Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanomaterials for miRNA detection: the hybridization chain reaction strategy
Brij
Mohan
 *a,
Sandeep
Kumar
b,
Suresh
Kumar
*a,
Sandeep
Kumar
b,
Suresh
Kumar
 *c,
Krunal
Modi
*c,
Krunal
Modi
 d,
Deependra
Tyagi
e,
Dimitri
Papukashvili
e,
Nino
Rcheulishvili
e and
Armando J. L.
Pombeiro
a
d,
Deependra
Tyagi
e,
Dimitri
Papukashvili
e,
Nino
Rcheulishvili
e and
Armando J. L.
Pombeiro
a
aCentro de Química Estrutural, Institute of Molecular Sciences, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001 Lisboa, Portugal. E-mail: brizharry17@gmail.com
bSchool of Science, Harbin Institute of Technology (Shenzhen), Shenzhen 518055, China
cDepartment of Chemistry, SUS Government PG College, Matak Majri, Karnal, 132041, Haryana, India. E-mail: sureshprocha@gmail.com
dDepartment of Humanity and Science, School of Engineering, Indrashil University, Mehsana-382740, Gujarat, India
eSouthern University of Science and Technology, Shenzhen 518000, China
First published on 8th December 2022
Abstract
MicroRNAs (miRNAs) with nucleotides are a class of endogenous small RNAs and can play crucial functions in diagnosing diseases. In particular, the group of miRNAs is responsible for information related to the cell and disease. Among various techniques for miRNA detection, the hybridization chain reaction (HCR) strategy shows potential and efficiency. This review summarizes and studies the most efficient HCR strategy for detecting miRNA using nanomaterials due to their ultrasensitive detection and excellent performances. In addition, signal amplification for the sensitive detection of miRNA due to the chain reaction has been studied. The key factors, such as limit of detection (LOD), linear range, the importance of the strategy, limitations or challenges, and future perspective are described. Finally, the study will provide new findings for developing a miRNA detection strategy applicable to disease diagnosis.
1. Introduction
Health is a natural global priority, but with the increasing health issues, different types of diseases, such as cancers, heart and kidney failure, HIV, and liver damage, have become a serious cause for millions of deaths.1–3 Also, it is difficult to monitor health checkups in many countries due to their high population and lack of resources. However, it can be achieved by developing simple techniques to detect microribonucleic acids (miRNAs) in various cell samples4–9 These are noncoding short-sequence RNAs that are helpful in the expression of a gene. Upon hybridization with messenger RNA, miRNAs cause a negative regulation of gene transcription. The expression of miRNAs is responsible for various diseases, most importantly cancers.10 Therefore, miRNAs can be used as exciting biomarkers to diagnose diseases. miRNA can be found in serum or plasma, and in circulating miRNA in the form of extracellular vesicle-derived miRNA. Spreading miRNAs are the best candidates as biomarkers, as extracellular vesicles increase the miRNA amount, making detection more accessible and reliable.11 Extracellular vesicles also protect miRNA from RNA degradation and increase its stability.12 It can thus be seen that miRNA is an essential biomolecule and can reveal important biological and biochemical information. Hence, there is a great need to develop new techniques for detecting miRNA to meet the disease diagnosis and public health needs.13,141.1 Importance of miRNA
miRNAs are from primary transcripts (pri-miRNAs); these can be controlled at the transcriptional and post-transcriptional levels. It is, however, challenging to collect the exact information for the regulation at multiple levels. Any changes in miRNA levels can symbolize a disorder in the body. The time course of mature and pri-miRNAs in production, their dynamic changes, and degradation can indicate a disorder. Also, nucleotide strands influence the stability of mature miRNAs, and their degradation can provide information about genes.15 In addition, miRNA is associated with cytokine signaling in the immune system and is essential for colorectal cancer, acute lymphoblastic leukemia, carcinoma, multiple myeloma, prostate, and ovarian cancers, etc. It can be determined by the expression level of miRNA in cells.16 In addition, miRNAs and noncoding RNAs help modify the autoimmune process in immune and nerve cells. Hence, miRNA can act as a biomarker for detection, diagnostic, and therapeutical applications for various diseases.17–191.2 Importance of the HCR strategy
The hybridization chain reaction (HCR) provides a promising way to detect miRNAs compared to other signal amplification methods. Recently, various detection methods have been used to develop multiple signal amplification strategies, but HCR has emerged as a powerful signal amplification technique. Moreover, it shows an enhanced application due to its nonenzymatic and isothermal features.20 One advanced feature in the HCR is a cross-opening that leads the initiator probe to trigger a pair of metastable hairpins. This then propagates an HCR event that results in a long-nicked double-stranded nucleic acid structure.21 Hence, it becomes an easy and advanced way to achieve excellent performance for the ultrasensitive detection of microRNA-21 based on the content of the hairpin probe.221.3 Detection techniques for miRNA and their scope
miRNA detection has experienced considerable advancements due to its high specificity and sensitivity in diagnosing diseases.23 Also, the abundant and circulating presence of miRNAs could affect their detection in tissues and biological fluids.24 The accurate detection of miRNA levels can help diagnose early diseases and thus enable effective therapeutic management.25,26 miRNA biosensing generally requires a biological system for recognizing and capturing target miRNA. Also, a transducer that can convert the capture reaction into an easily-triggered signal. Low miRNA expression in cells has resulted in the discovery of novel miRNA analysis methods with benefits of multiplexed detection, good reproducibility, low cost, and rapid detection.Recently, the ligation chain reaction with HCR for enzyme-free signal amplification strategies has been used to quantify microRNAs.27,28 Also, incorporating a 3D DNA layer on the electrode interface has been used to achieve ladder HCR and improved electrochemical signals for miRNA detection.29 However, it remains a challenge for researchers to ascertain the quantity of miRNA, mainly due to the short sequence and low abundance of miRNA.30 However, various techniques, such as microarrays, Northern blotting, and quantitative reverse transcription-polymerase chain reaction (qRT-PCR), are commonly employed for quantitative RNA measurement,31 but these typically all have limitations, such as lengthy procedures, the requirement for a large sample, and complicated and sophisticated instrumentation, that must be addressed.32
The most critical issues, such as the sensitivity and performances of biosensors for miRNA detection, have been resolved using Au-loaded nanoporous superparamagnetic Fe2O3, which can provide an active surface for miRNA adsorption.33 In addition, it has been used to develop rapid and inexpensive miRNA biosensor strategies.34 With the rapid growth in this research area, the hybridization chain reaction (HCR) provides an advanced, efficient, isothermal signal amplification that has been widely used in miRNA detection. It involves a linear elongation of double-stranded nicked DNA, single-stranded initiator DNA, and two hairpin fuel DNAs. Efforts have been made to explore HCR in recent times, given the utility of HCR in miRNA detection. Therefore, there is an urgent need to review all such articles for further improvement and for the utilization of the HCR in detecting miRNA.35,36
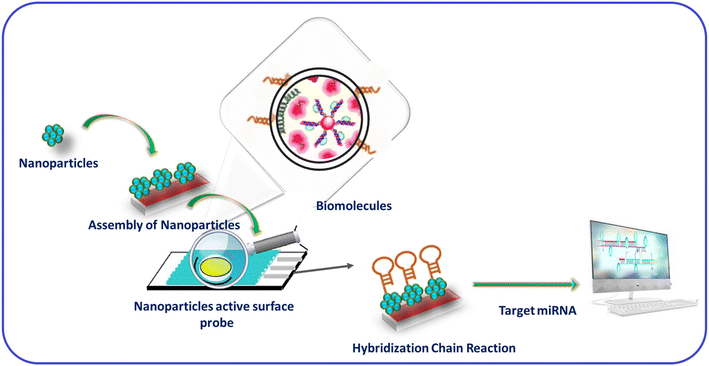
In recent years, various techniques, namely cyclic voltammetry (CV), Raman scattering, infrared, ultraviolet, visible, and fluorescence spectroscopy, mass spectrometry, surface plasmon resonance, and electrochemical-based approaches, have been used to detect biospecies.37–39 In addition, various inorganic and hybrid materials have been utilized for miRNA detection. In particular, nanomaterials have attracted considerable attention for detecting miRNAs among these materials due to their active surface area and robust sensing properties. In addition, the electropositive nature of metal-based nanomaterials (nanoparticles, nanorods, nanosheets, nanowires, nanoflares, nanotags, and nanoclusters) provides a suitable platform for miRNA recognition.40,41 Moreover, the two hairpin probes in miRNAs show interactions, resulting in the signal changes due to the HCR. In addition, biomolecules accumulation of nanomaterials will lead to signal amplification in miRNA detection. For example, AuNPs and magnetic 3D DNA walkers provided a solution for the colorimetric detection of miRNA. In addition, NPs with multiple metal ions have shown multianalyte detection with enhanced specificity and sensitivity.42 Hence, nanomaterials for HCR-based detection have allowed insights into the roles of various materials in signal amplification. In addition, it has created interest in researchers to collect information about miRNA binding to the unfolded H1 probe through SDR and then hybridization (Fig. 1).43
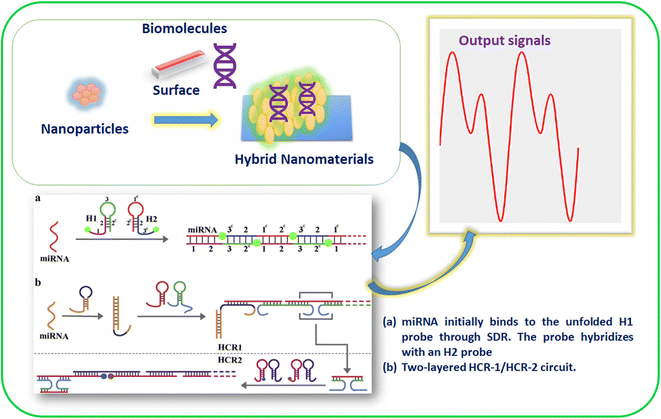 | ||
| Fig. 1 Materials for HCR-based miRNA detection and to gain working insights.43 | ||
Several review articles have been published in the past years summarizing materials for detecting miRNA.44 Despite these valuable studies, insights into the HCR-based signal amplification techniques have gained limited attention.45,46 Hence, studies into the insights provided by HCR-assisted signal amplification strategies are needed, mainly because of the extensive research in this field. Therefore, we have reviewed and analyzed HCR-assisted signal amplification strategies in this article to overview the new developments in this field for miRNA detection. Furthermore, we have explored several combined nanomaterials based on miRNA biosensors using HCR as representative examples, and finally, the future trends in miRNA detection are briefly discussed.
2. Development of fabricated platforms and HCR-active nanomaterial biosensors
Fabricating nanomaterials with the HCR can provide an active platform for detecting miRNA. The interactions through metal and electron-rich functionalities can allow signal amplification.47 For example, thiolated DNA-linked metal nanomaterials/NPs showed enhanced signals in spectroscopic and electrochemical studies.48 The fabrication of AuNPs with DNA molecules provided an active surface through Au–SH interactions.49 The designed nanocluster with an active surface could significantly detect miRNA in blood samples. It was found that the interaction significantly affected the photocurrent signal of the electrode while allowing detecting circulating miRNA-141 in whole blood samples.50 Developing a new fabricated platform for detecting miRNA needs to incorporate biorecognition modules with new materials.51 Various strategies have been developed to provide stable thermal, mechanical, and solution platforms for fabricating nanomaterials.Moreover, miRNA detection is based on the interactions for miRNA hybridization via signal changes and could lead to biorecognition and hairpin-shaped probes. The active surface in fabricated nanomaterials is helpful for a rapid HCR in miRNA recognition.52,53 The presence of functional sites in DNA-fabricated nanomaterials allows the hybridization of miRNA and follows a Watson–Crick base-pairing.54 This could result in rigid DNA–RNA or RNA–RNA hybrid structures. Therefore, it offers rapid and sensitive electrochemical, optical, or mechanical-based detection. Hence, biomolecule-fabricated NPs have advanced features for detecting miRNA with high sensitivity through hairpin-shaped nucleic acid probes. In addition, a loop is present that includes the target miRNA and an active surface acting as a stem in NPs.55 Therefore, the interactions between the active surfaces of fabricated nanomaterials and miRNA could result in loop dissociation, thus supporting the HCR. The presence of new functional sites in fabricated nanomaterials provides benefits in engineered nanomaterials biosensing.56 This strategy has thus emerged as a promising tool for HCR-based miRNA detection due to its active surface, nano-size surface![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) volume ratio, and enhanced optical-electronic properties.
volume ratio, and enhanced optical-electronic properties.
Immobilizing DNA molecules and amino acids could be useful for the fabrication or modulation of nanomaterials. Such fabricated nanomaterials (<10 nm) were found to exhibit better properties, such as water solubility, biocompatibility, photobleaching, and chemical resistance compared to traditional fluorophores.57 Also, nanomaterials with sizes of 1–100 nm showed applications in cancer detection due to their Tyndall effect or light-scattering properties. Furthermore, AgNPs, AuNPs, and magnetic nanocomposites have shown advanced applications in the colorimetric detection of miRNA.58
3. HCR-based sensing principles and techniques
The standard working principle of HCR-based sensing may follow two parts. In the first part, the chain reaction follows miRNA displacement amplification. Pierce and Dirks59 reported the mechanism for HCR for the first time, while it was further studied by Choi et al.60 The amplification of the miRNA could be responsible for the formation of a duplex and multi-way junction structures. This leads to the potential sensing of miRNA followed by the HCR on the biosensor surface. Also, the critical mechanism of HCR depends on the stored potential energy loops. The stored potential energy could employ the hairpin species. The employed hairpin system is followed by energy protection of the short loops by the long stems. The interaction of the molecular beacons with the nucleotide could lead to the HCR with kinetic trapping by the hairpin.59HCR-based miRNA detection produces double-strand molecules with overlapping hairpin pairs with partial complementarities. In addition, the detection process may include one DNA initiator and a pair of DNA hairpins (H1 and H2). The existing DNA hairpin pair gets opened in the solution with the initiator binding. It can be observed that the designed length plays a crucial role in the working and sensing mechanisms for HCR detection assembly. Therefore, using HCR to detect miRNA is a potential useful tool, and precise signals could be achieved by designing DNA hairpins.61
The electrochemical detection of miRNA offers a sensitive surface for electrochemical signals, which could be regarded as offering miRNA detection. However, photobleaching in the case of fluorescence and preparation of the electrochemical surface in detection can be complicated. Therefore, an appropriate hairpin design is crucial for miRNA's enhanced specific and sensitive detection.62 In addition, a fluorescence HCR assay to detect miRNA was designed by heating DNA H1 and DNA H2 at a particular temperature and then cooling them at room temperature, which led to activating the miRNA detection process.63 Upon miRNA addition with the designed hairpins, changes in the fluorescence intensity signals could be observed. A pair of hairpin DNA probes enriched with fluorophores was treated with miRNA in solution. Each miRNA molecule was triggered due to HCR between two hairpins, H1* and H2*, with accumulated changes in the signals.64
Various nanomaterials show excellent physicochemical properties and can be functionalized to develop fascinating biosensing platforms. Moreover, graphene-based nanomaterials have exhibited excellent detection for miRNA by forming π-conjugated supramolecular assemblies.65 For example, adding graphene oxide (GO) to the solution after the HCR resulted in the absorption of the two hairpins on the GO surface due to π–π stacking interactions.66 The sensitivity of this method was based mainly on the surface prepared for electrodes. The electrochemical HCR miRNA detection results could be read through the interaction with the target. In one study into the working mechanism of an HCR electrochemical sensor for miRNA detection, methylene blue (MB) and mesoporous silica containers (MSNs) were utilized for the signal release and helped in the sensitive electrochemical detection of miRNA. The DNA molecules acted as gate molecules to hold the hairpins for MB when miRNA was not present. In the presence of miRNA, hairpin 1 (H1) was released from the MSNs due to the miRNA interaction. Then MB was released due to the gate opening by the DNA. Moreover, the electrode surface was hybridized and resulted in an HCR amplification process (Fig. 2).67
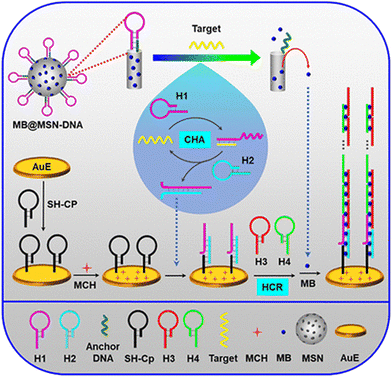 | ||
| Fig. 2 Silica containers and MB-based HCR electrochemical assay for miRNA detection; reproduced with permission; Copyright©2019, American Chemical Society.67 | ||
4. HCR strategy for miRNA detection
4.1 Signal amplification
The HCR can explain the signal amplification in biosensing because it is a kinetics-controlled reaction with excellent sensitivity and selectivity for detecting a target.68,69 Moreover, it does not require pH control, temperature, or buffer media.70 Miao et al. recently developed a dumbbell hybridization chain reaction (DHCR) combined with strand displacement amplification, in the developed sensing approach was based on two strategies. The first part involves the hybridization of miRNA with template DNA and a recycled polymerase catalyzed extension reaction and DNA nicking, resulting in many strands for the three-way junction (TWJ) structure marked as TWJ2 in Fig. 3. This demonstrated two dumbbell-shaped DNA DHP1 (dumbbell hybridization probe 1) and DHP2 (dumbbell hybridization probe 2) probes as fuel strands for HCR, having complementary sequences represented in Fig. 3 as b/b* and c/c*, which are helpful for dumbbell formation. DHP1 formed a three-way junction (TWJ1) with the electrodes and strands marked as TWJ2, created by strand displacement amplification.71 Next, DHP1 could be recognized, which helped the open the dumbbell structure and completed the three-way junction.72 Then, the complementary strand of DHP1 could activate the dumbbell structure of DHP2, resulting in a stacking of DHP1 and DHP2.73 Next, the complementary strand of DHP2 could start the dumbbell structure of a new probe (DHP1), resulting in recycled hybridization reactions, which is helpful for recording the electrochemical signal and evaluating miRNA levels.74 The combination of dual strategies significantly improved the sensitivity of the detection probe for miRNA with a low limit of detection (LOD) in the range of 7.3 aM, and was also applicable to analyzing samples from biological systems, such as human serum and cells, demonstrating their utility in point-of-care testing applications.71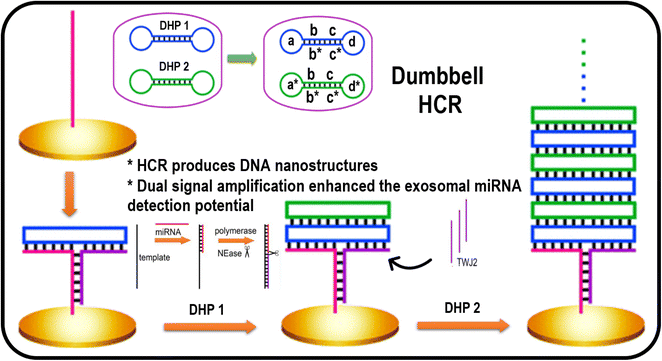 | ||
| Fig. 3 Representation of a biosensor based on DHCR for miRNA detection. Reproduced with permission Copyright©2020, American Chemical Society.74 | ||
Immobilized surface plasmon resonance and HCR can be utilized for an economical, robust, and straightforward biosensor for miRNA detection. The use of gold nanoparticles to detect miRNA (miR-17) is a method with good specificity for detection via hairpin probes, in combination with HCR to amplify the signal to extend the dynamic range of quantization. This process is rapid and required just 1 h for completion and displayed a very low LOD, nearly equal to 1 pM or 50 amol per measurement.75 A rapid electrochemical assay was developed for the HCR-based detection of microRNA-122. It was observed that hairpin DNA (hpDNA) gets opened on the gold electrode surface when miR-122 is present. The hpDNA helped to trigger HCR via the cross-opening of helper DNA hairpins and their hybridization (Fig. 4). The high density of hpDNA on the electrode resulted in HCR signal amplification. The LOD of the electrochemical assay for miR-122 detection was found to be 53 aM. The assay was also noted to selective and could detect exosomal miR-122 with high-valuable efficiency in cancer diagnostics.76
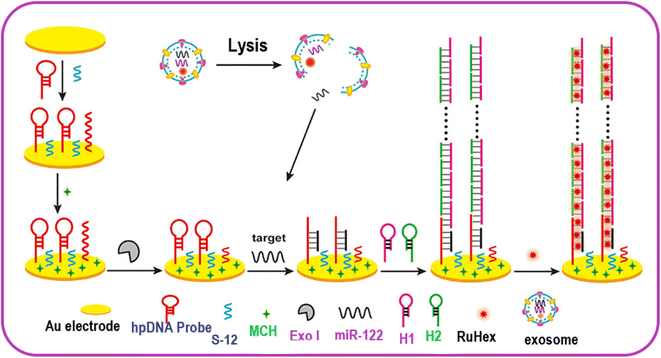 | ||
| Fig. 4 Electrochemical sensing of exosomal MicroRNA based on HCR signal amplification. Reproduced with permission Copyright©2020, American Chemical Society.76 | ||
4.2 Electrochemical and fluorescence signals
Moreover, the enzyme-free concatenated (EFC) detection of miRNA provided an advanced strategy in this area. For example, Wei et al. introduced an isothermal EFC hybridization chain reaction (C-HCR). This C-HCR was based on two different HCR layers (HCR-1 and HCR-2). In this system, the output from the HCR1 upstream layer is used as an intermediate input for activating the downstream layer of HCR-2. HCR-1 starts through the usual cross-opening of DNA hairpins to generate polymeric double-stranded DNA nanowires. These dsDNA nanowires are made up of various sequentially arranged triggers T in the form of an output. Reconstructed DNA segments T, termed amplicon T, are captured by HCR-2. Amplicon T helps in transduction for identifying analytes in the form of an amplified readout following the synchronization of both HCR layers (HCR-1 and HCR-2). In addition, it was observed by the authors that C-HCR occurred through the formation of leaf-like double-stranded DNA nanowires along with the production of amplified Förster resonance energy transfer (FRET) signals with the help of various analytical techniques.77In another example, Guao et al. recently described using time-gated Förster resonance energy transfer (TG-FRET) for microRNA analysis. The technique was used for terbium donors and dye acceptors in HCR for miR-20a and miR-21. The author's method provided excellent results for quantifying microRNA with a low LOD with a minimum of 240 amol of microRNA. The technique proved efficient at distinguishing between homologous microRNAs with high target specificity. The multiplexing of measurements in a FRET pair at excitation wavelength allowed the simultaneous quantification of miR-20a and miR-21, even at low concentrations of 30 and 300 pM. The developed HCR was applied equally to serum-free and serum-containing samples without RNase inhibitors. It can work in various living systems and can be applied for advanced nucleic acid biosensing.78
For example, MoS2 nanosheets enriched with MBs were used by Zhang et al. to develop hybridization chain reaction (HCR)-based miRNA detection. The authors used MoS2 nanosheets to capture MBs as an adsorption probe. In addition, the MoS2 nanosheets also worked as selective fluorescence quenching probes required for reducing background signals. The presence of target miRNAs triggered the HCR process, leading to many products. However, the products obtained from HCR as nanowires chains had a G-quadruplex abundance, which could not be adsorbed on the MoS2 surface and detached. Hence, Thioflavin T could be attached to the G-quadruplex and produced an electrochemical signal analyzed by fluorescence spectroscopy. This method could achieve a low detection for miRNA to a minimum of 4.2 pM in a wide linear range from 0.1 to 100 nM.79 In addition, Jia et al. used MoS2 quantum dots (QDs) in an HCR of G-quadruplex enzymatic catalysis for microRNA analysis via the inner filter effect. It was observed that the target microRNA triggered the HCR of two DNA probes and generated double-stranded DNA (dsDNA). The long dsDNA thus produced had many hemin/G-quadruplex enzymes with hemin. In the presence of hydrogen peroxide, these DNAzymes directly oxidized o-phenylenediamine into 2,3-diaminophenazine; this oxidation through the inner filter effect resulted in the fluorescence quenching of the MoS2 QDs. The electrochemical response of these QDs was directly proportional to the amount of miRNA. This approach was found to have a low LOD of 42 fM and can be used for practical applications.80 Ding et al. designed silicon nanoparticles (SiNPs) for detecting miRNA by fluorescence quenching due to the inner filter effects (IFEs). The HCR between DNA hairpin probes with G-quadruplex sequences led to horseradish peroxidase (HRP)-mimicking DNAzyme formation for targeting miRNA. This then catalyzed the H2O2-mediated oxidation of o-phenylenediamine to 2,3-diaminophenazine. The absorption and emission band overlapping resulted in fluorescence quenching.81 Also, Ying et al. demonstrated an HRP-based HCR strategy for detecting miRNA-155 with color change responses with an LOD of 31.8 fM. CP immobilized at the microplate played an active role in capturing miR-155 and the 3′ end of the reporter probe, while the 5′ end initiated the HCR. This resulted in signal amplification via dsDNA polymers having multiple fluorescein isothiocyanates. The observed colorimetric changes from colorless to blue color for miRNA detection could have been due to the antibody interactions on the microplate through the tetramethylbenzidine/H2O2 system.82 In another example, Feng et al. developed an HCR-based electrochemical miRNA-21 detection method by combining the DNA-generated current and target-triggered HCR. The immobilization of thiol-modified hairpin CP on a Au electrode resulted in miRNA-21 conformational change. This conformational change was responsible for HCR initiation for long DNA strands generated on the electrode surface. Also, the reaction between the DNA phosphate backbone and molybdate led to the redox probe molybdophosphate and generation of an electrochemical.83
miRNA detection in the presence of DNA has been widely explored and gained considerable attention.84 Zhou et al. developed a nonlinear HCR-based electrochemical method to detect microRNAs using Y-shaped DNA integration. Y-Shaped DNA consists of three sequences Y1, Y2, and Y3. These can act as stable and specific units for the detection of miRNA. It was reported that a competitive hybridization reaction occurs between miRNA and Y-shaped DNA when target miRNA is present. This was demonstrated by the freeing of the Y3 probe followed by the dissociation of the Y-shaped DNA structure. Subsequently, triggers blocked by Y3 become exposed, resulting in the onset of nonlinear HCR. This resulted in electrochemical changes in the signal through an amplification reaction, which could be recorded. The biosensor developed in this technique could detect microRNAs (miRNAs) up to an LOD of 0.3334 fM. The linear range was from 1 fM to 10 pM. Furthermore, the unique Y-shaped DNA turned out to be helpful to the biosensor for identifying single-base mutations.85
4.3 Electrochemical nanomaterials-based biosensors
For electrochemical nanomaterials-based biosensors to achieve high performance, surface fabrication with molybdenum disulfide (MoS2), GO, quantum dots (QDs), etc. can provide a unique electronic structure, which can help to improve the detection sensitivity and detection limits.86It is important to improve the reliability of miRNA for detecting progression-motivated disease diagnosis applications. Hence, developing an accelerated DNA nanoprobe for miRNA for the in situ monitoring of biofluids with a spatial strategy could provide a highly efficient and significant approach. In addition, a fast response would allow the nanoprobes to monitor the process of exosome endocytosis.87 For example, Zhuang et al. reported improved in situ hybridization methods for detecting miRNA through HCR. The in situ HCR method could detect miRNA even from mouse retinas. Furthermore, this process could be used to detect two microRNAs simultaneously, and even miRNA and mRNA could be detected simultaneously.88
Moreover, the combination of Fe magnetic nanoprobes with DNAs could be used for the HCR detection of microRNA-141 and microRNA-21 at a low level. For instance, SH-modified hairpin capture probes (CPs) were hybridized with miRNAs on an Au electrode, leading to conformation changes of the CPs. Furthermore, this triggered HCR to generate plentiful bonding sequences of the magnetic nanoprobes.89 Zhu et al. designed a Ru(bpy)32+-based ratiometric ECL–EC hybrid biosensor for the sensitive and low detection of miRNA-133a with an LOD of 12.17 aM. The Au–S bond in the DNA tetrahedron nanostructure and two Ru(bpy)32+-labeled H1 and H2 hairpins were utilized as ECL probes and fuel strands for an HCR. The MB with the 5′ end of the miRNA provided the internal reference signals.90 Extracellular vehicle (EV) cancer biomarkers can provide parent molecular information early on. For example, Wu et al. developed an EV-derived HCR-based strategy for detecting inherent in situ miRNAs. A modularized DNAzyme-amplified two-stage cascaded HCR circuit created the detector amplifier. This was followed by HCR1 as an analyte-generated output and HCR2 as a trigger input. Moreover, the designed modular CHCR–DNAzyme circuit acted as a “plug-and-play” sensing mode for detecting miRNA. Interestingly the developed model worked in vitro in different cells by the amplified detection of the miRNA biomarkers in the EVs.91
Au-based nanomaterials have demonstrated improved biosensing performance that would be helpful for disease identification.92 Recently, Yuan et al. reported a ratiometric electrochemical assay for detecting miRNA from thionine (Thi) and ferrocene (Fc). First, a Au electrode was developed through a Au–S reaction through a thiol-modified and ferrocene-labeled hairpin CP. Then, the HCR-based target detection of miRNA was accompanied by a hybridization of the chain probes and unfolding of the miRNA-DNA duplexes hairpin and Kamchatka crab duplex-specific, resulting in the miRNA release. The far electrode distance of Fc led to the Fc signal-off state, and the residual fragment process on the electrode surface was responsible for the HCR for generating dsDNA. The in situ HCR had a primer, HDNA, and HDNA′ probes for capturing numerous DNA/Au NPs/Thi, resulting in the signal-on state of Thi. The observed dual-amplification mechanism signal-off for Fc and signal-on for Thi provided a sensitive HCR technique for detecting miRNA-141 with an LOD of 11 aM.93
Yang et al. designed a strategy to introduce a branched HCR circuit. The use of a terbium(II) organic gel (TOG) electrode elevated the biosensor sensitivity by a bHCR circuit for miRNA-141 detection with a low LOD of 0.18 fM. The advancement in this work was a one-step approach to modify nucleic acids to electrodes to introduce the DNA structure. This method was reported to be more precise and can avoid errors that may arise in the stepwise modification methods due to the electrode's drawback of low-molecular-weight nucleic sequences.94 In another example, Zhang et al. designed polydopamine (PDA)-encapsulated photonic crystal (PhC) barcodes for target-triggering cycle amplification and HCR for detecting miRNA with an LOD of 8.0 fM. Moreover, the barcodes showed structural colors for different encoding miRNAs that could immobilize biomolecules for helping the reaction with amino-modified hairpin probes (H1) and could initiate HCR for cycle amplification.95
miRNAs as riveting RNAs have significance in gene regulation and specific roles in certain pathological and physiological or pathological processes. The use of Au nanomaterials-based sensors provides a tool for the rapid and sensitive detection of the miRNA assay.96 For example, Lu et al. designed an ECL biosensor by immobilizing a CP on Fe3O4@SiO2@AuNPs to detect femtomolar miRNA-141 with a low LOD of 0.03 fM. The HCR-assisted cascade amplification and Faraday cage-type strategy through GO and signal unit from the material (Ru(phen)32+-HCR/GO) were allowed via nucleic acid hybridization. The large surface area and electronic transport properties in this system were responsible for the enhanced signal amplification. In addition, GO concentration on the electrode surface resulted in the sensor's outer Helmholtz plane (OHP) extension.97 Also, Zheng et al. reported that Fe3O4@SiO2@AuNPs-cDNA nanomaterials coated by hairpin cDNA could be used to detect miRNA-126 with an LOD of 2 fM. The ECL signal unit was fixed through DNA and HCR-Ru(phen)32+ for target miRNA-126, which opened the stem-loop structure of cDNA.98 In another example, Fan et al. reported target HCR-based detection for microRNA with an LOD of 4.2 fM. The GO has far-reaching significance in signal change for HCR miRNA detection, which was achieved by a helicase-assisted GO-based reaction platform for microRNA (miRNA) detection.99
Nanomaterials-based electrochemical sensors hold excellent promise for fast miRNAs detection in real samples. Combining G-quadruplex DNA probes and single-stranded anchor DNA could help seal the pores.100 Furthermore, the combination of materials can provide new materials for biosensing applications. For example, a magnetic beads and duplex-specific nuclease enzyme combination showed an enhanced detection potential for miRNAs-21 with an LOD of 170 aM.101 Tang et al. designed DNA/Fe3O4 nanosheets as a triple-amplification assay for detecting miRNA let-7a with an LOD of 13 aM. The magnetic nanosheet networks were initiated by the target miRNA-associated HCR. Also, DNA-combined networks were reported to catalyze peroxidase for a colorimetric reaction.102 Nanomaterials with CdSe QDs showed enhanced signals for detecting miRNA. In addition, the hybrid materials were helpful for surface programmatic chain reactions and multiple amplification.103 In another example, a nucleic acid framework designed by Qu et al. acted as a multiple miRNAs sensor. The surface was modulated with DNA probes via lateral interactions that enabled a programmable tailoring of the enhanced kinetics and hybridization efficiency for sensing. The microassay framework combined with the HCR amplification strategy was used to detect miRNA (e.g., FNA-miR-652, FNA-miR-627, and FNA-miR-629) biomarkers in gastric cancer.104
In another example, Zhu et al. designed flower-like gold nanostructures (HFGNs) as an electrochemical sensor for detecting miRNA-21 with an LOD of 0.12 fM in a linear range from 1 fM to 1 nM. The HFGNs-deposited ITO first captured DNA (DNA-1) on its surface, and then HCR was attached to the electrode through a target miRNA-mediated sandwich hybridization for signal amplification.105 Xue et al. recently designed a label-free DNA dendrimers HCR-mediated multiple G-quadruplex to detect miRNAs. The hairpin switch probe was employed for enhancing the weak signals of the split G-quadruplex of double-stranded DNAs and nonlinear HCR assembly.106 Nanomaterials have been used for various biomedical faces due to their active biological immobilization properties. Hence, HCR-based miRNA detection provide an effective strategy for detecting miRNA, and the observed results can be evaluated with easy handling (Fig. 5). The current early detection of diseases is often ineffective due to the significant population needing testing. An HCR-based sensor could be helpful in this regard for the effective and early detection of miRNA to provide disease information. Moreover, HCR-based outcomes are effective with nanomaterials (Table 1).107
| Sr. no. | Materials/electrode | Target | LOD | Linear range | Ref. |
|---|---|---|---|---|---|
| 1 | Gold electrode surface | miR-122 | 53 aM | 76 | |
| 2 | MoS2 nanosheets | miRNAs | 4.2 pM | 0.1 to 100 nM | 79 |
| 3 | MoS2 quantum dots | miRNA | 42 fM | — | 80 |
| 4 | CP immobilized | miRNA-155 | 31.8 fM | — | 82 |
| 5 | Y-Shaped DNA integration | miRNAs | 0.3334 fM | 1 fM to 10 pM | 85 |
| 6 | Ru(bpy)32+-based ratiometric ECL–EC hybrid biosensor | miRNA-133a | 12.17 aM | — | 90 |
| 7 | DNA/Au electrode | miRNA-141 | 11 aM | — | 93 |
| 8 | Terbium(II) organic gel | miRNA-141 | 0.18 fM | — | 94 |
| 9 | Polydopamine (PDA) encapsulated photonic crystal | miRNAs | 8.0 fM | — | 95 |
| 10 | Fe3O4@SiO2@AuNPs | miRNA-141 | 0.03 fM | — | 97 |
| 11 | Fe3O4@SiO2@AuNPs-cDNA | miRNA-126 | 2 fM | — | 98 |
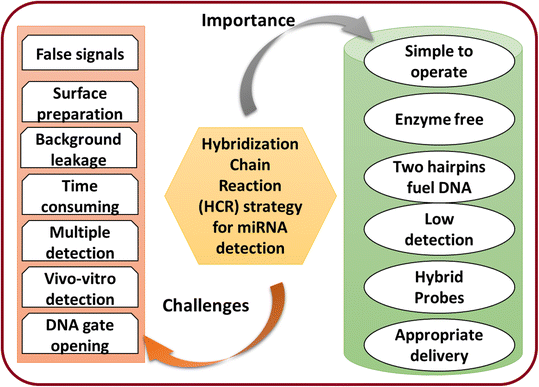
5. Importance and challenges
miRNA detection is critical due to miRNA's involvement in various biological processes. Different artificial materials and techniques can achieve the detection of miRNA. Therefore, there is a tremendous demand in academia and in clinical purposes for miRNA detection to address health issues. Currently, reusable and cost-effective portable devices are most important for detecting miRNA.108 Therefore, the recently developed materials have been utilized as an essential tool for miRNA detection from various cells. The critical feature of the detection technique is to perform well in detection with signal amplification. The advanced approaches combine the function of enhancing the trace target molecules, avoiding traditional labels, and reducing noise effectively. The common importance and challenges of the HCR strategy for miRNA detection to make the study more beneficial for researchers are illustrated in Fig. 6.Furthermore, HCR-based miRNA detection and the use of this technique for disease diagnosis will be highly beneficial for society. This can be achieved by using various newly designed material matrices. This approach will have significant scientific importance to aid developing a technology that can overcome several issues in diagnosis. The design of HCR-based detection for miRNA will help avoid the need for the use of expensive and complex techniques for clinical applications.109 In particular, while different amplification techniques based on enzymes and nuclease have been potentially utilized for miRNA detection, these generally still suffer some challenges and limitations, such as selection and the selectivity of the analyte. The feasible guidelines for real-world applications are still lacking. By solving these limitations, the materials science for detecting multiple miRNAs will be advanced and solutions could be utilized for real-world applications. The significant challenges are discussed as following.
1. Other issues, such as the concentrations of the hairpins and leakage, are still challenging and need to be resolved to address the background leakage. Moreover, DNA present during the HCR of the hairpin without miRNA could be subject to an unnecessary chain reaction. These unwanted factors could be responsible for false-positive signals. Various factors, such as temperature, concentration, solvent choice, and pressure, must be controlled for the HCR assay.
2. It was seen that the HCR-based biosensors follow the random diffusion of DNA and displacement. Therefore, it could take a longer time to overcome the slow kinetics. Thus, designing a model for rapid miRNA detection is still challenging.
3. The lack of sensitivity with current amplification technologies needs to be improved. In addition, the detection of multiple miRNAs is still a challenge. Therefore, these imitations need a combined detection technology with enhanced signals and probes.
4. The development of portable detection devices is an urgent need and a challenge for portable detection tools to meet real-world applications.
5. Also, the low concentration of miRNAs in vivo could limit the detection process. On the other hand, the in vitro detection of miRNA could lead to degradation and affect the efficiency of the detection techniques. Hence, there is an urgent need to develop potential and efficient detection methods for in vivo detection. This could be achieved by developing fabricated or hybrid materials.
6. Conclusions
In conclusion, HCR-based miRNA detection has been widely studied to develop biosensors. The signal amplification approaches were presented herein as an advanced feature of the HCR. The HCR strategy for detecting miRNA with selective electrochemical signal amplification provides a versatile platform with low cost and enhanced sensitivity. However, limitations like the negative charge, sufficient degradation, and centering DNA hairpin probes regarding miRNA detection must be overcome. Some innovative research discussed in this review provided directions for developing new HCR strategy-based miRNA detection materials. The miRNA detection performance acquired by nanomaterials has been explored for HCR targeting. In addition, immobilized nanocomposites with enhanced efficiency, sensitivity, and detection potentials for detecting miRNA have been studied. While studies and analyses have been primarily conducted in vitro, the biological effects in different aspects are encouraging. In vivo studies are critical to confirm the observed effects, assess the side effects, and make a comprehensive evaluation before achieving the ultimate clinical application of DNA-based nanostructures.7. Future prospectives
Although potential research has been devoted to the design of HCR-based miRNA detection, to better use the HCR strategy for miRNA detection, the design of new materials is crucial to enhance performance. Developing efficient software and calculations with enzyme-free amplification could improve the detection signal. Therefore, the HCR-based detection strategies will provide a practical approach for rapidly detecting miRNAs. In addition, using noble metal nanomaterials with the HCR strategy could be helpful for particular disease diagnoses. This should also overcome the high cost of imaging methods and make RNA accessible in detecting living cells. Consequently, the study of HCR-based nanomaterials miRNA detection strategies will also be an emerging research area to open new future research gates.Author contributions
Brij Mohan: conceptualization, methodology, data curation, writing – original draft preparation, software, Sandeep Kumar: software, writing – review & editing, Suresh Kumar: conceptualization, methodology, data curation writing – review & editing, Krunal Modi: writing – review & editing, Deependra Tyagi: review & editing, Dimitri Papukashvili: review & editing, Nino Rcheulishvili: review & editing, Armando J. L. Pombeiro: review & editing, software.Conflicts of interest
There are no conflicts of interest.Acknowledgements
The authors gratefully acknowledge the Fundação para a Ciência e Tecnologia (FCT), Portugal, for the Centro de Química Estrutural, Institute of Molecular Sciences projects UIDB/00100/2020, UIDP/00100/2020 and LA/P/0056/2020 projects.References
- F. A. Romero, C. T. Jones, Y. Xu, M. Fenaux and R. L. Halcomb, J. Med. Chem., 2020, 63, 5031–5073 CrossRef CAS PubMed.
- R. D. Crapnell, A. G.-M. Ferrari, N. C. Dempsey and C. E. Banks, Sens. Diagn., 2022, 1, 405–428 RSC.
- K. Pollet, N. Garnier, S. Szunerits, A. Madder, D. Hober and I. Engelmann, Sens. Diagn., 2023 10.1039/D2SD00140C.
- K. M. Koo, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, Anal. Chem., 2016, 88, 6781–6788 CrossRef CAS PubMed.
- J. M. Sasso, B. J. B. Ambrose, R. Tenchov, R. S. Datta, M. T. Basel, R. K. Delong and Q. A. Zhou, J. Med. Chem., 2022, 65, 6975–7015 CrossRef CAS PubMed.
- J. Zhang, S. Zhao, J. Wu, J. Zhang, S. Zhao and J. Wu, Biosensors - Current and Novel Strategies for Biosensing, 2022, DOI:10.5772/INTECHOPEN.93937.
- J. Liu, M. Shen, J. Talap, X. Shen, Z. Song, H. Hu, S. Zeng and S. Cai, Sens. Diagn., 2022, 1, 1063–1068 RSC.
- Z. Yao, P. Coatsworth, X. Shi, J. Zhi, L. Hu, R. Yan, F. Güder and H.-D. Yu, Sens. Diagn., 2022, 1, 312–342 RSC.
- A. Marín-Romero, M. Tabraue-Chávez, J. W. Dear, J. J. Díaz-Mochón and S. Pernagallo, Sens. Diagn., 2022, 1, 1243–1251 RSC.
- C. Costa, M. Teodoro, C. A. Rugolo, C. Alibrando, F. Giambò, G. Briguglio and C. Fenga, Toxicol. Rep., 2020, 7, 759–767 CrossRef CAS PubMed.
- R. Drula, L. F. Ott, I. Berindan-Neagoe, K. Pantel and G. A. Calin, Cancers, 2020, 12, 1–24 CrossRef PubMed.
- M. A. C. Pomatto, B. Bussolati, S. D'Antico, S. Ghiotto, C. Tetta, M. F. Brizzi and G. Camussi, Mol. Ther.--Methods Clin. Dev., 2019, 13, 133–144 CrossRef CAS PubMed.
- P. Khashayar, S. Al-Madhagi, M. Azimzadeh, V. Scognamiglio and F. Arduini, TrAC, Trends Anal. Chem., 2022, 156, 116706 CrossRef CAS.
- J. Sun and X. Sun, TrAC, Trends Anal. Chem., 2020, 127, 115900 CrossRef CAS.
- L. Zhou, M. Y. T. Lim, P. Kaur, A. Saj, D. Bortolamiol-Becet, V. Gopal, N. Tolwinski, G. Tucker-Kellogg and K. Okamura, eLife, 2018, 7, 38389 CrossRef PubMed.
- S. Kadkhoda and S. Ghafouri-Fard, Cancer Cell Int., 2022, 22, 1–16 CrossRef PubMed.
- H. Beyrampour-Basmenj, M. Rahmati, M. P. Moghamddam, M. E. Kalan, M. Alivand, Z. Aliyari-Serej, P. Nastarin, M. Omrani, S. Khodakarimi and A. Ebrahimi-Kalan, Gene Rep., 2022, 26, 101457 CrossRef CAS.
- R. Bhattacharjee, P. Mitra, N. Gupta, S. Sharma, V. K. Singh, N. Mukerjee, A. Dhasmana and R. Gundamaraju, Adv. Cancer Biol.: Metastasis, 2022, 5, 100050 CAS.
- M. Labib, E. H. Sargent and S. O. Kelley, Chem. Rev., 2016, 116, 9001–9090 CrossRef CAS PubMed.
- Y. Duan, Y. Li, C. Zhang, J. Chen, R. Sun, Z. Huang, Z. Luo, C. Zhou and M. Wu, ACS Sens., 2020, 5, 2977–3000 CrossRef PubMed.
- H. Li, X. Wang, S. Wei, C. Zhao, X. Song, K. Xu, J. Li, B. Pang and J. Wang, Anal. Chim. Acta, 2022, 1190, 338930 CrossRef CAS PubMed.
- Z. Li, B. Li, Y. Zhou, H. Yin, J. Wang and S. Ai, Anal. Biochem., 2017, 538, 20–25 CrossRef CAS PubMed.
- O. A. Goryacheva, A. S. Novikova, D. D. Drozd, P. S. Pidenko, T. S. Ponomaryeva, A. A. Bakal, P. K. Mishra, N. V. Beloglazova and I. Y. Goryacheva, TrAC, Trends Anal. Chem., 2019, 111, 197–205 CrossRef CAS.
- S. Chettimada, D. R. Lorenz, V. Misra, S. M. Wolinsky and D. Gabuzda, BMC Immunol., 2020, 21, 1–20 CrossRef PubMed.
- A. S. Silantyev, L. Falzone, M. Libra, O. I. Gurina, K. S. Kardashova, T. K. Nikolouzakis, A. E. Nosyrev, C. W. Sutton, P. D. Mitsias and A. Tsatsakis, Cells, 2019, 8(8), 863 CrossRef CAS PubMed.
- O. A. Goryacheva, A. M. Vostrikova, A. A. Kokorina, E. A. Mordovina, D. V. Tsyupka, A. A. Bakal, A. V. Markin, R. Shandilya, P. K. Mishra, N. V. Beloglazova and I. Y. Goryacheva, TrAC, Trends Anal. Chem., 2019, 119, 115613 CrossRef CAS.
- W. Fan, Y. Qi, X. Lu, W. Ren, C. Liu and Z. Li, Chem. Commun., 2020, 56, 7179–7182 RSC.
- Y. Qi, L. Qiu, W. Fan, C. Liu and Z. Li, Analyst, 2017, 142, 2967–2973 RSC.
- H. Chai, J. Zhu, Z. Guo, Y. Tang and P. Miao, Biosens. Bioelectron., 2023, 220, 114900 CrossRef CAS PubMed.
- M. G. M. Kok, M. W. J. de Ronde, P. D. Moerland, J. M. Ruijter, E. E. Creemers and S. J. Pinto-Sietsma, Biomol. Detect. Quantif., 2018, 15, 1–5 CrossRef CAS PubMed.
- Z. Cai, M. Zafferani, O. M. Akande and A. E. Hargrove, J. Med. Chem., 2022, 65, 7262–7277 CrossRef CAS PubMed.
- S. A. Razavi, M. Afsharpad, M. H. Modarressi, M. Zarkesh, P. Yaghmaei, S. Nasiri, S. M. Tavangar, H. Gholami, A. Daneshafrooz and M. Hedayati, Sci. Rep., 2019, 9, 1–11 CrossRef CAS PubMed.
- M. N. Islam, M. K. Masud, N. T. Nguyen, V. Gopalan, H. R. Alamri, Z. A. Alothman, M. S. Al Hossain, Y. Yamauchi, A. K. Y. Lam and M. J. A. Shiddiky, Biosens. Bioelectron., 2018, 101, 275–281 CrossRef CAS PubMed.
- N. B. Aziz, R. G. Mahmudunnabi, M. Umer, S. Sharma, M. A. Rashid, Y. Alhamhoom, Y. B. Shim, C. Salomon and M. J. A. Shiddiky, Analyst, 2020, 145, 2038–2057 RSC.
- R. M. Borum and J. V. Jokerst, Biomater. Sci., 2021, 9, 347–366 RSC.
- Y. Nie, M. Yang and Y. Ding, Microchim. Acta, 2018, 185, 1–7 CrossRef CAS PubMed.
- P. Ghorai, K. Pal, P. Karmakar and A. Saha, Dalton Trans., 2020, 49, 4758–4773 RSC.
- A. Kumar, P. Verma and P. Jindal, Microelectron. Eng., 2022, 111897 Search PubMed.
- C. Steiner, P. Lescuyer, P. Cutler, J.-C. Tille and A. Ducret, Mol. Cell. Proteomics, 2022, 21, 100416 CrossRef CAS PubMed.
- Y. L. Zhu, Y. M. Lian, J. K. Wang, Z. P. Chen and R. Q. Yu, Anal. Chem., 2021, 93, 5839–5848 CrossRef CAS PubMed.
- K. Singh, P. Kumar, C. Jagadeesh, M. Patel, D. Das and J. Saha, Adv. Synth. Catal., 2020, 362, 4130–4137 CrossRef CAS.
- S. Wang, R. Wang, D. Jiang, N. Zhang and W. Jiang, Sens. Actuators, B, 2022, 357, 131400 CrossRef CAS.
- C. Shu Zhu, L. Zhu, D. An Tan, X. Yuan Qiu, C. Yang Liu, S. Si Xie and L. Yun Zhu, Comput. Struct. Biotechnol. J., 2019, 17, 904–916 CrossRef PubMed.
- J. Wang, J. Wen and H. Yan, Chem. – Asian J., 2021, 16, 114–128 CrossRef CAS PubMed.
- Y. Wu, S. Cui, Q. Li, R. Zhang, Z. Song, Y. Gao, W. Chen and D. Xing, Biosens. Bioelectron., 2020, 165, 112449 CrossRef CAS PubMed.
- F. Li, Y. Zhou, H. Yin and S. Ai, Biosens. Bioelectron., 2020, 166, 112476 CrossRef CAS PubMed.
- Y. Cheng, L. Dong, J. Zhang, Y. Zhao and Z. Li, Analyst, 2018, 143, 1758–1774 RSC.
- L. He, J. Mu, O. Gang and X. Chen, Adv. Sci., 2021, 8, 2003775 CrossRef CAS PubMed.
- J. W. Oh, D. K. Lim, G. H. Kim, Y. D. Suh and J. M. Nam, J. Am. Chem. Soc., 2014, 136, 14052–14059 CrossRef CAS PubMed.
- N. Zhang, X. M. Shi, H. Q. Guo, X. Z. Zhao, W. W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2018, 90, 11892–11898 CrossRef CAS PubMed.
- L. Zhang, C. Gu, J. Wen, G. Liu, H. Liu and L. Li, Anal. Bioanal. Chem., 2021, 413, 83–102 CrossRef CAS PubMed.
- C. S. M. Martins, A. P. Lagrow and J. A. V. Prior, ACS Sens., 2022, 7, 1269–1299 CrossRef CAS PubMed.
- L. Zhao, F. Ahmed, Y. Zeng, W. Xu and H. Xiong, ACS Sens., 2022, 7(10), 2833–2856 CrossRef CAS PubMed.
- J. Stenvang and S. Kauppinen, Expert Opin. Biol. Ther., 2007, 8, 59–81 CrossRef PubMed.
- E. Cesewski and B. N. Johnson, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- M. T. Gabr and S. Brogi, J. Med. Chem., 2020, 63, 9695–9704 CrossRef CAS PubMed.
- Y. Jian, H. Wang, F. Lan, L. Liang, N. Ren, H. Liu, S. Ge and J. Yu, Microchim. Acta, 2018, 185, 1–8 CrossRef CAS PubMed.
- R. Shandilya, S. Ranjan, S. Khare, A. Bhargava, I. Y. Goryacheva and P. K. Mishra, Drug Discovery Today, 2021, 26, 1501–1509 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- H. M. T. Choi, V. A. Beck and N. A. Pierce, ACS Nano, 2014, 8, 4284–4294 CrossRef CAS PubMed.
- X. Wu, Y. Chai, R. Yuan, Y. Zhuo and Y. Chen, Sens. Actuators, B, 2014, 203, 296–302 CrossRef CAS.
- L. Lan, Q. Guo, H. Nie, C. Zhou, Q. Cai, J. Huang and X. Meng, Chem. Sci., 2019, 10, 2034–2043 RSC.
- W. Zhang, Z. Tian, S. Yang, J. Rich, S. Zhao, M. Klingeborn, P. H. Huang, Z. Li, A. Stout, Q. Murphy, E. Patz, S. Zhang, G. Liu and T. J. Huang, Microsyst. Nanoeng., 2021, 7, 63 CrossRef CAS PubMed.
- K. T. Kim, S. Angerani and N. Winssinger, Chem. Sci., 2021, 12, 8218–8223 RSC.
- C. Zhang, P. Miao, M. Sun, M. Yan and H. Liu, Small, 2019, 15, 1901867 CrossRef PubMed.
- L. Yang, C. Liu, W. Ren and Z. Li, ACS Appl. Mater. Interfaces, 2012, 4, 6450–6453 CrossRef CAS PubMed.
- H. Cheng, W. Li, S. Duan, J. Peng, J. Liu, W. Ma, H. Wang, X. He and K. Wang, Anal. Chem., 2019, 91, 10672–10678 CrossRef CAS PubMed.
- H. Ju, J. Anal. Test., 2017, 1, 1–18 CrossRef.
- T. Fozooni, H. Ravan and H. Sasan, Appl. Biochem. Biotechnol., 2017, 183, 1224–1253 CrossRef CAS PubMed.
- S. Delkhahi, M. Rahaie and F. Rahimi, J. Fluoresc., 2017, 27, 603–610 CrossRef CAS PubMed.
- P. Miao and Y. Tang, Anal. Chem., 2020, 92, 12026–12032 CrossRef CAS PubMed.
- B. N. M. Van Buuren, F. J. J. Overmars, J. H. Ippel, C. Altona and S. S. Wijmenga, J. Mol. Biol., 2000, 304, 371–383 CrossRef CAS PubMed.
- C. E. Carr and L. A. Marky, Biophys. J., 2017, 113, 529–539 CrossRef CAS PubMed.
- J. de Dieu Habimana, R. Huang, B. Muhoza, Y. N. Kalisa, X. Han, W. Deng and Z. Li, Biosens. Bioelectron., 2022, 114033 CrossRef PubMed.
- A. Miti, S. Thamm, P. Müller, A. Csáki, W. Fritzsche and G. Zuccheri, Biosens. Bioelectron., 2020, 167, 112465 CrossRef CAS PubMed.
- Q. Guo, Y. Yu, H. Zhang, C. Cai and Q. Shen, Anal. Chem., 2020, 92, 5302–5310 CrossRef CAS PubMed.
- J. Wei, X. Gong, Q. Wang, M. Pan, X. Liu, J. Liu, F. Xia and F. Wang, Chem. Sci., 2017, 9, 52–61 RSC.
- J. Guo, C. Mingoes, X. Qiu and N. Hildebrandt, Anal. Chem., 2019, 91, 3101–3109 CrossRef CAS PubMed.
- F. Zhang, S. Wang, J. Feng, R. Zou, L. Xiang and C. Cai, Talanta, 2019, 202, 342–348 CrossRef CAS PubMed.
- J. Ge, Z. Qi, L. Zhang, X. Shen, Y. Shen, W. Wang and Z. Li, Nanoscale, 2020, 12, 808–814 RSC.
- L. Ding, H. Liu, L. Zhang, L. Li and J. Yu, Sens. Actuators, B, 2018, 254, 370–376 CrossRef CAS.
- N. Ying, T. Sun, Z. Chen, G. Song, B. Qi, S. Bu, X. Sun, J. Wan and Z. Li, Anal. Biochem., 2017, 528, 7–12 CrossRef CAS PubMed.
- K. Feng, J. Liu, L. Deng, H. Yu and M. Yang, Microchim. Acta, 2018, 185, 28 CrossRef PubMed.
- A. Oleksi, A. G. Blanco, R. Boer, I. Usón, J. Aymamí, A. Rodger, M. J. Hannon and M. Coll, Angew. Chem., 2006, 118, 1249–1253 CrossRef.
- L. Zhou, Y. Wang, C. Yang, H. Xu, J. Luo, W. Zhang, X. Tang, S. Yang, W. Fu, K. Chang and M. Chen, Biosens. Bioelectron., 2019, 126, 657–663 CrossRef CAS PubMed.
- J. Dong, H. Yang, J. Zhao, L. Wen, C. He, Z. Hu, J. Li, D. Huo and C. Hou, Microchim. Acta, 2022, 189, 1–9 CrossRef PubMed.
- R. Isaac, F. C. G. Reis, W. Ying and J. M. Olefsky, Cell Metab., 2021, 33, 1744–1762 CrossRef CAS PubMed.
- P. Zhuang, H. Zhang, R. M. Welchko, R. C. Thompson, S. Xu and D. L. Turner, Sci. Rep., 2020, 10, 1–10 CrossRef PubMed.
- Y. H. Yuan, Y. Di Wu, B. Z. Chi, S. H. Wen, R. P. Liang and J. D. Qiu, Biosens. Bioelectron., 2017, 97, 325–331 CrossRef CAS PubMed.
- L. Zhu, J. Ye, S. Wang, M. Yan, Q. Zhu, J. Huang and X. Yang, Chem. Commun., 2019, 55, 11551–11554 RSC.
- Q. Wu, H. Wang, K. Gong, J. Shang, X. Liu and F. Wang, Anal. Chem., 2019, 91, 10172–10179 CrossRef CAS PubMed.
- Z. Xu, K. Zheng, Z. Du, J. Xin, M. Luo and F. Wang, Biotechnol. Appl. Biochem., 2022, 69, 974–980 CrossRef CAS PubMed.
- Y. H. Yuan, B. Z. Chi, S. H. Wen, R. P. Liang, Z. M. Li and J. D. Qiu, Biosens. Bioelectron., 2018, 102, 211–216 CrossRef CAS PubMed.
- Y. Li, C. Z. Huang and Y. F. Li, Anal. Chem., 2019, 91, 9308–9314 CrossRef CAS PubMed.
- D. Zhang, F. Bian, L. Cai, T. Wang, T. Kong and Y. Zhao, Biosens. Bioelectron., 2019, 143, 111629 CrossRef CAS PubMed.
- P. Miao, Y. Tang, Z. Mao and Y. Liu, Part. Part. Syst. Charact., 2017, 34, 1600405 CrossRef.
- J. Lu, L. Wu, Y. Hu, S. Wang and Z. Guo, Biosens. Bioelectron., 2018, 109, 13–19 CrossRef CAS PubMed.
- Y. Zheng, Y. Xu, L. Chen, X. Yin, F. Lin, S. Weng and X. Lin, J. Electrochem. Soc., 2020, 167, 167502 CrossRef CAS.
- X. Fan, Y. Qi, Z. Shi, Y. Lv and Y. Guo, Sens. Actuators, B, 2018, 255, 1582–1586 CrossRef CAS.
- X. Xie, Z. Wang, M. Zhou, Y. Xing, Y. Chen, J. Huang, K. Cai and J. Zhang, Small Methods, 2021, 5, 2101072 CrossRef CAS PubMed.
- K. Djebbi, B. Shi, T. Weng, M. Bahri, M. A. Elaguech, J. Liu, C. Tlili and D. Wang, ACS Omega, 2022, 7, 2224–2233 CrossRef CAS PubMed.
- S. Tang, Y. Li, A. Zhu, Y. Yao, J. Sun, F. Zheng, Z. Lin and W. Shen, Chem. Commun., 2019, 55, 8386–8389 RSC.
- J. Ge, C. Li, Y. Zhao, X. Yu and G. Jie, Chem. Commun., 2019, 55, 7350–7353 RSC.
- X. Qu, M. Xiao, F. Li, W. Lai, L. Li, Y. Zhou, C. Lin, Q. Li, Z. Ge, Y. Wen, H. Pei and G. Liu, ACS Appl. Bio Mater., 2018, 1, 859–864 CrossRef CAS PubMed.
- D. Zhu, W. Liu, W. Cao, J. Chao, S. Su, L. Wang and C. Fan, Electroanalysis, 2018, 30, 1349–1356 CrossRef CAS.
- Q. Xue, C. Liu, X. Li, L. Dai and H. Wang, Bioconjugate Chem., 2018, 29, 1399–1405 CrossRef CAS PubMed.
- M. A. Bustos, N. Rahimzadeh, S. Ryu, R. Gross, L. T. Tran, V. M. Renteria-Lopez, R. I. Ramos, A. Eisenberg, P. Hothi, S. Kesari, G. Barkhoudarian, Y. Takasumi, C. Cobbs, D. F. Kelly and D. S. B. Hoon, Lab. Invest., 2022, 2022, 1–11 Search PubMed.
- L. Wang, X. Wang, Y. Wu, M. Guo, C. Gu, C. Dai, D. Kong, Y. Wang, C. Zhang, D. Qu, C. Fan, Y. Xie, Z. Zhu, Y. Liu and D. Wei, Nat. Biomed. Eng., 2022, 2022, 1–10 Search PubMed.
- X. Liu, M. Zhang, Z. Chen, J. Cui, L. Yang, Z. Lu, F. Qi and H. Wang, Front. Bioeng. Biotechnol., 2021, 9, 1310 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |