Open Access Article
Open Access ArticleCanine parvovirus 2 detection using a LSPR biosensing method with gold nanoparticles†
Ana Carolina
Yamakawa
 *a,
Caroline Rodrigues
Basso
*a,
Caroline Rodrigues
Basso
 ab,
Valber de Albuquerque
Pedrosa
b and
João Pessoa
Araújo Júnior
*ab
ab,
Valber de Albuquerque
Pedrosa
b and
João Pessoa
Araújo Júnior
*ab
aBiotechnology Institute, São Paulo State University, Botucatu, SP 18607-440, Brazil. E-mail: a.yamakawa@unesp.br; joao.pessoa@unesp.br
bChemical and Biological Sciences Department, Bioscience Institute, São Paulo State University, Botucatu, SP 18618-000, Brazil
First published on 13th October 2022
Abstract
Canine parvovirus 2 (CPV-2) is responsible for one of the most common gastrointestinal diseases that affect mainly young dogs. This severe condition can lead to death, and the development of low-cost and fast diagnostic techniques is essential since only clinical diagnosis is not accurate. Furthermore, noble metal nanoparticles, such as gold nanoparticles (AuNPs), have dynamic physicochemical characteristics that allow the transduction of various signals, which makes them efficient biosensors. The present report describes a protocol to identify CPV-2 from stool samples using AuNPs modified with antibody deposition. The protocol was standardized using monoclonal and polyclonal antibodies, and a combination of both. A total of 60 positive and five negative stool samples for CPV-2 were subjected to the technique. There was a significant increase in wavelengths after the addition of the positive samples. The combination of both antibodies showed the greatest difference. In contrast, when adding the negative samples, there was no statistically significant difference in wavelengths compared to that in the step with the antibodies. Adenovirus and porcine circovirus 2 samples were also subjected to the technique, and no nonspecific binding was detected. This technique provides a quick (40 min), low cost ($2 per sample), and sensitive and specific diagnosis. The results found here are promising and may serve as a basis for future point of care testing AuNP protocols.
Introduction
Canine parvovirus 2 (CPV-2) is one of the smallest non-enveloped viruses, with a single-stranded DNA (ssDNA) genome of about 5 kb.1–3 CPV-2 has been distributed worldwide since the 80s, probably due to the lack of previous immunity.4 Right after the virus' global emergence, the first antigenic variant, CPV-2a, was identified.5,6 A few years later, in 1984, the second variant CPV-2b5 was reported and in 2000, the third CPV-2c was identified in Italy.7 Nowadays, all variants are distributed worldwide.8CPV-2 is one of the most important enteric pathogens in dogs, especially in high canine density populations such as in kennels and shelters.9 Characterized as an acute infectious disease, faecal–oronasal contact is the main transmission route with a significant spread rate.1,10,11 After oronasal contact, CPV-2 can replicate on cells in the dividing phase, especially on gastrointestinal and lymphoid tissue.1 Thus, infection of leucocytes and then the crypt cells of the small intestine leads to shortening and even necrosis.1 Along with these effects, they can induce severe gastrointestinal signs such as haemorrhagic diarrhoea, especially for puppies, and may lead to sepsis with high mortality rates.12,13
There are some available diagnosis techniques for CPV-2 identification from biological samples.9 The gold standard diagnosis uses molecular techniques, such as qPCR.9,14 Likewise, we can detect CPV-2 by viral isolation with cell culture,15,16 transmission electron microscopy,17 hemagglutination tests,18 and ELISA and immunochromatographic tests (IC).19–21 However, they can be expensive and time-consuming techniques.
Nanotechnology has been growing as a focus of research in the areas of biology and medicine since the beginning of the 21st century, due to the ability to manipulate the physical, chemical, and biological characteristics of nanoparticles.22 Biosensors use biological materials such as enzymes, antibodies, nucleic acids, and aptamers, among others, which, when connected to a transducer, will transform the target into an electrical, optical, gravimetric, or thermal signal that can be measurable for reading.23–25 Furthermore, the versatility in diagnostic tests has been proving advantageous for rapid and real-time analysis of in vivo samples.26,27
The use of gold nanoparticles (AuNPs) as biosensors has been gaining attention, already being reported in the detection of pesticides, hormones, viruses, and bacteria.23,28–34 Due to their unique characteristics, such as high biocompatibility, easy functionalization with proteins, and optical and electronic properties, they are ideal to produce analysis protocols.35–37
Another important property that allows the use of AuNPs in different biomedical applications is their scattering and absorption of light in the visible region by the oscillation of free electrons on their surface, an event known as localized surface plasmon resonance (LSPR).38 This resonance is directly affected by the size and shape of the AuNPs and can be assessed by the wavelength of the solution.38 Consequently, the deposition of biomolecules in AuNPs affects their resonance, and consequently their wavelength, and it is possible to follow their changes by the LSPR technique.39 Therefore, this report proposes a parvovirus detection protocol using AuNPs and LSPR analyses to establish a fast, accurate and inexpensive diagnosis.
Experimental methods
Chemicals
The chemicals used in the present experiments were gold(III) chloride trihydrate 99% (HAuCl4); sodium citrate dihydrate 99%, 11-mercaptoundecanoic acid 95% (MUA), N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), absolute ethanol 99%, and phosphate-buffered saline (PBS, pH 7.4), and were purchased from Sigma-Aldrich (Saint Louis, MO, USA).Samples and CPV-2 diagnosis
A total of 60 positive and five negative to CPV-2 stool samples were tested. The samples were kindly provided from partnerships with the Laboratory of Veterinary Molecular Diagnosis (LDMVET) and with the infectious diseases sector from the veterinary hospital of São Paulo State University, Botucatu. The samples were stored in 15 mL conical centrifuge tubes at −20 °C until processed.CPV-2 presence in the feces was confirmed by a quantitative polymerase chain reaction (qPCR), the gold standard diagnosis. DNA extraction was performed with an adapted protocol using magnetic beads (Sera-Mag™ Magnetic SpeedBeads™ Carboxylate-Modified Dia.: 1 μm; 3 EDAC/PA5; 15 mL) (GE Healthcare, Chicago, Illinois, EUA).40 For the reaction, 10 μL of GoTaq® qPCR Master Mix (Promega, Madison, USA) was used, along with 0.8 μL of each specific primer14 (10 pmol μL−1) and 4.4 μL nuclease-free water. The cycling consisted of five minutes of 95 °C denaturation, followed by 40 cycles of 95 °C per 15 seconds and 60 °C per minute, and a melting curve. The reaction was conducted on an AriaMX real-time PCR System (Agilent, Santa Clara, CA, EUA). All reactions were performed with nuclease-free water as negative control and a previous positive control.
Hemagglutination tests (HA) to determine the viral titer were performed using fresh swine blood as previously described.18 The feces were serially diluted in borate buffer (pH 8.7) with a factor of two and starting on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution.
20 dilution.
Next-generation sequencing of seven samples was also performed to identify the antigenic variants (CPV-2a, CPV-2b, and CPV-2c). A library was prepared with an Illumina DNA Prep kit (Illumina Inc., CA, USA) and a pool of all samples was processed on a 300 cycle micro flow cell on a MiSeq equipment (Illumina Inc., CA, USA). The sequences were analysed with the software Geneious Prime (Biomatters, Auckland, New Zealand) and aligned to a reference sequence of the CPV-2 complete genome (GenBank NC_001539.1).
AuNP synthesis and self-assembled monolayer (SAM) modification
For the AuNP synthesis, a chloroauric acid (HAuCl4) solution (1 mM) was heated until its boiling point, and then the solution was reduced by adding sodium citrate (30 mM), as previously described.28,29 Sodium citrate reduces the gold ions into metallic gold nanoparticles, and the excess citrate anions in the solution maintain the metallic surface of gold by giving a negative charge to each nanoparticle.41 Once at room temperature, the solution was stored in an amber bottle. The synthesis success was confirmed by localized surface plasmon resonance (LSPR) analyses using a Biochrom Lira S11 ultraviolet-visible (UV-vis) spectrophotometer (Biochrom Ltd., Cambridge, UK); transmission electron microscopy (TEM) images analysed with the software ImageJ (version 1.8.0_172); and dynamic light scattering (DLS) on a Dyna PRO Titan (Wyatt Technology, Califórnia, EUA) equipment at 30 °C and 100 acquisitions of 10 seconds each. The LSPR and DLS data were analysed with the software OriginPro 8.5.0 and DYNAMICS 6.10, respectively.Self-assembled monolayers (SAMs) were arranged by the addition of 100 μL of 11-mercaptoundecanoic acid (MUA) (0.018 M) in 1 mL of AuNPs, followed by a 40 minute incubation. The thiol group present in MUA binds irreversibly to the AuNP structure. Then, 50 μL of 0.1 M N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and 0.05 M N-hydroxysuccinimide (NHS) were added, followed by a 4 °C overnight incubation. The SAMs are responsible for mediating the attachment of the antibodies under the gold nanoparticles, through the interaction of their non-active functional group with the thiol group of the monolayer. To characterize the SAM and AuNP complex, the LSPR and TEM techniques were performed.
CPV-2 detection methodology
The experiments were performed with polyclonal and monoclonal antibodies. Hyperimmune serum containing the polyclonal immunoglobulin produced in guinea pigs was kindly given by MSc Marcela Ribeiro. Immunoglobulin viability analyses and titration were performed by hemagglutination tests and by hemagglutination inhibition (CARMICHAEL; JOUBERT; POLLOCK, 1980). The estimated globulin concentration was 21.7 mg ml−1. The value was determined from the albumin/globulin ratio and total proteins present in the serum. Monoclonal antibodies (ab140431; Abcam, UK) were purchased commercially and presented a stock concentration of 2.17 mg mL−1. The combination of both immunoglobulins in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was also tested.
1 ratio was also tested.
To evaluate the technique specificity and selectivity, biological samples containing porcine circovirus 2 (stool and serum) and a porcine adenovirus (serum) were also tested. The families Circoviridae and Adenoviridae were preferred since they have similarities to the family Parvoviridae, like the absence of a viral envelope.2,42,43 In addition, the Circoviridae and Parvoviridae families also present a round capsid with a T = 1 icosahedral symmetry, besides having a similar size (around 18 nm) and ssDNA.2,42
Data analyses
To analyse the wavelength changes in the solution with the immunoglobulins and after the addition of the positive and negative samples, the Wilcoxon test was used. The comparison of the wavelength results of the positive and negative samples was performed by the Mann–Whitney test. To evaluate the wavelength difference values of each immunoglobulin before and after the addition of positive samples, Friedman's test was used. Descriptive analyses, such as mean, standard deviation (±) and 95% confidence interval (95% IC) were also determined for the wavelength data. The correlation of qPCR and hemagglutination results with the wavelength increase data from the positive samples was performed with Spearman's test. All statistical analyses were performed with a 95% significance level in GraphPad Prism 8.0 software.Ethical statement
The project was approved under protocol 0163/2020 of the Ethics Committee on Animal Use of the College of Veterinary Medicine and Animal Sciences, São Paulo State University.Results and discussion
Molecular diagnosis and hemagglutination tests
The presence of CPV-2 in the 60 stool samples was confirmed by qPCR. Of these, 49 presented a hemagglutination titer. Regarding the sequencing analyses, the three variants' data were identified: one CPV-2a (sample 20), four CPV-2b (samples 8, 21, 29, and 30), and two CPV-2c (samples 5 and 12). It was possible to identify the variant of the sequences of the seven samples, by the analysis of the codons, genes, and amino acids from the VP2 protein. Region 426 is the most relevant for the differentiation of the variants, with PVC-2a characteristically having the presence of Asn, PVC-2b having Asp, and PVC-2c having Glu.3,7Characterization of AuNP synthesis and self-assembled monolayers
The AuNP synthesis was satisfactory. AuNP LSPR analyses showed a peak as displayed in Fig. 1A at 524 nm, similar to results described in other studies.28,34,44 Also, in Fig. 1A, we have the absorption spectra of the modification steps, evidencing the wavelength shift of the peak between each step. With the addition of MUA, we have the formation of the self-organized monolayers with a peak at 527 nm and then after the addition of EDC/NHS, the formation of N-hydroxysuccinimide groups with a peak at 529 nm, revealing that with each modification, the pattern of light absorption by the AuNPs changed.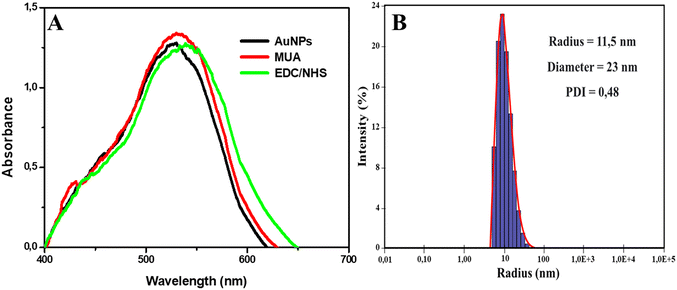 | ||
| Fig. 1 AuNP characterization. A) UV-vis spectra of AuNPs synthesized and the formation of the self-organized monolayers. B) DLS graphical analysis of AuNP size and polydispersity. | ||
The DLS analysis was also satisfactory. In Fig. 1B, we can observe that the AuNPs presented an average radius and diameter of 11.5 and 23 nm, respectively, with a polydispersity index (PDI) of 0.48. The PDI represents the size heterogeneity within a sample, with the higher the index, the greater the size variation. Previous studies found similar PDI values in experiments with AuNPs.45,46
Analysis of the TEM images corroborate the spectrophotometry and DLS results, about the success of synthesis and diameter of AuNPs, respectively. In Fig. S1A and B,† it is possible to observe the TEM image and histogram of the diameter distribution of AuNPs, with an average diameter of 23.64 nm ± 5.41 nm, agreeing with the value found by DLS of 23 nm.
Immunoglobulin ideal concentration
Binding analyses of both monoclonal and polyclonal immunoglobulins were also performed. Concentrations of 0.5, 1.5, 2 and 2.5 μg mL−1 of the monoclonal immunoglobulin were tested. All presented a wavelength increase, confirming the binding on the self-organized monolayers, as shown in Fig. 2. The concentration with the greatest wavelength difference from the EDC/NHS step was 2.5 μg mL−1, which was then used in the following experiments.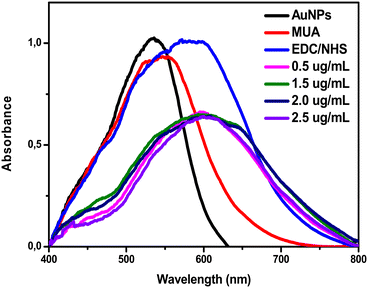 | ||
| Fig. 2 UV-vis spectrum plots of monoclonal antibodies at different concentrations (0.5, 1.5, 2 and 2.5 μg mL−1) diluted in PBS. | ||
The binding of the amine groups of the polyclonal antibodies present in the hyperimmune serum with the N-hydroxysuccinimide groups was tested with pure serum and that diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, and 1
200, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 in PBS. The 1
400 in PBS. The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution was the most satisfactory, showing an increase in wavelength compared to that in the EDC/NHS step with less noise, as shown in the graphs present in Fig. 3.
100 dilution was the most satisfactory, showing an increase in wavelength compared to that in the EDC/NHS step with less noise, as shown in the graphs present in Fig. 3.
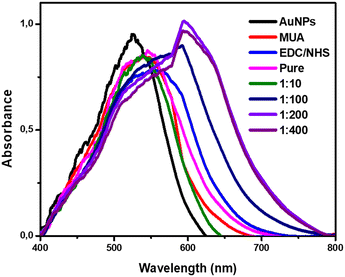 | ||
Fig. 3 UV-vis spectrum plots of the polyclonal antibodies with different dilutions (pure, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1 10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1 100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, and 1 200, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) in PBS. 400) in PBS. | ||
Technique standardization
Tests with the monoclonal immunoglobulin at 2.5 μg mL−1, polyclonal immunoglobulin diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and the combination of both at a 1
100, and the combination of both at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, were performed with a CPV-2 viral isolate in cell culture. In Fig. S2,† a slight increase in wavelength can be seen after the addition of the viral isolate. With the use of the monoclonal immunoglobulin, an increase from 531.04 nm to 533.97 nm was obtained after the addition of the isolate, with the polyclonal immunoglobulin, an increase from 532.40 nm to 534.80 nm, and with the combination of both, an increase from 531.45 nm to 534.59 nm, confirming the viral binding with all the complexes.
1 ratio, were performed with a CPV-2 viral isolate in cell culture. In Fig. S2,† a slight increase in wavelength can be seen after the addition of the viral isolate. With the use of the monoclonal immunoglobulin, an increase from 531.04 nm to 533.97 nm was obtained after the addition of the isolate, with the polyclonal immunoglobulin, an increase from 532.40 nm to 534.80 nm, and with the combination of both, an increase from 531.45 nm to 534.59 nm, confirming the viral binding with all the complexes.
To exclude possible nonspecific interactions with bacteria, proteins, and/or salts present in the stool, five CPV-2 negative stool samples were diluted in PBS (pH 7.4) at the ratios 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150, 1
150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, 1
200, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, and 1
500, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. The monoclonal and polyclonal antibodies, and the combination of both antibodies, have not presented any nonspecific interactions at 1
1000. The monoclonal and polyclonal antibodies, and the combination of both antibodies, have not presented any nonspecific interactions at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
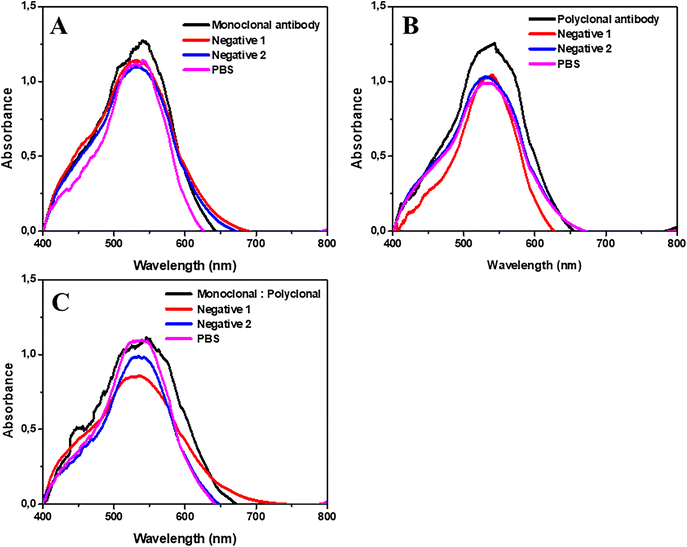
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 dilution as presented in Fig. 4. As presented in Fig. 4A, the monoclonal antibodies presented a wavelength of 531.25 nm, and after the addition of the negatives 1 and 2, and only PBS, the wavelength became 529.34 nm, 529.24 nm, and 530.16 nm, respectively. In Fig. 4B, we have the polyclonal antibody wavelength values, being 532.41 nm and 531.58 nm after the addition of negative 1, 529.82 nm after adding negative 2, and 530.07 nm for the PBS only. The combination of both immunoglobulins is represented in Fig. 4C, with a wavelength of 531.26 nm, and after the addition of negatives 1 and 2, and PBS, it became 528.42 nm, 530.49 nm, and 528.98 nm, respectively. Therefore, the experiments with all positive and negative samples were performed at 1
150 dilution as presented in Fig. 4. As presented in Fig. 4A, the monoclonal antibodies presented a wavelength of 531.25 nm, and after the addition of the negatives 1 and 2, and only PBS, the wavelength became 529.34 nm, 529.24 nm, and 530.16 nm, respectively. In Fig. 4B, we have the polyclonal antibody wavelength values, being 532.41 nm and 531.58 nm after the addition of negative 1, 529.82 nm after adding negative 2, and 530.07 nm for the PBS only. The combination of both immunoglobulins is represented in Fig. 4C, with a wavelength of 531.26 nm, and after the addition of negatives 1 and 2, and PBS, it became 528.42 nm, 530.49 nm, and 528.98 nm, respectively. Therefore, the experiments with all positive and negative samples were performed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 dilution in PBS.
150 dilution in PBS.
CPV-2 detection methodology
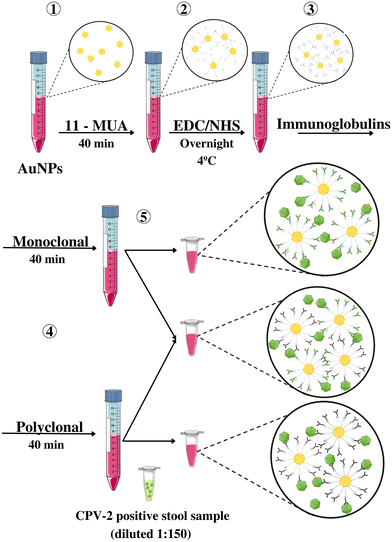
The 60 positive and 5 negative stool samples for CPV-2 were subjected to the protocol with the monoclonal and polyclonal antibodies, and a combination of both in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The experiments were performed as shown in Fig. 5.
1 ratio. The experiments were performed as shown in Fig. 5.
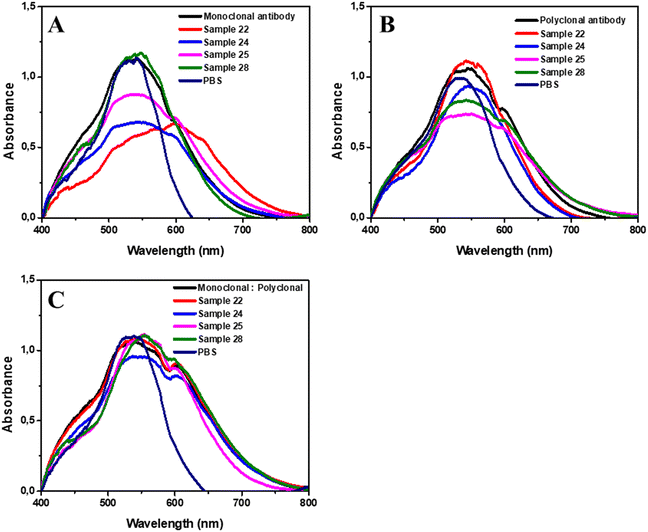
Further, the UV-vis spectrophotometry analyses confirmed the viral binding in the AuNP complex by the positive sample wavelength increase, as shown in Fig. 6. In Table S1,† we have the wavelength values of the positive and negative samples, and the respective differences compared to that in the antibody step. Regarding the detection of the three antigenic variants (PVC-2a, PVC-2b, and PVC-2c), the protocol was also proved satisfactory. The three variants PVC-2a (A20), PVC-2b (A8, A21, A29, and A30), and PCV-2c (A5 and A12) all showed an increase in their wavelengths compared to the respective immunoglobulin steps, as presented in Table S1.†
The mean wavelength after binding of the monoclonal antibodies in the complex was 531.1 ± 0.16 nm (95% CI, 531.1–531.2), the polyclonal antibodies showed a mean of 532.8 ± 0.43 nm (95% Cl, 532.7–532.9) and the combination of both in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio showed a mean of 532.2 ± 0.76 nm (95% IC, 532–532.4). The wavelength after the addition of the positive samples and incubation showed a mean of 537.2 ± 4.46 nm (95% IC, 536.1–538.4) with the use of monoclonal immunoglobulin, with polyclonal immunoglobulin, the mean was 536.9 ± 2.21 nm (95% IC, 536.3–537.5) and the combination of both showed the highest mean with 538.4 ± 3.73 nm (95% IC, 537.4–539.4).
1 ratio showed a mean of 532.2 ± 0.76 nm (95% IC, 532–532.4). The wavelength after the addition of the positive samples and incubation showed a mean of 537.2 ± 4.46 nm (95% IC, 536.1–538.4) with the use of monoclonal immunoglobulin, with polyclonal immunoglobulin, the mean was 536.9 ± 2.21 nm (95% IC, 536.3–537.5) and the combination of both showed the highest mean with 538.4 ± 3.73 nm (95% IC, 537.4–539.4).
Regarding the addition of the negative samples in the complex with the monoclonal antibodies, the mean wavelength of the samples was 529.9 ± 0.85 nm (95% IC, 528.8–530.9), with the polyclonal antibodies, the mean was 531.2 ± 0.87 nm (95% IC, 530.1–532.2) and using the combination of both showed a mean of 529.8 ± 0.97 nm (95% IC, 528.6–531).
The difference between the immunoglobulin's wavelength and that after the addition of positive samples had a mean increase of 6.05 ± 4.43 nm (95% IC, 4.91–7.2; p < 0.0001) with the monoclonal antibodies and 4.07 ± 2.02 nm (95% IC, 3.55–4.59; p < 0.0001) with the polyclonal antibodies, and the combination of both presented a mean increase of 6.21 ± 3.59 nm (95% IC, 5.29–7.14; p < 0.0001). The three groups showed a statistically significant wavelength increase compared to those in the previous step. However, the combination of monoclonal and polyclonal antibodies obtained the greatest difference. Furthermore, the use of monoclonal antibodies and the combination of monoclonal and polyclonal antibodies showed a statistically significant difference compared to that of polyclonal antibodies (p = 0.014 and p = 0.0002, respectively). However, the use of the monoclonal antibodies and the combination of both antibodies had no statistical difference (p = 0.706).
Regarding the negative samples, there was no increase in the wavelength compared to the AuNPs@MUA@EDC/NHS@Immunoglobulin step for all groups. With the monoclonal antibodies, the mean difference was −1.32 ± 0.88 nm (95% IC, −2.42–−0.23; p = 0.0625), that with the polyclonal antibodies was −1.29 ± 0.85 nm (95% IC, −2.3–−0.24; p = 0.0625) and that with the combination of both was −1.55 ± 0.94 nm (95% IC, −2.71–−0.38; p = 0.0625). No statistical difference was found between the wavelengths of the negative samples with the immunoglobulins. Regardless of the antibody, all negative samples showed a lower wavelength compared to those in the previous step, and similar results to those found in previous studies.23,29,30 The difference between the median wavelengths of the positive and negative samples are proved statistically significant with p < 0.0001, for all antibodies (monoclonal, polyclonal, and combination).
The TEM analysis corroborates the UV-visible results. In Fig. S3A,† an increase in the average diameter of the AuNPs is noticed, previously from 23.64 nm to 29.24 nm after the formation of the complex AuNPs@MUA@EDC/NHS@Immunoglobulins. After the addition of a positive sample, an increase in the average diameter to 32.73 nm is observed (Fig. S3B†). Similar results were found in previous experiments,30,47 demonstrating an increase in the diameter of AuNPs from biomolecule deposition.
In Fig. S4,† we have TEM images, where we can observe, in Fig. S4A,† the AuNPs at a 500 nm scale with a uniform size and dispersed. In Fig. S4B,† we have the formation of a monolayer around the AuNPs and in Fig. S4C,† the complex with the binding of the combination of monoclonal and polyclonal antibodies. Further, in Fig. S4D,† we have the addition of a positive sample, and it is possible to observe a subtle shadow around the AuNPs and an increase in the nanoparticle aggregates. The results are similar to those of previous studies with AuNPs.30,48
Limit of detection
To identify the limit of detection, we diluted the positive sample 12 at ratios 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150, 1
150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, 1
500, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103, 1
103, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104, 1
104, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105, 1
105, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 and 1
106 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 (Fig. 7). The limit of detection was 1
107 (Fig. 7). The limit of detection was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105, since it was at a higher dilution that an increase in wavelength (2.02 nm) is observed compared to that in the antibody step. In Fig. 7B, we have the comparison of the wavelength values with log2 of the respective dilutions, with an R2 = 0.97.
105, since it was at a higher dilution that an increase in wavelength (2.02 nm) is observed compared to that in the antibody step. In Fig. 7B, we have the comparison of the wavelength values with log2 of the respective dilutions, with an R2 = 0.97.
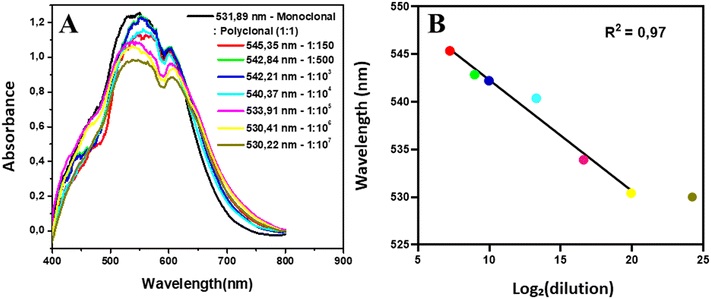 | ||
| Fig. 7 Protocol limit of detection. A. UV-vis graphs of diluted sample 12. B. Comparison between the wavelength values and log2 of the respective dilutions. | ||
To evaluate the selectivity of the protocol, serum samples positive for adenovirus and porcine circovirus 2 (PCV-2) and one stool sample positive for PCV-2 were subjected to the technique. Both viral families, Adenoviridae and Circoviridae, comprise DNA viruses with the absence of a viral envelope, similar to parvoviruses.2,42,43 In Fig. 8, we have the UV-vis analysis for evaluation of possible cross-linking with other viral samples. The step with the combination of both antibodies showed a wavelength of 531.89 nm, which, with the addition of the adenovirus and CVS-2 serum sample, changed to −1.43 and −2.92 nm, respectively. The PCV-2 stool sample also showed a wavelength reduction of −2.23 nm, as well as with the addition of PBS (−3.92 nm). No nonspecific binding of the AuNP complex to the parvovirus-like viral samples was evident.
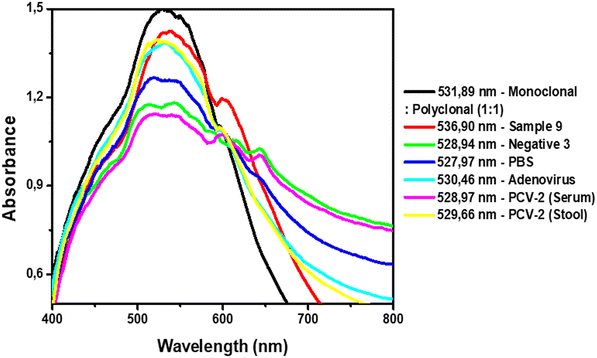 | ||
| Fig. 8 UV-vis analysis of the AuNPs with complexes and after addition of positive, negative, PBS and interfering samples. | ||
The use of AuNPs for diagnostic methods has already been proven advantageous and effective, with a combination of low cost, ease of conjugation, and unique optical properties.23,29 The use of AuNPs as biosensors to detect CPV-2 by LSPR analysis proved to be a fast, reliable, and inexpensive technique compared to the gold standard test qPCR. Here in the protocol, there is no necessity for previous steps, such as DNA extraction,14 which can delay and increase the diagnosis cost.
The combination of monoclonal and polyclonal antibodies showed a more satisfactory result, especially when compared to the use of the polyclonal antibodies alone. It was possible to detect the three variants using the protocol, without cross-referencing with other similar viral samples, demonstrating the satisfactory selectivity of the immunosensor. Furthermore, the sensitivity was confirmed to be remarkable when compared to the qPCR results, and all the 60 CPV-2 positive samples presented an increase in the wavelength. Similar results were found in previous experiments performed by our research group for dengue virus detection using AuNPs.29
Despite the absence of an association of the wavelength value with the viral concentration in the biological sample, for canine parvovirus, only qualitative diagnosis (positive or negative) is enough to continue the treatment and subsequent outbreak control.
The protocol also proved to be more financially advantageous and fast, with an execution time of approximately 40 minutes. With the reagents used, the cost was around USD 2.00 per sample, which is more affordable compared to molecular methods and fast diagnostic kits available.
Some technique limitations are that LSPR can have less efficient and sensitive results in comparison to the conventional surface plasmon resonance (SPR) and surface-enhanced Raman scattering (SERS), due to interferences in the peak shape from a diversity of parameters (shape, size, medium/solution).25,49–51 However, LSPR has advantages as well, such as the possibility of NP solutions being easy and practical to transport around locations. Further, LSPR usage with metallic nanoparticles in the detection of several compounds is widely described in the literature as reported by Basso et al.,28–30,44 which can be measured on an ultraviolet-visible (UV-vis) spectrophotometer showing results with higher sensitivity and lower detection limits, due to collective interactions between NPs, altering the optical response of the overall NP assembly.
To date, there have been no studies in the literature using AuNPs in solution with the LSPR technique for CPV-2 diagnosis. This new methodology involves low cost, easy manipulation, and simple operational procedures with good bioconjugation yield. Along with the potential to be used as an alternative to traditional detection tests, the methodology can also be a start to establishing a point of care testing diagnosis as well. Besides, AuNP diagnostic techniques already have been related to fast colorimetric diagnostic protocols;30,34,44,52–55 the present experiments did not have a colorimetric change after virus binding on the AuNP complex, similar to a previous report on dengue virus detection.29 However, when the AuNPs were combined with surface active maghemite nanoparticles and specific aptamers, a colorimetric change was observed.44 AuNP use in immunochromatographic (IC) strips, also known as lateral flow assays, is another possibility.46,55 Most IC commercial tests are based on the use of only one or two monoclonal antibodies (capture and detection).46 A previous immunochromatographic strip test study, using a monoclonal antibody specific to CPV-2 as a detection antibody and a polyclonal antibody as capture antibody, has established a high specificity protocol.46 Further, the combined use of monoclonal and polyclonal antibodies as detectors in IC tests may increase the sensitivity and reduce the costs, since polyclonal antibodies are less expensive. Accordingly, the results herein can benefit and give guidance to a future point of care testing protocol using AuNPs combining monoclonal and polyclonal antibodies.
Conclusions
The present report described a successful protocol to identify CPV-2 presence in stool samples, using AuNPs with antibody deposition. The use of AuNPs with monoclonal and polyclonal antibodies, and the combination of both antibodies with LSPR analyses provides a reliable diagnosis when compared to usual molecular techniques. The combination of both immunoglobulins presented better results. Further, the protocol promotes a fast and affordable alternative to CPV-2 diagnosis. Besides, the results found herein can help to establish point of care testing diagnostics in the future.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to the Laboratory of Veterinary Molecular Diagnosis (LDMVET) and the infectious diseases sector from the veterinary hospital of São Paulo State University, Botucatu, for all the help. This work was only possible thankfully due to the financial support from FAPESP (process: 2020/01629-3), CAPES Brazil, and Fundibio/UNESP.Notes and references
- N. Decaro and C. Buonavoglia, Vet. Microbiol., 2012, 155, 1–12 CrossRef CAS PubMed.
- M. Mietzsch, J. J. Pénzes and M. Agbandje-Mckenna, Viruses, 2019, 11, 4 CrossRef PubMed.
- J. Tsao, M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans and C. R. Parrish, Science, 1991, 251, 1456–1464 CrossRef CAS.
- L. E. Carmichael and L. N. Binn, Adv. Vet. Sci. Comp. Med., 1981, 25, 1–37 CAS.
- C. R. Parrish, P. Have, W. J. Foreyt, J. F. Evermann, M. Senda and L. E. Carmichael, J. Gen. Virol., 1988, 69, 1111–1116 CrossRef PubMed.
- C. R. Parrish, P. H. O'Connell, J. F. Evermann and L. E. Carmichael, Science, 1985, 230, 1046–1048 CrossRef CAS.
- C. Buonavoglia, V. Martella, A. Pratella, M. Tempesta, A. Cavalli, D. Buonavoglia, G. Bozzo, G. Elia, N. Decaro and L. Carmichael, J. Gen. Virol., 2001, 82, 3021–3025 CrossRef CAS.
- W. de Oliveira Santana, V. P. Silveira, J. M. Wolf, D. Kipper, S. Echeverrigaray, C. W. Canal, U. Truyen, V. R. Lunge and A. F. Streck, Infect., Genet. Evol., 2022, 98 Search PubMed.
- R. Khatri, Poonam, H. Mohan, Minakshi and C. S. Pundir, J. Vet. Sci. Med. Diagn., 2017, 6, 3 Search PubMed.
- A. Behdenna, T. Lembo, O. Calatayud, S. Cleaveland, J. E. B. Halliday, C. Packer, F. Lankester, K. Hampson, M. E. Craft, A. Czupryna, A. P. Dobson, E. J. Dubovi, E. Ernest, R. Fyumagwa, J. G. C. Hopcraft, C. Mentzel, I. Mzimbiri, D. Sutton, B. Willett, D. T. Haydon and M. Viana, Proc. R. Soc. B, 2019, 286, 1–10 Search PubMed.
- I. C. V. P. Gogone, F. R. O. de Barros, F. Possatti, A. A. Alfieri and E. Takiuchi, Can. J. Microbiol., 2020, 66, 138–143 CrossRef CAS PubMed.
- E. Kilian, J. S. Suchodolski, K. Hartmann, R. S. Mueller, G. Wess and S. Unterer, PLoS One, 2018, 13, 1–11 CrossRef.
- C. G. Lamm and G. B. Rezabek, Vet. Clin. North Am. Small Anim. Pract., 2008, 38, 837–850 CrossRef.
- N. Decaro, G. Elia, V. Martella, C. Desario, M. Campolo, L. Di Trani, E. Tarsitano, M. Tempesta and C. Buonavoglia, Vet. Microbiol., 2005, 105, 19–28 CrossRef CAS PubMed.
- R. A. Crandell, C. G. Fabricant and W. A. Nelson-Rees, In Vitro, 1973, 9, 176–185 CrossRef CAS PubMed.
- G. Kaur, M. Chandra, P. N. Dwivedi and N. S. Sharma, Vet. World, 2015, 8, 52–56 CrossRef PubMed.
- G. Burtonboy, F. Coignoul, N. Delferriere and P. P. Pastoret, Arch. Virol., 1979, 61, 1–11 CrossRef CAS PubMed.
- L. E. Carmichael, J. C. Joubert and R. V. Pollock, Am. J. Vet. Res., 1980, 41, 784–791 CAS.
- N. Decaro, C. Desario, M. Billi, E. Lorusso, M. L. Colaianni, V. Colao, G. Elia, G. Ventrella, I. Kusi, S. Bo and C. Buonavoglia, Vet. J., 2013, 198, 504–507 CrossRef CAS.
- H. Y. Esther Yip, A. Peaston, L. Woolford, S. J. Khuu, G. Wallace, R. S. Kumar, K. Patel, A. A. Azari, M. Akbarzadeh, M. Sharifian, R. Amanollahi, R. J. Jozani, A. Khabiri and F. Hemmatzadeh, Viruses, 2020, 12, 1–13 Search PubMed.
- S. Schmitz, C. Coenen, M. Konig, H.-J. Thiel and R. Neiger, J. Vet. Diagn. Invest., 2009, 21, 344–345 CrossRef PubMed.
- S. E. McNeil, J. Leukocyte Biol., 2005, 78, 585–594 CrossRef CAS.
- C. R. Basso, C. De Camargo Tozato, M. C. Mendes Ribeiro, J. P. Araujo Junior and V. A. Pedrosa, Anal. Methods, 2013, 5, 5089–5095 RSC.
- Z. Xi, H. Ye and X. Xia, Chem. Mater., 2018, 30, 8391–8414 CrossRef CAS.
- A. M. Shrivastav, U. Cvelbar and I. Abdulhalim, Commun. Biol., 2021, 4, 1–12 CrossRef.
- B. D. Malhotra and A. Chaubey, Sens. Actuators, B, 2003, 91, 117–127 CrossRef CAS.
- J. Ou, Z. Zhou, Z. Chen and H. Tan, Int. J. Mol. Sci., 2019, 20, 18 Search PubMed.
- C. R. Basso, C. C. Tozato, J. P. A. Junior and V. A. Pedrosa, Anal. Methods, 2015, 7, 2264–2267 RSC.
- C. R. Basso, C. C. Tozato, B. P. Crulhas, G. R. Castro, J. P. A. Junior and V. A. Pedrosa, Virology, 2018, 513, 85–90 CrossRef CAS.
- C. R. Basso, A. C. Yamakawa, V. A. Pedrosa, M. Magro and F. Vianello, Pathogens, 2022, 11, 570 CrossRef CAS.
- B. Negahdari, M. Darvishi and A. A. Saeedi, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 469–474 CrossRef CAS PubMed.
- K. Omidfar, S. Kia, S. Kashanian, M. Paknejad, A. Besharatie, S. Kashanian and B. Larijani, Appl. Biochem. Biotechnol., 2010, 160, 843–855 CrossRef CAS PubMed.
- A. Razmi, A. Golestanipour, M. Nikkhah, A. Bagheri, M. Shamsbakhsh and S. Malekzadeh-Shafaroudi, J. Virol. Methods, 2019, 267, 1–7 CrossRef CAS.
- N. Verdoodt, C. R. Basso, B. F. Rossi and V. A. Pedrosa, Food Chem., 2017, 221, 1792–1796 CrossRef CAS PubMed.
- D. Lin, J. Wu, H. Ju and F. Yan, Biosens. Bioelectron., 2013, 45, 195–200 CrossRef CAS PubMed.
- L. S. Selvakumar, K. V. Ragavan, K. S. Abhijith and M. S. Thakur, Anal. Methods, 2013, 5, 1806–1810 RSC.
- N. Seow, P. S. Lai and L. Y. L. Yung, Anal. Biochem., 2014, 451, 56–62 CrossRef CAS PubMed.
- I. Fratoddi, Nanomaterials, 2018, 8, 1 Search PubMed.
- E. A. Smith and R. M. Corn, Appl. Spectrosc., 2003, 57, 320A–332A CrossRef CAS PubMed.
- P. Oberacker, P. Stepper, D. M. Bond, S. Höhn, J. Focken, V. Meyer, L. Schelle, V. J. Sugrue, G. J. Jeunen, T. Moser, S. R. Hore, F. Meyenn, K. Hipp, T. A. Hore and T. P. Jurkowski, PLoS Biol., 2019, 17, 1–16 CrossRef.
- C. Wang and J. Irudayaraj, Small, 2008, 4, 2204–2208 CrossRef CAS.
- D. Gava, V. H. B. Serrão, L. T. Fernandes, M. E. Cantão, J. R. Ciacci-Zanella, N. Morés and R. Schaefer, Braz. J. Microbiol., 2018, 49, 351–357 CrossRef CAS PubMed.
- S. Kulanayake and S. K. Tikoo, Viruses, 2021, 13, 388 CrossRef CAS.
- C. R. Basso, B. P. Crulhas, M. Magro, F. Vianello and V. A. Pedrosa, Talanta, 2019, 197, 482–490 CrossRef CAS.
- S. Ghosh, S. C. Dasgupta, A. K. Dasgupta, A. Gomes and A. Gomes, J. Nanosci. Nanotechnol., 2020, 20, 3404–3414 CrossRef CAS PubMed.
- C. Sharma, M. Singh, V. Upmanyu, V. Chander, S. Verma, S. Chakrovarty, G. K. Sharma, H. Dhanze, P. Singh, S. Shrivastava, J. Kumar, T. K. Goswami and V. K. Gupta, Arch. Virol., 2018, 163, 2359–2368 CrossRef CAS PubMed.
- M. Reth, Nat. Immunol., 2013, 14, 765–767 CrossRef CAS PubMed.
- R. Mradula, S. Devi Raj and S. Mishra, Anal. Sci., 2020, 36, 799–806 CrossRef CAS PubMed.
- S. Balbinot, A. M. Srivastav, J. Vidic, I. Abdulhalim and M. Manzano, Trends Food Sci. Technol., 2021, 111, 128–140 CrossRef CAS.
- A. J. Haes, C. L. Haynes, A. D. McFarland, G. C. Schatz, R. P. Van Duyne and S. Zou, MRS Bull., 2005, 30, 368–375 CrossRef CAS.
- V. Shvalya, G. Filipič, J. Zavašnik, I. Abdulhalim and U. Cvelbar, Appl. Phys. Rev., 2020, 7, 3 Search PubMed.
- C. R. Basso, C. C. Tozato, J. P. A. Junior and V. A. Pedrosa, Anal. Methods, 2015, 7, 2264–2267 RSC.
- L. Gao, M. Liu, G. Ma, Y. Wang, L. Zhao, Q. Yuan, F. Gao, R. Liu, J. Zhai, Z. Chai, Y. Zhao and X. Gao, ACS Nano, 2015, 9, 10979–10990 CrossRef CAS.
- G. Liu, M. Lu, X. Huang, T. Li and D. Xu, Sensors, 2018, 18, 1–16 Search PubMed.
- M. Marin, M. V. Nikolic and J. Vidic, Compr. Rev. Food Sci. Food Saf., 2021, 1–21 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00114d |
| This journal is © The Royal Society of Chemistry 2023 |