, DE-97080 Wuerzburg, Germany
First published on 7th June 2023
Abstract
Periodontitis is a chronic inflammatory disease that affects an estimated 20–50% of the world's population and can lead to high follow-up costs for the healthcare system. The disease is closely associated with a dysbiotic, pro-inflammatory change of the oral microbiota. As the disease progresses, the inflammation affects the periodontium with subsequent tooth mobility and tooth loss as clinical outcomes. Early diagnosis and early treatment are vital and can leverage overall clinical success, resulting in possible preservation. Therefore, periodontal inflammation should be detected as soon as possible. Unfortunately, early stages of periodontitis are mostly asymptomatic and unnoticed by affected patients or only noticed at advanced stages, making diagnosis impossible without medical consultation. Ideally, highly intuitive diagnostic tools would be available to anyone, anywhere and anytime for personal check-ups. Therefore, we developed peptide sensors, that are cleaved by upregulated matrix metalloproteinases (MMP) as present in oral inflammation. This leads to the release of a denatonium-based flavor substance, providing a rapid diagnostic indicator for further professional clarification. Two distinct techniques were employed to enhance sensor specificity. A peptide sensor was developed for disease-specific protease (MMP) digest using previously published information and proteome wide screening techniques, followed by optimization against nonspecific proteases present in the oral cavity. Finally, the selected sensor was tested in patient saliva, and the sensor cleavage correlated with MMP concentration and activity. Furthermore, the sensors significantly differentiated patient pools and pools of healthy volunteers, and cleavage was linked to pocket probing depths, a clinical sign of periodontal diseases.
Introduction
Chronic inflammations of the periodontium, such as gingivitis and periodontitis, are among the most common oral diseases worldwide. If left untreated, they can lead to tooth loss. This is accompanied by significant functional limitations and considerable impairment of aesthetics and phonetics, leading to exclusion and other adverse effects on social life.1,2 Worldwide, periodontitis affects an estimated 20–50% of the global population and around 40% of the adult population in the US.3,4 The consequential costs of late or undetected cases for the healthcare system are high and will likely rise with an aging population.5,6Periodontitis is a chronic inflammatory process fueled by microbial dysbiosis, dental plaque formation, genetic predisposition, systemic diseases, smoking, or lifestyle.7 Progressing inflammation drives irreversible damage to the tooth-supporting tissues and, ultimately, tooth loss.8 Therefore, therapy aims to start in the early stages of the disease.9 The sooner inflammation in the oral cavity is detected and treated, the greater the chance of preventing the occurrence of irreversible damage and maintaining oral health into old age.
Self-diagnosis is impossible. The clinical dental examination includes measuring pocket probing depth, recession, bleeding on probing (BoP), tooth mobility, furcation, and radiographic findings of bone loss.9 Unfortunately, periodontitis often starts symptom-free, stays unnoticed, and is only noticed in an advanced state by affected patients. Thus patient compliance, even where patients are entitled to use the offer of an annual dental check-up of their oral health, is often insufficient, and an early diagnosis can usually not be guaranteed.9,10 That gap defines the central challenge for this manuscript.
Saliva offers easy, non-invasive diagnostic access to multiple diseases.11 A variety of methods have been described targeting saliva, e.g., proteomics, point of care testing (POCT), or electronic tongues.12–14 While proteomics and electronic tongues provide broader information, ranging from information of whole salivary proteomes to phonetic interpretation of complex liquids, POCT is mainly focused on defined analytes and simple readouts.12,15 Here, promising saliva biosensing platforms can quantify analytes like virus particles to screen for infectious diseases or address non-communicable diseases such as cancer by microRNAs.16,17 A well-known example of a US Food and Drug Administration (FDA)- and EU-approved PoC test kit that uses mouth rinse samples rather than saliva is the commercially available aMMP-8-based lateral flow immunoassay (e.g., PerioSafe®, Implant Safe®, Oral Risk Indicator®).18 Using digital reader systems, quantifiability of the results and high sensitivity is possible, allowing the mapping of the different stages of periodontitis and the selection of appropriate therapeutic approaches.19,20 In previous studies, we could achieve good results in the release of the bitter flavor with the electronic tongue using the precursor model, a particle-based sensor.21 With our new system presented here, we aim to achieve a simple, fast, and ubiquitous readout in complex fluids, i.e., translating differences (healthy and periodontitis patients) in matrix metalloproteinase activity in saliva, a surrogate marker for inflammation, into a bitter substance and finally into taste.22,23 The taste is supposed to motivate the patients for professional consultation. Therefore, this system complements current pull strategies (patients comply with regular visits for check-ups) with push strategies (the sensory chewing gum pushes patients into the office).
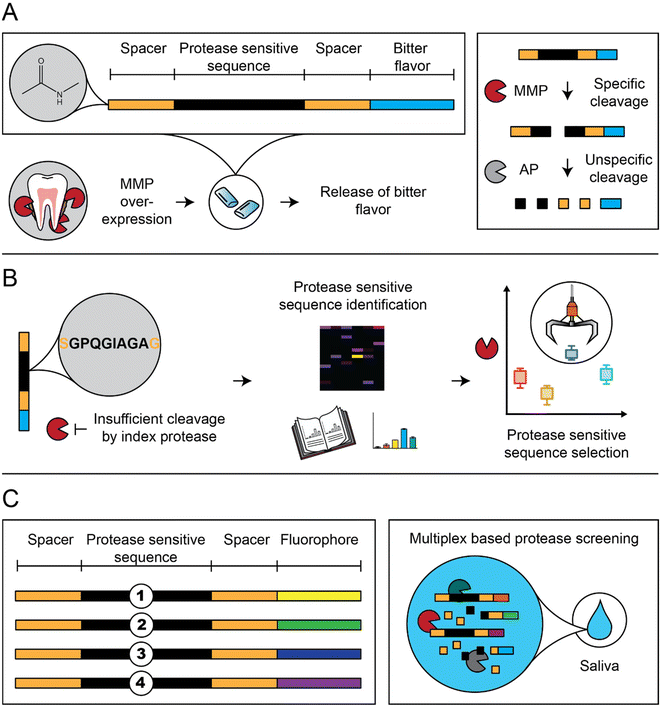
The peptide-based sensors disintegrate by upregulated MMPs, surrogate markers of oral inflammation, and lead to the release of a bitter-tasting denatonium derivative (Fig. 1A).21,24 First, we screened our sensors with MMP-8, a collagenolytic protease correlating with oral inflammation, including periodontitis.25,26 We deployed proteome-wide search strategies (proteomic identification of protease cleavage sites; PICS) to screen for new protease-sensitive sequences (PSS; Fig. 1B).27 In addition, selected sensors were N-acetylated to prevent unspecific proteolytic digestion by salivary aminopeptidases (AP). Finally, sensor selection was performed in patient saliva to include the full periodontitis orchestra (Fig. 1C).28–33
Material and methods
Proteomic identification of cleavage sites (PICS)
Identification of protease cleavage sites was performed with minor changes according to PICS protocols described by Schilling et al.34 Three different PICS libraries were generated, namely human embryonic kidney (HEK) 293 (ATCC-Number CRL-1573, ATCC, Manassas, VA,) cell based, Escherichia coli (E. coli) (Thermo Fisher Scientific, Waltham, USA) 14N and E. coli15N. A detailed method section is provided in the ESI.†Peptide synthesis
All peptides described in this manuscript were synthesized using fluorenylmethoxycarbonyl protecting group (Fmoc) based solid phase peptide synthesis, following established protocols using a polypropylene-reactor (MultiSynTech, Witten, Germany). A detailed method section is provided in the ESI.†Liquid chromatography-mass spectrometry (LC-MS)
Peptide mass spectra were acquired via a Shimadzu LC-MS system (Shimadzu Scientific Instruments, Columbia, MD, USA) equipped with a DGU-20A3R degassing unit, a LC20AB liquid chromatograph using a Synergi 4 μm fusion-RP column (150 × 4.6 mm) (Phenomenex lnc., Torrance, CA), SPD-20A UV/vis detector, coupled with a single quadrupole LCMS-2020. Mobile phase A was 0.1% (v/v) FA in Millipore water. Mobile phase B was methanol with 0.1% (v/v) FA, flow was set to 1 ml min−1, the wavelength of the detector was set to λ = 214 nm and the injected volume of the sample was 20 μL. The gradient was increased in 8 min from 5 to 90% B, then held at 90% B for 5 min, reduced to 5% B within 1 min, and held at 5% B for 4 min. Detection was done at λ = 214 nm. MS-detection range was set to 60 to 1000 (m/z).MMP activation
Pro-Matrix-metalloproteinases (MMP-1, MMP-8, and MMP-9) (Merck Millipore, Burlington, MA, USA) were activated prior usage. To activate Pro-MMPs a fresh 50 mM 4-aminophenylmercuric acetate (APMA) stock solution in 0.1 M NaOH was prepared. The stock solution was added in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio to Pro-MMPs (Pro-MMPs
1 (v/v) ratio to Pro-MMPs (Pro-MMPs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) APMA) to reach a final concentration of 5 mM APMA. The solution was incubated for 3 h at 37 °C without agitation. Following incubation, the activated MMPs were stored in aliquots at −80 °C.
APMA) to reach a final concentration of 5 mM APMA. The solution was incubated for 3 h at 37 °C without agitation. Following incubation, the activated MMPs were stored in aliquots at −80 °C.
Peptide cleavage
Lyophilized peptides were solubilized in MMP buffer (200 mM NaCl, 50 mM Tris-HCl, 5 mM CaCl2, 1 μM ZnCl2, 0.05% Brij35 at pH 6.8–7.0) to 1 mM stock solutions. Cleavage was performed at 37 °C under rigorous shaking in 30 μL MMP buffer containing 0.1 mM of the respective peptide and 900 ng mL−1 MMP-8. The reaction was stopped by addition of 5 mM EDTA final concentration. Iterative improvement and sensor stage experiments were performed n = 3, and experiments comparing Y1–8 and X3 were performed n = 5.Analytical HPLC was carried out via an Agilent 1260 infinity II HPLC (Agilent Technologies Inc., Waldbronn, Germany) using a Zorbax 300SB-CN 5 μm analytical 4.6 × 150 mm LC column (Agilent Technologies Inc., Waldbronn, Germany) The device was equipped with a diode array detector (G7115A, Agilent), a fluorescence detector (G7121A, Agilent), an automatic vial sampler (G7129C, Agilent), flexible pump (G7104C, Agilent), and multicolumn oven (G7116A, Agilent). Mobile phase A was 0.1% TFA in Millipore water. Mobile phase B was ACN with 0.1% TFA, flow was set to 1 ml min−1, injection volume was 15 μl, and the wavelength of the detector was set to λ = 214 nm for PICS derived peptides.
The gradient was held at 15% B for 1 minute, then increasing to 40% within 7 minutes, increased again for 0.5 minutes to 95% B, held for 0.5 min, then back to 15% B within 0.5 minute, and held for 5.5 minutes.
Cleavage percent was analyzed by comparing the area under the curve (AUC) of the main peak from the compound incubated with MMP and in absence of MMP.
Sensor synthesis
Denatonium labeled sensors were synthesized via coupling of purified, side-chain-protected peptides with 2 molar equivalents of denatonium–CH2–NH2 and 4 molar equivalents of DIPEA in DMF. The reaction was performed at RT for 16 h under rigorous shaking. The sensors were then precipitated with DEE as described above.DBCO-functionalized dyes were incubated with 1.5 molar excess of the respective azide-functionalized peptides in 50/50 DMSO/water (v/v) for 16 h under shaking at room temperature. The sensors were then purified by reverse phase purification as described before.
Alkyne-functionalized dyes were incubated with 1.5 molar excess of the respective azide-functionalized peptides in water with 500 μM tris-hydroxypropyltriazolylmethylamine, 100 μM CuSO4 and 5 mM sodium ascorbate. The reaction was incubated for 1 h at room temperature and stopped by adding 500 μM ethylenediaminetetraacetic acid (EDTA). The sensors were then purified via anion exchange chromatography via fast protein liquid chromatography on a GE ÄKTA pure (GE Healthcare, Chalfont St Giles, UK) system with a Hitrap Q FF (GE Healthcare, Chalfont St Giles, UK) column, with eluant A (20 mM Tris HCL pH 8) and eluent B (20 mM Tris HCL pH 8, 1 M NaCl) from 0% B to 100% B in 60 minutes. Afterwards, the sensor was purified via reverse phase purification as described before.
Sensors were freeze-dried (as described above) and stored at −80 °C until further usage. Subsequently, sensors were characterized by HPLC and LC-MS.
Sensor cleavage
Lyophilized sensors and Y3 were solubilized in MMP buffer (200 mM NaCl, 50 mM Tris-HCl, 5 mM CaCl2, 1 μM ZnCl2, 0.05% Brij35 at pH 6.8–7.0) to 1 mM stock solutions. Cleavage was performed at 37 °C under rigorous shaking in 30 μL MMP buffer containing 0.1 mM of the respective sensor/peptide and 900 ng mL−1 MMP-8, or a molar equivalent of MMP-1 or MMP-9 for single cleavage conditions. Cleavage of sensors in mixture was performed with sensor concentrations of 25 μM for each sensor. The reaction was stopped via addition of 5 mM EDTA final concentration. Experiments were performed n = 5. Cleavage was analyzed via fluorescent detection via HPLC with following wavelengths: SP3 (absorption: 646 nm; emission: 661 nm), SP5 (absorption: 494 nm; emission: 517 nm), SP7 (absorption: 553 nm; emission: 566 nm) and SN3 (absorption: 280 nm; emission: 350 nm). Y3 cleavage was analyzed as described before. Cleavage percent was analyzed by comparing the AUC of the main peak from the compound incubated with MMP and in absence of MMP. Cleavage percent was then normalized by multiplying cleavage results with a correction factor. The correction factor was determined individually for every experiment/MMP by dividing Y3 cleavage results acquired in protease sensitive linker identification study by cleavage results of Y3 in individual sensor cleavage experiments (Fig. 3D–F).Sensor cleavage in human saliva
Lyophilized sensors were solubilized in MMP buffer to 0.05 mM each and mixed with saliva 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) to reach a final concentration of 25 μM per sensor. The solution was incubated for 15 min at 37 °C under rigorous shaking and the reaction was stopped via addition of 5 mM EDTA final concentration. The samples were diluted 1
1 (v/v) to reach a final concentration of 25 μM per sensor. The solution was incubated for 15 min at 37 °C under rigorous shaking and the reaction was stopped via addition of 5 mM EDTA final concentration. The samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 with acetonitrile, 1
2 with acetonitrile, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 with water (1
5 with water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 total dilution) and analyzed via HPLC as described before. Cleavage efficiency was analyzed by comparing the AUC of the newly formed peaks compared to the total AUC of the combined peaks. Peaks with LU < 1 were considered below the limit of detection and set to 0.5 LU for further analysis.
10 total dilution) and analyzed via HPLC as described before. Cleavage efficiency was analyzed by comparing the AUC of the newly formed peaks compared to the total AUC of the combined peaks. Peaks with LU < 1 were considered below the limit of detection and set to 0.5 LU for further analysis.
Aminopeptidase cleavage
Lyophilized sensors were solubilized in AP buffer (50 mM Tris-HCl, 1 mM CaCl2, 150 mM NaCl at pH 7.0) to 1 mM stock solutions. Cleavage was performed at 37 °C under rigorous shaking in 30 μL AP buffer containing 0.1 mM of the respective sensor and 0, 0.87, 8.7, 39 or 78 μg mL−1 AP for 1 h for concentration-based cleavage. For time resolved cleavage of S1c, cleavage was performed at 37 °C under rigorous shaking in 30 μL AP buffer containing 0.1 mM of the respective sensor and 0.87 μg mL−1 AP for 0, 5, 10, 20, 30 and 60 min. The reaction was stopped via 15 min incubation at 95 °C and analyzed by HPLC described before. Cleavage was calculated as the decrease of the main peak in relation to a negative control, indicating cleavage as the cleavage of one or more amino acids from the N-terminus. All experiments were performed n = 3.Active MMP-8 concentration determination
Saliva samples were thawed at 4 °C, overnight, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 and analyzed by MMP-8 activity assay (Quickzyme, Leiden, Netherlands) following provided protocols. In brief, reagents were prepared as described by protocols. A 96 well plate was prepared: anti-MMP-8 antibody solution was added, allowed to bind at 37 °C for 2 h, followed by 4 wash steps, incubation with assay buffer and then addition of either samples (100 μL of diluted saliva) or standards as supplied. The plate was covered and incubated at 4 °C overnight and washed 4 times with wash buffer. 1.5 mM APMA solution was freshly prepared and added to wells containing standards. Samples were treated with assay buffer to measure endogenous MMP-8 levels. The plate was covered and incubated at 37 °C for 1 h, detection reagent freshly prepared and added. Absorbance was measured at 405 nm in a microplate reader (Tecan Infinite F50, Crailsheim, Germany) at 0, 2, 4 and 6 h. Between measurements, the plate was incubated at 37 °C.
200 and analyzed by MMP-8 activity assay (Quickzyme, Leiden, Netherlands) following provided protocols. In brief, reagents were prepared as described by protocols. A 96 well plate was prepared: anti-MMP-8 antibody solution was added, allowed to bind at 37 °C for 2 h, followed by 4 wash steps, incubation with assay buffer and then addition of either samples (100 μL of diluted saliva) or standards as supplied. The plate was covered and incubated at 4 °C overnight and washed 4 times with wash buffer. 1.5 mM APMA solution was freshly prepared and added to wells containing standards. Samples were treated with assay buffer to measure endogenous MMP-8 levels. The plate was covered and incubated at 37 °C for 1 h, detection reagent freshly prepared and added. Absorbance was measured at 405 nm in a microplate reader (Tecan Infinite F50, Crailsheim, Germany) at 0, 2, 4 and 6 h. Between measurements, the plate was incubated at 37 °C.
Statistical analysis
Comparison between two groups and correlations were performed using MEDAS (C. Grund, Margetshöchheim, Germany). For comparisons between two groups, Student's t-test was performed when Gaussian distribution was provided and Mann–Whitney U test was used when data was not normally distributed. Correlations were described by analysis of Spearman's rank correlation coefficient (ρ/rho) when data had no ties and not normally distributed. When data had ties and not normally distributed, Kendall rank correlation (τ/tau) coefficient was used to describe correlations. P-Values <0.05 were considered significant and displayed as * for values <0.05, ** for values <0.005 and *** for values <0.001.Results
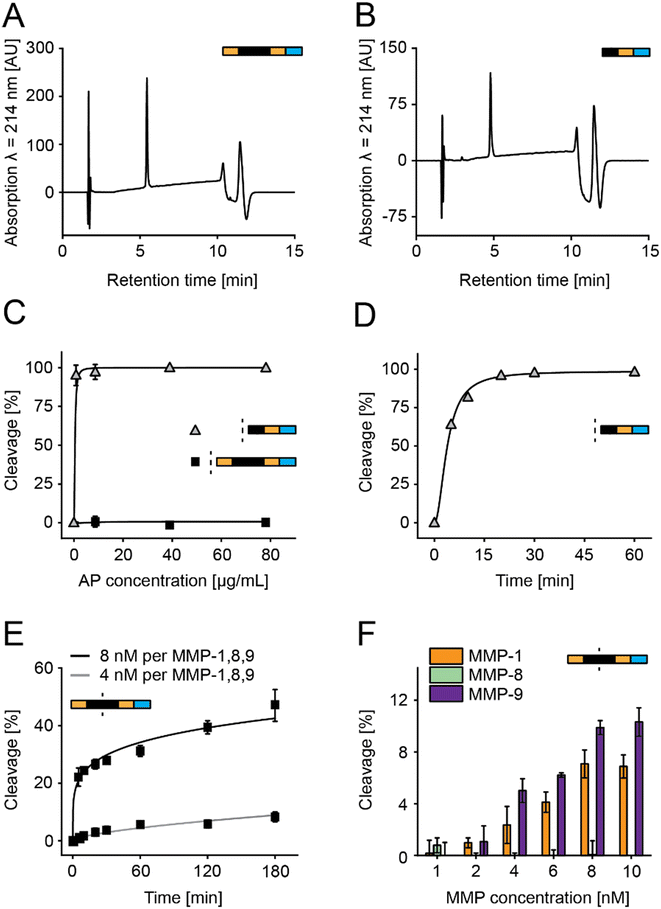
In this study, we optimized peptide-based sensors, which release a bitter substance (denatonium derivative) after MMP-specific cleavage (Fig. 1A).21,35,36 This design simplified previous designs using beads and removed previous challenges on salivary stability and storage stability by introducing acetylated N-termini and a storage stable C terminal denatonium modification.21,36 We optimized sensors in vitro using MMP-8, a key surrogate marker in periodontitis (Fig. 1B), but selected the final sensors from responses observed in a multiplex based MMP-screening in saliva samples collected from diagnosed patients (Fig. 1C).To allow the C-terminal coupling of denatonium, an amine-functionalized denatonium derivative (Den–CH2–NH2) was synthesized and characterized (Fig. S1†).35 Sensor S1 and a cut variant (S1c, representing the C-terminal fragment after MMP cleavage) were synthesized and characterized by HPLC and LC-MS, showing purity of >95% and the predicted molecular mass, as well as long term storage stability (Fig. 2A and B and S2A–C†).
Aminopeptidase stability of S1 and S1c was studied for 1 h with aminopeptidase concentrations ranging from 0 to 78 μg mL−1, showing no cleavage for all tested concentrations for S1 and a cleavage rate of 95% at the lowest concentration tested for S1c (Fig. 2C). Cleavage over time was evaluated for S1c with 0.87 μg mL−1 aminopeptidase over 1 h, indicating a cleavage rate of 95% after 20 min (Fig. 2D). This confirmed that unspecific aminopeptidase digest of the sensor occurs only after the specific cleavage event and finally leads to the release of the bitter substance.
S1 was then incubated with either 4 nM or 8 nM MMP mix (a mixture of MMPs 1, 8, and 9), resulting in cleavage rates of <1% and >20% after 5 min, respectively (Fig. 2E). To test the cleavage of individual MMPs, the sensor was incubated with MMP-1, MMP-8, and MMP-9 for 15 min with concentrations ranging from 1 to 10 nM (Fig. 2F). Cleavage was observed for MMP-1 and MMP-9 but not for MMP-8, with a maximum cleavage of 7% for MMP-1 and 10% for MMP-9. Therefore, we had to redesign the PSS to address the low overall MMP sensitivity and the lack of MMP-8 susceptibility.21
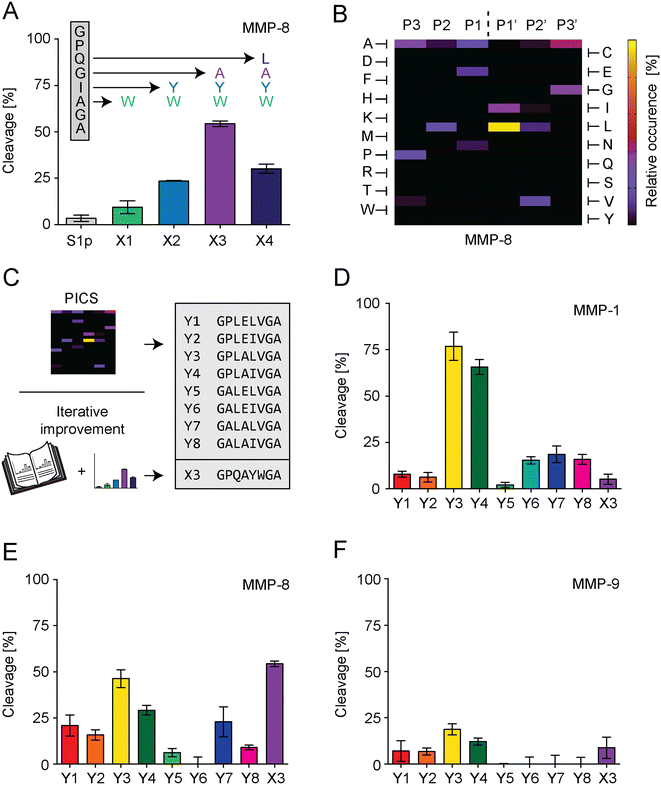
For S1 improvement, two different strategies were followed to optimize the PSS of S1 (Fig. 3C). The first strategy exchanged individual amino acids of the S1-PSS in an iterative process based on previously published information reporting increased MMP-8 specificity (Fig. 3A).37 These reports rated tryptophan in position P2′ as the most effective means of increasing MMP-8 cleavage rate and suggested placing tyrosine in P1′, alanine in P1, and leucine in P2. Consequently, we replaced alanine in the S1-PSS with tryptophan (referred to as PSS-X1) and observed an increase in cleavage. PSS-X1 was further improved by introducing tyrosine (PSS-X2), and the PSS-X2 was further enhanced by introducing alanine (PSS-X3).37 Leucine introduction, resulting in PSS-X4, reduced cleavage compared to PSS-X3. Therefore, PSS-X3 was selected. Our second strategy searched alternative PSSs by proteomic identification of protease cleavage sites (PICS; Fig. 3B and S6–S8†).38 We generated chemically modified proteome libraries from E. coli, 15N labeled E. coli and HEK-293 cells. These libraries were incubated with MMP-8, and resulting cleavage products were isolated, enriched, and analyzed by MS. PICS indicated that P3 should be an alanine or proline, P2 a leucine, P1 a glutamic acid or alanine, P1′ a leucine or isoleucine, P2′ a valine, and P3′ an alanine or glycine. These amino acids were selected by considering their relative occurrence, as determined through WebPICS.39 We set the relative occurrence threshold at 11.5% or more (average of at least three independent PICS results) to limit the number of possible candidates for peptide design.38 We chemically synthesized eight PSSs based on these insights, excluding the P3′ glycine variant to avoid structural repetitions. Furthermore, none of the hits exceeded the threshold of 11.5% for P4 or P4′, so we selected glycine and alanine for these positions, respectively. In addition, glycine and alanine are in positions P4 and P4′ in the S1-PSS (Table S1†). The PICS-derived PSSs are called PSS-Y1 to PSS-Y8 (Fig. 3C), were characterized, and tested in comparison to PSS-X3 for cleavage rates at 17.5 nM MMP-1, MMP-8, or MMP-9 with an incubation time of 15 minutes (Fig. S9 and S10† and 3D–F). We selected three PSSs for further testing. PSS-Y3 was chosen for its high cleavage rate by all MMPs. PSS-Y5 and PSS-Y7 were selected based on cleavage by MMP-8 while not being cleaved by MMP-9. PSS-X3 (see above; optimized from previously reported insights37) was chosen due to its high cleavage rate for MMP-8 and low cleavage rate for MMP-1 and MMP-9.
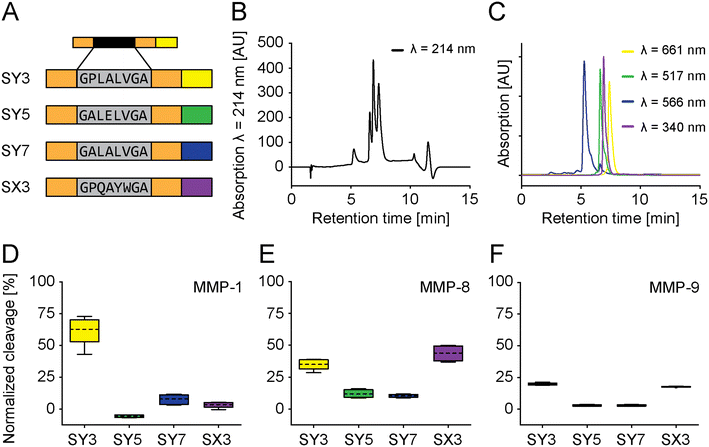
These four selected PSSs were then assembled into fully functional sensors using four different fluorophores and referred to as SY3, SY5, SY7, and SX3 in reference to PSS-Y3, PSS-Y5, PSS-Y7, and PSS-X3 (vide supra). SY3, SY5, and SY7 were synthesized with C-terminal L-azidohomoalanine (Aha) and “clicked” to different fluorescent dyes at their respective C-termini. SX3 was modified with the (fluorescent) denatonium derivative. The fluorescence spectra did not overlap. Therefore, we could simultaneously multiplex all sensors under identical conditions (Fig. 4A–C and S11–S13†). Multiplexing was tested against MMP-1, MMP-8, or MMP-9 (Fig. S15D–F,† 17.5 nM; incubation 15 minutes). The cleavage outcome of the sensors generally confirmed the effect observed for the PSS alone (Fig. 3D–F). SY3 was cleaved most effectively by MMP-1 and MMP-8 and, to a lesser extent, MMP-9. SY5 responded to MMP-8 but not MMP-1 or MMP-9, with generally low cleavage rates. SY7 had a low general cleavage rate and responded to all MMPs other than MMP-9. SX3 showed high cleavage rates for MMP-8, low cleavage rates for MMP-1, and medium cleavage rates for MMP-9. Further modifications of the fluorophore distant from the PSS did not impact cleavage performances (e.g., PSS, spacer-PSS-spacer, or spacer-PSS-spacer-fluorophore, Fig. S3, and S14†).
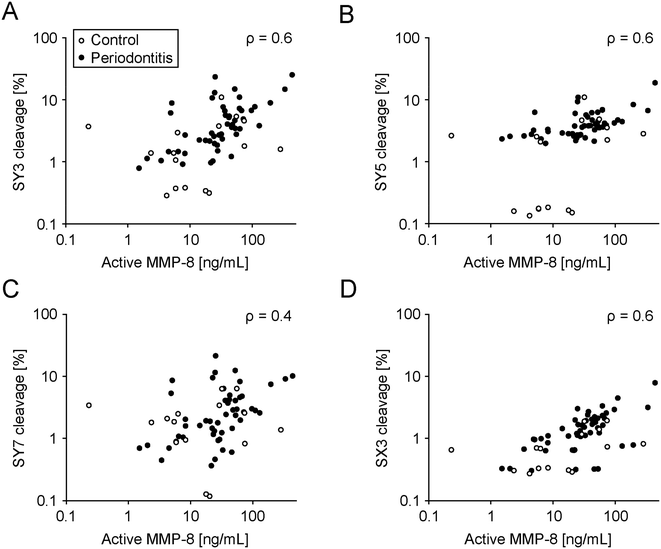
These sensors were profiled in saliva samples from 16 healthy subjects and 50 established periodontitis patients. The saliva was characterized for total MMP-8 and active MMP-8 and correlated with cleavage rates of the sensors (Fig. 5 and S16, Tables S2–S4†). We collected further patient information, including age, smoking status, BoP, the number of missing teeth, recession, plaque, tooth mobility, furcation involvement, gingival pocket depth, clinical attachment level, periodontal epithelial surface area (PESA), and periodontal inflamed surface area (PISA) separately. The selected cohort consisted of treated and stable patients, and for example a chewing gum as a future medical product could be profiled in this group for follow-up checks at home. We chose these patients as we wanted to establish a saliva library with different MMP-8 profiles and test our sensors on that library accordingly. Sensor cleavage significantly correlated with total and active MMP-8 concentration (Fig. 5; Table S4†). However, another arguably more relevant patient pool than the one used here (to establish a proof of mechanism) are naïve periodontitis patients to facilitate diagnosis in early stages. Future studies should address this patient pool. Furthermore, all sensors except SY7 were significantly better cleaved in saliva from patients compared to saliva from healthy donors (Table S5†). Lastly, gingival pocket depths significantly correlated with sensor cleavage, except for SY7 (Table S6†).
Discussion
The peptide-based sensors were stable against unspecific salivary aminopeptidases. Following MMP-cleavage of their PSS, they rapidly disintegrated, releasing a bitter flavor (Fig. 2C and D). The new soluble design replaces previous particle-based sensors.21,36 One challenge during replacement was that the PSS, used in the previous particle-based design, was unresponsive to MMP-8 when integrated into the new and soluble sensors. Therefore, we initiated a search for novel MMP-8 responsive PSSs compatible with the new and soluble setup (Fig. 3). Identified hits (PSS) were assessed, individually encoded with fluorescent dyes (sensor) and multiplexed in patient and control saliva. Sensor cleavage correlated with MMP-8 (total and active) levels in vitro and MMP-8 levels measured in patient saliva (Fig. 5). Furthermore, cleavage rates were significantly higher in patient saliva compared to saliva from healthy donors (Table S5†). Further adaptations became necessary. For example, the particle in the old design protected the N-terminus of the sensor from unspecific cleavage by salivary aminopeptidases. The same was achieved in the new soluble sensor by N-terminal acetylation. As a result, replacing the old, particle-based design will reduce the costs of goods for this high volume, commodity product, facilitates manufacturing, including the integration into the development of new oral thin films or chewing gums (for example), and reduces the effort for quality control reflecting the soluble state of the new sensors.Contrasting previous studies linking salivary MMP-8 to periodontitis, our patient pool did not show these correlations (Fig. S18, Table S5†).40 These differences arguably root in our inclusion/exclusion criteria, recruiting treated periodontitis patients in regular supportive aftercare with comparatively low BoP-values. Treatment reduces acute inflammatory processes and MMP levels.41,42 Furthermore, our criteria did not gate salivary collection regiments, such as the time of the day or last food/drink intake. MMP-8 is not the only marker correlating with periodontitis.28–31 Therefore, the possible promiscuity of our sensors might be advantageous, which is why we screened in patient saliva to reflect the complexity in periodontitis. Some sensors which were not exceptionally responsive to MMP-8 (SY3, SY5, and SX3) discriminated against periodontitis, supporting final screenings in biological fluids. Our patient pool donated saliva with a relevant range of active inflammation, but this patient pool is not the one targeted by the product. The future target profile is the broad, un-diagnosed population to early segment subjects at risk of developing periodontitis. The clinical correlation demonstrated further exciting potential. For example, sensor cleavage correlated with pocket probing depth, an important factor for disease progression.43 If corroborated by future clinical studies using pocket depth as the primary endpoint, this evidence further strengthens the approach for community screening, targeted dental diagnosis and dental care in prescreened and enriched patient pools.
MMP-8 is a significant biomarker for a spectrum of physiological processes ranging from inflammation, wound healing, and immune responses to malignant tissue and tissue remodeling, highlighting its potential as a versatile diagnostic and therapeutic target.44 As a diagnostic marker, MMP-8 is already used in various rapid test systems in the field of periodontitis (vide supra). Our sensor does not offer the possibility of quantifying periodontitis but allows a simple, fast and ubiquitous use without a professional background. Moreover, the sensors are modular. The PSS is readily replaced by other PSSs, paving the way for new products, for example, marking bacterial infection (strep throat) or viral challenge (flue).45,46 We also currently explore use in spotting malignancies like squamous cell carcinomas of the head and neck.47 The rapid PICS-identification tools for new PSSs leverage modular flexibilities and, when combined with multiplexing provide paired, head-to-head comparison within each biological sample. Nevertheless, there is bias in our studies. We rely on previous information, such as the link between MMP-8 and periodontitis.22,23 However, MMP-8 levels correlate with periodontitis but not necessarily vice versa.23 Consequently, our approach is burdened by the limitations associated with MMP-8 as a biomarker for periodontitis.
In summary, this platform technology holds the potential to leverage active community screening for various challenges. Future designs may target viral outbreaks. The modular design and the rapid and proteome-wide search tools for new PSSs suggest quick response rates, easy mass-production, and distribution at affordable costs.
Conflicts of interest
L. M. is an inventor of related patents/patent applications (EP13710333.9, EP17758824.1, EP17762078.8, and EP19708959.2A) and states a possible conflict of interest. M. R. is an inventor of a related patent application (EP19708959.2A). The remaining authors declare no competing interests.Acknowledgements
We gratefully acknowledge the support of Constantin Wiesner for MMP characterization in saliva, the help of Sophie Wiebecke with patient diagnosis, and Dr. Lukas Hahn for critical review. The research was supported by the Federal Ministry of Education and Research, grant # 03ZZF75B, grant # 13GH0478C, and grant # 03COV18A.References
- R. Brignardello-Petersen, Tooth loss, periodontal disease, and dental caries may be associated with decreased oral health–related quality of life, but there is no evidence about the magnitude of this association, J. Am. Dent. Assoc., JADA, 2017, 148(10), e150 CrossRef PubMed.
- E. D. Roumanas, The social solution-denture esthetics, phonetics, and function, J. Prosthodontics, 2009, 18(2), 112–115 CrossRef PubMed.
- M. A. Nazir, Prevalence of periodontal disease, its association with systemic diseases and prevention, Int. J. Health Sci., 2017, 11(2), 72–80 Search PubMed.
- P. I. Eke, W. S. Borgnakke and R. J. Genco, Recent epidemiologic trends in periodontitis in the USA, Periodontol. 2000, 2020, 82(1), 257–267 CrossRef PubMed.
- W. He, D. Goodkind and P. R. Kowal, An aging world: 2015, United States Census Bureau, Washington, DC, 2016 Search PubMed.
- P. Madianos, W. Papaioannou, D. Herrera, M. Sanz, A. Baeumer and A. Bogren, et al., EFP Delphi study on the trends in Periodontology and Periodontics in Europe for the year 2025, J. Clin. Periodontol., 2016, 43(6), 472–481 CrossRef PubMed.
- P. M. Bartold and T. E. Van Dyke, An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis, J. Clin. Periodontol., 2019, 46(1), 6–11 CrossRef PubMed.
- A. Savage, K. A. Eaton, D. R. Moles and I. Needleman, A systematic review of definitions of periodontitis and methods that have been used to identify this disease, J. Clin. Periodontol., 2009, 36(6), 458–467 CrossRef PubMed.
- D. F. Kinane, P. G. Stathopoulou and P. N. Papapanou, Periodontal diseases, Nat. Rev. Dis. Primers, 2017, 3(1), 17038 CrossRef PubMed.
- A. Saito, M. Kikuchi, F. Ueshima, S. Matsumoto, H. Hayakawa and H. Masuda, et al., Assessment of oral self-care in patients with periodontitis: a pilot study in a dental school clinic in Japan, BMC Oral Health, 2009, 9(1), 27 CrossRef PubMed.
- C.-Z. Zhang, X.-Q. Cheng, J.-Y. Li, P. Zhang, P. Yi and X. Xu, et al., Saliva in the diagnosis of diseases, Int. J. Oral Sci., 2016, 8(3), 133–137 CrossRef CAS PubMed.
- E. Pappa, K. Vougas, J. Zoidakis and H. Vastardis, Proteomic advances in salivary diagnostics, Biochim. Biophys. Acta, Proteins Proteomics, 2020, 1868(11), 140494 CrossRef CAS PubMed.
- M. Podrażka, E. Bączyńska, M. Kundys, P. S. Jeleń and N. E. Witkowska, Electronic Tongue—A Tool for All Tastes?, Biosensors, 2018, 8(1) DOI:10.3390/bios8010003.
- K. Aro, F. Wei, D. T. Wong and M. Tu, Saliva Liquid Biopsy for Point-of-Care Applications, Front. Public Health, 2017, 5, 77 Search PubMed.
- J. Liu, S. Liu, X. Lian, X. Li, S. S. Low and Z. Zhao, et al., Taste Analog Perception System Based on Impedance Spectrum Sensor Array and Human-Like Fuzzy Evaluation Cloud Model, IEEE Sens. J., 2022, 22(20), 19513–19523 CAS.
- S. Shin Low, Y. Pan, D. Ji, Y. Li, Y. Lu and Y. He, et al., Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept, Sens. Actuators, B, 2020, 308, 127718 CrossRef CAS.
- L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen and C. Zhao, et al., One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device, Biosens. Bioelectron., 2021, 171, 112685 CrossRef CAS PubMed.
- T. Sorsa, S. O. Nwhator, D. Sakellari, A. Grigoriadis, K. A. Umeizudike and E. Brandt, et al., aMMP-8 Oral Fluid PoC Test in Relation to Oral and Systemic Diseases, Front. Oral Health, 2022, 3, 897115 CrossRef PubMed.
- T. Sorsa, V. Sahni, N. Buduneli, S. Gupta, I. T. Räisänen and L. M. Golub, et al., Active matrix metalloproteinase-8 (aMMP-8) point-of-care test (POCT) in the COVID-19 pandemic, Expert Rev. Proteomics, 2021, 18(8), 707–717 CrossRef CAS PubMed.
- M. Keskin, J. Rintamarttunen, E. Gülçiçek, I. T. Räisänen, S. Gupta and T. Tervahartiala, et al., A Comparative Analysis of Treatment-Related Changes in the Diagnostic Biomarker Active Metalloproteinase-8 Levels in Patients with Periodontitis, Diagnostics, 2023, 13(5), 903 CrossRef CAS PubMed.
- J. Ritzer, T. Lühmann, C. Rode, M. Pein-Hackelbusch, I. Immohr and U. Schedler, et al., Diagnosing peri-implant disease using the tongue as a 24/7 detector, Nat. Commun., 2017, 8, 264 CrossRef CAS PubMed.
- S. Kc, X. Z. Wang and J. E. Gallagher, Diagnostic sensitivity and specificity of host-derived salivary biomarkers in periodontal disease amongst adults: Systematic review, J. Clin. Periodontol., 2020, 47(3), 289–308 CrossRef PubMed.
- N. Arias-Bujanda, A. Regueira-Iglesias, C. Balsa-Castro, L. Nibali, N. Donos and I. Tomás, Accuracy of single molecular biomarkers in saliva for the diagnosis of periodontitis: A systematic review and meta-analysis, J. Clin. Periodontol., 2020, 47(1), 2–18 CrossRef CAS PubMed.
- A. Brockhoff, M. Behrens, A. Massarotti, G. Appendino and W. Meyerhof, Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium, J. Agric. Food Chem., 2007, 55(15), 6236–6243 CrossRef CAS PubMed.
- V. Rangbulla, A. Nirola, M. Gupta, P. Batra and M. Gupta, Salivary IgA, Interleukin-1β and MMP-8 as Salivary Biomarkers in Chronic Periodontitis Patients, Chin. J. Dent. Res., 2017, 20(1), 43–51 Search PubMed.
- P. H. M. Hernandez-Rios, M. Garrido, T. Tervahartiala, J. Leppilahti, H. Kuula, A. M. Heikkinen, P. Mäntylä, N. Rathnayake, S. Nwhator and T. Sorsa, Oral fluid matrix metalloproteinase (MMP)-8 as a diagnostic tool in chronic periodontitis, Metalloproteinases Med., 2016, 3, 11–18 Search PubMed.
- O. Schilling and C. M. Overall, Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites, Nat. Biotechnol., 2008, 26(6), 685–694 CrossRef CAS PubMed.
- I. Luchian, A. Goriuc, D. Sandu and M. Covasa, The Role of Matrix Metalloproteinases (MMP-8, MMP-9, MMP-13) in Periodontal and Peri-Implant Pathological Processes, Int. J. Mol. Sci., 2022, 23(3), 1806 CrossRef CAS PubMed.
- M. R. Guimaraes-Stabili, M. C. de Medeiros, D. Rossi, A. C. Camilli, C. F. Zanelli and S. R. Valentini, et al., Silencing matrix metalloproteinase-13 (Mmp-13) reduces inflammatory bone resorption associated with LPS-induced periodontal disease in vivo, Clin. Oral Investig., 2021, 25(5), 3161–3172 CrossRef PubMed.
- I. Luchian, M. Moscalu, A. Goriuc, L. Nucci, M. Tatarciuc and I. Martu, et al., Using Salivary MMP-9 to Successfully Quantify Periodontal Inflammation during Orthodontic Treatment, J. Clin. Med., 2021, 10(3), 379 CrossRef CAS PubMed.
- W. T. Y. Loo, M. Wang, L. J. Jin, M. N. B. Cheung and G. R. Li, Association of matrix metalloproteinase (MMP-1, MMP-3 and MMP-9) and cyclooxygenase-2 gene polymorphisms and their proteins with chronic periodontitis, Arch. Oral Biol., 2011, 56(10), 1081–1090 CrossRef CAS PubMed.
- W. Nesse, F. Abbas, I. Van Der Ploeg, F. K. L. Spijkervet, P. U. Dijkstra and A. Vissink, Periodontal inflamed surface area: quantifying inflammatory burden, J. Clin. Periodontol., 2008, 35(8), 668–673 CrossRef PubMed.
- J. Geiger, S. Both, S. Kircher, M. Neumann, A. Rosenwald and R. Jahns, Hospital-integrated Biobanking as a Service – The Interdisciplinary Bank of Biomaterials and Data Wuerzburg (ibdw), Open J. Bioresour., 2018, 5, 6 CrossRef.
- O. Schilling, U. auf dem Keller and C. M. Overall, Protease Specificity Profiling by Tandem Mass Spectrometry Using Proteome-Derived Peptide Libraries, in Gel-Free Proteomics: Methods and Protocols, ed. K. Gevaert and J. Vandekerckhove, Humana Press, Totowa, NJ, 2011, pp. 257–272 Search PubMed.
- L. Meinel, T. Luehmann and R. Raschig, Aminomethyl-functionalized denatonium derivatives, their preparation and use, EP3755685B1, 2019.
- M. Schnabelrauch, Chewing gum for the diagnosis of inflammatory tissues in dental applications patent, EP2822602B1, 2013.
- H. Nagase and G. B. Fields, Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides, Pept. Sci., 1996, 40(4), 399–416 CrossRef CAS.
- U. Eckhard, P. F. Huesgen, O. Schilling, C. L. Bellac, G. S. Butler and J. H. Cox, et al., Active site specificity profiling datasets of matrix metalloproteinases (MMPs) 1, 2, 3, 7, 8, 9, 12, 13 and 14, Data Brief, 2016, 7, 299–310 CrossRef PubMed.
- O. Schilling, U. auf dem Keller and C. M. Overall, Factor Xa subsite mapping by proteome-derived peptide libraries improved using WebPICS, a resource for proteomic identification of cleavage sites, Biol. Chem., 2011, 392(11), 1031–1037 CrossRef CAS PubMed.
- L. Zhang, X. Li, H. Yan and L. Huang, Salivary matrix metalloproteinase (MMP)-8 as a biomarker for periodontitis: A PRISMA-compliant systematic review and meta-analysis, Medicine, 2018, 97(3), e9642 CrossRef CAS PubMed.
- C. M. Cobb, Non-surgical pocket therapy: mechanical, Ann. Periodontol., 1996, 1(1), 443–490 CrossRef CAS PubMed.
- A. M. Marcaccini, C. A. Meschiari, L. R. Zuardi, T. S. De Sousa, M. Taba Jr and J. M. Teofilo, et al., Gingival crevicular fluid levels of MMP-8, MMP-9, TIMP-2, and MPO decrease after periodontal therapy, J. Clin. Periodontol., 2010, 37(2), 180–190 CrossRef CAS PubMed.
- G. Matuliene, B. E. Pjetursson, G. E. Salvi, K. Schmidlin, U. Brägger and M. Zwahlen, et al., Influence of residual pockets on progression of periodontitis and tooth loss: Results after 11 years of maintenance, J. Clin. Periodontol., 2008, 35(8), 685–695 CrossRef PubMed.
- E. Dejonckheere, R. E. Vandenbroucke and C. Libert, Matrix metalloproteinase8 has a central role in inflammatory disorders and cancer progression, Cytokine Growth Factor Rev., 2011, 22(2), 73–81 CrossRef CAS PubMed.
- C.-C. Wang, H.-C. Houng, C.-L. Chen, P.-J. Wang, C.-F. Kuo and Y.-S. Lin, et al., Solution Structure and Backbone Dynamics of Streptopain, J. Biol. Chem., 2009, 284(16), 10957–10967 CrossRef CAS PubMed.
- G. M. Air, Influenza neuraminidase, Influenza Other Respir. Viruses, 2012, 6(4), 245–256 CrossRef CAS PubMed.
- D. E. Bassi, H. Mahloogi, L. Al-Saleem, R. Lopez De Cicco, J. A. Ridge and A. J. P. Klein-Szanto, Elevated furin expression in aggressive human head and neck tumors and tumor cell lines, Mol. Carcinog., 2001, 31(4), 224–232 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00031a |
| This journal is © The Royal Society of Chemistry 2023 |