Open Access Article
Open Access ArticleDearomative intermolecular [2 + 2] photocycloaddition for construction of C(sp3)-rich heterospirocycles on-DNA†
Longbo
Li
 a,
Bianca
Matsuo
a,
Bianca
Matsuo
 a,
Guillaume
Levitre
a,
Edward J.
McClain‡
bd,
Eric A.
Voight
a,
Guillaume
Levitre
a,
Edward J.
McClain‡
bd,
Eric A.
Voight
 b,
Erika A.
Crane‡
*c and
Gary A.
Molander
b,
Erika A.
Crane‡
*c and
Gary A.
Molander
 *a
*a
aDepartment of Chemistry, Roy and Diana Vagelos Laboratories, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104-6323, USA. E-mail: gmolandr@sas.upenn.edu
bDrug Discovery Science & Technology, Discovery Research & Development, AbbVie, Inc., 1 North Waukegan Rd, North Chicago, Illinois 60064-1802, USA
cDrug Hunter, Inc., 13203 SE 172nd Ave, Suite 166 PMB 2019, Happy Valley, Oregon 97086, USA. E-mail: erika@drughunter.com
dDepartment of Chemistry, University of Wisconsin−Madison, Madison, Wisconsin 53706, USA
First published on 9th February 2023
Abstract
DNA-encoded library (DEL) screens have significantly impacted new lead compound identification efforts within drug discovery. An advantage of DELs compared to traditional screening methods is that an exponentially broader chemical space can be effectively screened using only nmol quantities of billions of DNA-tagged, drug-like molecules. The synthesis of DELs containing diverse, sp3-rich spirocycles, an important class of molecules in drug discovery, has not been previously reported. Herein, we demonstrate the synthesis of complex and novel spirocyclic cores via an on-DNA, visible light-mediated intermolecular [2 + 2] cycloaddition of olefins with heterocycles, including indoles, azaindoles, benzofurans, and coumarins. The DNA-tagged exo-methylenecyclobutane substrates were prepared from easily accessible alkyl iodides and styrene derivatives. Broad reactivity with many other DNA-conjugated alkene substrates was observed, including unactivated and activated alkenes, and the process is tolerant of various heterocycles. The cycloaddition was successfully scaled from 10 to 100 nmol without diminished yield, indicative of this reaction's suitability for DNA-encoded library production. Evaluation of DNA compatibility with the developed reaction in a mock-library format showed that the DNA barcode was maintained with high fidelity, with <1% mutated sequences and >99% amplifiable DNA from quantitative polymerase chain reaction (PCR) and next generation sequencing (NGS).
Introduction
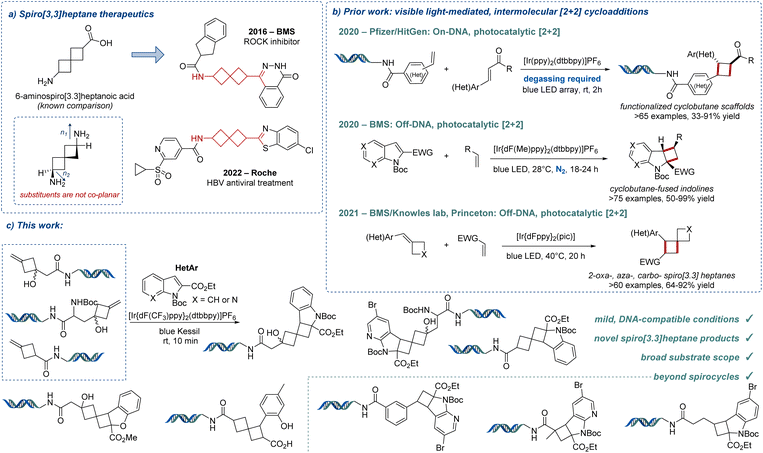
Spirocycles are important structural motifs in drug discovery, as their three-dimensionality provides an opportunity to orient functional groups in different spatial directions, avoiding the restrictions of planarity that are commonly encountered with highly unsaturated, sp2-rich arene cores.1–3 In particular, spiro[3.3]heptanes represent a unique class of spirocyclic compounds with two cyclobutane rings that project functional groups into non-coplanar space (Fig. 1a).4 Because of their novel 3D properties, spiro[3.3]heptanes have become a point of interest for medicinal chemists.5–9 Syntheses of spiro[3.3]heptanes have been mostly achieved via photochemical [2 + 2] cycloaddition.10–12 However, to our knowledge, a general method for the preparation of diverse spiro[3.3]heptanes on-DNA has yet to be reported.DNA-encoded library (DEL) technology has emerged as a powerful tool for hit identification in modern drug discovery.13–21 DELs can be readily assembled to afford exceptionally large combinational libraries that can be efficiently screened in a cost-efficient manner, as only nanomolar quantities of both drug-like molecules and the biomolecular targets are required.21 Furthermore, prior successes in the identification of valuable hits for challenging therapeutic targets via DEL chemistry have attracted increasing attention in academia and industry.22,23 Solution-based chemical syntheses on-DNA must be carried out within strict reaction parameters, as the transformations must be tolerant of aqueous conditions and the basic DNA backbone.24,25 To this end, visible light-mediated transformations have attracted significant attention in DEL chemistry for the ability to generate novel C–C bonds26–41 while expanding chemical space under extremely mild conditions.42–44 For example, Kölmel and coworkers demonstrated that this mild reactivity paradigm could be utilized for the synthesis of strained cyclobutane scaffolds on-DNA via visible light-mediated [2 + 2] photocycloaddition between cinnamate substrates and styrene derivatives (Fig. 1b).41 Additional relevant transformations have been performed off-DNA, including a highly diastereoselective dearomative [2 + 2] photocycloaddition of indoles and diverse alkenes to afford cyclobutene-fused indolines, also requiring inert conditions.45 New heteroarene dearomative functionalization strategies have recently demonstrated great value, particularly those enabled by visible light, enhancing the molecular complexity and three-dimensionality of these venerable products.46 Additionally, BMS in collaboration with the Knowles laboratory reported a different [2 + 2] photocycloaddition to access polysubstituted (hetero)spiro[3.3]heptane products.12
A recent collaborative study from our groups focused on unlocking the radical reactivity of 1,3-disubstituted [1.1.1]bicyclopentyl (BCP) halides for on-DNA reactions.33 Enabled by visible-light photochemistry, the method allowed the preparation of diverse, sp3-rich chemical matter on-DNA. As part of our continued study of BCP reactivity, we sought to develop a method that utilized the exo-methylenecyclobutanol products derived from the strain-driven hydrolytic ring opening of DNA-conjugated BCP halides (Fig. 1c).47,48 With well-documented, mild, DNA-compatible reactivity, we envisioned that visible-light photochemistry could be leveraged to engage these and other olefins in a triplet-sensitized [2 + 2]-cycloaddition with indoles and other heterocycles, providing access to novel spirocyclic substructures on-DNA.12,45,49 As an extension of this chemistry, we also explored additional DNA-conjugated olefin substrates, including exo-methylenespiro[2.3]hexanes,50 unactivated alkenes, styrene derivatives, and acrylamides. The latter three have revealed significant reactivity toward indoles and other heterocycles to form fused ring systems.45 Finally, we focused on important and abundant scaffolds in drug discovery as the [2 + 2] coupling partners, including indoles, azaindoles, benzofurans, and coumarins, generating complex spiro- and fused polycyclic ring systems.
Results and discussion
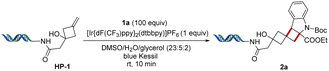
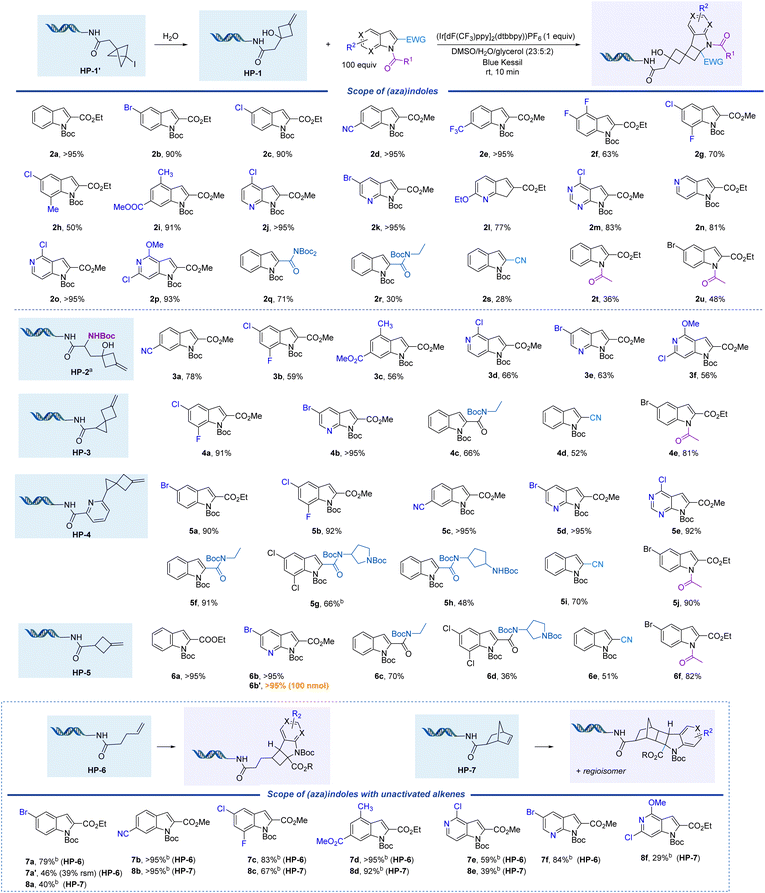
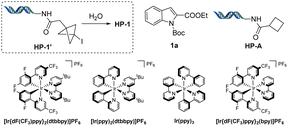
The initial exo-methylene cyclobutanol HP-1 was prepared by solvolysis of 1-iodo-bicyclo[1.1.1]pentane (BCP-I) on-DNA (HP-1′, Table 1). HP-1′ and other derivatives were prepared by reaction of [1.1.1]propellane with abundant alkyl- or aryl iodides via atom transfer radical addition (ATRA).33,51 The reaction of HP-1 and ethyl indole-2-carboxylate (1a) was investigated to determine suitable reaction parameters (Table 1). Quantitative conversion was observed when using one equivalent of photosensitizer [Ir{dF(CF3)ppy}2(dtbbpy)]PF6 and a small amount of glycerol as cosolvent in under 10 minutes of blue Kessil lamp irradiation at room temperature (>95%, entry 1, Table 1). Control experiments demonstrated that the reaction requires photosensitizer (entry 2), light (entry 3), and glycerol (entry 4), which likely acts as radical scavenger.41 Other results indicated that the reaction is not complete after 5 min (entry 5), and fewer equivalents (25 and 50 equivalents, respectively) of indole 1a result in slightly lower yields (entries 6 and 7). Interestingly, by increasing the number of equivalents of indole to 200, a lower reaction yield was also observed (67%) with recovery of starting material (entry 8). We expect this to be caused by the dimerization of indole 1a,45 as the probability of self-reaction increases with indole concentration under the fixed catalyst concentration, illumination conditions, and reaction duration. Solvents other than DMSO, such as methanol and DMA were investigated, but gave lower yields (entries 9 and 10). Unsurprisingly, other iridium complexes with lower triplet state energies (EnT) such as [Ir(ppy)2(dtbbpy)]PF6, which was used in the previous on-DNA photocatalytic [2 + 2] cycloaddition,41 as well as Ir(ppy)3 did not yield any product (entries 11 and 12). [Ir{dF(CF3)ppy}2(bpy)]PF6 proved to be a viable alternative photosensitizer, providing the desired product in 83% yield (entry 13). A control experiment with HP-A (entry 14) demonstrated that the double-stranded DNA tag is not participating in the [2 + 2] photocycloaddition. Overall, quantitative yields were achieved within 10 min without any degassing, providing an expedient and convenient access to a unique class of molecules on-DNA.| Entry | Deviations from standard conditions | Yield (%) |
|---|---|---|
| a Standard conditions: DNA (2 mM in H2O, 10 nmol, 1 equiv.), 1a (100 mM in DMSO, 100 equiv.), [Ir{dF(CF3)ppy}2(dtbbpy)]PF6 (2 mM in DMSO, 1 equiv.), glycerol (2 μL in 8 μL of DMSO), rt, 10 min, blue Kessil. Work-up: ethanol precipitation. n.d. = product not detected. rsm = recovered starting material. | ||
| 1 | None | >95 |
| 2 | No photocatalyst | n.d. (>95 rsm) |
| 3 | No light | n.d. (>95 rsm) |
| 4 | No glycerol | Degradation |
| 5 | 5 min instead of 10 min | 75 (25 rsm) |
| 6 | 25 equiv. of indole | 88 |
| 7 | 50 equiv. of indole | 93 |
| 8 | 200 equiv. of indole | 67 (19 rsm) |
| 9 | MeOH instead of DMSO | 15 (68 rsm) |
| 10 | DMA instead of DMSO | 44 |
| 11 | Ir(ppy)2(dtbbpy)]PF6 | n.d. (71 rsm) |
| 12 | Ir(ppy)3 | n.d. (70 rsm) |
| 13 | [Ir{dF(CF3)ppy}2(bpy)]PF6 | 83 |
| 14 | HP-A instead of HP-1 | n.d. (71 rsm) |
|
||
To probe the regio- and stereoselectivity of the reaction on-DNA, off-DNA reactions were conducted between 1a and ethyl 3-methylenecyclobutane-1-carboxylate (see ESI Section 3.1† for details). Only one regioisomer was observed, which is consistent with previous off-DNA reports.45 However, four stereoisomers (two sets of diastereomeric enantiomers) were generated, in a 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio by chiral supercritical fluid chromatography (SFC). Despite the low stereoselectivity for this cycloaddition, the unprecedented novelty of these products covers uncharted chemical space, and methods to deconvolute DEL hits as stereoisomeric mixtures of comparable complexity are well established.52
1 ratio by chiral supercritical fluid chromatography (SFC). Despite the low stereoselectivity for this cycloaddition, the unprecedented novelty of these products covers uncharted chemical space, and methods to deconvolute DEL hits as stereoisomeric mixtures of comparable complexity are well established.52
We evaluated the scope of cycloaddition partners, reacting many diverse indoles and azaindoles with HP-1 (Fig. 2). Indoles with various functional groups at different positions about the aromatic ring resulted in good to excellent yields (2b–2i). Functional groups including halogens (2b–2c), a nitrile (2d), and CF3 (2e) were well tolerated with yields >80%. Functional handles at the 5-, 6-, 7-, and 8-positions of the indole ring including F and Cl substituents, as well as methyl and ester groups, also performed well under the standard conditions (2f–2i). Azaindoles and pyrrolo[2,3-d]pyrimidines with additional functional groups (Cl, Br, or ether substituents) gave good to excellent yields (2j–2p). Various electron-withdrawing groups at the 2-position of the indole were tolerated in this reaction. Indole-2-carboxamides provided moderate to good yields of cycloaddition products (2q, 2r). A 2-cyano-substituted indole provided a lower yield of 28% (as compared to the corresponding ester 2s). Even further, N-acetyl-protected indoles afforded moderate yields of products (2t, 2u).
Commercially available Boc-3-iodo-L-alanine methyl ester was used to prepare methylenecyclobutanol substrate HP-2 utilizing the same strategy as used with HP-1. Diverse indoles 3a–3c and azaindoles 3d–3f performed well in reactions with this headpiece, indicating that the cycloaddition is not limited to DNA-conjugated exo-methylenecyclobutanol HP-1.
We expanded this reaction further to afford dispiro[2.1.3.1]nonane ring systems. The DNA-conjugated exo-methylene spiro[2.3]hexane substrates were prepared using a nickel-catalyzed cyclopropanation of [1.1.1]propellane and readily accessible styrenes or acrylates.50HP-3 demonstrated good reactivity with indoles containing various groups at the 2-position, including an ester (4a), amide (4c), and nitrile (4d). Azaindole 4b and N-Ac indole 4e also worked well with HP-3. Related HP-4, prepared from the corresponding vinyl pyridine, performed well with indoles 5a–5e in >90% yield. Indoles with 2-substituted amides gave moderate to excellent yields (5f, 5g, and 5h). 2-Cyano and N-Ac indoles afforded products 5i and 5j in 70% and 90% yields, respectively. HP-5 was prepared from commercially available 3-exo-methylenecyclobutanecarboxylic acid and performed well with various indoles (6a, 6c–6f) and azaindoles (6b). Even further, we successfully scaled this reaction from 10 nmol (6b) to 100 nmol (6b′) with no loss in yield, revealing excellent promise for subsequent functionalization and implementation in DNA-encoded library production.
The reactivity of unactivated alkene headpieces was also investigated (Fig. 2). For HP-6, 50 equivalents of indole gave higher yields (e.g., 7a, 79% yield) than 100 equivalents (e.g., 7a′, 46% yield + 39% starting material) after irradiation for 10 min. This is likely caused by the lower reactivity of HP-6 compared to exo-methylenecyclobutane headpieces HP-1–HP-4. The indole at a higher concentration competes with DNA-conjugated alkenes and reacts with the excited triplet 1,2-diradical indole species to form a dimer, which leads to a lower yield of product in the same reaction time. Similar products have been observed in analogous off-DNA [2 + 2] photocycloaddition studies.45 Various indoles (7a–7d) and azaindoles (7e and 7f) performed well under this set of conditions. We expect high regioselectivity of 7a–7f as indicated by a similar reaction off-DNA.45 Another headpiece, HP-7, was also investigated with indoles (8a–8d) and azaindoles (8e and 8f) (Fig. 2). The complex polycycles obtained are unique, with no reported precedent in the literature.
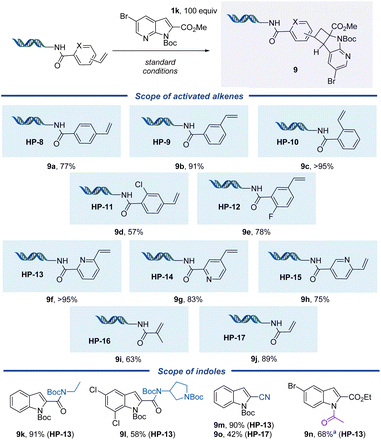
Activated alkenes, including styrene derivatives HP-8–HP-15 and acrylamides HP-16 and HP-17, reacted smoothly with azaindole 1k with yields >50% (Fig. 3). The regioselectivity of a similar product off-DNA (indole 1a and styrene) has been reported in the literature.45para-(HP-8), meta-(HP-9), and ortho-substituted styrenes (HP-10) performed well with bromoindole 1k in 77%, 91%, and >95% yields, respectively. Chloro-substituted HP-11, primed for post-functionalization, provided a 57% yield of product. Fluoride-substituted HP-12 reacted well in 78% yield, and vinyl pyridine headpieces (HP-13–HP-15) provided polycyclic products in yields >70%. Acrylamide HP-17 showed higher reactivity than methyl acrylamide HP-16, providing the desired cycloaddition products in 89% and 63% yields, respectively. We explored the reactivity of other indoles toward the vinyl pyridine-derived substrates further. Vinyl pyridine HP-13 performed well with Boc-protected indole-2-carboxamides (9k and 9l) and indole-2-carbonitrile (9m), as well as N-acylindole-2-carboxy ester (9n), with yields >50%. Acrylamide HP-17 reacted with indole-2-carbonitrile to afford 9o in a moderate 42% yield.
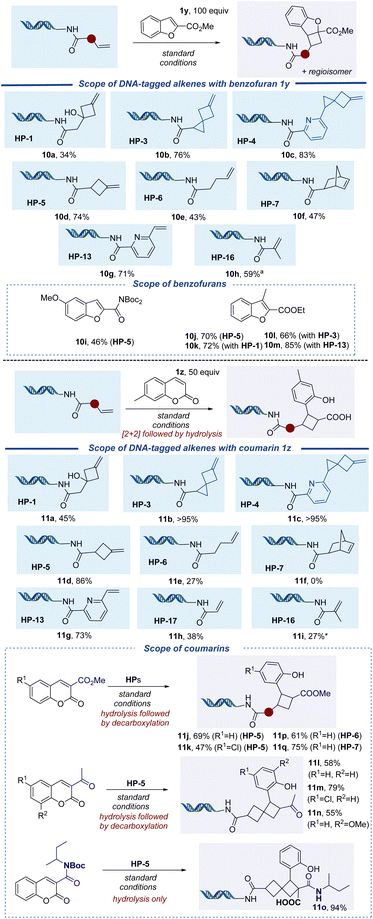
Given the prevalence of diverse heterocyclic compounds in the medicinal chemistry field, additional heterocyclic coupling partners were also explored, including benzofuran and coumarin derivatives. Methyl benzofuran-2-carboxylate (1y) reacted smoothly with olefin headpieces 10a–10h to afford products in good yield (Fig. 4). The off-DNA reaction of benzofuran and exo-methylenecyclobutanol proceeds in a ∼1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dr and ∼3
1 dr and ∼3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 regioisomeric ratio (see ESI Section 3.2† for details). This regioisomeric ratio is consistent with what has been reported off-DNA.45 A variety of benzofurans were reacted with HP-5. The 2-carboxamide afforded 10i in a moderate 46% yield and, surprisingly, 3-methylbenzofuran-2-carboxylate afforded 10j in a good 70% yield of the cycloaddition product. This substrate showed good reactivity with other DNA-conjugated olefin substrates, including exo-methylenecyclobutanes HP-1 (10k) and HP-3 (10l), as well as vinyl pyridine HP-13 (10m) to give >60% yield in each case. Benzofurans with a Cl and/or methyl ether substitution both proceeded in low yields (see ESI Section 7, Fig. S4†), as did the corresponding 2-nitrile substrate.
1 regioisomeric ratio (see ESI Section 3.2† for details). This regioisomeric ratio is consistent with what has been reported off-DNA.45 A variety of benzofurans were reacted with HP-5. The 2-carboxamide afforded 10i in a moderate 46% yield and, surprisingly, 3-methylbenzofuran-2-carboxylate afforded 10j in a good 70% yield of the cycloaddition product. This substrate showed good reactivity with other DNA-conjugated olefin substrates, including exo-methylenecyclobutanes HP-1 (10k) and HP-3 (10l), as well as vinyl pyridine HP-13 (10m) to give >60% yield in each case. Benzofurans with a Cl and/or methyl ether substitution both proceeded in low yields (see ESI Section 7, Fig. S4†), as did the corresponding 2-nitrile substrate.
We investigated the reactivities of coumarin derivatives with different DNA-conjugated alkenes, including exo-methylenecyclobutanes, unactivated alkenes, styrene derivatives, and acrylamides (Fig. 4). Visible light-mediated intermolecular [2 + 2] cycloadditions of coumarin-3-carboxylates and acrylamide analogs off-DNA have been previously reported.53 We conducted these analogous off-DNA reactions to confirm the regio- and diastereoselectivity of the [2 + 2] cycloaddition products (see ESI Section 3.3† for details). Initial on-DNA experiments revealed that a small percentage of lactone hydrolysis product was observed under standard conditions, along with the expected [2 + 2] cycloaddition adduct as the major product. We found that the [2 + 2] product could be funneled to hydrolyzed product in aqueous solution by heating at 70 °C for 20 min. Overall, 7-methylcoumarin (1z) showed good reactivity with methylenecyclobutanes HP-1, HP-3, HP-4, and HP-5, affording spiro[3.3]heptanes 11a–11d in good to excellent yields. Unactivated alkene substrate HP-6 resulted in a low 27% yield (11e), while HP-7 did not afford any desired product. For the activated alkene substrates, vinyl pyridine HP-13 and acrylamide HP-17 reacted to provide 11g and 11h in 73% and 38% yields, respectively. The reactivity of coumarin derivatives containing an ester, ketone, or amide was evaluated with exo-methylenecyclobutane HP-5. For coumarin-containing esters and ketones, hydrolysis followed by decarboxylation yielded spiro[3.3]heptane products 11j–11n in >45% yield. For a coumarin-containing amide, only the hydrolysis product without decarboxylation was observed, affording 11o in 94% yield. Furthermore, unactivated alkenes HP-6 and HP-7 reacted to provide 11p and 11q in good yields. Control experiments with a headpiece that lacked an alkene functional group were conducted under the standard reaction conditions with benzofuran and coumarin (HP-A, see Table 1 and ESI Section 6† for details). We observed benzofuran and coumarin adducts in 10% and 7% yields, respectively, which indicates the potential addition of benzofuran and coumarin to the DNA tag. Therefore, actual yields for 10a–10m and 11a–11n are slightly lower than reported.
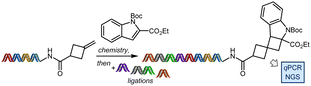
To ensure that this method would be compatible with DNA-encoded library production, exo-methylenecyclobutane substrate HP-5 was prepared on an elongated DNA tag and then subjected to the reaction with indole 1a or other control conditions (Table 2). Upon isolation of the single constructs by ethanol precipitation and filtration, a series of ligations were performed, mimicking the library production process, followed by quantitative polymerase chain reaction (qPCR) and next-generation sequencing (NGS) of the full-length tag (see ESI Section 8† for details). The ligations for all six samples proceeded with good efficiencies (80–100%), as confirmed by gel electrophoresis. Amplification efficiency of the full-length sequence by qPCR was comparable for all six samples (93–98%) and the reaction maintained 99% amplifiable DNA, as compared to the no light, no reagents control (entry 6, Table 2). In contrast, some of the most often used non-photonic on-DNA chemistries often have only 30–50% amplifiable DNA remaining.54 Finally, NGS analysis revealed that all samples as compared to the control had <1% mutated sequences.
| Entry | Conditions | Amplifiable DNA (%) | Mutated sequences (%) |
|---|---|---|---|
| 1 | Standard reaction conditions | 99 | 0.5 |
| 2 | Standard conditions under N2 | 100 | 0.1 |
| 3 | Reagents with no irradiation control | 100 | 0.2 |
| 4 | Irradiation with no reagents control | 99 | 0.4 |
| 5 | Standard conditions, no BCP-I | 100 | 0.2 |
| 6 | No irradiation, no reagents control | 100 | 0 |
Conclusions
In conclusion, a versatile and operationally simple dearomative [2 + 2] cycloaddition is described that provides access to diverse DNA-encoded libraries of novel and compact hetero-spirocycles with high Fsp3 content. This protocol utilizes abundant heterocycles including indoles, azaindoles, benzofurans, and coumarins, which readily react with various easily accessible DNA-conjugated exo-methylenecyclobutanes. In addition, we demonstrated the versatility of this method for building diverse cyclobutane-fused scaffolds from unactivated and activated DNA-tagged alkenes. In most cases, the reaction of the various alkene headpieces and indoles proceeded in high yields. The reaction could be scaled up ten-fold while maintaining >95% yield, which will render it useful in library production, allowing further functionalization in subsequent chemistry cycles. Through the developed method, vast new DEL chemical space can be accessed. The prepared compounds possess unique chemical structures bearing different functional groups and reactive handles, which is key for further functionalization strategies aimed at the construction of structurally diversified libraries. Furthermore, the integrity of the DNA tag was maintained under the mild reaction conditions, indicating that this method could be applied to DNA-encoded library production to afford libraries with previously inaccessible hetero-spirocycles, utilizing abundant and diverse indoles, azaindoles, benzofurans, or coumarins.Data availability
General considerations, preparation of indoles, headpieces, general procedure for the cycloadditions, limitations of the reaction, and characterization of all compounds synthesized are available in the ESI.†Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are thankful for financial support provided by NIGMS (R35 GM 131680 to GAM) and the National Science Foundation (CHE-1952583 to GAM). Financial support for this research was also provided in part by AbbVie. The NSF Major Research Instrumentation Program (award NSF CHE-1827457), the NIH supplements awards 3R01GM118510-03S1 and 3R01GM087605-06S1, as well as the Vagelos Institute for Energy Science and Technology supported the purchase of the NMRs used in this study. We thank Dr Charles W. Ross, III (UPenn) for mass spectral data and Kessil for donation of LED lamps. We thank Gary O'Donovan (AbbVie) for his participation in the AbbVie-Molander collaboration. We thank Dr Gaonan Wang and the WuXi AppTec HitS Unit for experimental execution of ligations, qPCR, NGS, and analysis for the DNA damage assessment.Notes and references
- F. Lovering, J. Bikker and C. Humblet, J. Med. Chem., 2009, 52, 6752–6756 CrossRef CAS PubMed.
- F. Lovering, Med. Chem. Commun., 2013, 4, 515–519 RSC.
- K. Hiesinger, D. Dar’in, E. Proschak and M. Krasavin, J. Med. Chem., 2021, 64, 150–183 CrossRef CAS PubMed.
- D. S. Radchenko, S. O. Pavlenko, O. O. Grygorenko, D. M. Volochnyuk, S. V. Shishkina, O. V. Shishkin and I. V. Komarov, J. Org. Chem., 2010, 75, 5941–5952 CrossRef CAS PubMed.
- X. Lin, H. Yun, B. Zhang and X. Zheng, WO2022112207, 2022.
- X. Lin, J. Wang, H. Yun and X. Zheng, WO2022112188, 2022.
- X. Lin, H. Yun, B. Zhang and X. Zheng, WO2022112140, 2022.
- V. Ladziata, P. W. Glunz, Z. Hu and Y. Wang, US20160016914A1, 2016.
- M. Hagihara, Y. Tsuzaki, K. Komori, H. Nishida, K. Kido, T. Fujimoto, T. Matsugi and A. Shimazaki, WO2007142323, 2007.
- O. P. Demchuk, O. V. Hryshchuk, B. V. Vashchenko, A. V. Kozytskiy, A. V. Tymtsunik, I. V. Komarov and O. O. Grygorenko, J. Org. Chem., 2020, 85, 5927–5940 CrossRef CAS PubMed.
- X. Gu, Y. Wei and M. Shi, Org. Chem. Front., 2021, 8, 6823–6829 RSC.
- P. R. D. Murray, W. M. M. Bussink, G. H. M. Davies, F. W. van der Mei, A. H. Antropow, J. T. Edwards, L. A. D'Agostino, J. M. Ellis, L. G. Hamann, F. Romanov-Michailidis and R. R. Knowles, J. Am. Chem. Soc., 2021, 143, 4055–4063 CrossRef CAS PubMed.
- S. Brenner and R. A. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381–5383 CrossRef CAS PubMed.
- M. W. Kanan, M. M. Rozenman, K. Sakurai, T. M. Snyder and D. R. Liu, Nature, 2004, 431, 545–549 CrossRef CAS PubMed.
- W. Decurtins, M. Wichert, R. M. Franzini, F. Buller, M. A. Stravs, Y. Zhang, D. Neri and J. Scheuermann, Nat. Protoc., 2016, 11, 764–780 CrossRef CAS PubMed.
- J. Ottl, L. Leder, J. V. Schaefer and C. E. Dumelin, Molecules, 2019, 24, 1629 CrossRef CAS PubMed.
- F. V. Reddavide, M. Cui, W. Lin, N. Fu, S. Heiden, H. Andrade, M. Thompson and Y. Zhang, Chem. Commun., 2019, 55, 3753–3756 RSC.
- M. Song and G. T. Hwang, J. Med. Chem., 2020, 63, 6578–6599 CrossRef CAS PubMed.
- N. Favalli, G. Bassi, C. Pellegrino, J. Millul, R. De Luca, S. Cazzamalli, S. Yang, A. Trenner, N. L. Mozaffari, R. Myburgh, M. Moroglu, S. J. Conway, A. A. Sartori, M. G. Manz, R. A. Lerner, P. K. Vogt, J. Scheuermann and D. Neri, Nat. Chem., 2021, 13, 540–548 CrossRef CAS PubMed.
- A. L. Satz, A. Brunschweiger, M. E. Flanagan, A. Gloger, N. J. V. Hansen, L. Kuai, V. B. K. Kunig, X. Lu, D. Madsen, L. A. Marcaurelle, C. Mulrooney, G. O'Donovan, S. Sakata and J. Scheuermann, Nat. Rev. Methods Primers, 2022, 2, 3 CrossRef CAS.
- R. A. Goodnow, C. E. Dumelin and A. D. Keefe, Nat. Rev. Drug Discovery, 2017, 16, 131–147 CrossRef CAS PubMed.
- D. T. Flood, C. Kingston, J. C. Vantourout, P. E. Dawson and P. S. Baran, Isr. J. Chem., 2020, 60, 268–280 CrossRef CAS.
- Y. Shi, Y. Wu, J. Yu, W. Zhang and C. Zhuang, RSC Adv., 2021, 11, 2359–2376 RSC.
- M. L. Malone and B. M. Paegel, ACS Comb. Sci., 2016, 18, 182–187 CrossRef CAS PubMed.
- P. R. Fitzgerald and B. M. Paegel, Chem. Rev., 2021, 121, 7155–7177 CrossRef CAS PubMed.
- D. K. Kölmel, R. P. Loach, T. Knauber and M. E. Flanagan, ChemMedChem, 2018, 13, 2159–2165 CrossRef PubMed.
- R. Chowdhury, Z. Yu, M. L. Tong, S. V. Kohlhepp, X. Yin and A. Mendoza, J. Am. Chem. Soc., 2020, 142, 20143–20151 CrossRef CAS PubMed.
- S. O. Badir, J. Sim, K. Billings, A. Csakai, X. Zhang, W. Dong and G. A. Molander, Org. Lett., 2020, 22, 1046–1051 CrossRef CAS PubMed.
- H. Wen, R. Ge, Y. Qu, J. Sun, X. Shi, W. Cui, H. Yan, Q. Zhang, Y. An, W. Su, H. Yang, L. Kuai, A. L. Satz and X. Peng, Org. Lett., 2020, 22, 9484–9489 CrossRef CAS PubMed.
- R. Wu, T. Du, W. Sun, A. Shaginian, S. Gao, J. Li, J. Wan and G. Liu, Org. Lett., 2021, 23, 3486–3490 CrossRef CAS PubMed.
- J. Shan, X. Ling, J. Liu, X. Wang and X. Lu, Bioorg. Med. Chem., 2021, 42, 116234 CrossRef CAS PubMed.
- S. O. Badir, A. Lipp, M. Krumb, M. J. Cabrera-Afonso, L. M. Kammer, V. E. Wu, M. Huang, A. Csakai, L. A. Marcaurelle and G. A. Molander, Chem. Sci., 2021, 12, 12036–12045 RSC.
- E. Yen-Pon, L. Li, G. Levitre, J. Majhi, E. J. McClain, E. A. Voight, E. A. Crane and G. A. Molander, J. Am. Chem. Soc., 2022, 144, 12184–12191 CrossRef CAS PubMed.
- J. P. Phelan, S. B. Lang, J. Sim, S. Berritt, A. J. Peat, K. Billings, L. Fan and G. A. Molander, J. Am. Chem. Soc., 2019, 141, 3723–3732 CrossRef CAS PubMed.
- D. K. Kölmel, J. Meng, M.-H. Tsai, J. Que, R. P. Loach, T. Knauber, J. Wan and M. E. Flanagan, ACS Comb. Sci., 2019, 21, 588–597 CrossRef PubMed.
- D. K. Kölmel, A. S. Ratnayake and M. E. Flanagan, Biochem. Biophys. Res. Commun., 2020, 533, 201–208 CrossRef PubMed.
- Y. Zhang, H. Luo, H. Ma, J. Wan, Y. Ji, A. Shaginian, J. Li, Y. Deng and G. Liu, Bioconjugate Chem., 2021, 32, 1576–1580 CrossRef CAS PubMed.
- M. Krumb, L. M. Kammer, S. O. Badir, M. J. Cabrera-Afonso, V. E. Wu, M. Huang, A. Csakai, L. A. Marcaurelle and G. A. Molander, Chem. Sci., 2022, 13, 1023–1029 RSC.
- Y. Shen, G. Yang, W. Huang, A. Shaginian, Q. Lin, J. Wan, J. Li, Y. Deng and G. Liu, Org. Lett., 2022, 24, 2650–2654 CrossRef CAS PubMed.
- S. R. N. Kolusu and M. Nappi, Chem. Sci., 2022, 13, 6982–6989 RSC.
- D. K. Kölmel, A. S. Ratnayake, M. E. Flanagan, M.-H. Tsai, C. Duan and C. Song, Org. Lett., 2020, 22, 2908–2913 CrossRef PubMed.
- S. Patel, S. O. Badir and G. A. Molander, Trends Chem., 2021, 3, 161–175 CrossRef CAS PubMed.
- V. M. Lechner, M. Nappi, P. J. Deneny, S. Folliet, J. C. K. Chu and M. J. Gaunt, Chem. Rev., 2022, 122, 1752–1829 CrossRef CAS PubMed.
- R. J. Fair, R. T. Walsh and C. D. Hupp, Bioorg. Med. Chem. Lett., 2021, 51, 128339 CrossRef CAS PubMed.
- M. S. Oderinde, A. Ramirez, T. G. M. Dhar, L. A. M. Cornelius, C. Jorge, D. Aulakh, B. Sandhu, J. Pawluczyk, A. A. Sarjeant, N. A. Meanwell, A. Mathur and J. Kempson, J. Org. Chem., 2021, 86, 1730–1747 CrossRef CAS PubMed.
- N. Kratena, B. Marinic and T. J. Donohoe, Chem. Sci., 2022, 13, 14213–14225 RSC.
- K. B. Wiberg and V. Z. Williams, J. Am. Chem. Soc., 1967, 89, 3373–3374 CrossRef CAS.
- E. Della and D. Taylor, Aust. J. Chem., 1990, 43, 945–948 CrossRef CAS.
- M. R. Becker, E. R. Wearing and C. S. Schindler, Nat. Chem., 2020, 12, 898–905 CrossRef CAS PubMed.
- S. Yu, A. Noble, R. B. Bedford and V. K. Aggarwal, J. Am. Chem. Soc., 2019, 141, 20325–20334 CrossRef CAS PubMed.
- J. Nugent, C. Arroniz, B. R. Shire, A. J. Sterling, H. D. Pickford, M. L. J. Wong, S. J. Mansfield, D. F. J. Caputo, B. Owen, J. J. Mousseau, F. Duarte and E. A. Anderson, ACS Catal., 2019, 9, 9568–9574 CrossRef CAS.
- M. V. Westphal, L. Hudson, J. W. Mason, J. A. Pradeilles, F. J. Zécri, K. Briner and S. L. Schreiber, J. Am. Chem. Soc., 2020, 142, 7776–7782 CrossRef CAS PubMed.
- Q. Liu, F.-P. Zhu, X.-L. Jin, X.-J. Wang, H. Chen and L.-Z. Wu, Chem.–Eur. J., 2015, 21, 10326–10329 CrossRef CAS PubMed.
- B. Sauter, L. Schneider, C. Stress and D. Gillingham, Bioorg. Med. Chem., 2021, 52, 116508 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and characterization. CCDC 2215502. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3sc00144j |
| ‡ EAC and EJM are former employees of AbbVie. |
| This journal is © The Royal Society of Chemistry 2023 |