Open Access Article
Open Access ArticlePhoto-caged 2-butene-1,4-dial as an efficient, target-specific photo-crosslinker for covalent trapping of DNA-binding proteins†‡
Jiahui
Li
,
Zenghui
Cui
,
Chaochao
Fan
 ,
Yifei
Zhou
,
Mengtian
Ren
and
Chuanzheng
Zhou
,
Yifei
Zhou
,
Mengtian
Ren
and
Chuanzheng
Zhou
 *
*
State Key Laboratory of Elemento-Organic Chemistry, Frontiers Science Center for New Organic Matter, Department of Chemical Biology, College of Chemistry, Nankai University, Tianjin 300071, China. E-mail: chuanzheng.zhou@nankai.edu.cn
First published on 19th September 2023
Abstract
Covalent trapping of DNA-binding proteins via photo-crosslinking is an advantageous method for studying DNA–protein interactions. However, traditional photo-crosslinkers generate highly reactive intermediates that rapidly and non-selectively react with nearby functional groups, resulting in low target-capture yields and high non-target background capture. Herein, we report that photo-caged 2-butene-1,4-dial (PBDA) is an efficient photo-crosslinker for trapping DNA-binding proteins. Photo-irradiation (360 nm) of PBDA-modified DNA generates 2-butene-1,4-dial (BDA), a small, long-lived intermediate that reacts selectively with Lys residues of DNA-binding proteins, leading in minutes to stable DNA–protein crosslinks in up to 70% yield. In addition, BDA exhibits high specificity for target proteins, leading to low non-target background capture. The high photo-crosslinking yield and target specificity make PBDA a powerful tool for studying DNA–protein interactions.
Introduction
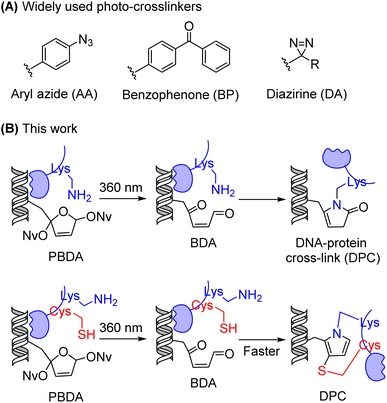
The maintenance and function of DNA in cells depend on its interactions with various proteins, including but not limited to histones,1 epigenetic writer/reader/eraser proteins,2 DNA repair enzymes,3 and transcription factors.4 DNA–protein interactions are generally non-covalent and transient, and therefore the identification of DNA-binding proteins by many of the routinely used technologies is tedious and difficult.5 Photo-crosslinkers have attracted considerable attention as a potential solution to this problem.6,7 Photo-irradiation of DNA modified with a photo-crosslinker can covalently trap DNA-binding proteins, thus making it possible to lock transient interactions with high spatiotemporal resolution in vitro and in vivo.8–12 To date, three types of photo-crosslinkers have been extensively used for this purpose: aryl azides (AA), benzophenones (BP), and diazirines (DA) (Fig. 1A).13,14 However, photo-irradiation of these compounds generates highly reactive intermediates that react non-selectively with nearby functional groups, resulting in low target-capture yields and high non-target background capture.15 Although several new photo-crosslinkers,13 including 2-aryl-5-carboxytetrazole,16,17 photo-caged quinone methide,18 and o-nitrobenzyl alcohol,19 exhibit interesting features, the development of photo-crosslinkers that can trap DNA-binding proteins with both high specificity and high yield remains a challenge.DNA-binding proteins are generally rich in Lys residues,20 and modifying DNA with Lys-reactive functionality has been reported as a new strategy for trapping DNA-binding proteins.19,21–23 We have previously demonstrated that a C4′-oxidized abasic site (C4-AP) reacts with primary amines to form stable conjugates.24–28 In the present study, we designed and synthesized an analogue of C4-AP, photo-caged 2-butene-1,4-dial (PBDA, Fig. 1B), as a new photo-crosslinker for identifying DNA-binding proteins. The irradiation (360 nm) of this crosslinker generates 2-butene-1,4-dial (BDA),29–33 a small, relatively long-lived intermediate that reacts selectively with Lys residues on DNA-binding proteins, leading to stable DNA–protein crosslinks in yields up to 70%. If there is a Cys residue close to the Lys, the Cys can participate in and speed up crosslink formation.
Results and discussion
Synthesis of PBDA
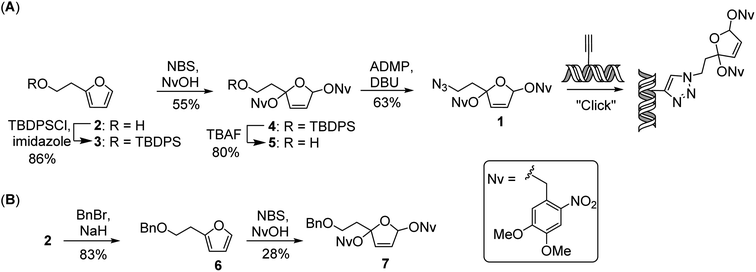
As a versatile tool for introducing the PBDA moiety into DNA, we designed and synthesized azide 1, which can be used for postsynthetic labeling of DNA via click chemistry (Scheme 1A). The synthesis of 1 started from 2-furanethanol (2). The hydroxyl group of 2 was protected as a t-butyldiphenylsilyl ether, and the resulting ether compound (3) could be treated either with 4,5-dimethoxy-2-nitrobenzyl alcohol in the presence of N-bromosuccinimide34 or with 2,3-dichloro-5,6-dicyanobenzoquinone35,36 to afford photo-caged compound 4 in 55% or 28% yield, respectively. After removal of the t-butyldiphenylsilyl protecting group, the resulting alcohol was converted to an azide by treatment with 2-azido-1,3-dimethylimidazolinium hexafluorophosphate in the presence of diazabicycloundecene to afford 1 in 63% yield. Using the same strategy, we also synthesized PBDA analogue 7 (Scheme 1B), which was used for assessing the reactivity of PBDA with model peptides.Mechanistic study of photo-reactions of PBDA analogue 7 with model peptides
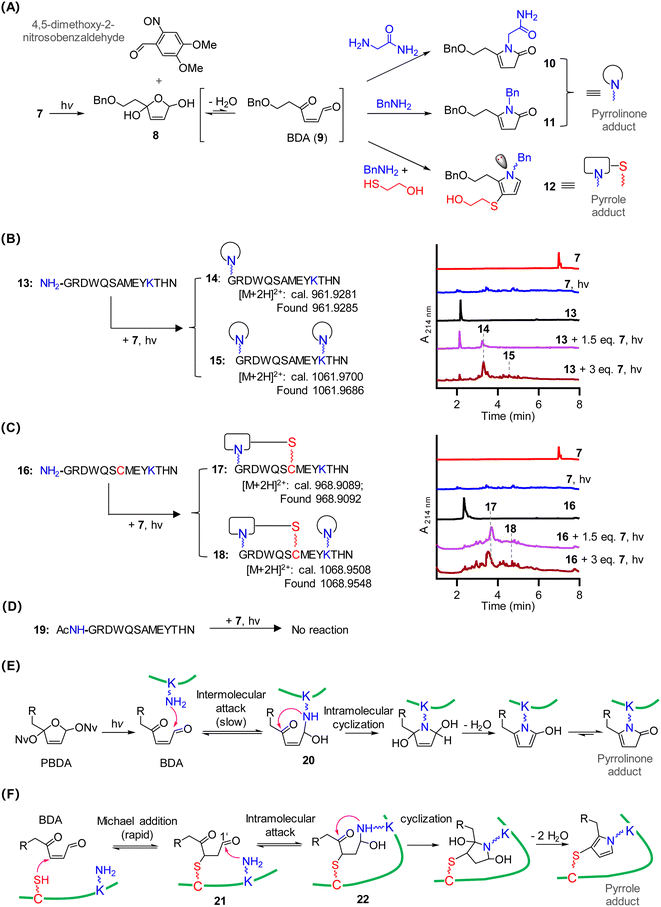
To characterize the reactivity of PBDA with amino acids, we photo-irradiated 7 at 365 nm in the presence of glycinamide, which removed the Nv protecting group to generate intermediate 8 (Fig. 2A). This intermediate could be detected by ultra-performance liquid chromatography mass spectrometry (UPLC-MS), suggesting that it was long-lived. Ring-opening of 8 in aqueous solution afforded highly electrophilic BDA 9, which reacted with glycinamide to give pyrrolinone adduct 10 as the predominant product. Similarly, photo-irradiation of 7 in the presence of BnNH2 gave pyrrolinone adduct 11. In contrast, photo-irradiation of 7 in the presence of BnNH2 and 2-mercaptoethanol yielded pyrrole adduct 12 as the predominant product (Fig. S1‡). Because the nitrogen atom in 12 is a chiral center, two stable isomers were formed (12-I and 12-II), and they could be separated into their pure forms. Pyrrolinone adduct 10 and pyrrole adducts 12-I and 12-II were unambiguously characterized by means of various one- and two-dimensional NMR spectroscopy techniques (see the ESI‡). Neither 10 nor 12-II decomposed when incubated for 12 h in neutral, acidic, or basic buffers, suggesting that both adducts were stable (Fig. S2‡). Of note, photo-irradiation of 7 generates another active electrophile, 4,5-dimethoxy-2-nitrosobenzaldehyde (Fig. 2A), which could also react with amines. However, we did not observe side products formed by the reaction of 4,5-dimethoxy-2-nitrosobenzaldehyde with glycinamide/BnNH2. Thus, BDA outcompetes 4,5-dimethoxy-2-nitrosobenzaldehyde on reacting with amines. We think two factors may account for the result: first, 4,5-dimethoxy-2-nitrosobenzaldehyde, as an aromatic aldehyde, is much less electrophilic than the aliphatic aldehyde BDA 9;37 Second, the reaction of 4,5-dimethoxy-2-nitrosobenzaldehyde with amines is reversible, affording unstable products.38 In contrast, the reaction of BDA with amines generates rather stable products.Next, we designed and carried out experiments to assess the reactivity of 7 with polypeptides 13, 16, and 19 (Fig. 2B–D). After a mixture of peptide 13 and 7 (1.5 equiv.) in phosphate buffer (pH 7.4) was irradiated and then incubated for 20 min, UPLC-MS showed that roughly 40% of peptide 13 had been transformed to a pyrrolinone monoadduct with an m/z value of 961.9285 ([M + 2H]2+, Fig. 2B). MS/MS analysis of the adduct revealed that the pyrrolinone moiety was on the N-terminal α-amino group (14, Fig. S3‡). Increasing the amount of 7 to 3 equiv. led to the formation of adduct 15, which had pyrrolinone moieties both on the ε-amino group of the Lys residue and on the N-terminal α-amino group (Fig. S4‡). These results indicate that the α-amino group was more reactive than the Lys side chain, which is consistent with previous observations that N-terminal α-amino groups outcompete the ε-amino group of Lys in reactions with electrophiles in neutral buffer.39,40
The incubation of peptide 16, which has Cys instead of Ala at position 7, with 1.5 equiv. of 7 completely transformed 16 to cyclic pyrrole adduct 17 in 20 min (Fig. 2C and S5‡). This result suggests that the formation of the Cys-involved pyrrole adduct was much faster than pyrrolinone adduct formation. Product 18 (calc m/z of [M + 2H]2+, 1068.9508; found 1068.9548), which contains both a pyrrole adduct and a pyrrolinone adduct (Fig. S6‡), was obtained when the amount of 7 was increased to 3 equiv.
No reaction occurred upon photo-irradiation of a mixture of 7 and peptide 19 which contains no Cys, Lys, and free N-terminal α-amino functional groups (Fig. 2D and S7‡). In addition, the reaction of photo-decaged 7 with a peptide containing one Cys reside but no Lys residues and no free N-terminal α-amino group also proceeded smoothly, but no stable product could be isolated, indicating that reversible crosslinks could be formed (data not shown).
On the basis of the above-described results and our previous work on peptides with a C4′-oxidized abasic site (BDA analogues),24–27 we propose the following mechanism for the reaction of PBDA with peptides. For peptides containing Lys but not Cys, the ε-amino group of Lys reacts with BDA to afford 20. Once 20 forms, intramolecular cyclization and elimination of water occur to generate a pyrrolinone adduct (Fig. 2E). For peptides containing both Cys and Lys, Michael addition of Cys to BDA to generate 21 is much more rapid than the reaction between BDA and the ε-amine group of Lys.41,42 After the formation of 21, the Lys ε-amino group intramolecularly attacks the C1′ aldehyde to generate intermediate 22. The subsequent cyclization and elimination of water afford a pyrrole adduct (Fig. 2F). For peptides with a Cys residue but no Lys residue and free N-terminal α-amino group, the adduct formed by reaction between Cys and BDA is not stable, and decomposition occurs via the retro-Michael addition reaction. Taken together, our results indicate that PBDA is a Lys-specific photo-crosslinker and that a Cys residue near Lys can speed up crosslink formation.
Intramolecular photo-crosslinking of PBDA-modified DNA with histones in nucleosome core particles
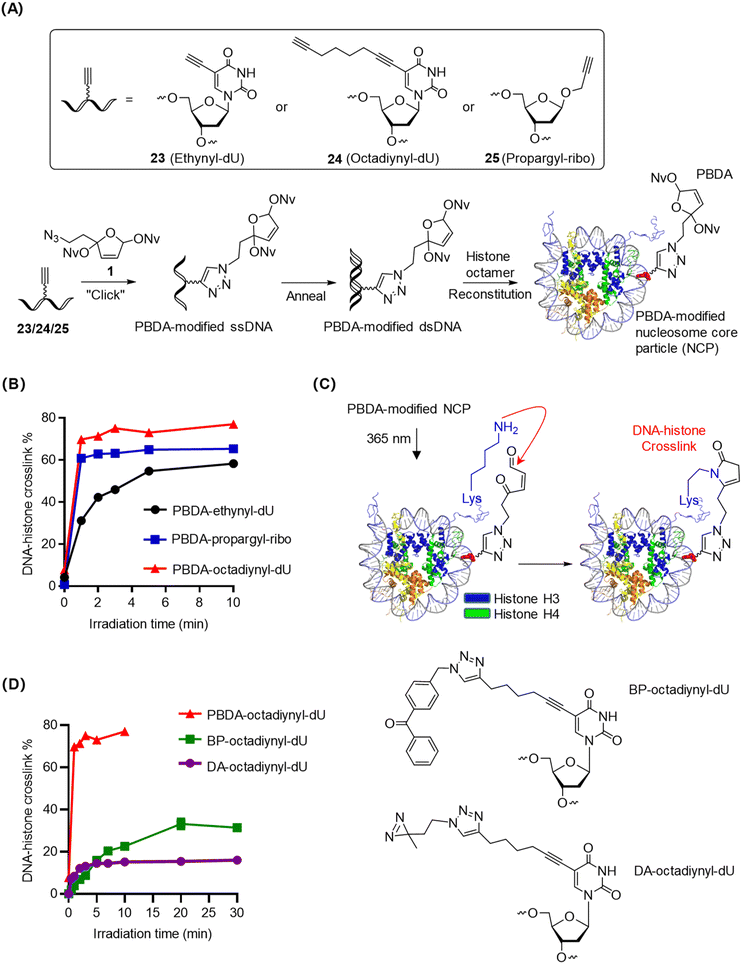
Nucleosome core particles (NCPs), the fundamental unit of chromatin, are compact and stable DNA–histone complexes.1 We prepared PBDA-modified NCPs as intramolecular models for optimizing the conditions for photo-crosslinking of PBDA with DNA-binding proteins. Specifically, single-stranded DNA with a 5-ethynyl-2′-deoxyuridine (ethynyl-dU), 5-(1,7-octadiynyl)-2′-deoxyuridine (octadiynyl-dU), or 1′-propargyl-2′-deoxyribose (propargyl-ribo) modification was coupled with PBDA (1) in the presence of Cu(I), and the obtained product was annealed with the complementary DNA strand to afford PBDA-modified double-stranded DNA (dsDNA). Finally, PBDA-modified NCPs were prepared by reconstitution of the modified dsDNA with a histone octamer (Fig. 3A, S8, and S9, Table S1‡).28,39,43PBDA-modified NCPs were irradiated for various durations, and the DNA–histone crosslink yields were quantified by SDS-PAGE (Fig. S10‡). As shown in Fig. 3B, photo-crosslinking of PBDA to histones was clearly affected by the nature of the linker between the DNA strand and the PBDA moiety. The most flexible linker (the 1,7-octadiynyl linker) gave the best results: the DNA–histone crosslink yield reached 70% within 2 min.
The flexible N-terminal tails of histones H3 and H4 are in proximity to the PBDA-octadiynyl-dU moiety on the NCPs (Fig. 3C), and these histone tails contain many Lys residues but no Cys.44 Thus, we reasoned that DNA–histone crosslinking occurred via the formation of pyrrolinone adducts between photo-decaged PBDA and Lys residues on these histone tails. After photo-crosslink formation, further incubation of the reaction mixture for up to 12 h resulted in no changes in the crosslink yields (Fig. S11‡), confirming that PBDA-modified-DNA–protein crosslinks were stable.
To compare the photo-crosslinking efficiency of PBDA with that of widely used photo-crosslinkers, we prepared benzophenones (BP)-octadiynyl-dU- and diazirine (DA)-octadiynyl-dU-modified NCPs and studied their photo-crosslinking kinetics. Photo-crosslinking of DA to histones within NCPs was rapid, yielding DNA–histone crosslinking in up to 15% yield within 3 min (Fig. 3D and S12‡). Photo-crosslinking of BP to histones occurred more slowly but gave a slightly higher crosslinking yield, reaching a plateau (30%) at 20 min (Fig. S12‡). Under the same conditions, PBDA yielded 70% DNA–histone crosslinking in 2 min. Taken together, PBDA is more rapid and efficient than classical photo-crosslinkers for the covalent trapping of DNA-binding proteins.
Intermolecular photo-crosslinking of PBDA-modified DNA with DNA-binding proteins
Because the above-described results demonstrated that PBDA-octadiynyl-dU was an excellent photo-crosslinker, rapidly yielding DNA–protein crosslinks (>70% yield) in DNA–protein complexes, we next prepared dsDNA 26 (Fig. 4A), which was modified with both PBDA-octadiynyl-dU and 5-formyl-2′-deoxycytidine (5fC), an epigenetic modification that can be recognized and removed by the base excision repair enzyme thymine DNA glycosylase (TDG, Fig. 4B).45,46 Once prepared, dsDNA 26 was used as a substrate to study intermolecular photo-crosslinking of DNA with a DNA-binding protein. Specifically, 26 was preincubated with 10 equiv. of human TDG and bovine serum albumin (BSA, 0.05 mg mL−1, Fig. S13‡), and the mixture was photo-irradiated for 7 min. Tricine-SDS-PAGE analysis of the reaction mixture showed a DNA–TDG crosslink yield of 63% (Fig. 4C, lane 4). No DNA–BSA crosslinks (Fig. 4C, lane 2 and Fig. S14‡) or DNA interstrand crosslinks (Fig. S15‡) were observed. Two pathways can account for the DNA–TDG crosslinking: (1) the formation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds between the formyl group of 5fC and Lys residues on TDG39 and (2) the reactions of photo-decaged PBDA with Lys (and Cys) on TDG. We concluded that the contribution of the former pathway was negligible, because dsDNA–TDG crosslinking was negligible in the absence of photo-irradiation (Fig. 4C, lane 3).
N bonds between the formyl group of 5fC and Lys residues on TDG39 and (2) the reactions of photo-decaged PBDA with Lys (and Cys) on TDG. We concluded that the contribution of the former pathway was negligible, because dsDNA–TDG crosslinking was negligible in the absence of photo-irradiation (Fig. 4C, lane 3).
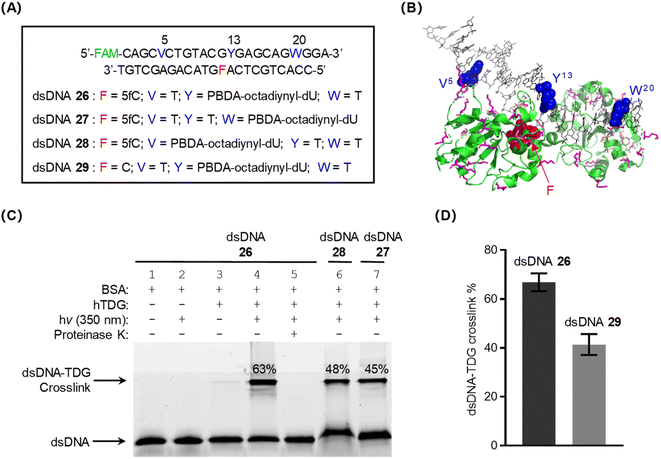 | ||
| Fig. 4 Intermolecular photo-crosslinking of PBDA-modified DNA with the DNA base excision repair enzyme hTDG. (A) PBDA-octadiynyl-dU- and 5fC-modified dsDNA used in this study. (B) Crystal structure of the dsDNA–TDG complex (PDB ID: 3UO7). The PBDA-octadiynyl-dU and 5fC modification sites are indicated by blue and red spheres, respectively. Lys residues and Cys residues are indicated by pink and orange sticks, respectively. (C) Tricine-SDS-PAGE (15.5%) analysis of photo-crosslinking of dsDNA with TDG. The gel was visualized by means of fluorescence (fluorescein, FAM) imaging. BSA, bovine serum albumin. (D) Comparison of photo-crosslink yields for dsDNA 26 and dsDNA 29. | ||
When dsDNA 27 or 28, in which the PBDA-octadiynyl-dU modification is farther from the TDG-binding region, was used as the substrate, decreased crosslink yields were observed (Fig. 4C). TDG has been shown to have less binding affinity for native dsDNA than for 5fC-modified dsDNA.46 Consistent with this fact, we found that the crosslink yield for the combination of TDG and dsDNA 29 (which does not contain 5fC) was clearly lower than that for the TDG/dsDNA 26 combination (Fig. 4D and S16‡). These results confirm that PBDA is an excellent photo-crosslinker for covalently trapping DNA-binding proteins and that it exhibits rather good specificity for target protein.
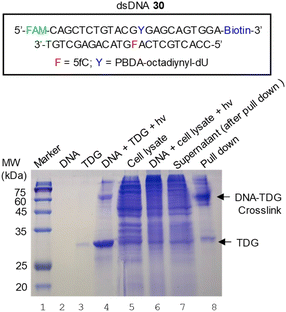
Pull down of DNA-binding proteins from cell lysate using PBDA-modified DNA
Escherichia coli cells that overexpress human TDG were harvested, lysed, and centrifuged; and the supernatant was collected for a pull-down assay. Because of serious sedimentation, the concentration of expressed human TDG in the supernatant was very close to the concentrations of the endogenous proteins of E. coli (Fig. 5, lane 5), making this supernatant an ideal sample for determining whether 5fC- and PBDA-modified dsDNA 30 could be used to pull down hTDG from a protein mixture via photo-crosslinking.Biotinylated dsDNA 30 and cell lysate were mixed and incubated at 4 °C for 2.5 h to allow for DNA–protein binding. After photo-irradiation for 7 min, the mixture was incubated at 37 °C for 30 min (Fig. 5, lane 6). Then streptavidin-coated magnetic beads were added to capture the biotinylated dsDNA. After removal of the supernatant (lane 7), the beads were successively washed with 1 M NaCl, 0.1% SDS, 6 M urea, and phosphate-buffered saline. Finally, the trapped DNA and the crosslinked DNA–protein complexes were eluted with SDS loading buffer at 70 °C and analyzed with 15% SDS-PAGE (Fig. 5, lane 8). The major product was identified as crosslinked DNA–TDG on the basis of the following observations: (1) the product co-migrated with the authentic sample of DNA–TDG in the gel (Fig. 5, lane 4); (2) the band was visible during fluorescence imaging (Fig. S17A‡), which suggests that the product contained fluorescein-labeled dsDNA 30; (3) the band was visible during anti-TDG western blot analysis (Fig. S17B‡), suggesting the presence of TDG; (4) in-gel trypsin digestion followed by MS/MS analysis revealed that hTDG was abundant in this band (Table S2‡). Of note, unexpected proteins were also identified by MS/MS. Some of them, such as human hornerin and serum albumin, could be contaminants introduced during sample handling,47,48 whereas some others could be ascribed to non-specific trapping of DNA-binding proteins.
Conclusions
In this study, we demonstrated that PBDA is an efficient, target-specific photo-crosslinker for trapping and identification of DNA-binding proteins. Compared with traditional photo-crosslinkers such as aryl azide, benzophenone, and diazirine, PBDA has three obvious advantages. First, it gives higher crosslink yields (up to 70%). Two factors may account for the high yields: (i) the intermediate generated by photo-decaging of PBDA (BDA) is less reactive than radicals, carbenes, and nitrenes, and thus its background quenching is negligible, and (ii) the small size and linearity of BDA (in situ generated by photo-irradiation of PBDA) facilitate its approach to and reaction with DNA-binding proteins. Second, the photo-reaction of PBDA with DNA-binding proteins was Lys-specific and was complete in minutes. The high residue specificity and moderate reaction rate allowed photo-decaged PBDA to selectively trap target proteins, minimizing non-target background trapping. Third, given that MS/MS analysis has become the predominant method for protein identification, the Lys-specific reactivity of PBDA makes MS/MS data analysis less complicated than when a residue-nonspecific crosslinker is employed.In addition to being abundant in nucleic-acid-binding proteins, Lys is present in almost all proteins, especially in regions with relatively high solvent accessibility.49–51 Thus, PBDA may find broad applications for investigating RNA–protein and protein–protein interactions in addition to DNA–protein interactions.
Data availability
Experimental procedures and all relevant data are available in ESI‡ and from the authors.Author contributions
J. L. and Z. C. performed the experiments. C. F., Y. Z. and M. R. provided guidance on some experimental details and contributed new reagents/analytic tools. C. Z. designed and supervised the project. The initial draft of the manuscript was written by J. Li and C. Z. All authors discussed the results and have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 22377059 and 91953115), the Natural Science Foundation of Tianjin City (no. 20JCZDJC00830), and the Frontiers Science Center for New Organic Matter, Nankai University (no. 63181206).Notes and references
- R. K. McGinty and S. Tan, Chem. Rev., 2015, 115, 2255–2273 CrossRef CAS PubMed.
- R. Ren, J. R. Horton, X. Zhang, R. M. Blumenthal and X. Cheng, Curr. Opin. Struct. Biol., 2018, 53, 88–99 CrossRef CAS PubMed.
- N. C. Bauer, A. H. Corbett and P. W. Doetsch, Nucleic Acids Res., 2015, 43, 10083–10101 CAS.
- D. Yesudhas, M. Batool, M. A. Anwar, S. Panneerselvam and S. Choi, Genes, 2017, 8, 192 CrossRef PubMed.
- M. F. Carey, C. L. Peterson and S. T. Smale, Cold Spring Harb. Protoc., 2012, 1, 18–33 Search PubMed.
- Y. Tian and Q. Lin, Chimia, 2018, 72, 758–763 CrossRef CAS PubMed.
- G. W. Preston and A. J. Wilson, Chem. Soc. Rev., 2013, 42, 3289–3301 RSC.
- J. J. Tate, J. Persinger and B. Bartholomew, Nucleic Acids Res., 1998, 26, 1421–1426 CrossRef CAS PubMed.
- U. K. Shigdel, J. L. Zhang and C. He, Angew. Chem., Int. Ed., 2008, 47, 90–93 CrossRef CAS PubMed.
- M. Winnacker, S. Breeger, R. Strasser and T. Carell, ChemBioChem, 2009, 10, 109–118 CrossRef CAS PubMed.
- Y. Liu, W. Zheng, W. Zhang, N. Chen, Y. Liu, L. Chen, X. Zhou, X. Chen, H. Zheng and X. Li, Chem. Sci., 2015, 6, 745–751 RSC.
- J. Zhang, J. Peng, Y. Huang, L. Meng, Q. Li, F. Xiong and X. Li, Angew. Chem., Int. Ed., 2020, 59, 17525–17532 CrossRef CAS PubMed.
- J. P. Holland, M. Gut, S. Klingler, R. Fay and A. Guillou, Chem.–Eur. J., 2020, 26, 33–48 CrossRef CAS PubMed.
- M. M. Hassan and O. O. Olaoye, Molecules, 2020, 25, 2285 CrossRef CAS PubMed.
- P. Kleiner, W. Heydenreuter, M. Stahl, V. S. Korotkov and S. A. Sieber, Angew. Chem., Int. Ed., 2017, 56, 1396–1401 CrossRef CAS PubMed.
- A. Herner, J. Marjanovic, T. M. Lewandowski, V. Marin, M. Patterson, L. Miesbauer, D. Ready, J. Williams, A. Vasudevan and Q. Lin, J. Am. Chem. Soc., 2016, 138, 14609–14615 CrossRef CAS PubMed.
- Y. Tian, M. P. Jacinto, Y. Zeng, Z. Yu, J. Qu, W. R. Liu and Q. Lin, J. Am. Chem. Soc., 2017, 139, 6078–6081 CrossRef CAS PubMed.
- J. Liu, L. Cai, W. Sun, R. Cheng, N. Wang, L. Jin, S. Rozovsky, I. B. Seiple and L. Wang, Angew. Chem., Int. Ed., 2019, 58, 18839–18843 CrossRef CAS PubMed.
- W. Hu, Y. Yuan, C.-H. Wang, H.-T. Tian, A.-D. Guo, H.-J. Nie, H. Hu, M. Tan, Z. Tang and X.-H. Chen, Chem, 2019, 5, 2955–2968 CAS.
- J. Zhang, Z. Ma and L. Kurgan, Briefings Bioinf., 2019, 20, 1250–1268 CrossRef CAS PubMed.
- C. B. Rosen, L. B. KodalAnne, J. S. Nielsen, D. H. Schaffert, C. Scavenius, A. H. Okholm, N. V. Voigt, J. J. Enghild, J. Kjems, T. Tørring and K. V. Gothelf, Nat. Chem., 2014, 6, 804–809 CrossRef CAS PubMed.
- V. Raindlova, R. Pohl and M. Hocek, Chem.–Eur. J., 2012, 18, 4080–4087 CrossRef CAS PubMed.
- M. Hocek, Acc. Chem. Res., 2019, 52, 1730–1737 CrossRef CAS PubMed.
- C. Zang, H. Wang, T. Li, Y. Zhang, J. Li, M. Shang, J. Du, Z. Xi and C. Zhou, Chem. Sci., 2019, 10, 8973–8980 RSC.
- Y. Zhang, X. Zhou, Y. Xie, M. M. Greenberg, Z. Xi and C. Zhou, J. Am. Chem. Soc., 2017, 139, 6146–6151 CrossRef CAS PubMed.
- C. Z. Zhou, J. T. Sczepanski and M. M. Greenberg, J. Am. Chem. Soc., 2013, 135, 5274–5277 CrossRef CAS PubMed.
- Y. Zhang, C. Zang, G. An, M. Shang, Z. Cui, G. Chen, Z. Xi and C. Zhou, Nat. Commun., 2020, 11, 1015 CrossRef CAS PubMed.
- M. Ren, M. M. Greenberg and C. Zhou, Acc. Chem. Res., 2022, 55, 1059–1073 CrossRef CAS PubMed.
- L. A. Peterson, Chem. Res. Toxicol., 2013, 26, 6–25 Search PubMed.
- J. Nunes, I. L. Martins, C. Charneira, I. P. Pogribny, A. de Conti, F. A. Beland, M. M. Marques, C. C. Jacob and A. M. M. Antunes, Toxicol. Lett., 2016, 264, 106–113 CrossRef CAS PubMed.
- L. L. G. Carrette, T. Morii and A. Madder, Bioconjugate Chem., 2013, 24, 2008–2014 CrossRef CAS PubMed.
- J. S. Jha, J. Yin, T. Haldar, Z. Yang, Y. Wang and K. S. Gates, J. Am. Chem. Soc., 2022, 144, 10471–10482 CrossRef CAS PubMed.
- A. Manicardi, E. Cadoni and A. Madder, Chem. Sci., 2020, 11, 11729–11739 RSC.
- S. Caddick, S. Khan, L. M. Frost, N. J. Smith, S. Cheung and G. Pairaudeau, Tetrahedron, 2000, 56, 8953–8958 CrossRef CAS.
- C. Gatanaga, B. Yang, Y. Inadomi, K. Usui, C. Ota, T. Katayama, H. Suemune and M. Aso, ACS Chem. Biol., 2016, 11, 2216–2221 CrossRef CAS PubMed.
- C. Paolella, D. D'Alonzo, G. Palumbo and A. Guaragna, Org. Biomol. Chem., 2013, 11, 7825–7829 RSC.
- R. Appel and H. Mayr, J. Am. Chem. Soc., 2011, 133, 8240–8251 CrossRef CAS PubMed.
- M. E. Belowich and J. F. Stoddart, Chem. Soc. Rev., 2012, 41, 2003–2024 RSC.
- F. Li, Y. Zhang, J. Bai, M. M. Greenberg, Z. Xi and C. Zhou, J. Am. Chem. Soc., 2017, 139, 10617–10620 CrossRef CAS PubMed.
- J. Bai, Y. Zhang, Z. Xi, M. M. Greenberg and C. Zhou, Chem. Res. Toxicol., 2018, 31, 1364–1372 Search PubMed.
- A. Gennari, J. Wedgwood, E. Lallana, N. Francini and N. Tirelli, Tetrahedron, 2020, 76, 131637 CrossRef CAS.
- C. Z. Zhou, J. T. Sczepanski and M. M. Greenberg, J. Am. Chem. Soc., 2012, 134, 16734–16741 CrossRef CAS PubMed.
- M. Ren, M. Shang, H. Wang, Z. Xi and C. Zhou, Nucleic Acids Res., 2021, 49, 257–268 CrossRef PubMed.
- D. Vasudevan, E. Y. D. Chua and C. A. Davey, J. Mol. Biol., 2010, 403, 1–10 CrossRef CAS PubMed.
- A. Maiti and A. C. Drohat, J. Biol. Chem., 2011, 286, 35334–35338 CrossRef CAS PubMed.
- L. Zhang, X. Lu, J. Lu, H. Liang, Q. Dai, G.-L. Xu, C. Luo, H. Jiang and C. He, Nat. Chem. Biol., 2012, 8, 328–330 CrossRef CAS PubMed.
- B. O. Keller, J. Sui, A. B. Young and R. M. Whittal, Anal. Chim. Acta, 2008, 627, 71–81 CrossRef CAS PubMed.
- K. Hodge, S. T. Have, L. Hutton and A. I. Lamond, J. Proteomics, 2013, 88, 92–103 CrossRef CAS PubMed.
- B. Dubreuil, O. Matalon and E. D. Levy, J. Mol. Biol., 2019, 431, 4978–4992 CrossRef CAS PubMed.
- E. D. Levy, J. Mol. Biol., 2010, 403, 660–670 CrossRef CAS PubMed.
- J. Warwicker, S. Charonis and R. A. Curtis, Mol. Pharm., 2014, 11, 294–303 CrossRef CAS PubMed.
Footnotes |
| † Dedication to Professor Zhen Xi on the occasion of his 60th birthday. |
| ‡ Electronic supplementary information (ESI) available: General experimental methods, spectroscopic data and supplementary figures and tables (PDF). See DOI: https://doi.org/10.1039/d3sc03719c |
| This journal is © The Royal Society of Chemistry 2023 |