Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Stepwise conversion of the Cys6[4Fe–3S] to a Cys4[4Fe–4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase†
Andrea
Schmidt‡
a,
Jacqueline
Kalms‡§
a,
Christian
Lorent‡
 b,
Sagie
Katz‡
b,
Stefan
Frielingsdorf‡
b,
Sagie
Katz‡
b,
Stefan
Frielingsdorf‡
 b,
Rhiannon M.
Evans
b,
Rhiannon M.
Evans
 c,
Johannes
Fritsch¶
b,
Elisabeth
Siebert
b,
Christian
Teutloff
d,
Fraser A.
Armstrong
c,
Johannes
Fritsch¶
b,
Elisabeth
Siebert
b,
Christian
Teutloff
d,
Fraser A.
Armstrong
 c,
Ingo
Zebger
c,
Ingo
Zebger
 *b,
Oliver
Lenz
*b,
Oliver
Lenz
 *b and
Patrick
Scheerer
*b and
Patrick
Scheerer
 *a
*a
aCharité – Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Institute of Medical Physics and Biophysics (CC2), Group Structural Biology of Cellular Signaling, Charitéplatz 1, 10117 Berlin, Germany. E-mail: patrick.scheerer@charite.de
bInstitut für Chemie, Biophysical Chemistry, Technische Universität Berlin, Straße des 17. Juni 135, 10623 Berlin, Germany. E-mail: oliver.lenz@tu-berlin.de; ingo.zebger@tu-berlin.de
cDepartment of Chemistry, University of Oxford, OX1 3QR Oxford, UK
dDepartment of Physics, Freie Universität Berlin, Arnimallee 14, 14195 Berlin, Germany
First published on 20th September 2023
Abstract
The membrane-bound [NiFe]-hydrogenase of Cupriavidus necator is a rare example of a truly O2-tolerant hydrogenase. It catalyzes the oxidation of H2 into 2e− and 2H+ in the presence of high O2 concentrations. This characteristic trait is intimately linked to the unique Cys6[4Fe–3S] cluster located in the proximal position to the catalytic center and coordinated by six cysteine residues. Two of these cysteines play an essential role in redox-dependent cluster plasticity, which bestows the cofactor with the capacity to mediate two redox transitions at physiological potentials. Here, we investigated the individual roles of the two additional cysteines by replacing them individually as well as simultaneously with glycine. The crystal structures of the corresponding MBH variants revealed the presence of Cys5[4Fe–4S] or Cys4[4Fe–4S] clusters of different architecture. The protein X-ray crystallography results were correlated with accompanying biochemical, spectroscopic and electrochemical data. The exchanges resulted in a diminished O2 tolerance of all MBH variants, which was attributed to the fact that the modified proximal clusters mediated only one redox transition. The previously proposed O2 protection mechanism that detoxifies O2 to H2O using four protons and four electrons supplied by the cofactor infrastructure, is extended by our results, which suggest efficient shutdown of enzyme function by formation of a hydroxy ligand in the active site that protects the enzyme from O2 binding under electron-deficient conditions.
Introduction
Molecular hydrogen has been almost inexhaustible on the ancient earth and is supposed to represent one of the first energy sources for early microbes.1 Biocatalytic splitting of H2 into protons and electrons occurs at transition metal centers that constitute the active sites of hydrogenases. Depending on the metal center composition, these metalloenzymes are classified as either [FeFe]- or [NiFe]-hydrogenases.2,3 Consistent with their anaerobic origin, hydrogenases are commonly highly susceptible to molecular oxygen, which either inactivates or even destroys the enzymes. Thus, the emergence of the oxygenic atmosphere posed evolutionary pressure towards a microbial metabolism that also functions under oxic conditions. Although the availability of free H2 declined dramatically during these times, some bacterial species evolved O2-tolerant [NiFe]-hydrogenases that feed the H2-derived electrons into the quinone pool of the aerobic respiratory chain.4 A prominent member of these so-called Knallgas bacteria is Cupriavidus necator H16 (formerly Ralstonia eutropha H16), which employs four different O2-tolerant [NiFe]-hydrogenases for generating cellular energy from the controlled oxidation of H2 in the presence of O2.5 Among them, only the tripartite membrane-bound [NiFe]-hydrogenase (MBH) is directly linked to the respiratory chain.6 The MBH large subunit contains the catalytic center composed of a nickel and an iron ion, which are coordinated to the protein by four Cys-derived thiolates. One CO and two cyanide ligands additionally ligate the iron (Fig. 1). This architecture provides a substrate/inhibitor-binding site located between the two metal ions.7,8 Notably, the first and second coordination spheres of the MBH catalytic center are identical to those of O2-sensitive [NiFe]-hydrogenases, suggesting that the O2 tolerance of the enzyme originates from structural features furtheraway from the active site.9 Besides variations in their gas tunnel networks,10,11 a major difference in cofactor structure/composition has been found in the small, electron-transferring subunit of the C. necator MBH and closely related enzymes compared to O2-sensitive hydrogenases.9,12 Typically, membrane-bound [NiFe]-hydrogenases contain a relay of three iron–sulfur clusters that connect the active site with the third subunit, a cytochrome b. The most distal cluster relative to the active site is a [4Fe–4S] cluster coordinated by three cysteines and a histidine, while the medial cluster is a [3Fe–4S] species with three cysteines as protein anchors.2,3 However, in the proximal position, instead of a standard Cys4[4Fe–4S] cluster, the O2-tolerant MBHs possess a [4Fe–3S] cluster coordinated by six cysteine residues (Fig. 1 and S1†).13–16 The unique architecture of the [4Fe–3S] cluster is associated with a redox-dependent structural flexibility that allows two redox-transitions within a physiological potential region, i.e., the cofactor can assume a super-oxidized, an oxidized, and a reduced state.13,15,17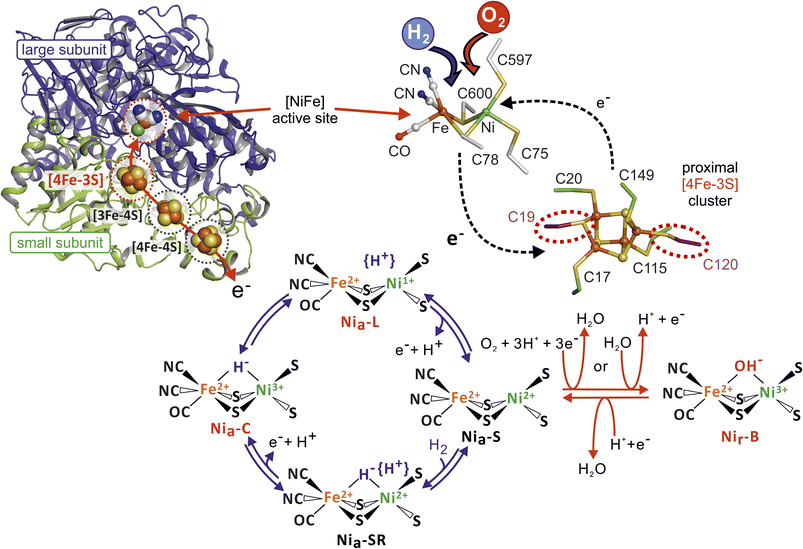 | ||
| Fig. 1 Overview of oxygen-tolerant membrane-bound [NiFe] hydrogenase from Cupriavidus necator (MBH). Left: Cartoon representation of the MBH heterodimer (large subunit HoxG in blue, small subunit HoxK in green) in the reduced state (PDB entry 3RGW). The [NiFe] active site in the large subunit and the proximal [4Fe–3S] cluster (both highlighted by dashed red circles) as well as the remaining Fe–S clusters of the small subunit (marked by dashed black circles) are shown in a ball representation. The red arrow illustrates the electron pathway between the active site and the iron–sulfur clusters. Right: Close up view of the [NiFe]-active site and the unique [4Fe–3S] cluster with its two supernumerary cysteines (dashed red circles). Depending on the presence of oxygen or hydrogen, the electron exchange between the two metal-centers occurs in a different direction. Bottom: Illustration of the various redox states of the [NiFe]-active site during catalytic H2 oxidation/proton reduction (blue arrows) and the proposed in-/re-activation process (red arrows) upon O2 exposure. Active site states (Niu-A, Niia-S), which are discussed in the text but whose structures are unknown and presumably formed only under in vitro conditions are not shown. | ||
This unusual trait is supported by a plethora of biochemical, electrochemical, spectroscopic, computational, and crystallographic results obtained for MBH enzymes from Aquifex aeolicus, Escherichia coli, Hydrogenovibrio marinus, Cupriavidus necator, and Salmonella enterica.12–17 Moreover, a functional [4Fe–3S] cluster is essential for O2 tolerance, as the replacement of both or just one of the two additional cysteine residues results in the loss of the capability to sustain H2 oxidation at high O2 concentrations. This requirement has been demonstrated for the MBH from C. necator and the closely related Hyd-1 from E. coli based on bacterial growth and protein film electrochemistry (PFE) experiments.9 According to a current model, the proximal [4Fe–3S] cluster contributes to efficient O2 detoxification in the catalytic center, i.e., rapid and complete reduction of O2 to 2H2O with 4e− and 4H+ previously obtained from H2 oxidation (Fig. 1).9,15 In fact, the production of water as a result of O2 reduction with H2-derived electrons has already been reported for another O2-tolerant [NiFe]-hydrogenase from C. necator18 and Hyd-1 from E. coli.19 Catalytic intermediates of the H2/H+ cycling process at [NiFe]-hydrogenase active sites are well-established (Fig. 1). Notably, the only well-defined, oxygen-containing intermediate is an active site featuring Ni3+ and Fe2+ ions bridged by an OH− ligand.3 This so-called Nir-B state is considered to represent a resting state of the enzyme that is rapidly re-activated upon reduction, although there is no experimental evidence that O2 is the origin of the bridging hydroxy ligand.20,21 Further O2-mediated modifications of the catalytic center, which could also be caused by X-ray damage, include oxygenated cysteines observed in a number of crystal structures of both O2-sensitive and O2-tolerant [NiFe]-hydrogenases.13,15,22–24 Such oxygenated cysteine thiolates are considered to impede catalytic activity either temporarily or even permanently, and are associated with the inactive state referred to as Niu-A (catalytically defined as an ‘unready’ state).3,22,25
Prevention of any oxidative damage of the active site is a prerequisite for sustained H2 oxidation activity under aerobic conditions, and the proximal [4Fe–3S] cluster appears to play a crucial role in this protective mechanism. In this context, C. necator MBH variants bearing cysteine exchanges at the proximal [4Fe–3S] cluster were analyzed under various oxidizing and reducing conditions by X-ray crystallography, biochemistry, protein film electrochemistry, spectroscopy, and spectro-electrochemistry to investigate the role of the two supernumerary cysteines at the proximal cluster. The results are of great importance for a generalization on the origin of O2-tolerance of [NiFe]-hydrogenases.
Results
In order to obtain insight into the individual role of the supernumerary cysteines in the coordination and function of the [4Fe–3S] cluster, we purified C. necator MBH derivatives carrying exchanges of cysteine for glycine at the positions 19 and/or 120 in the small subunit HoxK, which are hereafter referred to as MBHC19G, MBHC120G and MBHC19G/C120G. While the native MBH and the MBHC19G protein variants showed similar yields, approx. half the amount was obtained for MBHC19G/C120G (Table S1†). Only a fractional amount of pure protein per g of cells was collected for MBHC120G. The H2-driven methylene blue reduction activities of the purified MBHC19G, MBHC120G and MBHC19G/C120G proteins were 53%, 13%, and 52%, respectively, of the activity of native MBH (140 U mg−1 ref. 9) (Table S1†).To obtain a detailed view of the proximal cluster architecture, all three engineered MBH variants were analyzed by protein X-ray crystallography. Additionally, the two MBHC19G and MBHC120G variants were investigated by PFE and surface-enhanced infrared absorption (SEIRA) spectro-electrochemistry as well as infrared (IR), resonance Raman (RR) and electron paramagnetic resonance (EPR) spectroscopy, to correlate activities with the structural/electronic properties of the metal centers.
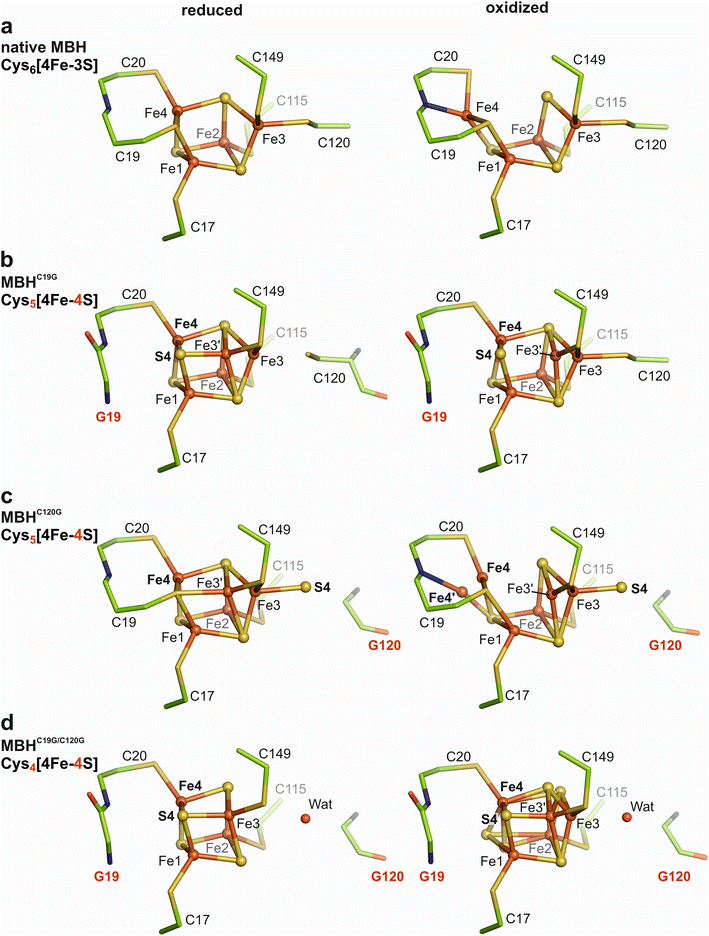
Cysteine exchanges lead to major changes in the composition and architecture of the proximal Fe–S cluster
The MBHC19G, MBHC120G and MBHC19G/C120G variants were crystallized under either aerobic/oxidizing or anaerobic/H2-reducing conditions. The crystal structures were solved by molecular replacement, and the models were refined to resolutions ranging from 1.60 Å to 1.93 Å (PDB entries 8POU-8POZ). To qualify positive peaks in the mFo-DFC electron density maps, anomalous diffraction data were collected.The electron density maps (Fig. S3†) of oxidized MBHC120G crystals (at 1.93 Å resolution) revealed the proximal Fe–S cluster in a similar structure/composition as in the reduced protein, except that Fe4 showed conformational flexibility (Fig. 2c). The electron density distribution was interpreted with 50% of Fe4 residing in a comparable position as in H2-reduced MBHC120G (Fig. 2c). However, an Fe4–S3 distance of 2.9 Å indicates the absence of a covalent bond (Fig. 2c). The remaining 50% of Fe4′ in oxidized MBHC120G were shifted by 1.2 Å to form a covalent bond with the backbone nitrogen of Cys20, bringing Fe4′ into hydrogen bonding distance (2.8 Å) with Glu76 of the MBH small subunit. This particular Fe4′ position differs by 0.6 Å from the classical Fe4 position of native MBH in the super-oxidized state, and the distance between Fe4′ and Sγ-Cys19 and Sγ-Cys20 is 2.9 Å and 2.7 Å, respectively. As observed for all single cluster variants of MBH, the Fe3 was found in a double conformation. Approximately 75% resided in the native Fe3 position and formed a covalent bond to S4 (Fe3–S4 distance 2.4 Å), consistent with the S4 occupancy. The remaining 25% of the Fe3 were shifted by 1.1 Å toward a more cubic conformation (Fe3′). However, a covalent binding to Sγ-Cys19 was not established (Fe3′ – Sγ-Cys19 distance 3.2 Å), resulting in a three-fold ligation of Fe3′ (Fig. S3b†).
Structure of the [NiFe]-active site in the [4Fe–3S] cluster variants
The active site structures of the MBH variants carrying modified proximal iron–sulfur clusters were found in the same conformations as previously described for native MBH (Fig. S7†).14,17 The identity includes the positions of the two metal atoms as well as the CO and the two CN− ligands of the iron, which were modelled into the structure according to previous QM/MM calculations.17 The oxidized structures exhibited an electron density between the Ni and Fe ions (Ni–Fe) distance, 2.9 Å; (Fig. S7†), which is consistent with the presence of a hydroxyl group occupying the bridging position between the two metals corresponding to the resting state known as Nir-B (Fig. 1). The reduced structures exhibited a lower Ni–Fe distance of 2.6 Å, which is typical of catalytic cycle intermediates26 and consistent with our spectroscopic studies (see below). One exception was found for H2-reduced MBHC19G, which contained a positive mFo-DFc electron density between nickel and iron (Fig. S7†). We interpreted this density as a mono-oxygen species with an occupancy of about 50%, causing a slightly enlarged Ni–Fe distance of 2.7 Å. A very minor positive electron density located between nickel and iron was also observed for H2-reduced MBHC19G/C120G. This small density was interpreted as a mono-oxygen species with an occupancy below 10%. Furthermore, the oxidized MBHC19G variant contained a sulfoxygenated (occupancy of 20%), nickel-coordinating cysteine (Cys597). Such modification was less evident in the structure of H2-reduced MBHC19G (occupancy below 10%) and can therefore be considered as partly reversible. Notably, we found no evidence of oxygenation at the coordinating cysteine thiolates or of further damage in any of the other crystal structures.Spectroscopic properties of the metal cofactors in the MBH variants
To investigate changes in the redox properties for the [NiFe]-active site and the proximal Fe–S cluster of MBH caused by the C19G and C120G exchanges, we carried out IR and EPR spectroscopic experiments in solution, complemented by resonance Raman data obtained from single crystals. Goris et al.9 already described the IR/EPR-spectroscopic characterization of the MBHC19G/C120G variant, and we will refer to these data mainly in the discussion.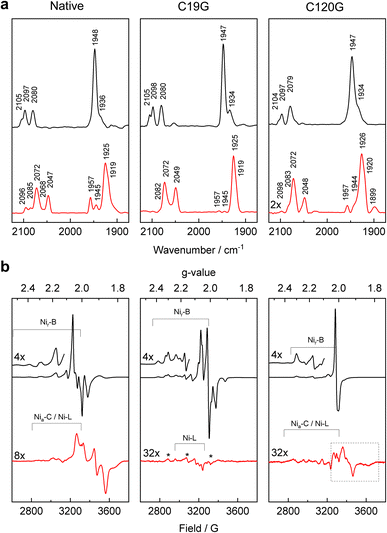 | ||
| Fig. 3 IR and EPR spectroscopic investigation of the MBH variants. (a) IR absorbance spectra of oxidized (black) and H2-reduced (red) samples of native MBH (left), MBHC19G (middle), and MBHC120G (right), recorded at 10 °C. The band assignment is summarized in Table S3.† (b) EPR spectra of oxidized (black) and H2-reduced (red) samples of native MBH (left), MBHC19G (middle), and MBHC120G (right) measured at 20 K. Signals marked with an asterisk (gx = 2.31, gy = 2.15, gz = 2.00) correspond to a so far unknown paramagnetic state of the active site. The signals in the dashed box can be assigned to different reduced Fe–S clusters (see Fig. S11†). | ||
The IR spectrum of the reduced MBHC19G variant (Fig. 3a) was dominated by one of the EPR-silent, fully reduced states, namely Nia-SR′, with characteristic modes at 1925, 2049 and 2072 cm−1. A shoulder at 1919 cm−1 also indicated the presence of the Nia-SR′′ species (Table S3†). In addition to these states, the corresponding IR spectrum of native MBH contained bands at 1945 and 1957 cm−1 (Fig. 3a). This observation implies that a fraction of the active site in native enzyme resides in the Nia-SR state (∼5%) and the paramagnetic Nia-C state (∼10%). The spectrum of the reduced MBHC19G variant showed only traces of these reduced states, suggesting different steady state equilibria caused by the modified proximal cluster.
EPR measurements revealed that H2-treated MBHC19G was mainly EPR-silent. Only trace amounts of Nia-L were observed. In contrast, H2-reduced native MBH showed a distinct signal typical for the reduced proximal [4Fe–3S] cluster in O2-tolerant MBHs from different organisms (Fig. 3b, red traces). It should be noted that clear indications for the reduced distal [4Fe–4S] cluster have not been observed by EPR.9,28–30
To investigate whether the reduction of MBHC19G is reversible, we exposed the initially H2-reduced enzyme to air. According to EPR, this re-oxidation should have led to a diminishment of both the EPR signal of the proximal cluster and its magnetic coupling to the active site in the Nir-B state (Fig. S9a,† dotted lines). Instead, a large uncoupled axial signal appeared (g⊥ = 2.017; g∥ = 2.001), which was assigned to the oxidized medial [3Fe–4S]+ cluster. A similar observation was obtained for native MBH (Fig. S9a†).9 The corresponding IR spectrum of re-oxidized MBHC19G protein was dominated by signals related to the Nir-B state (Fig. S9b†), while ∼20% of the protein molecules resided in the inactive Niia-S state (Table S3†), which should be irreversible under physiological conditions but has eluded structural characterization to date.29 Again, native MBH showed a similar behavior (Fig. S9b†). These results suggest that isolated native MBH and MBHC19G suffered from the re-oxidation treatment in the same way. Notably, the oxidative inactivation was completely reversible when the same procedure was performed with membrane fragments (Fig. S9a,† solid lines). This observation shows that the results obtained with the isolated heterodimeric enzyme have to be treated with caution.29
In conclusion, the absence of any Fe–S cluster-related EPR signal in the reduced MBHC19G variant can be interpreted as the presence of a diamagnetic proximal cluster. This result suggests that the new Cys5[4Fe–4S] cluster in MBHC19G undergoes only one redox transition, corresponding to a reductive conversion from a paramagnetic, super-oxidized state into an EPR-silent oxidized state. The second reduction step towards a fully reduced state appears to be beyond physiological redox potentials. This assumption is supported by the incomplete disappearance of characteristic Fe–S bands (at ca. 340 cm−1) in the resonance Raman spectrum of H2-reduced MBHC19G crystals when compared to the corresponding spectra of native MBH and MBHC120G (Fig. S10†).
The observed bands in the IR spectrum of H2-reduced MBHC120G correspond to the catalytically relevant, fully reduced sub-states Nia-SR, Nia-SR′ and Nia-SR′′, in addition to the partially reduced Nia-C state and a small amount of the Nia-L species (Fig. 3a and Table S3†). The same bands, though in different relative ratios, were observed for samples of reduced native MBH (Fig. 3a). The lower intensities of the absorption bands of the H2-reduced MBHC120G variant, however, indicate protein instability caused by the C120G exchange, which was not observed for the MBHC19G variant (Fig. 3a).
The EPR spectrum of H2-reduced MBHC120G lacks the characteristic Fe–S cluster-derived signal present in reduced native MBH (Fig. 3b). Instead, very weak signals occurred in the g-value range from 2.05 to 1.80, which we attribute to traces of one or more reduced proximal [4Fe–4S] cluster species. EPR measurements at various temperatures and microwave powers indeed revealed different spin relaxation properties (Fig. S11†), suggesting the presence of at least two electronically and structurally different species. This is consistent with the crystallographic data, which suggest multiple conformations of the reduced proximal cluster (Fig. 2c). The corresponding resonance Raman spectra revealed the disappearance of all cluster-related Fe–S modes in H2-reduced MBHC120G crystals, indicating the complete reduction of the Fe–S cluster relay (Fig. S10†).
Re-oxidation of H2-reduced MBHC120G did not result in the re-appearance of Nir-B-associated EPR signals with the same intensity as initially observed in the oxidized enzyme (Fig. S9a†). In addition, the amount of Niia-S species (∼45%) was almost as large as that of Nir-B (∼55%) as revealed by IR spectroscopy (Fig. S9b†). These results support the inherent instability of the solubilized MBHC120G variant upon repeated redox treatments.
H2 turnover activity of the MBH variants in the presence of O2
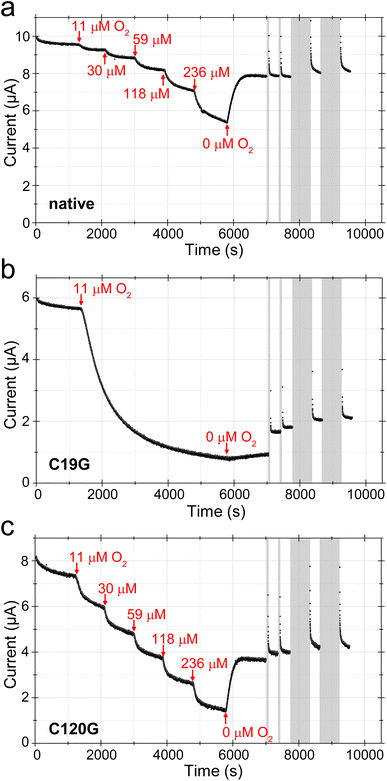
Protein film electrochemistry (PFE) is an established method to investigate the H2/H+-cycling properties of (bio-)catalysts immobilized onto conducting surfaces such as pyrolytic graphite electrodes or metal (oxides) with biocompatible coatings.31–34 The measured currents are proportional to enzymatic turnover making the technique particularly useful for testing the influence of inhibitory gases such as O2 during H2 activation. Here, it is important that the gas composition is tightly controlled during the time the catalytic current is monitored, since changes in activity in response to a particular gas may be slow. The electrochemical cell is thus sealed, and equipped to allow a controlled mixture of gases to flow through the headspace above the solution while the electrode is rotated. Gases to be tested are introduced or removed by adjusting a mass flow controller that rapidly mixes the different components in a precise manner (H2, Ar, O2etc.).35 PFE has already been employed to investigate the MBHC19G/C120G variant on a bare pyrolytic graphite electrode.9 The protein rapidly lost its H2-oxidizing activity as soon as O2 was added to the assay solution, which was in deep contrast to the observations for native MBH.9 Here, we tested the capabilities of the MBHC19G and MBHC120G proteins to sustain H2 conversion in the prolonged presence of O2. To avoid irreversible ‘loss’ of protein film from the electrode, seen as a continuous loss of current over time,9 film stability can be improved by covalently attaching the enzyme onto pyrolytic graphite electrodes coated with pyrene-butyric acid-modified multi-walled carbon nanotubes (MWNT).36 The activity of MBHC19G and MBHC120G was followed by cyclic voltammetry (CV) in the presence of 80% H2 (606 μM) under otherwise anoxic conditions (Fig. S12†). Both variants showed the typical overpotential of about 80 mV for the onset of catalysis and similar inactivation kinetics under oxidizing conditions at higher potential. However, they reacted differently to short pulses of O2 (Fig. S12†), prompting us to investigate the effect of prolonged O2 exposure on the H2 oxidation activity of the MBH variants in chronoamperometric experiments (Fig. 4). The potential was set to 0 V to avoid direct O2 reduction by the electrode. Native MBH showed the typical behavior upon O2 treatment.37,38 Each O2 addition led to a distinct current drop (Fig. 4a). The resulting steady state current derives from active enzyme, the level of which is determined by the rate of O2-mediated inactivation and the rate of re-activation. A prerequisite for this behavior is that unready states of the active site are not formed upon O2 incubation, which would lead to a constant decline of the current. An important feature of the native MBH is its capability to regain a significant portion of activity as soon as O2 is removed from the cell headspace (Fig. 4a).39 Stepwise addition of O2 to H2-oxidizing MBHC120G variant revealed a comparable current progression as observed for native MBH (Fig. 4b). However, at the highest O2 concentration, MBHC120G lost almost 80% of its initial activity, while corresponding loss of native MBH was just ∼50%. As with native MBH, flushing O2 from the cell resulted in rapid spontaneous recovery of catalytic activity. In contrast to native MBH, however, a further current increase was observed upon application of a series of short (60 s) and extended (600 s) low potential pulses (−0.659 V). Nevertheless, the current increase was rather small compared to the recovery observed after O2 removal (Fig. 4b).The MBHC19G variant showed a more pronounced response toward O2 exposure. Addition of just 1% O2 (11 μM) resulted in a continuous drop in H2 oxidation current (Fig. 4c). After 4600 s of low-level O2 exposure, the current reached a stable level corresponding to less than 20% of the initial activity. In contrast to native MBH and MBHC120G, the removal of O2 from the solution, resulted in only a marginal recovery of the current. A more significant recovery was achieved, however, after the application of low potential pulses (Fig. 4c).
To obtain insight into the nature and distribution of the active site redox states during catalysis, we employed a setup allowing surface-enhanced IR absorption (SEIRA) spectroscopy under electrochemical control (Fig. 5). The MBH variants were immobilized onto a nanostructured Au electrode coated with a self-assembled monolayer, and we carried out a chronoamperometric study by employing similar, but not identical conditions as used for the PFE experiments described above (see Fig. S13† for details). IR spectra were recorded during H2 conversion in the absence and presence of O2, as well as after flushing out the O2 followed by reductive reactivation at negative potentials (Fig. 5 and S13†). Under pure H2, the spectra of the native MBH and the variants C19G and C120G were dominated by the catalytic Nia-C and Nia-SR′ states. Furthermore, two inactive states (Nir-B and Niia-S) were observed in all variants. The amount of the Niia-S state (CO peak at 1930 cm−1) seemed to increase continuously during the chronoamperometric measurements, irrespectively of the gas composition. As the quantity of immobilized hydrogenase did not change during the course of the spectro-electrochemical measurements (Fig. S14†), we interpret the intrinsic decline of the catalytic current (often stated as film loss) with the accumulation of the apparently irreversibly inactive Niia-S state. The signal attributed to the Nir-B state (CO peak at 1948 cm−1), which accumulates in the presence of the H2/O2 mixture, disappeared completely in native MBH after removal of oxygen. In contrast, the same treatment did not result in the complete elimination of the Nir-B signal for the MBHC120G variant and especially for the MBHC19G variant (Fig. 5b and S13†). Only the application of low-potential pulses under anaerobic conditions resulted in lowering of the Nir-B signals in favor of the population of signals resulting from reduced states. This result is consistent with the level of current recovery during chronoamperometry, which was highest for the MBHC19G variant (Fig. 4c and S13†). Notably, IR bands of other inactive species such as Niu-A were not found in detectable amounts in any of the immobilized MBH variants.
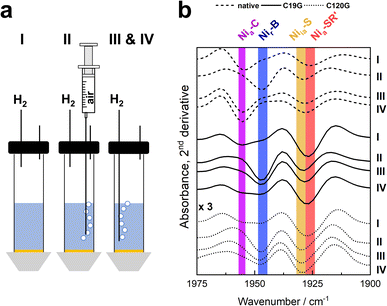 | ||
| Fig. 5 SEIRA spectroscopy of the MBH variants under electrochemical control. Proteins were immobilized on bio-compatibly coated Au electrodes, and spectroscopic changes were followed during chronoamperometry at 0 mV (vs. SHE). (a) Experimental set-up and procedure of the experiment. In phase I, the protein solution was subjected to a pure H2 atmosphere. Phase II included the stepwise addition of air to the buffer reservoir which was subsequently removed in phase III by flushing with pure H2. In phase IV, two low-potential pulses were applied at −400 mV, and the potential was then set back to 0 mV. (b) Second derivatives of the SEIRA spectra in the region characteristic for the CO absorptions recorded at the end of each phase. Spectra of MBHC120G are enlarged by a factor of three (Fig. S14†). The redox states corresponding to specific absorption bands are marked with coloured bars. For further details, see Fig. S13.† | ||
Discussion
Substitution of auxiliary cysteines changes iron–sulfur cluster types with unprecedented structural features
Exchanges of the supernumerary cysteines of the proximal [4Fe–3S] cluster had distinct consequences regarding the yield and catalytic activity of all MBH variants. In fact, the single C120G exchange caused the most drastic effect, as both the protein yield and the H2-mediated methylene blue reduction capacity (under anoxic conditions) were only about 10% of the corresponding values for the native protein. Despite the low yield, we were able to solve the crystal structures of the MBHC120G variant of the oxidized and H2-reduced forms. Among the MBH variants, the MBHC120G protein showed the highest structural plasticity with at least two different states of the proximal Fe–S cluster for each of the two redox conditions. In approximately 50% of the H2-reduced MBHC120G crystals, this new cluster resembles the structure of the reduced native [4Fe–3S] cluster, except that the Cys120 thiolate is partially replaced by an exogenous sulfur species (S4) that coordinates Fe3, resulting in a [4Fe–4S] cluster in a non-cubic form. The other 50% does not contain the S4, making Fe3 part of a cubane, one corner of which is occupied by the Cys19-derived sulfur. To our knowledge, such a novel cubic cluster in which one of the inorganic sulfides is replaced by a cysteine sulfur remains unprecedented. The replacement of Cys120 results in a double conformation of Fe3 that correlates with the exo-sulfur species in terms of the occupancy. The proximal Cys5[4Fe–4S] cluster in oxidized MBHC120G is even more flexible, forming a new cluster scaffold with altered redox properties and electronic structure. In one fraction of the protein, Fe4 is bound to the backbone nitrogen of Cys20 (Fe4′ position), as observed for the native super-oxidized [4Fe–3S] cluster. The other fraction (Fe4) remains in the equivalent position to that found for the reduced state (Fig. 2c). The observations clearly indicate that Cys19 is the crucial ligand that allows the movement of Fe4 to move in the native proximal [4Fe–3S] cluster of MBH. However, the overall coordination of Fe4 in MBHC120G differs from that in the native [4Fe–3S] cluster. In conclusion, our results underline the stabilizing role of Cys120 in the plasticity and redox function of the Cys6[4Fe–3S] cluster. In addition to a change in redox properties, removal of Cys120 resulted in an inappropriate flexibility of the proximal cluster in MBHC120G, which, in turn, caused decreased protein stability, as manifested by low protein yield and low abundance of this MBH variant in C. necator cells.9The crystal structures of H2-reduced MBHC19G and MBHC19G/C120G revealed almost cubic [4Fe–4S] clusters coordinated by the four canonical cysteine residues, which had been expected in case of the C19G/C120G exchange, but experimental evidence was lacking.9,40 In MBHC19G, however, the presence of Cys120 and the new sulfide (S4) resulted in a certain redox-dependent flexibility of Fe3, which is comparable but not identical to the situation in MBHC120G. In H2-reduced MBHC19G crystals, a major fraction of Fe3 is not covalently bound to Sγ-Cys120, which is in contrast to the reduced [4Fe–3S] cluster of native MBH. This result can be explained by the fact that sulfide (S4) is a more tightly bound ligand than a thiolate, which leads to coordinative bonding to the shifted Fe3′. The corresponding cubic conformation increases the stability of the cluster. As expected, the Fe4 shift does not occur in the oxidized [4Fe–4S] cluster of MBHC19G. Here, most of Fe3 is covalently bound to Cys120 and not to S4, which is similar to the cluster structure of the C19G variant of E. coli Hyd-1 resulting in a less negative reduction potential.41 Overall, our data show that the covalent bond between Cys120 and Fe3 is crucial for stabilizing the super-oxidized state and thus for the physiological function of the proximal cluster.
Even in the MBHC19G/C120G protein, which contains a four Cys-coordinated [4Fe–4S] species, oxidation resulted in cluster flexibility, indicating that the binding pocket is optimized for an Fe–S cluster of larger dimensions, i.e., the Cys6[4Fe–3S] cluster (Fig. 2). Thus, the Cys4[4Fe–4S] cluster in MBHC19G/C120G is similar but not identical to the classical Cys4[4Fe–4S] cluster of O2-sensitive [NiFe]-hydrogenases.
Insight into the maturation of the proximal Cys6[4Fe–3S] cluster
The different crystal structures of the MBH variants prompted us to propose a possible scheme of the transformation process from a [4Fe–4S] cluster to a [4Fe–3S] species (Fig. 6). The first step is presumably the IscU-assisted insertion of a preformed [4Fe–4S] cluster,42 which becomes attached to the canonical cysteines Cys17, 20, 115 and 149 (also present in all periplasmic, membrane-associated [NiFe]-hydrogenases carrying a [4Fe–4S] cluster in the proximal position) of the MBH small subunit. The [4Fe–4S] cluster as present in the H2-reduced MBHC19G/C120G may represent this maturation intermediate, as this species is the most stable among the cluster variants. However, this hypothesis may be supported by future studies on MBH variants in which the canonical cysteine residues were exchanged. We also hypothesize that cluster rearrangement is subsequently triggered by one of the two additional cysteines, Cys19 or Cys120. In the first scenario, Cys19 would initiate the rearrangement, and its thiolate takes over the coordination of Fe1 and Fe4, replacing sulfide S4. The released S4 could either be delivered to the solvent (assuming that the binding pocket of the proximal cluster is exposed to the solvent during maturation) or swings over towards the position of the thiol group of Cys120. Either of these steps would be accompanied by a shift of Fe3 from the corner position of the cubane. These transformation steps would result in the Cys5[4Fe–4S] cluster as present in the reduced MBHC120G variant. In the second scenario, the Cys120 would first covalently bind Fe3, which should lead to a non-tetrahedral coordination of S4 (represented by the oxidized Cys5[4Fe–4S] cluster of MBHC19G) and its subsequent release. Both scenarios would eventually lead to covalent binding of Cys19 and Cys120 to the corresponding iron ions of the mature Cys6[4Fe–3S] cluster (Fig. 6). Under aerobic conditions, this transformation process appears to require specific auxiliary and protective proteins42,43 because the proximal Fe–S cluster in the isolated small subunit is solvent-exposed and therefore susceptible to O2.6 Interestingly, the EPR spectrum of the oxidized C19G variant of MBH (Fig. 3b) shows striking similarities to that of MBH isolated from a C. necator mutant strain in which the rubredoxin-like maturase-encoding gene hoxR was deleted.44 This is the first indication that HoxR enables the correct bridging coordination of Cys19 to Fe1 and Fe4 under aerobic conditions for both of the proposed maturation pathways.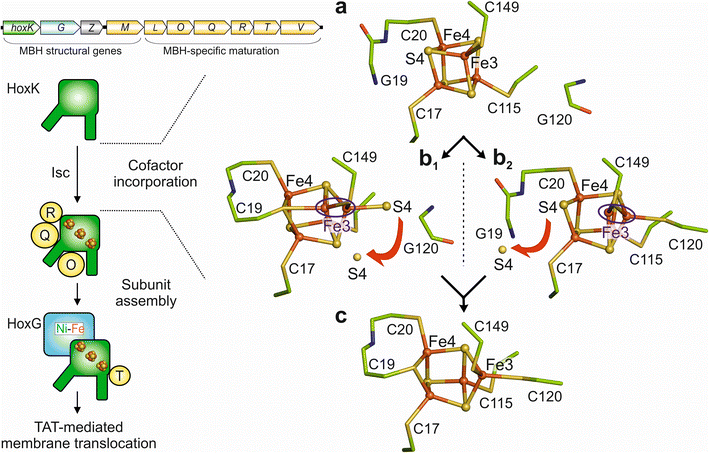 | ||
| Fig. 6 Possible maturation pathway(s) of the proximal [4Fe–3S] cluster. (Left) Schematic overview of the MBH maturation, which requires dedicated maturases (yellow circles) encoded in the MBH gene cluster (top).42 (Right) (a) The first step would be the insertion of a standard cubane [4Fe–4S] cluster, followed by either (b1) covalent binding of Cys19 to Fe4 and Fe1 or (b2) binding of Cys120 to Fe3. Binding of the first additional cysteine is expected to lead to a destabilization of the cubane and subsequent release of S4. This would allow covalent binding of the second additional cysteine, leading to stabilization of the classical [4Fe–3S] cluster conformation (c). The proximal cluster, selected amino acids and water molecules are depicted as ball/sticks, sticks and red spheres respectively. The destabilization at Fe3 is highlighted by a blue circle. Red arrows indicate the release of S4. | ||
Cysteine exchanges restrict the number of redox transitions mediated by the proximal cluster
IR spectroscopy revealed that all three oxidized MBH derivatives investigated in this study form predominantly the Nir-B state, which is consistent with the crystal structure and EPR data. The sulfoxygenated cysteine observed in oxidized MBHC19G crystals is not reflected by the spectroscopic data, probably because of its low occupancy, which is even lower in H2-reduced crystals, indicating that this modification is slowly reactivatable rather than irreversibly inactive. In addition to MBHC19G and MBHC120G, the oxidized MBHC19G/C120G protein9 forms preferentially the Nir-B state, showing that the manifold engineered proximal clusters does not necessarily lead to accumulation of Niu-A.Concerning the EPR analysis, the simplest EPR spectrum was observed for oxidized MBHC120G. It consists of an axial signal attributed to the medial [3Fe–4S]1+ cluster in addition to the typical rhombic Nir-B signal (Ni3+). In contrast to the corresponding spectrum of the native MBH, the proximal cluster of the oxidized MBHC120G remains diamagnetic, as no additional signals and no splitting or broadening of the signals for Nir-B and the medial cluster were observed. The same observation has been made for the oxidized C19G/C120G exchange variants of MBH and E. coli Hyd-1.9,30 In addition, H2-reduced MBHC19G/C120G and Hyd-1C19G/C120G, were EPR-silent, suggesting a fully reduced active site and a diamagnetic medial [3Fe–4Fe]0 cluster. This observation, in turn, is consistent with the EPR spectrum of reduced MBHC120G, which contained only very weak signals interpretable as reduced Fe–S cluster species. The spectrum is also consistent with the predominant formation of EPR-silent, fully reduced Nia-SR sub-states as observed by IR. Although MBHC120G, MBHC19G/C120G, and Hyd-1C19G/C120G showed catalytic activity, their proximal cluster remains non-detectable by EPR after reduction, possibly caused by the presence of an integer spin or an unusual non-integer high spin configuration that is not detectable under the applied measurement conditions. We conclude that the Cys5[4Fe–4S] cluster of MBHC120G and most likely also the proximal Cys4[4Fe–4S] clusters of MBHC19G/C120G and Hyd-1C19G/C120G perform only one low-potential electron transition.15,17,45 This is in accordance with our protein tunneling calculations of the corresponding crystal structures that the exchange of Cys120 with a smaller Gly residue increases the solvent accessibility of the proximal Fe–S cluster in MBHC120G and MBHC19G/C120G crystals (Fig. S6†). This shifted solvent accessibility could contribute to confer a single low redox potential transition.46
In contrast, a major fraction of the Cys5[4Fe–4S] cluster of the MBHC19G variant was EPR-active under oxidizing conditions, whereas no Fe–S cluster-related EPR signal was observed under reducing conditions. Thus, the proximal Cys5[4Fe–4S] cluster of the MBHC19G variant mediates a 3+ to 2+ transition at higher potentials, while the 2+ to 1+ transition does not take place within a physiological potential range.46 This conclusion is consistent with data obtained for Hyd-1C19G,41 for which a redox titration in combination with EPR spectroscopy and computational studies revealed that the proximal [4Fe–4S] cluster is capable of undertaking the “high-potential” transition of the two redox transitions mediated by the native [4Fe–3S] site.41 The associated redox potential of +240 mV was very close to that of the super-oxidized/oxidized couple of the [4Fe–3S] cluster (230 mV).30 This result agrees with our observations for the proximal cluster of the MBHC19G variant, which also seems to allow only the high-potential redox transition.
Accumulation of inactive states during H2 cycling under oxygenic conditions
According to both the cyclic voltammetry and the chronoamperometric PFE experiments, all MBH variants lost a certain amount of activity upon O2 incubation, with native MBH being least and MBHC19G most affected. In the case of MBHC19G and partially also for MBHC120G, O2 exposure led to the accumulation of inactive enzyme species that could in part be reactivated by applying a very negative electrode potential. Our PFE results are in line with those obtained for E. coli Hyd-1 variants carrying equivalent exchanges of the supernumerary cysteines.40 Accompanying spectro-electrochemical SEIRA studies indicate that the current decrease upon O2 incubation is related to the accumulation of two inactive states of the catalytic center in all MBH variants. The first one is the inactive Niia-S state, the amount of which increased steadily during the experiment, regardless of which gas mixture was present, indicating no direct correlation between Niia-S state accumulation and O2-induced inactivation. Besides Niia-S (and the reduced states), all three investigated MBH variants formed the Nir-B state during H2/H+-cycling in the presence of O2. The fact that for the MBHC19G protein in the Nir-B state, current recovery occurred mainly after applying negative potential pulses indicates major kinetic limitations regarding its reactivation ability, which resulted in only a marginal current recovery upon O2 removal. In fact, the redox potential of the Nir-B state of the C. necator MBH lies at −110 mV.47 Reactivation of the Nir-B state requires one proton and one electron (Fig. 1), the latter of which must be supplied in a reverse fashion by the iron–sulfur cluster chain of the MBH small subunit. With the medial [3Fe–4S] cluster and the proximal Cys5[4Fe–4S] cluster, the MBHC19G protein carries two rather high-potential clusters in a row, which creates a more severe kinetic barrier for the efficient reactivation of Nir-B.Conclusions
Re-interpretation of the oxygen tolerance mechanism conferred by the [4Fe–3S] cluster
The oxidized active site of all MBH derivatives seems to reside predominantly in the Nir-B state, which we consider as a very important observation. The bridging hydroxy ligand prevents the active site from interacting with other substrates. Our results indicate that generation of the Nir-B state in oxygen-tolerant MBH is not intimately linked to the two-electron transfer capacity of the unique [4Fe–3S] cluster, which is actually impeded in all proximal cluster variants of the MBH. This proposal is supported by previous results from ascorbate-treated, partially reduced native MBH, which resulted in the formation of a mixture of super-oxidized and oxidized forms of the [4Fe–3S] cluster, while the catalytic center remained in the Nir-B state.17 Similarly, the oxidized crystal structures of Hyd-1 from E. coli and MBH of H. marinus contained an active site fully occupied with a hydroxide ligand, in combination with mixtures of oxidized/reduced and super-oxidized [4Fe–3S] clusters. Only treatment with the strong oxidant ferricyanide resulted in exclusive binding of Fe4 to the backbone nitrogen of Cys20.13,15 These observations suggest that formation of the super-oxidized [4Fe–3S] cluster is mainly triggered by the redox status of the iron–sulfur relay, which interacts with the quinone pool of the respiratory chain in the native environment.29 This assumption implies that in vivo only a completely oxidized iron–sulfur cluster chain (i.e. lacking any reducing capacity as might occur during H2 oxidation under aerobic conditions) leads to the formation of the super-oxidized state of the [4Fe–3S] cluster, as suggested previously45 and supported by the fact that O2-mediated full oxidation to the super-oxidized state exclusively takes place if the MBH is still coupled to the cellular quinone pool (Fig. S9†).Removal of the active site OH ligand under H2/electron-limiting conditions, regenerating the Nia-S state, leaves the active site unprotected and vulnerable to O2-mediated inactivation. Reduction of the [4Fe–3S] cluster by one electron (originating from the Fe–S cluster chain), however, results in the movement of Fe4 back into the Fe–S cluster core. As this reorganization is time-consuming,45 the [4Fe–3S]5+ cluster functions as an implemented time-delay preventing premature reactivation of the Nir-B state in the case of H2 scarcity under otherwise oxic conditions. Importantly, the active site protection mechanism described above does not preclude the previously described model of O2 detoxification by four electrons and four protons delivered to the active site in the reverse direction to reduce O2 to harmless water.9,14,18,19,37,48 In the proposed new mechanism, activity can be switched off under electron-deficient conditions even before O2 would gain access to the catalytic center. In fact, Knallgas bacteria such as C. necator usually face an (micro)aerobic environment in which H2 occurs transiently and only in traces.4
Experimental section
Material and methods
Construction of MBHC19G and MBHC120G. The amino acid exchanges C19G and C120G in the small MBH subunit HoxK were introduced by the QuikChange (Agilent) method using primers #793/#794 for C19G and #795/#796 for C120G9 on plasmid pCH1229.37 The modified plasmids were digested with SpeI/SmaI and the resulting 1171 bp fragments were transferred to SpeI/SmaI-cut pJH2430 (Litmus28 containing PMBH-hoxKStrep-hoxV) resulting in plasmids pJH5116 (C19G) and pJH5127 (C120G). Plasmids pJH5116 and pJH5127 were digested with SpeI/BstZ17I and the resulting 2581 bp fragments were ligated into SpeI/BstZ17I-cut pCH785 (ref. 49) resulting in pJH5143 (C19G) and pJH5148 (C120G). These plasmids were digested with SpeI/XbaI and the resulting 21.5 kbp fragments were ligated into XbaI-cut pEDY309 (ref. 50) resulting in pJH5175 (C19G) and pJH5179 (C120G). pJH5175 and pJH5179 were conjugated using E. coli S17-1 (ref. 51) into C. necator HF210 resulting in strains HP83 (C19G) and HP84 (C120G).
Construction of MBHC19G/C120G. The double amino acid exchange C19G/C120G in the small MBH subunit HoxK was introduced by digesting pCH1487 9 with SpeI/BstZ17I and ligating the resulting 2581 bp fragment into SpeI/BstZ17I-cut pCH785 resulting in plasmid pJH4052. The latter was digested with SpeI/XbaI and the resulting 21.5 kbp fragment was ligated into XbaI-cut pEDY309 resulting in plasmid pJH4081 which was conjugated into C. necator HF210 resulting in strain HP29. Recombinant C. necator strains were cultivated and the MBH variants purified as described elsewhere.51
Diffraction data were collected at the synchrotron facilities BESSY II (Berlin, Germany) and ESRF (Grenoble, France). The best diffracting data for H2-reduced MBHC19G/C120G was collected at BESSY II, beamline BL14.1 (ref. 52) with a Rayonix MX-225 detector at a wavelength of 0.918 Å (PDB entry 8POV). For oxidized MBHC19G/C120G, data collection was executed at ESRF, beamline ID29 (ref. 53) with a PILATUS M6-F detector at a wavelength of 0.976 Å (PDB entry: 8POU). For anomalous data collection, the wavelength was shifted towards 1.734 Å. The best diffracting data for oxidized and H2-reduced MBHC19G/C120G were collected at ESRF, beamline ID29 (ref. 53) with a PILATUS M6-F detector at a wavelength 0.969 Å and 1.00 Å, respectively (PDB entries 8POW and 8POX). Data for oxidized and H2-reduced MBHC120G crystals were collected at the ESRF, beamline ID29 (ref. 53) with a PILATUS M6-F detector at a wavelength of 0.969 Å (PDB entries: 8POY and 8POZ). Additionally, anomalous data at 1.734 Å was recorded for MBHC120G. Images were indexed, integrated and scaled using the XDS program package54 and the CCP4 program SCALA.55,56 All crystals showed the orthorhombic space group P212121. Table S2† summarizes the statistics for crystallographic data collection and structural refinement. Initial phases for the MBH structures were obtained by molecular replacement using crystal structures of H2-reduced native (reduced) MBH (PDB entry 3RGW14) and as-isolated (super-oxidized) native MBH (PDB entry 4IUB17) as the initial search models for the MBH variants. Molecular replacement was achieved using the CCP4 program PHASER.57 Phasing and heavy atom search for analyzing anomalous data were performed by using the SHELX package.58 Statistics for anomalous crystallographic data collection are provided in Table S2.† The structures of the MBH variants were refined with the programs PHENIX and CCP4 program REFMAC5 using refinement strategies (inter alia real-space refinement, B-factor refinements) and simulated-annealing (slow cooling protocol, maximum likelihood target function, energy minimization) to overcome potential phase biases.55,59–61 First water molecules were depicted with the automatic search method in PHENIX.60 Manual model building and electron density interpretation were performed in COOT.62 After every round of manual model building, energy minimization and B-factor refinement were performed with REFMAC5.61,63 The final crystal structures were validated using the RCSB PDB Validation Server,64MolProbity server65 and WHAT IF server.66 Solvent accessibility was calculated in PyMOL using the program CAVER3.0.67 All molecular graphics representations were created in PyMOL.68
Covalent immobilization of MBH on electrodes was performed as described previously.69 In each case, a dispersion of multi-walled carbon nanotubes (MWNT) in DMF (16 μL of 1 mg mL−1) was placed on the surface of a PGE-disk electrode and allowed to dry overnight. The MWNT-coated PGE electrodes (geometric area 0.2–0.28 cm2) were then coated with a solution of 1-pyrenebutyric acid (Py) in DMF (10 mM) for 1 h to obtain strong π–π stacked MWNT-Py assemblies on PGE (PGE/MWNT/Py). The electrodes were rinsed in purified water and the free carboxylic groups of Py were activated for 20–40 min using a freshly prepared solution of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, 0.4 M) + N-hydroxy sulfosuccinimide (NHSS, 0.1 M) before treating with MBH variant solutions at room temperature for 60 min to make covalent attachments via the surface lysine residues (∼33 for MBH from C. necator). This procedure resulted in modified electrodes denoted PGE/MWNT/Py-MBH. The electrodes were then rinsed in purified water and used immediately or stored at room temperature in 50 mM potassium phosphate, 100 mM NaCl buffer, pH 5.5.
Protein film electrochemistry experiments were carried out using an electrochemical analyzer (Autolab PGSTAT128N) controlled by a PC operating NOVA software (EcoChemie) in an anaerobic glovebox (M. Braun) comprising a N2 atmosphere (O2 <2 ppm). The enzyme-coated electrodes were placed in enzyme-free buffered electrolyte (50 mM potassium phosphate, 100 mM NaCl, pH 5.5) so that all enzyme molecules addressed in the experiments were subjected to the same regime of strict potential control. In all experiments the electrode was rotated at a constant rate (>2500 rpm) to provide efficient supply of substrate and removal of product. The electrode rotator (EcoChemie) was placed in a specially designed, gas-tight, glass electrochemical cell incorporating a Pt wire counter electrode. A saturated calomel reference electrode (SCE) was contained in a sidearm containing 100 mM NaCl, separated from the main cell compartment by a Luggin capillary. Potentials (E) are quoted with respect to the standard hydrogen electrode (SHE) using the conversion ESHE = ESCE + 0.241 V at 25 °C.70
Experiments were performed in phosphate buffer (50 mM; Fisher Scientific) containing NaCl (100 mM; Analytical Grade, Fisher Scientific) and titrated to the desired pH at the experimental temperature. Experiments were performed under gas mixtures of H2 (Premier Grade, Air Products), Ar, and O2 (BOC gases) created using mass flow controllers (Sierra Instruments).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 ratio, with initial concentrations of 1 mM. Thereby, a mixed self-assembled monolayer (SAM) was formed on the electrode surface. Subsequently, the MBH variants were immobilized according to ref. 33 and 71. The spectro-electrochemical measurements were performed using a three-electrode setup that employed the electroless-deposited gold film as a working electrode (geometric area of 0.79 cm2), a Pt mesh as a counter electrode, and an Ag/AgCl 3 M KCl (Dri-Ref, WPI) reference electrode. A schematic representation of the homemade SEIRA spectro-electrochemical cell used in this work is depicted elsewhere.72 Electrochemical control was achieved using a Metrohm μAutolab potentiostat.
9 ratio, with initial concentrations of 1 mM. Thereby, a mixed self-assembled monolayer (SAM) was formed on the electrode surface. Subsequently, the MBH variants were immobilized according to ref. 33 and 71. The spectro-electrochemical measurements were performed using a three-electrode setup that employed the electroless-deposited gold film as a working electrode (geometric area of 0.79 cm2), a Pt mesh as a counter electrode, and an Ag/AgCl 3 M KCl (Dri-Ref, WPI) reference electrode. A schematic representation of the homemade SEIRA spectro-electrochemical cell used in this work is depicted elsewhere.72 Electrochemical control was achieved using a Metrohm μAutolab potentiostat.
Data availability
All experimental data and crystallographic analyses are available in the ESI† file and the raw data are available from the authors.Author contributions
P. S., O. L and I. Z. conceived and supervised the project. J. F. and O. L. constructed the MBH variants. S. F. and J. F. purified the MBH samples and analyzed them biochemically. A. S., J. K., S. F., J. F., and P. S. managed the MBH crystallization. A. S., J. K., and P. S. collected the X-ray and anomalous diffraction data, performed data processing and solved the MBH structures. A. S., J. K. and P. S. refined and interpreted the MBH structures. C. L. recorded and analyzed EPR data supported by C. T. IR and RR and spectro-electrochemistry data were obtained and analyzed by S. K., C. L. and I. Z. R. M. E. and F. A. A. recorded and analyzed electrochemistry data; A. S., J. K., S. F., C. L., S. K., I. Z., O. L. and P. S. wrote the paper with contributions from all co-authors.Conflicts of interest
The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to P. S. (E-mail: patrick.scheerer@charite.de; ), O. L. (oliver.lenz@tu-berlin.de; ), and I. Z. (ingo.zebger@tu-berlin.de).Acknowledgements
We are grateful to Manfred Weiss and the scientific staff of the BESSY-MX/Helmholtz Zentrum Berlin für Materialien und Energie at beamlines BL14.1, BL14.2 and BL14.3 operated by the Joint Berlin MX-Laboratory at the BESSY II electron storage ring (Berlin-Adlershof, Germany) and the scientific staff of the European Synchrotron Radiation Facility (ESRF, Grenoble) at beamlines ID30A-1-3, ID30B, ID23-1, ID23-2 and ID29, where the data were collected, for continuous support. We are indebted to Friedhelm Lendzian for preliminary EPR measurements. This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through grants CRC 1423, project number 421152132, (subproject Z03) to P. S., CRC 1078, project number 221545957, (subproject B06) to P. S., the SPP 1927 “Iron–sulfur for Life” – ZE 510/2-2 (to I. Z.) as well as LE 2934/1-2 (to O. L.) and the cluster of excellence “UniSysCat” under Germany's Excellence Strategy-EXC2008/1-390540038 to A. S., S. F., C. T., I. Z., O. L., and P. S. Research in Oxford was supported by the Biological and Biotechnological Sciences Research Council (Grants BB/I022309 and BB/L009722). We also thank the Einstein Center of Catalysis (EC2) funded by the Einstein Foundation Berlin for support. O. L. and I. Z. are also grateful for funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no 810856.References
- W. F. Martin, Biol. Direct, 2011, 6, 1–25 CrossRef.
- P. M. Vignais and B. Billoud, Chem. Rev., 2007, 107, 4206–4272 CrossRef CAS.
- W. Lubitz, H. Ogata, O. Ru and E. Reijerse, Chem. Rev., 2014, 114, 4081–4148 CrossRef CAS.
- E. Schwartz, J. Fritsch and B. Friedrich, in The Prokaryotes, ed. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson, Springer Berlin Heidelberg, 4th edn, 2013, pp. 119–199 Search PubMed.
- O. Lenz, L. Lauterbach, S. Frielingsdorf and B. Friedrich, in Biohydrogen, ed. M. Rögner, Walter de Gruyter GmbH & Co. KG, Berlin, Boston, 2015, pp. 61–88 Search PubMed.
- O. Lenz, M. Ludwig, T. Schubert, I. Bürstel, S. Ganskow, T. Goris, A. Schwarze and B. Friedrich, ChemPhysChem, 2010, 11, 1107–1119 CrossRef CAS PubMed.
- M. Bruschi, M. Tiberti, A. Guerra and L. De Gioia, J. Am. Chem. Soc., 2014, 136, 1803–1814 CrossRef CAS PubMed.
- Y. Ilina, C. Lorent, S. Katz, J. H. Jeoung, S. Shima, M. Horch, I. Zebger and H. Dobbek, Angew. Chem., Int. Ed., 2019, 58, 18710–18714 CrossRef CAS PubMed.
- T. Goris, A. F. Wait, M. Saggu, J. Fritsch, N. Heidary, M. Stein, I. Zebger, F. Lendzian, F. A. Armstrong, B. Friedrich and O. Lenz, Nat. Chem. Biol., 2011, 7, 310–318 CrossRef CAS PubMed.
- J. Kalms, A. Schmidt, S. Frielingsdorf, P. van Der Linden, D. von Stetten, O. Lenz, P. Carpentier and P. Scheerer, Angew Chem. Int. Ed. Engl., 2016, 38043, 5586–5590 CrossRef PubMed.
- J. Kalms, A. Schmidt, S. Frielingsdorf, T. Utesch, G. Gotthard, D. von Stetten, A. Royant, M. A. Mroginski, P. Carpentier, O. Lenz and P. Scheerer, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E2229–E2237 CrossRef CAS PubMed.
- M. Pandelia, W. Nitschke, P. Infossi, M.-T. Giudici-Orticoni, E. Bill and W. Lubitz, Proc. Natl. Acad. Sci. U.S.A., 2011, 1, 1–6 Search PubMed.
- Y. Shomura, K. S. Yoon, H. Nishihara and Y. Higuchi, Nature, 2011, 479, 253–256 CrossRef CAS PubMed.
- J. Fritsch, P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz and C. M. T. Spahn, Nature, 2011, 479, 249–252 CrossRef CAS PubMed.
- A. Volbeda, P. Amara, C. Darnault, J.-M. Mouesca, A. Parkin, M. M. Roessler, F. A. Armstrong and J. C. Fontecilla-Camps, Proc. Natl. Acad. Sci. U.S.A., 2012, 109, 1–6 CrossRef PubMed.
- L. Bowman, L. Flanagan, P. K. Fyfe, A. Parkin, W. N. Hunter and F. Sargent, Biochem. J., 2014, 458, 449–458 CrossRef CAS PubMed.
- S. Frielingsdorf, J. Fritsch, A. Schmidt, M. Hammer, J. Löwenstein, E. Siebert, V. Pelmenschikov, T. Jaenicke, J. Kalms, Y. Rippers, F. Lendzian, I. Zebger, C. Teutloff, M. Kaupp, R. Bittl, B. Friedrich, O. Lenz and P. Scheerer, Nat. Chem. Biol., 2014, 5, 378–385 CrossRef PubMed.
- L. Lauterbach and O. Lenz, J. Am. Chem. Soc., 2013, 135, 17897–17905 CrossRef CAS PubMed.
- P. Wulff, C. C. Day, F. Sargent and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6606–6611 CrossRef CAS PubMed.
- M. Carepo, D. L. Tierney, C. D. Brondino, T. C. Yang, A. Pamplona, J. Telser, I. Moura, J. J. G. Moura and B. M. Hoffman, J. Am. Chem. Soc., 2002, 124, 281–286 CrossRef CAS PubMed.
- J. W. van der Zwaan, J. M. Coremans, E. C. Bouwens and S. P. Albracht, Biochim. Biophys. Acta, 1990, 1041, 101–110 CrossRef CAS PubMed.
- A. Volbeda, L. Martin, E. Barbier, O. Gutierrez-Sanz, A. L. De Lacey, P.-P. Liebgott, M. Rousset and J. C. Fontecilla-Camps, J. Biol. Inorg. Chem., 2015, 20, 11–22 CrossRef CAS PubMed.
- H. Ogata, S. Hirota, A. Nakahara, H. Komori, N. Shibata, T. Kato, K. Kano and Y. Higuchi, Structure, 2005, 13, 1635–1642 CrossRef CAS.
- H. Ogata, P. Kellers and W. Lubitz, J. Mol. Biol., 2010, 402, 428–444 CrossRef CAS PubMed.
- J. C. Fontecilla-Camps, A. Volbeda, C. Cavazza and Y. Nicolet, Chem. Rev., 2007, 107, 4273–4303 CrossRef CAS.
- H. Ogata, K. Nishikawa and W. Lubitz, Nature, 2015, 520, 571–574 CrossRef CAS PubMed.
- E. Siebert, Y. Rippers, S. Frielingsdorf, J. Fritsch, A. Schmidt, J. Kalms, S. Katz, O. Lenz, P. Scheerer, L. Paasche, V. Pelmenschikov, U. Kuhlmann, M. A. Mroginski, I. Zebger and P. Hildebrandt, J. Phys. Chem. B, 2015, 119, 13785–13796 CrossRef CAS PubMed.
- A. Parkin, L. Bowman, M. M. Roessler, R. A. Davies, T. Palmer, F. A. Armstrong and F. Sargent, FEBS Lett., 2012, 586, 536–544 CrossRef CAS PubMed.
- M. Saggu, I. Zebger, M. Ludwig, O. Lenz, B. Friedrich, P. Hildebrandt and F. Lendzian, J. Biol. Chem., 2009, 284, 16264–16276 CrossRef CAS PubMed.
- M. M. Roessler, R. M. Evans, R. A. Davies, J. Harmer and F. A. Armstrong, J. Am. Chem. Soc., 2012, 134, 15581–15594 CrossRef CAS PubMed.
- M. Sensi, M. del Barrio, C. Baffert, V. Fourmond and C. Léger, Curr. Opin. Electrochem., 2017, 5, 135–145 CrossRef CAS.
- D. Millo, P. Hildebrandt, M.-E. Pandelia, W. Lubitz and I. Zebger, Angew Chem. Int. Ed. Engl., 2011, 50, 2632–2634 CrossRef CAS PubMed.
- N. Heidary, T. Utesch, M. Zerball, M. Horch, D. Millo, J. Fritsch, O. Lenz, R. von Klitzing, P. Hildebrandt, A. Fischer, M. A. Mroginski and I. Zebger, PLoS One, 2015, 10, 1–9 CrossRef PubMed.
- F. A. Armstrong, R. M. Evans and C. F. Megarity, in Fe–S Cluster Enzymes PartB, ed. S. S. David, Academic Press, 2018, vol. 599, pp. 387–407 Search PubMed.
- R. M. Evans, S. E. Beaton, P. R. Macia, Y. Pang, K. L. Wong, L. Kertess, W. K. Myers, R. Bjornsson, P. A. Ash, K. A. Vincent, S. B. Carr and F. A. Armstrong, Chem. Sci., 2023, 14, 8531–8551 RSC.
- R. M. Evans and F. A. Armstrong, in Metalloproteins, ed. J. C. Fontecilla-Camps and Y. Nicolet, Humana Press, Totowa, NJ, 2014, pp. 73–94 Search PubMed.
- M. Ludwig, J. A. Cracknell, K. A. Vincent, F. A. Armstrong and O. Lenz, J. Biol. Chem., 2009, 284, 465–477 CrossRef CAS PubMed.
- K. A. Vincent, A. Parkin, O. Lenz, S. P. J. Albracht, J. C. Fontecilla-Camps, R. Cammack, B. Friedrich and F. A. Armstrong, J. Am. Chem. Soc., 2005, 127, 18179–18189 CrossRef CAS.
- K. A. Vincent, J. A. Cracknell, O. Lenz, I. Zebger, B. Friedrich and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16951–16954 CrossRef CAS PubMed.
- M. J. Lukey, M. M. Roessler, A. Parkin, R. M. Evans, R. A. Davies, O. Lenz, B. Friedrich, F. Sargent and F. A. Armstrong, J. Am. Chem. Soc., 2011, 133, 16881–16892 CrossRef CAS PubMed.
- A. Volbeda, J. M. Mouesca, C. Darnault, M. M. Roessler, A. Parkin and F. A. Armstrong, Chem. Commun., 2018, 54, 7175–7178 RSC.
- J. Fritsch, O. Lenz and B. Friedrich, Nat. Rev. Microbiol., 2013, 11, 106–114 CrossRef CAS PubMed.
- J. Fritsch, O. Lenz and B. Friedrich, J. Bacteriol., 2011, 193, 2487–2497 CrossRef CAS PubMed.
- J. Fritsch, E. Siebert, J. Priebe, I. Zebger, F. Lendzian, C. Teutloff, B. Friedrich and O. Lenz, J. Biol. Chem., 2014, 289, 7982–7993 CrossRef CAS PubMed.
- V. Radu, S. Frielingsdorf, O. Lenz and L. J. C. Jeuken, Chem. Commun., 2016, 52, 2632–2635 RSC.
- A. Dey, F. E. Jenney Jr, M. W. W. Adams, E. Babini, Y. Takahashi, K. Fukuyama, K. O. Hodgson, B. Hedman and E. I. Solomon, Science, 2007, 318, 1464–1469 CrossRef CAS PubMed.
- K. Knuttel, K. Schneider, A. Erkens, W. Plass, A. Müller, E. Bill and A. Trautwein, Bull. Pol. Acad. Sci., 1994, 42, 495–511 Search PubMed.
- J. A. Cracknell, A. F. Wait, O. Lenz, B. Friedrich and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(49), 20681–20686 CrossRef PubMed.
- O. Lenz, A. Gleiche, A. Strack and B. Friedrich, J. Bacteriol., 2005, 187, 6590–6595 CrossRef CAS PubMed.
- L. Kleihues, O. Lenz, M. Bernhard, T. Buhrke and B. Friedrich, J. Bacteriol., 2000, 182, 2716–2724 CrossRef CAS PubMed.
- O. Lenz, L. Lauterbach and S. Frielingsdorf, in Enzymes of Energy Technology, ed. F. A. Armstrong, Academic Press, 2018, vol. 613, pp. 117–151 Search PubMed.
- U. Mueller, N. Darowski, M. R. Fuchs, R. Förster, M. Hellmig, K. S. Paithankar, S. Pühringer, M. Steffien, G. Zocher and M. S. Weiss, J. Synchrotron Radiat., 2012, 19, 442–449 CrossRef CAS PubMed.
- D. De Sanctis, A. Beteva, H. Caserotto, F. Dobias, J. Gabadinho, T. Giraud, A. Gobbo, M. Guijarro, M. Lentini, B. Lavault, T. Mairs, S. McSweeney, S. Petitdemange, V. Rey-Bakaikoa, J. Surr, P. Theveneau, G. A. Leonard and C. Mueller-Dieckmann, J. Synchrotron Radiat., 2012, 19, 455–461 CrossRef CAS PubMed.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- Collaborative Computational Project Number 4, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1994, D50, 760–763 CrossRef PubMed.
- P. Evans, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 72–82 CrossRef PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 479–485 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- P. D. Adams, V. Pavel, V. B. Chen, W. Ian, N. Echols, N. W. Moriarty, R. J. Read, D. C. Richardson, S. Jane and C. Thomas, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 213–221 CrossRef CAS PubMed.
- A. A. Vagin, R. A. Steiner, A. A. Lebedev, L. Potterton, S. McNicholas, F. Long and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2184–2195 CrossRef PubMed.
- P. Emsley and B. Lohkamp, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, D66, 486–501 CrossRef PubMed.
- M. D. Winn, M. Isupov and G. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2001, D57, 122–133 CrossRef CAS PubMed.
- H. M. Berman, T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne, K. Burkhardt, Z. Feng, G. L. Gilliland, L. Iype, S. Jain, P. Fagan, J. Marvin, D. Padilla, V. Ravichandran, B. Schneider, N. Thanki, H. Weissig, J. D. Westbrook and C. Zardecki, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 899–907 CrossRef PubMed.
- I. W. Davis, A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall, J. Snoeyink, J. S. Richardson and D. C. Richardson, Nucleic Acids Res., 2007, 35, 375–383 CrossRef PubMed.
- R. Rodriguez, G. Chinea, N. Lopez, T. Pons and G. Vriend, Bioinformatics, 1998, 14, 523–528 CrossRef CAS PubMed.
- E. Chovancova, A. Pavelka, P. Benes, O. Strnad, J. Brezovsky, B. Kozlikova, A. Gora, V. Sustr, M. Klvana, P. Medek, L. Biedermannova, J. Sochor and J. Damborsky, PLoS Comput. Biol., 2012, 8, 23–30 CrossRef PubMed.
- Version 2.5.5 Schrödinger.
- S. Krishnan and F. A. Armstrong, Chem. Sci., 2012, 3, 1015–1023 RSC.
- A. J. Bard and L. R. Faulkner, Russ. J. Electrochem., 2001, 38, 1364–1365 Search PubMed.
- N. Wisitruangsakul, O. Lenz, M. Ludwig, B. Friedrich, F. Lendzian, P. Hildebrandt and I. Zebger, Angew. Chem., Int. Ed., 2009, 48, 611–613 CrossRef CAS PubMed.
- T. G. A. A. Harris, N. Heidary, J. Kozuch, S. Frielingsdorf, O. Lenz, M.-A. Mroginski, P. Hildebrandt, I. Zebger and A. Fischer, ACS Appl. Mater. Interfaces, 2018, 10, 23380–23391 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03739h |
| ‡ These authors contributed equally to this work. |
| § Current address: UCB Pharmaceutical Manufacturing, 208 Bath Road, Slough, SL1 3WE, UK. |
| ¶ Current address: Leopoldina, Nationale Akademie der Wissenschaften, c/o ABC Business Center, Friedrichstraβe 79, 10117 Berlin, Germany. |
| This journal is © The Royal Society of Chemistry 2023 |