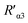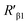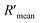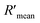Open Access Article
Open Access ArticleSingle-nucleobase resolution of a surface energy transfer nanoruler for in situ measurement of aptamer binding at the receptor subunit level in living cells†
Yu
Zhang‡
a,
Mengke
Su‡
 a,
Xingru
Fang
a,
Wenwen
Huang
a,
Hao
Jiang
a,
Qi
Li
a,
Nisar
Hussain
a,
Mao
Ye
b,
Honglin
Liu
a,
Xingru
Fang
a,
Wenwen
Huang
a,
Hao
Jiang
a,
Qi
Li
a,
Nisar
Hussain
a,
Mao
Ye
b,
Honglin
Liu
 *ab and
Weihong
Tan
*ab and
Weihong
Tan
 *bcd
*bcd
aChina Light Industry Key Laboratory of Meat Microbial Control and Utilization, School of Food and Biological Engineering, Engineering Research Center of Bio-Process, Ministry of Education, Hefei University of Technology, Hefei 230601, P. R. China. E-mail: liuhonglin@mail.ustc.edu.cn; liuhonglin@hfut.edu.cn
bMolecular Science and Biomedicine Laboratory, State Key Laboratory for Chemo/Bio-Sensing and Chemometrics, College of Material Science and Engineering, College of Chemistry and Chemical Engineering, College of Biology, Hunan University, Changsha 410082, China. E-mail: tan@hnu.edu.cn
cThe Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences, Hangzhou, Zhejiang 310022, China
dInstitute of Molecular Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, College of Chemistry and Chemical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China
First published on 23rd August 2023
Abstract
In situ identification of aptamer-binding targets on living cell membrane surfaces is of considerable interest, but a major challenge, specifically, when advancing recognition to the level of membrane receptor subunits. Here we propose a novel nanometal surface energy transfer (NSET) based nanoruler with a single-nucleobase resolution (SN-nanoruler), in which FAM-labeled aptamers and single-sized gold nanoparticle (GNP) antibody conjugates act as a donor and an acceptor. A single nucleobase resolution of the SN-nanoruler was experimentally illustrated by molecular size, orientation, quenching nature, and other dye-GNP pairs. The SN-nanoruler provides high reproducibility and precision for measuring molecule distance on living cell membranes at the nanometer level owing to only the use of single-sized antibody-capped GNPs. In situ identification of the aptamer binding site was advanced to the protein subunit level on the living cell membrane for the utilization of this SN-nanoruler. The results suggest that the proposed strategy is a solid step towards the wider application of optical-based rulers to observe the molecular structural configuration and dynamic transitions on the membrane surface of living cells.
Introduction
Cell systematic evolution of ligands by exponential enrichment (cell-SELEX) is an exciting method that enables the creation of cell-specific aptamers without prior identification of a specific target and expands the various applications of aptamers in diagnostics, drug delivery, therapeutics, etc.1 Precise identification of desired aptamer binding sites is more important and still considered the main challenging scientific issue, the solution of which will greatly promote further aptamer development in the future. To address this issue, various analytical methods are conducted. For example, chromatography2 and mass spectroscopy3,4 have evidenced that aptamer Sgc8 optimally binds to membrane protein tyrosine kinase 7,5 AS1411 recognizes nucleoli proteins,6,7 SYL3C recognizes the epithelial cell adhesion molecule,8,9 XQ-P3 binds to programmed death ligand 1, etc. Additionally, XQ-2d, a pancreatic ductal adenocarcinoma PL45 cell-binding aptamer, has been confirmed to bind the transferrin receptor protein (CD71).10,11 Interestingly, the aforementioned methods are complicated, tedious, labor-intensive, costly, and all performed in vitro and failed to accurately determine the specific protein subunit at the aptamer recognition site due to the limitations of the measuring principle. Thus, the measurement of aptamer binding sites on the surface of intact living cells in situ is necessary for understanding the exact biological functions of aptamers. There is an urgent need to develop in situ strategies for living cell systems, particularly when advancing recognition to the protein subunit level.Optical-based distance measurements at the nanoscale, referred to as “nanoruler”, are one of the key topics in cell biology, promising a potential tool for in situ determination of ligand-binding sites. The most commonly used nanoruler is Förster resonance energy transfer (FRET), as proposed by Förster et al. in 1948, but it is limited to a scale of less than 10 nm. Quenching efficiency (Φ) of FRET follows the law Φ = 1/[1 + (r/r0)6].12 Alternatively, nanometal surface energy transfer (NSET) is an energy transfer process between a nanometal surface and fluorescent dyes, as proposed by T. L. Jennings et al. in 2005. The Φ of NSET is speculated to follow the law Φ = 1/[1 + (r/r0)4].13 Similar to FRET, Φ is the energy transfer efficiency, r0 is the distance at which a specific dye displays equal probabilities for energy transfer and spontaneous emission, and r is the separation distance from the fluorophore to the surface of GNPs or from the donor dye to the receptor dye.14,15 The angstrom resolution of FRET has been shown by rigorously considering molecular distances and orientations at the single-molecule level.16 In principle, NSET can measure much longer distances than FRET13 and could become an essential nanoscale analytical tool.17 For instance, NSET has been innovatively designed to detect the conformational changes in RNA binding18 and to explore the interaction between DNA and fluorescent dyes,19,20 but the resolution limits of its distance measurements have not been addressed experimentally. Therefore, developing new models of the NSET nanoruler with a known-resolution is crucially needed to expand its potential for in situ accurate measurement on living cells, especially for the scalability and possibility of measuring more refined spatial configuration.
We herein propose a novel in situ NSET-based nanoruler with single-nucleobase resolution (hereinafter shortened to SN-nanoruler), composed of an FAM-labeled aptamer and single-sized GNP-antibody conjugates as the donor and acceptor, respectively, on the living cell membrane surface. Taking into account molecular distance, orientation, quenching nature, etc., single-nucleobase resolution of the SN-nanoruler is evidenced by both fluorescence intensity (FLI) and lifetime (FLT) experiments. By inserting an “n TA” base spacer between the dye and 5′ end of the XQ-2d aptamer, the Φ values gradually increase by approximately 6% for lengthening of each per base. Application of single-sized antibody-capped GNPs enables ultra-high reproducibility and accuracy during distance measurements. Size of antibodies here is considered by three methods: theoretical calculations, TEM and AFM measurements, respectively. The error coming from antibody measurements by the three methods causes a non-significant difference on further distance measurements, which evidences the feasibility of theoretical calculations on antibody size. High single-nucleobase resolution of the SN-nanoruler enables advancing aptamer binding site in situ identification to the protein subunit level on living cell membranes.
Results
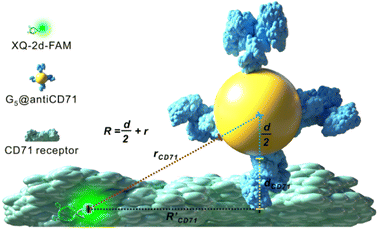
Design principle of the SN-nanoruler
The principle of the SN-nanoruler can be illustrated by selecting an XQ-2d aptamer and its target CD71 receptor as a model system on the PL45 cell membrane surface (Scheme 1). Although GNPs with a larger size could accept energy more efficiently, GNPs with a smaller size were reported to be the least intrusive for living cells.14 Also, GNPs with a diameter of 5 nm (G5NPs) have simplicity and reproducibility in both chemical synthesis and surface modification procedures.21,22 Importantly, the r0 has already been determined to be 13 nm for the G5NP and FAM dye pair with a high agreement between theoretical predictions and experimental data.15 Thus, XQ-2d labeled with FAM dye (XQ-2d-FAM) acts as an energy transfer donor, while single-sized G5NPs act as energy transfer acceptors. Notably, FAM is far from an ideal fluorophore in the living cell system; its emission might be influenced by surrounding factors. Nevertheless, more information on the NSET of GNP-FAM pairs could be referred to in many previous studies; more importantly, the emission of FAM completely overlaps with the absorption of G5NPs which is an important parameter in the NSET model.15,23,24 Hence, here FAM was selected for constructing the SN-nanoruler. As evidence, a very stable but expensive dye, Alexa488, was labelled on the 5′ end of XQ-2d and showed similar quenching behaviors in the SN-nanoruler model although its r0 value is unknown to calculate the exact distance, which will be discussed later.Accordingly, G5NPs were functionalized with saturated antiCD71 on their surface (G5@antiCD71), and the 5′ end of XQ-2d was labeled with the FAM fluorophore. After XQ-2d and antiCD71 respectively carried the FAM fluorophore and G5NPs to their binding sites on the CD71 receptor protein, the energy transfer was monitored by flow cytometric analysis on FLI and time-correlated single photon counting measurement on FLT, respectively. A single nucleobase resolution of the SN-nanoruler was experimentally illustrated by molecular size, orientation, quenching nature, and other dye-GNP pairs. Here, antibody size (dAb) was estimated by TEM, AFM and theoretical prediction which will be discussed in detail later, and was taken into account by assuming a spatial configuration of a right triangle.
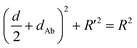 | (1) |
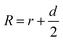 | (2) |
Importantly, the SN-nanoruler working model is the interaction process of a point dipole and surface dipole,25 where the donor (dye) is viewed as the point dipole and the acceptor (GNP) models as a collection of infinite point dipoles,26 and consequently, the equation of separation distance-dependent quenching efficiency proposed by Yun et al. is as follows:13
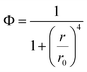 | (3) |
Eqn (3) exhibits the relationship between Φ and r. In particular, for the FAM-G5NP pair, Φ could be calculated as r0 has been determined to be 13 nm,15 which means that these parameters can be acquired according to the change in the FLI of XQ-2d-FAM before and after the binding of G5@antiCD71.
The anisotropy of ligand binding is an important parameter affecting the NSET process. A C6 chain is introduced as a spacer between the FAM dye and 5′ end of each aptamer (Table S1†). This flexible spacer offers a cone of probability for randomizing the dipole vector relative to the nanometal surface, assures an isotropic distribution of electronic dipoles, and guarantees that the measurement result is not affected by dye orientation.14
FLI-based quenching could be dynamic quenching or static quenching. So, it is necessary to find a way to examine the nature of fluorescence quenching. Here, the Stern–Volmer quenching constant (Ksv) was examined that enjoy the relationship as follows:27
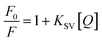 | (4) |
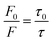 | (5) |
Single-nucleobase spatial resolution of the SN-nanoruler on living cell membranes
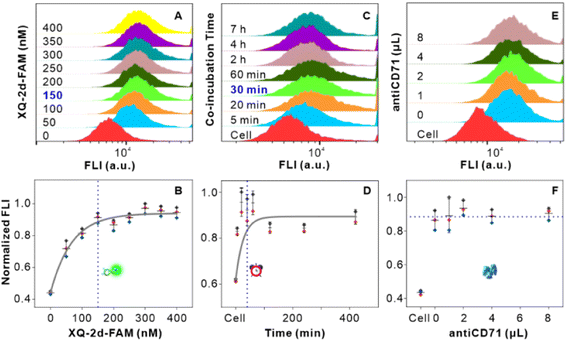
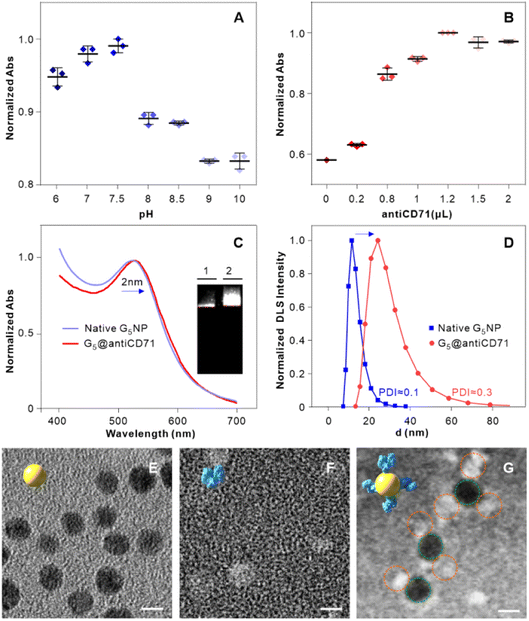
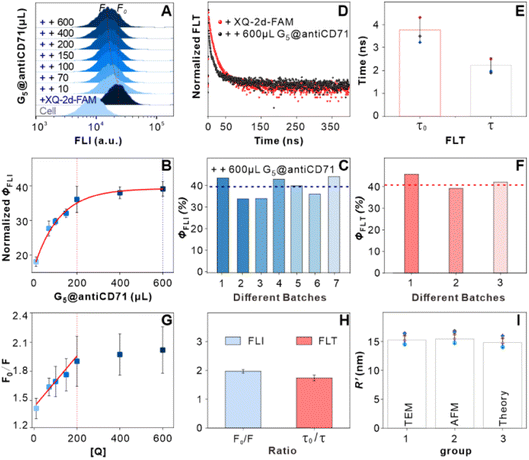
Firstly, we examined the binding features of two ligands on PL45 cells. PL45 cells were uncontaminated with high integrity of the cell nucleus as evidenced by confocal imaging (Fig. S1†). PL45 cells were treated with a series of concentrations of XQ-2d-FAM, examined by flow cytometry (FCM) (Fig. 1A and B). The result indicated that the binding saturation concentration of XQ-2d-FAM is 150 nM. Notably, although the used concentration of XQ-2d-FAM is saturated, which is much less than the self-quenching concentration,28,29 it effectively avoids the self-quenching that happened. Then, the optimal incubation time for XQ-2d-FAM was verified to be 30 min through incubating 150 nM XQ-2d-FAM with these cells for different times (Fig. 1C and D). And the calculated equilibrium dissociation constant (Kd) is of ∼11 nM, consistent with our previous report, indicating that the labeling of the XQ-2d aptamer by the FAM fluorophore does not affect its high binding affinity on the PL45 cell membrane (Fig. S2†). Also, competitive binding between the aptamer XQ-2d-FAM and the monoclonal antibody antiCD71 was further excluded by FCM (Fig. 1E and F). Increasing amounts of antiCD71 were incubated with PL45 cells for CD71 receptor protein recognition to compete with saturated XQ-2d-FAM, but no decrease in the FLI of FAM was observed, demonstrating non-competition between XQ-2d-FAM and antiCD71. Moreover, another competitive test was carried out by incubating PL45 cells first with different amounts of antiCD71 and then with saturated XQ-2d-FAM (Fig. S3A and B†). It also evidenced that XQ-2d-FAM and antiCD71 binding to two distinct sites of the CD71 receptor protein, i.e., the CD71 receptor, antiCD71 and XQ-2d-FAM could act as ideal components on the living cell membrane to examine the validity of the SN-nanoruler.Secondly, we precisely conjugated G5NPs and antiCD71. Antibody is amphoteric, pH impacts the antibody adsorbed onto GNPs.30 Thus, we explored the pH effect on conjugating G5NPs with antiCD71, which evidenced a good stability and high conjugation efficiency at pH 7.5 (Fig. 2A). Then, the saturated amounts of antiCD71 were incubated with 200 μL of G5NPs to maximize the conjugation of antibody on the GNP surface (Fig. 2B), which indicated that 1.2 μL of antiCD71 could reach saturation binding in 200 μL of the G5NP system. The successful conjugation of G5NPs with antiCD71 was evidenced by UV-VIS absorbance and agarose gel electrophoresis (Fig. 2C), as well as zeta potential analysis (Fig. S4†). Compared with native G5NPs, G5@antiCD71 had a larger diameter of hydration dynamics and the poly-dispersity index (PDI) of DLS assays increased from 0.1 to 0.3 after conjugation (Fig. 2D); nevertheless, the results demonstrated a good dispersibility of G5@antiCD71. GNPs, antibody, and aptamer are the key components in the calculation of separation distance. In the SN-nanoruler, sizes of G5NPs and antibody were considered when calculating the distance between the energy donor and receptor. TEM, AFM, and theoretical calculations were respectively used to estimate the sizes of antiCD71 and G5@antiCD71. Negative-staining TEM analysis evidenced that the diameters of G5NPs, antiCD71, and G5@antiCD71 were 5.5 ± 0.4 nm, 5.2 ± 0.2 nm, and 10.0 ± 1.0 nm, respectively (Fig. 2E–G and S5†). Notably, although the coating of antiCD71 has heterogeneity, the protein corona does not affect the properties of GNPs, such as dispersion and stability.31 Therefore, antiCD71 antibody protein corona coating on GNPs can carry the GNPs to the site on the CD71 receptor but does not affect the reliability and accuracy of this SN-nanoruler function. AFM observed that the sizes of G5NP, antiCD71 and G5@antiCD71 were 5.3 ± 0.6 nm, 4.9 ± 0.6 nm, and 9.0 ± 1.2 nm, respectively (Fig. S6A–D†). Notably, Stokes has proposed a theoretical calculation of protein sizes according to eqn (6) (ref. 32).
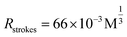 | (6) |
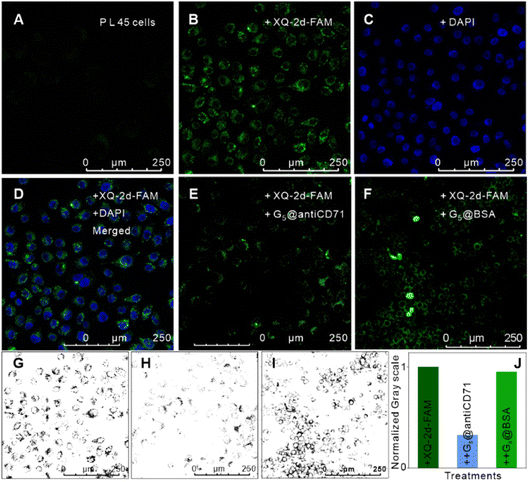
Thirdly, we measured the specificity and validity of the SN-nanoruler at the CD71 receptor protein level in living PL45 cells. We verified the incubation time of G5@antiCD71 coupling with PL45 cells. FCM analysis showed that the FLI of PL45 cells gradually dropped and reached a plateau after co-incubating with G5@antiCD71 for 3 h (Fig. S8†), which was chosen as the co-incubation time of G5@antiCD71 in the following assays. Moreover, confocal imaging visualized the NSET effect of XQ-2d-FAM on G5@antiCD71. XQ-2d-FAM clearly bound to the membrane of PL45 cells (Fig. 3A and B). The staining of the cell nucleus with 4,6-diamidino-2-phenyl indole (DAPI) dye further evidenced the membrane surface binding of XQ-2d-FAM (Fig. 3C and D). Next, a significant fluorescence quenching was observed after adding G5@antiCD71 (Fig. 3E), compared to a negative control of G5@BSA conjugates (Fig. 3F), which verified the high specific quenching from G5@antiCD71. Furthermore, the FLI of XQ-2d-FAM before and after the binding of G5@antiCD71 was quantitatively analyzed using grayscale maps (Fig. 3G–J), indicating a fluorescence quenching of about over 60%. Confocal imaging showed unavoidable large errors in FLI analysis but clearly evidenced the specificity and validity of the SN-nanoruler.
We further deduced the feasibility and specificity of the SN-nanoruler by FLI and FLT related experiments. Quantitated FLI was estimated by FCM. During the whole assay, PL45 cells had good monodispersities before and after the binding of XQ-2d-FAM and different amounts of G5@antiCD71, respectively (Fig. S9†), and the FCM charts are presented in Fig. 4A. The Φ could be written as
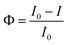 | (7) |
 was 15.22 ± 0.78 nm, 15.43 ± 0.77 nm, and 14.81 ± 0.74 nm, respectively, based on the parameters determined by TEM, AFM, and theoretical calculation, and statistical analysis showed no significant difference among the three methods for determining
was 15.22 ± 0.78 nm, 15.43 ± 0.77 nm, and 14.81 ± 0.74 nm, respectively, based on the parameters determined by TEM, AFM, and theoretical calculation, and statistical analysis showed no significant difference among the three methods for determining 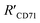 (Fig. 4I), which were all similar to the previously reported.11 Interestingly, the measurement error in
(Fig. 4I), which were all similar to the previously reported.11 Interestingly, the measurement error in 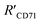 by the SN-nanoruler was about 4.29% from TEM analysis, 4.75% from AFM analysis, and 4.53% from the theoretical calculation, all smaller than previously reported errors of at least 10%.11 About 5% measurement error in R′ can be considered acceptable random fluctuations,33 so the SN-nanoruler is a relatively accurate spectral measurement tool in the field of chemo-metrics. Therefore, these results not only evidenced the specificity and validity of the SN-nanoruler, but also indicated that the antibody size could be theoretically estimated without laborious TEM or AFM experiments in the future.
by the SN-nanoruler was about 4.29% from TEM analysis, 4.75% from AFM analysis, and 4.53% from the theoretical calculation, all smaller than previously reported errors of at least 10%.11 About 5% measurement error in R′ can be considered acceptable random fluctuations,33 so the SN-nanoruler is a relatively accurate spectral measurement tool in the field of chemo-metrics. Therefore, these results not only evidenced the specificity and validity of the SN-nanoruler, but also indicated that the antibody size could be theoretically estimated without laborious TEM or AFM experiments in the future.
A nanoscale ruler with high spatial resolution is essential in materials engineering, genetics, biochemistry, and medicine,34 while the spatial resolution of the NSET effect has not been experimentally explored before. Here the SN-nanoruler model enables its spatial resolution to be examined by lengthening the “n TA” spacer between the 5′ end of XQ-2d and FAM dye, i.e., XQ-2d-n TA-FAM where n is the number of TA bases (hereinafter shortened to 0 TA, 1 TA, 2 TA, and 3 TA, Fig. 5A). Importantly, the 0 TA, 1 TA, 2 TA, and 3 TA binding to PL45 cells generated similar fluorescence features (Fig. 5B), and their Kd values were all lower than 15 nM (Fig. S10A–F†), indicating a strong binding affinity of 0 TA, 1 TA, 2 TA, and 3 TA to PL45 cells, and the slight difference did not affect the FLI under saturation binding. Previous reports also found that terminal modification did not alter the binding characteristics of the aptamer.35,36
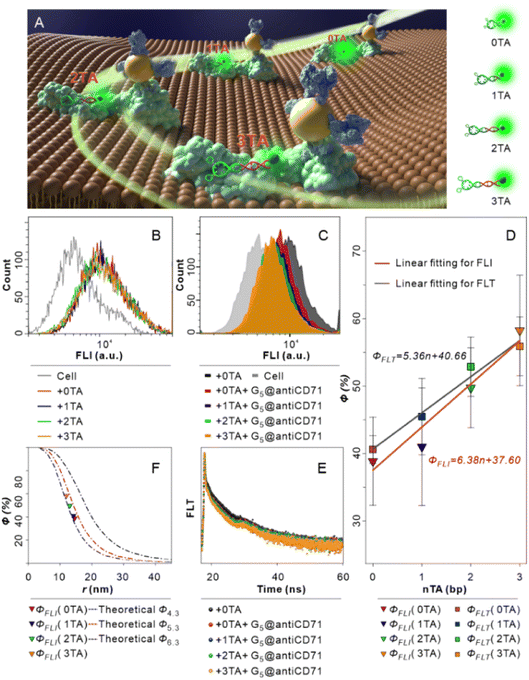 | ||
| Fig. 5 SN-nanoruler with a single-nucleobase resolution identified by FLI and FLT experiments. (A) Single-nucleobase resolution model by lengthening the “n TA” spacer, i.e., one TA base pair at a time, between the 5′ end of XQ-2d and FAM, where n is the number of TA base pairs. (B) FCM analysis of native PL45 cells (cell), cells incubated with saturated XQ-2d-FAM (0 TA), XQ-2d-1TA-FAM (1 TA), XQ-2d-2TA-FAM (2 TA), and XQ-2d-3TA-FAM (3 TA), respectively. (C) FCM analysis of cells, cells incubated with saturated 0 TA, and cells incubated with saturated n TA and then saturated G5@antiCD71, respectively. (D) Linear fitting relationship between the number of TA and ΦFLI (red line) and ΦFLT (black line), respectively, calculated according to the change in FAM FLI and FLT. (E) FLT analysis of cells incubated with saturated 0 TA and cells incubated with saturated n TA and then co-incubated with excess saturated G5@antiCD71, respectively. (F) Comparisons of our experimental data with the theoretical simulation curves. Theoretical curves show the r-dependent Φ of GNPs with diameters of 4.3 nm (blue line), 5.3 nm (red line), and 6.6 nm (black line), respectively, adopted from the literature.15 The color triangles indicate experimental Φ values calculated from FLI. | ||
The change in FLI was monitored after insertion of 1, 2, and 3 TA bases. In detail, PL45 cells were incubated with saturated 0 TA, 1 TA, 2 TA, and 3 TA, subsequently co-incubated with G5@antiCD71, washed, then re-suspended with binding buffer, and detected by FCM. The ΦFLI of the 0 TA group is approximately 39%, the same as the results in the above section. Then, the ΦFLI gradually increased to 41%, 50%, and 58% for 1 TA, 2 TA, and 3 TA groups, respectively (Fig. 5C). Clearly, the insertion of the “n TA” spacer decreased the separation distance between the FAM fluorophore and the metal surface, n TA-dependent ΦFLI fitting showed a good linear relationship, i.e., ΦFLI = 6.38n + 37.60, and the slope of linear fitting is 6.38% (red line in Fig. 5D), indicating that the ΦFLI value increased by 6.38% when lengthening every TA base pair.
Similarly, the change in the FLT was further monitored after the insertion of 1, 2, and 3 TA base pairs (Fig. 5E). By co-incubation with the saturated G5@antiCD71, the mean FLT of 0 TA, 1 TA, 2 TA, and 3 TA groups gradually dropped, from 3.77 to 2.23, 2.04, 1.76, and 1.65 ns, respectively (Fig. S11†). Correspondingly, the ΦFLT gradually increased from 41% that is similar to the result in the above section, to 46%, 53%, and 56%, respectively, following an “n TA”-dependent linear relationship, i.e., ΦFLT = 5.39n + 40.66 (black line in Fig. 5D), which is very similar to the results of FLI. There is a slight difference in the FLI and FLT results which may originate from operation and/or different instrumentation.
Three theoretical r-dependent Φ curves of GNPs with diameters of 4.3 nm (blue line), 5.3 nm (red line), and 6.6 nm (black line) adopted from the literature15 were plotted and compared with the experimental data corresponding to different “n TA” spacers (color triangle), and these data were overall located between the G4.3NP and G5.3NP theoretical curves (Fig. 5F), although ΦFLI (0 TA) with a value of 39% has a slight larger deviation. Actually, NSET has the highest sensitivity when the Φ is around 50%. When hypothesizing that r is nearly the value of r0 with a slight variation of Δr, Φ will be around 50%, and r can be written as
| r = r0 + Δr | (8) |
According to eqn (3),
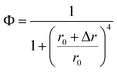 | (9) |
If Δr is considered as the change of r when lengthening every TA base pair, the insertion of 0, 1, 2 and 3 TA base pairs exactly induced the changes of Φ around 50%, and the corresponding r is 14.56 nm, 14.24 nm, 13.03 nm, and 11.96 nm, respectively, with a mean Δr of ∼0.87 nm. As reported, the pitch of the double DNA helix is 0.34 nm per base pair because of the double chain rigidity, while the extension of single-stranded DNA on the GNP surface is reported to be 0.15 nm per base because the orientation of ssDNA was more flexible compared to that of dsDNA.37 The length per base for oligonucleotides was estimated to be about 0.43 nm,38 while the calculated Δr is larger than the reported extension length per base pair.
Two factors may contribute to the above deviation: one factor may be the inaccurate estimation of previously reported r0, especially in the living cell system. Ignoring higher order terms, eqn (9) can be approximately written as
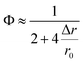 | (10) |
The reconfigured equation is as follows:
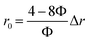 | (11) |
Obviously, r0 is directly proportional to Δr when Φ is a known value around 50%. Actually, the r0 could be affected by many factors as proposed by C. J. Breshike et al.15
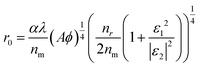 | (12) |
The other factor is the Φ(r) power law relationship. As discussed in the above section, when Φ = 1/[1 + (r/r0)4] is applied, the donor is seen as a point dipole while the GNP is seen as an infinite surface and a film mirror.14 C. J. Breshike et al. still thought that Φ and r exhibited a four-power law behaviour after redefining r0 according to the complex dielectric function (ε) and absorptivity (A).15 In fact, the FAM-labeled aptamer is not a perfect point dipole, and the G5NP acceptor is neither an infinite surface nor a perfect mirror, but a sphere with a curved surface and size only a few times larger than the dye. Thus, the four-power law of Φ(r) relationship may overestimate the interaction distance between the G5NP acceptor and the FAM dye donor. Nevertheless, all of the above data evidenced the capability of the SN-nanoruler for distinguishing single-nucleobase resolution measurement, even under the conditions of the living cell environment and the fluidity of the cell membrane.
Just in case, other dye-GNP pairs were considered. Alexa488, a quite stable dye commonly used for in situ analysis, was further used as the energy donor to verify the resolution of the SN-nanoruler. As shown in Fig. S12A,† the insertion of the “n TA” spacer did not alter the binding affinity of XQ-2d-Alexa488 to PL45 cells. After the saturated binding of XQ-2d-nTA-Alexa488, PL45 cells showed a similar FLI (Fig. S12B†). Subsequently, when co-incubated with saturated G5@antiCD71, the insertion of the “n TA” spacer caused the ΦFLI to increase from 34% to 40%, 53%, and 55%, respectively (Fig. S12C and D†), following an “n TA”-dependent linear relationship, i.e., ΦFLI = 7.56n + 33.82. Obviously, the quenching feature curve of Alexa488 dye on G5NPs is different from that of the FAM dye, i.e., the ΦFLI value increased approximately 7.56% when lengthening every TA base pair. So far, the r0 value of Alexa488-G5NP is unknown, but could be deduced from the n TA-dependent Φ values if assuming that the base-pair lengthening exactly equals the distance separating. It may pave a new way to determine the unknown r0 value of other dye-GNP pairs, which will be discussed in our on-going work. High precision, i.e., single-nucleobase spatial resolution of this SN-nanoruler exhibited high potential for in situ identification of aptamer-binding sites and advanced the molecular distance measurement further to the level of protein subunit domains in the living cell system.
In situ measurement of aptamer binding sites at the protein subunit level in living cells
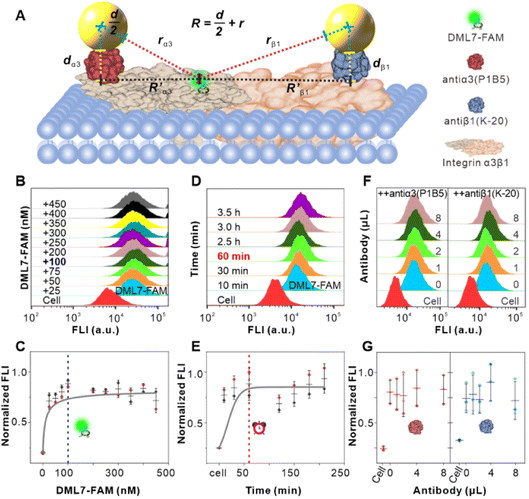
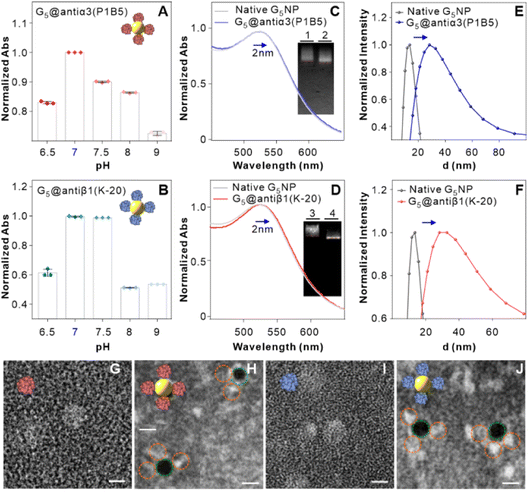
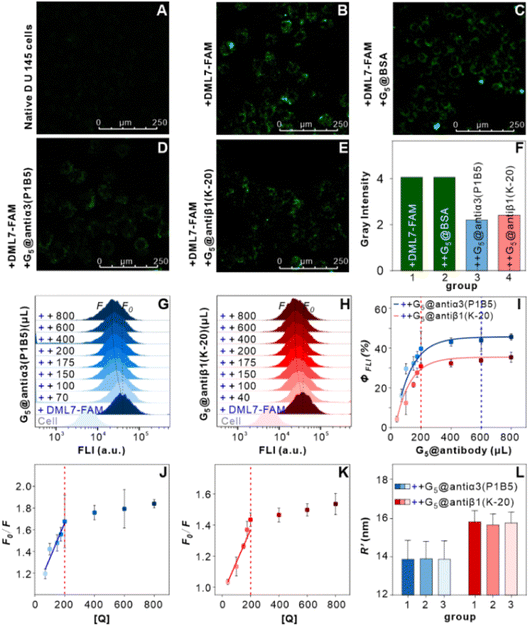
The above results of high single-nucleobase resolution inspired us to advance the in situ identification of aptamer binding sites to a much deeper level, e.g., protein subunits, in living cells. Recently, our group selected new aptamer DML7 that binds to DU145 cells.39 Preliminary data of SDS-PAGE, as well as pull-down and confocal analysis, hinted that its potential target might be integrin α3β1,40 a transmembrane heterodimer protein consisting of the α subunit domain and the β subunit domain.41 However, further distinguishing the protein subunit is very challenging with current technology. In this context, the integrin α3β1 receptor protein, antiα3 (P1B5) antibody, antiβ1 (K-20) antibody, and DML7-FAM aptamer were used for conducting the SN-nanoruler to in situ measure the separation distance on the DU145 cell membrane (Fig. 6A). It may provide a clue to the probable binding subunit of integrin α3β1. Notably, we first determined the saturation concentration of DML7-FAM binding to DU145 cells. DU145 cells were cultivated and shown to have good integrity of cell morphology and the nucleus by confocal microscopy (Fig. S13†). DU145 cells were then treated with different concentrations of DML7-FAM, followed by washing with washing buffer, suspending in binding buffer, and testing through FCM. The results indicated that the saturation concentration of DML7-FAM binding to DU145 cells is 100 nM (Fig. 6B and C). Similar to the above studies, we further determined the optimal time of DML7-FAM incubation with DU145 cells. DU145 cells were co-incubated with 100 nM DML7-FAM at different times. The saturation of FLI suggested that the optimal time for DML7-FAM binding with DU145 cells is 60 min (Fig. 6D and E).An important foundation of the SN-nanoruler is the non-competitive binding between two ligands when observing the distance between their binding sites. For this purpose, competition assays between the DML7-FAM aptamer and antiα3 (P1B5) or antiβ1 (K-20) antibody were respectively monitored by FCM. After saturation binding with the saturated DML7-FAM aptamer, DU145 cells were continuously incubated with various amounts of unlabeled antiα3 (P1B5) antibodies. It showed no obvious decline of FLI (left panel of Fig. 6F and G), demonstrating no competition between DML7-FAM and antiα3 (P1B5) antibody. Similarly, the results showed no competition between DML7-FAM and antiβ1 (K-20) antibody (right panel of Fig. 6F and G). To further exclude the possibility of co-recognition between DML7-FAM and antiα3 (P1B5) or antiβ1 (K-20) antibodies, DU145 is first incubated with antibodies and then with the aptamer, i.e., changing the incubation order of DML7-FAM and antibodies (Fig. S14A–D†). The incubation sequence did not affect the FLI of DML7-FAM bound to DU145 cells, indicating that DML7-FAM and the two antibodies have distinctly different binding sites on the DU145 cell membrane. Moreover, the competitive assay was examined by simultaneously adding DML7-FAM and each antibody to the cell system (Fig. S15A–D†). All these results indicated no competition between DML7-FAM and each antibody, further excluding nonspecific interference between different ligands. Similarly, the optimal pH value is near 7.0 for the coupling of G5@antiα3 (P1B5) (Fig. 7A) and G5@antiβ1 (K-20) (Fig. 7B). The UV-VIS absorbance spectroscopy, agarose gel electrophoresis (Fig. 7C and D), and zeta potential (Fig. S16A and B†) results evidenced the successful coupling of each G5@antibody. Furthermore, 0.2 μL of antiα3 (P1B5) and 0.4 μL of antiβ1 (K-20) were determined to be excess in 200 μL of G5NP solution to make a saturated conjugation (Fig. S17A and B†), and the DLS results demonstrated a good dispersibility of G5@antiα3 (P1B5) and G5@antiβ1 (K-20), respectively (Fig. 7E and F).
In particular, negative-staining TEM images confirmed that native antiα3 (P1B5), G5@antiα3 (P1B5), native antiβ1 (K-20), and G5@antiβ1 (K-20) have sizes of 7.1 ± 0.8 nm, 11.9 ± 1.2 nm, 6.8 ± 1.1 nm, and 12.5 ± 0.2 nm (Fig. 7G–J), respectively, and AFM images evidenced their sizes of 6.9 ± 0.6 nm, 12.9 ± 1.9 nm, 6.6 ± 0.8 nm, and 13.0 ± 2.4 nm, respectively (Fig. S18A–F†). According to eqn (6), the theoretical sizes of antiα3 (P1B5) and antiβ1 (K-20) with an M of 150 KD and 138 KD were 7.0 nm and 6.8 nm, respectively. Related sizes of native antibody and G5@antibody and the R′ value are summarized in Table 1. Statistical analysis showed no obvious difference among the measurement results by these three measurement methods (Fig. S19A and B†).
Confocal imaging first visualized the variation of NSET efficiency between DML7-FAM and different G5NP-antibody conjugates. Fig. 8A–C illustrate the binding of DML7-FAM on the surface of DU145 cells, but a significant fluorescence quenching of DML7-FAM was observed when adding saturated G5@antiα3 (P1B5) (Fig. 8D) or G5@antiβ1 (K-20) (Fig. 8E). G5@BSA conjugates as the negative control did not alter the FLI of DML7-FAM on DU145 cells which clearly excluded the nonspecific effects of G5@antibodies (Fig. 8F). Next, FCM was performed to quantitatively determine the initial FLI (I0) of the saturated DML7-FAM aptamer, quenched FLI(I) in the presence of saturated G5@antiα3 (P1B5) or G5@antiβ1 (K-20), and the background FLI (blank) of native DU145 cells. The population of the DU145 cells incubated with different ligands all had good monodispersities (Fig. S20A–D†). Next, the quenching effects of DML7-FAM by different amounts of G5@antiα3 (P1B5) or G5@antiβ1 (K-20) are exhibited in Fig. 8G and H, respectively. A plateau was reached after adding over 200 μL of G5@antiα3 (P1B5) (blue line) or G5@β1 (K-20) (red line) (Fig. 8I). Accordingly, the stable maximum of Φ values was produced to be nearly 44% by G5@antiα3 (P1B5) and 34% by G5@β1 (K-20).
According to eqn (4), F0/F follows a [Q]-dependent linear relationship for both G5@antiα3 (P1B5) and G5@β1 (K-20), rather than a rising curve bent to the y-axis (Fig. 8J and K), which indicated that the quenching from FLI is dynamic quenching. According to the parameters obtained from TEM, AFM, and theoretical calculations (Table 1), the calculated 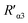 value was 14.09 ± 1.54 nm, 14.12 ± 1.45 nm, and 14.10 ± 1.49 nm, respectively, at a stable maximum of Φ in the presence of saturated G5@antiα3 (P1B5) (blue bars, Fig. 8L). Similarly, the calculated
value was 14.09 ± 1.54 nm, 14.12 ± 1.45 nm, and 14.10 ± 1.49 nm, respectively, at a stable maximum of Φ in the presence of saturated G5@antiα3 (P1B5) (blue bars, Fig. 8L). Similarly, the calculated 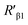 value was 16.13 ± 0.72 nm, 16.00 ± 0.69 nm, and 16.08 ± 0.69 nm, respectively, at a stable maximum of Φ in the presence of saturated G5@β1 (K-20) (red bars, Fig. 8L). Statistical analysis exhibited no significant difference among the three methods, i.e., TEM, AFM, and theoretical calculations, when calculating R′ (Fig. S21A and B†). Hence, the data obtained from the three methods were averaged, and the averaged values of
value was 16.13 ± 0.72 nm, 16.00 ± 0.69 nm, and 16.08 ± 0.69 nm, respectively, at a stable maximum of Φ in the presence of saturated G5@β1 (K-20) (red bars, Fig. 8L). Statistical analysis exhibited no significant difference among the three methods, i.e., TEM, AFM, and theoretical calculations, when calculating R′ (Fig. S21A and B†). Hence, the data obtained from the three methods were averaged, and the averaged values of 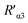 and
and 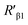 were 14.1 nm and 16.1 nm, in which the difference was greater than the resolution of the SN-nanoruler and thus had high reliability, i.e., DML7-FAM is much closer to G5@antiα3 (P1B5) rather than G5@antiβ1 (K-20).
were 14.1 nm and 16.1 nm, in which the difference was greater than the resolution of the SN-nanoruler and thus had high reliability, i.e., DML7-FAM is much closer to G5@antiα3 (P1B5) rather than G5@antiβ1 (K-20).
The measurement results of the SN-nanoruler strongly implied integrin α3β1 acting as a target of the DML7 aptamer and further hints that the binding site of the DML7 aptamer is closer to the α3 subunit domain, rather than to the β1 subunit domain. However, the SN-nanoruler may not accurately pinpoint the binding site of the aptamer, because the working model is highly related to selection of the antibody, and the selection is usually random experimentally. With this in mind, another antibody of the integrin α3 subunit, antiα3 (VM-2), was examined for competitive binding with DML7-FAM. Coincidentally, a significant competition was observed between DML7-FAM and native antiα3 (VM-2) (Fig. S22A and B†). This result not only provided full proof that the integrin α3 subunit domain precisely acted as the binding target of DML7-FAM, but also evidenced that the binding site of the DML7 aptamer overlapped with that of antiα3(VM-2) on the integrin α3 subunit domain.
Discussion
Our study developed a SN-nanoruler with a single-nucleobase resolution by virtue of a dye-labeled aptamer and single-sized GNP-antibody complex as probes to deduce the molecular distance between two binding ligands at the CD71 protein subunit domain level on living cell membranes. By lengthening the “n TA” spacer, i.e., one TA base at a time, between the end of the aptamer and dye, the single-nucleobase resolution of the SN-nanoruler was verified. The SN-nanoruler possessed a high precision, thus exhibiting high potential for in situ identification of aptamer-binding sites in living systems, and further advanced the molecular distance measurement down to the receptor protein subunit domain level in living cells. The SN-nanoruler can be a powerful complement or/and replacement to traditional energy transfer nanorulers for dynamically changing molecular event measurement at the high spatial resolution scale in biochemical fields and takes a step forward toward much wider applications of optical-based rulers in observing molecular structure transformation in living systems. For example, the SN-nanoruler is expected to in situ project each base of an aptamer on a specific plane, e.g., the GNP surface, in living systems, from the perspective of molecular distance, that enjoy great significance for establishing sensitive, reliable, and convenient aptamer-based diagnostic methods.Also, our study inspires several key issues that could be explored in the future. (1) The choice of fluorescent dyes is still very limited because of the absence of r0 values for different dyes and GNP pairs. There is an urgent need to develop novel strategies to easily determine more dyes and GNP pairs' r0 values, which would carry significance in biochemical sensing. The single-nucleobase resolution-based SN-nanoruler will pave a new way to determine the unknown r0 value of different GNP-dye pairs in living cell systems. (2) The R′ value is defined as the distance between the FAM fluorophore and antibody binding sites, and the fluorophore is labeled on the 5′ end of the aptamer. On the molecular scale, the spatial location of the fluorophore should not be identical to that of the aptamer site with specific three-dimensional scaffolds. However, a dye-labelled aptamer is considered a point dipole, which enables the fluorophore location to be the same as the aptamer site experimentally in this paper. Notably, 3′-end modifications of fluorophores in aptamers might produce different results, which could be deduced using the single-nucleobase resolution-based SN-nanoruler as well. (3) Application of the NSET effect is focused on monitoring the ensemble state of the aptamer these days. However, imaging of the transient state, e.g., single events on the living cell membrane, will expand our understanding of how a cell finely regulates molecular events on its membrane.42,43 Observing dynamically changing molecular events related to bio/chem-molecules and the transitions within the living biological system is significant to further understanding life processes and diagnosis of disease. In the future, the SN-nanoruler would be a nice spectral measurement tool in the chemo-metrics field, which could be applied for in situ monitoring of single event dynamic transitions, bimolecular structure changes, aptamer-protein or protein–protein interaction, cell–cell communication and the “on/off” state of an ion channel.
Data availability
Details of methods, experimental procedures, and characterization data are available in the ESI.† Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Honglin Liu (E-mail: liuhonglin@mail.ustc.edu.cn).Author contributions
Yu Zhang: investigation, conceptualization, methodology, formal analysis, data curation, visualization, writing – original draft, review, and editing. Mengke Su: formal analysis, data curation, writing – review and editing. Xingru Fang: investigation. Wenwen Huang: investigation. Hao Jiang: investigation. Qi Li: investigation. Nisar Hussain: writing – review & editing. Mao Ye: writing – review, editing and supervision. Honglin Liu: conceptualization, methodology, formal analysis, data curation, resources, writing – review, editing and supervision. Weihong Tan: writing – review, editing and supervision.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was funded by the National Natural Science Foundation of China (22274034, 21922403, and 21874034), the Key Research and Development Project of Anhui Province (202104a07020013), the Natural Science Research Project of Anhui Province (2108085QB84) and the Fundamental Research Funds for the Central Universities (JZ2022HGPB0307 and JZ2023HGTB0230).References
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Aptamers evolved from live cells as effective molecular probes for cancer study, Proc. Natl. Acad. Sci. U.S.A., 2006, 103(32), 11838–11843, DOI:10.1073/pnas.0602615103.
- A. Drabik, J. Ner-Kluza, P. Mielczarek, L. Civit, G. Mayer and J. Silberring, Advances in the Study of Aptamer-Protein Target Identification Using the Chromatographic Approach, J. Proteome Res., 2018, 17(6), 2174–2181, DOI:10.1021/acs.jproteome.8b00122.
- D. Shangguan, Z. Cao, L. Meng, P. Mallikaratchy, K. Sefah, H. Wang, Y. Li and W. Tan, Cell-specific aptamer probes for membrane protein elucidation in cancer cells, J. Proteome Res., 2008, 7(5), 2133–2139, DOI:10.1021/pr700894d.
- J. Tan, M. Zhao, J. Wang, Z. Li, L. Liang, L. Zhang, Q. Yuan and W. Tan, Regulation of Protein Activity and Cellular Functions Mediated by Molecularly Evolved Nucleic Acids, Angew Chem. Int. Ed. Engl., 2019, 58(6), 1621–1625, DOI:10.1002/anie.201809010.
- X. Miao, Z. Li, A. Zhu, Z. Feng, J. Tian and X. Peng, Ultrasensitive electrochemical detection of protein tyrosine kinase-7 by gold nanoparticles and methylene blue assisted signal amplification, Biosens. Bioelectron., 2016, 83, 39–44, DOI:10.1016/j.bios.2016.04.032.
- J. Guo, X. Gao, L. Su, H. Xia, G. Gu, Z. Pang, X. Jiang, L. Yao, J. Chen and H. Chen, Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery, Biomaterials, 2011, 32(31), 8010–8020, DOI:10.1016/j.biomaterials.2011.07.004.
- R. Yazdian-Robati, P. Bayat, F. Oroojalian, M. Zargari, M. Ramezani, S. M. Taghdisi and K. Abnous, Therapeutic applications of AS1411 aptamer, an update review, Int. J. Biol. Macromol., 2020, 155, 1420–1431, DOI:10.1016/j.ijbiomac.2019.11.118.
- T. Zheng, Q. Zhang, S. Feng, J. J. Zhu, Q. Wang and H. Wang, Robust nonenzymatic hybrid nanoelectrocatalysts for signal amplification toward ultrasensitive electrochemical cytosensing, J. Am. Chem. Soc., 2014, 136(6), 2288–2291, DOI:10.1021/ja500169y.
- Y. Song, Z. Zhu, Y. An, W. Zhang, H. Zhang, D. Liu, C. Yu, W. Duan and C. J. Yang, Selection of DNA Aptamers against Epithelial Cell Adhesion Molecule for Cancer Cell Imaging and Circulating Tumor Cell Capture, Anal. Chem., 2013, 85(8), 4141–4149, DOI:10.1021/ac400366b.
- X. Wu, Z. Zhao, H. Bai, T. Fu, C. Yang, X. Hu, Q. Liu, C. Champanhac, I. T. Teng and M. Ye, et al., DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition, Theranostics, 2015, 5(9), 985–994, DOI:10.7150/thno.11938.
- X. Wu, H. Liu, D. Han, B. Peng, H. Zhang, L. Zhang, J. Li, J. Liu, C. Cui and S. Fang, et al., Elucidation and Structural Modeling of CD71 as a Molecular Target for Cell-Specific Aptamer Binding, J. Am. Chem. Soc., 2019, 141(27), 10760–10769, DOI:10.1021/jacs.9b03720.
- T. Förster, Zwischenmolekulare energiewanderung und fluoreszenz, Ann. Phys., 1948, 437, 55–75, DOI:10.1002/andp.19484370105.
- C. S. Yun, A. Javier, T. Jennings, M. Fisher, S. Hira, S. Peterson, B. Hopkins, N. O. Reich and G. F. Strouse, Nanometal surface energy transfer in optical rulers, breaking the FRET barrier, J. Am. Chem. Soc., 2005, 127(9), 3115–3119, DOI:10.1021/ja043940i.
- T. L. Jennings, M. P. Singh and G. F. Strouse, Fluorescent lifetime quenching near d = 1.5 nm gold nanoparticles: probing NSET validity, J. Am. Chem. Soc., 2006, 128(16), 5462–5467, DOI:10.1021/ja0583665.
- C. J. Breshike, R. A. Riskowski and G. F. Strouse, Leaving Förster Resonance Energy Transfer Behind: Nanometal Surface Energy Transfer Predicts the Size-Enhanced Energy Coupling between a Metal Nanoparticle and an Emitting Dipole, J. Phys. Chem. C, 2013, 117(45), 23942–23949, DOI:10.1021/jp407259r.
- A. Gansen, A. Valeri, F. Hauger, S. Felekyan, S. Kalinin, K. Toth, J. Langowski and C. A. M. Seidel, Nucleosome disassembly intermediates characterized by single-molecule FRET, Proc. Natl. Acad. Sci. U.S.A., 2009, 106(36), 15308–15313, DOI:10.1073/pnas.090300510.
- H. Zhang, Z. Wang, L. Xie, Y. Zhang, T. Deng, J. Li, J. Liu, W. Xiong, L. Zhang and L. Zhang, et al., Molecular Recognition and In-Vitro-Targeted Inhibition of Renal Cell Carcinoma Using a DNA Aptamer, Mol. Ther.–Nucleic Acids, 2018, 12, 758–768, DOI:10.1016/j.omtn.2018.07.015.
- T. L. Jennings, J. C. Schlatterer, M. P. Singh, N. L. Greenbaum and G. F. Strouse, NSET molecular beacon analysis of hammerhead RNA substrate binding and catalysis, Nano Lett., 2006, 6(7), 1318–1324, DOI:10.1021/nl052458a.
- M. P. Singh, T. L. Jennings and G. F. Strouse, Tracking Spatial Disorder in an Optical Ruler by Time-Resolved NSET, J. Phys. Chem. B, 2009, 113(2), 552–558, DOI:10.1021/jp808997h.
- R. A. Riskowski, R. E. Armstrong, N. L. Greenbaum and G. F. Strouse, Triangulating Nucleic Acid Conformations Using Multicolor Surface Energy Transfer, ACS Nano, 2016, 10(2), 1926–1938, DOI:10.1021/acsnano.5b05764.
- E. Boisselier and D. Astruc, Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity, Chem. Soc. Rev., 2009, 38(6), 1759–1782, 10.1039/B806051G.
- P. Singh, S. Pandit, V. Mokkapati, A. Garg, V. Ravikumar and I. Mijakovic, Gold Nanoparticles in Diagnostics and Therapeutics for Human Cancer, Int. J. Mol. Sci., 2018, 19(7), 1979, DOI:10.3390/ijms19071979.
- X. Zhang, C. A. Marocico, M. Lunz, V. A. Gerard, Y. K. Gun'ko, V. Lesnyak, N. Gaponik, A. S. Susha, A. L. Rogach and A. L. Bradley, Wavelength, Concentration, and Distance Dependence of Nonradiative Energy Transfer to a Plane of Gold Nanoparticles, ACS Nano, 2012, 6(10), 9283–9290, DOI:10.1021/nn303756a.
- J. K. Vaishnav and T. K. Mukherjee, Long-Range Resonance Coupling-Induced Surface Energy Transfer from CdTe Quantum Dot to Plasmonic Nanoparticle, J. Phys. Chem. C, 2018, 122(49), 28324–28336, DOI:10.1021/ACS.JPCC.8B08757.
- C. Chen and N. Hildebrandt, Resonance energy transfer to gold nanoparticles: NSET defeats FRET, TrAC, Trends Anal. Chem., 2020, 123, 115748 CrossRef CAS.
- C. Chen, C. Midelet, S. Bhuckory, N. Hildebrandt and M. H. V. Werts, Nanosurface Energy Transfer from Long-Lifetime Terbium Donors to Gold Nanoparticles, J. Phys. Chem. C, 2018, 122(30), 17566–17574, DOI:10.1016/j.trac.2019.115748.
- S. Pushpam, M. Kottaisamy and V. Ramakrishnan, Dynamic quenching study of 2-amino-3-bromo-1,4-naphthoquinone by titanium dioxide nano particles in solution (methanol), Spectrochim. Acta, Part A, 2013, 114, 272–276, DOI:10.1016/j.saa.2013.05.038.
- W. Bae, T. Y. Yoon and C. Jeong, Direct evaluation of self-quenching behavior of fluorophores at high concentrations using an evanescent field, PLoS One, 2021, 16(2), e0247326, DOI:10.1371/journal.pone.0247326.
- J. R. Shao, S. J. Zhu, H. W. Liu, Y. B. Song, S. Y. Tao and B. Yang, Full-Color Emission Polymer Carbon Dots with Quench-Resistant Solid-State Fluorescence, Adv. Sci., 2017, 4(12), 1700395, DOI:10.1002/advs.201700395.
- G. Ruiz, K. Tripathi, S. Okyem and J. D. Driskell, pH Impacts the Orientation of Antibody Adsorbed onto Gold Nanoparticles, Bioconjugate Chem., 2019, 30(4), 1182–1191, DOI:10.1021/acs.bioconjchem.9b00123.
- A. M. Davidson, M. Brust, D. L. Cooper and M. Volk, Sensitive Analysis of Protein Adsorption to Colloidal Gold by Differential Centrifugal Sedimentation, Anal. Chem., 2017, 89(12), 6807–6814, DOI:10.1021/acs.analchem.7b01229.
- G. M. Rothe, Determination of molecular mass, Stokes' radius, frictional coefficient and isomer-type of non-denatured proteins by time-dependent pore gradient gel electrophoresis, Electrophoresis, 1988, 9(7), 307–316, DOI:10.1002/elps.1150090705.
- Y. Choi, Y. Park, T. Kang and L. P. Lee, Selective and sensitive detection of metal ions by plasmonic resonance energy transfer-based nanospectroscopy, Nat. Nanotechnol., 2009, 4(11), 742–746, DOI:10.1038/NNANO.2009.258.
- J. Zhou, B. Del Rosal, D. Jaque, S. Uchiyama and D. Jin, Advances and challenges for fluorescence nanothermometry, Nat. Methods, 2020, 17(10), 967–980, DOI:10.1038/s41592-020-0957-y.
- F. Zhou, T. Fu, Q. Huang, H. Kuai, L. Mo, H. Liu, Q. Wang, Y. Peng, D. Han and Z. Zhao, et al., Hypoxia-Activated PEGylated Conditional Aptamer/Antibody for Cancer Imaging with Improved Specificity, J. Am. Chem. Soc., 2019, 141(46), 18421–18427, DOI:10.1021/jacs.9b05063.
- Y. Jiang, X. Pan, J. Chang, W. Niu, W. Hou, H. Kuai, Z. Zhao, J. Liu, M. Wang and W. Tan, Supramolecularly Engineered Circular Bivalent Aptamer for Enhanced Functional Protein Delivery, J. Am. Chem. Soc., 2018, 140(22), 6780–6784, DOI:10.1021/jacs.8b03442.
- J. Guo, X. Qiu, C. Mingoes, J. R. Deschamps, K. Susumu, I. L. Medintz and N. Hildebrandt, Conformational Details of Quantum Dot-DNA Resolved by Forster Resonance Energy Transfer Lifetime Nanoruler, ACS Nano, 2019, 13(1), 505–514, DOI:10.1021/acsnano.8b07137.
- J. H. Hwang, S. Park, J. Son, J. W. Park and J. M. Nam, DNA-Engineerable Ultraflat-Faceted Core-Shell Nanocuboids with Strong, Quantitative Plasmon-Enhanced Fluorescence Signals for Sensitive, Reliable MicroRNA Detection, Nano Lett., 2019, 21(5), 2132–2140, DOI:10.1021/acs.nanolett.0c04883.
- M. Duan, Y. Long, C. Yang, X. Wu, Y. Sun, J. Li, X. Hu, W. Lin, D. Han, Y. Zhao and J. Liu, et al., Selection and characterization of DNA aptamer for metastatic prostate cancer recognition and tissue imaging, Oncotarget, 2016, 7(24), 36436–36446, DOI:10.18632/oncotarget.9262.
- W. Zhibo, Study on the Mechanism of Aptamer-Binding Targets of Prostatic Carcinoma Cells, CNKI, 2019 Search PubMed.
- S. W. Park, J. S. Bae, K. S. Kim, S. H. Park, B. H. Lee, J. Y. Choi, J. Y. Park, S. W. Ha, Y. L. Kim and T. H. Kwon, Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin, Exp. Mol. Med., 2004, 36(3), 211–219, DOI:10.1038/emm.2004.29.
- J. K. Jaiswal and S. M. Simon, Imaging single events at the cell membrane, Nat. Chem. Biol., 2007, 3(2), 92–98, DOI:10.1038/nchembio855.
- Y. Sako and T. Yanagida, Single-molecule visualization in cell biology, Nat. Rev. Mol. Cell Biol., 2003, SS1–SS5, DOI:10.1038/nrm1193.
Footnotes |
| † Electronic supplementary information (ESI) available: Full experimental procedures, Table S1, and Fig. S1–S22. See DOI: https://doi.org/10.1039/d3sc01244a |
| ‡ These authors contributed equally: Yu Zhang and Mengke Su. |
| This journal is © The Royal Society of Chemistry 2023 |