Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comprehensive structural, infrared spectroscopic and kinetic investigations of the roles of the active-site arginine in bidirectional hydrogen activation by the [NiFe]-hydrogenase ‘Hyd-2’ from Escherichia coli†
Rhiannon M.
Evans
 a,
Stephen E.
Beaton
a,
Patricia
Rodriguez Macia
a,
Stephen E.
Beaton
a,
Patricia
Rodriguez Macia
 a,
Yunjie
Pang
cd,
Kin Long
Wong
a,
Yunjie
Pang
cd,
Kin Long
Wong
 ab,
Leonie
Kertess
ab,
Leonie
Kertess
 a,
William K.
Myers
a,
William K.
Myers
 a,
Ragnar
Bjornsson
a,
Ragnar
Bjornsson
 de,
Philip A.
Ash
de,
Philip A.
Ash
 f,
Kylie A.
Vincent
f,
Kylie A.
Vincent
 a,
Stephen B.
Carr
a,
Stephen B.
Carr
 *ab and
Fraser A.
Armstrong
*ab and
Fraser A.
Armstrong
 *a
*a
aUniversity of Oxford, Department of Chemistry, South Parks Road, Oxford, UK. E-mail: stephen.carr@rc-harwell.ac.uk; fraser.armstrong@chem.ox.ac.uk
bResearch Complex at Harwell, Rutherford Appleton Laboratory, Harwell Campus, Didcot, UK
cCollege of Chemistry, Beijing Normal University, 100875, Beijing, China
dDepartment of Inorganic Spectroscopy, Max Planck Institute for Chemical Energy Conversion, Stiftstraße 34-36, 45470 Mülheim an der Ruhr, Germany
eUniv Grenoble Alpes, CNRS, CEA, IRIG, Laboratoire Chimie et Biologie des Métaux, 17 Rue Des Martyrs, F-38054 Grenoble Cedex, France
fSchool of Chemistry, The University of Leicester, University Road, Leicester, LE1 7RH, UK
First published on 25th July 2023
Abstract
The active site of [NiFe]-hydrogenases contains a strictly-conserved pendant arginine, the guanidine head group of which is suspended immediately above the Ni and Fe atoms. Replacement of this arginine (R479) in hydrogenase-2 from E. coli results in an enzyme that is isolated with a very tightly-bound diatomic ligand attached end-on to the Ni and stabilised by hydrogen bonding to the Nζ atom of the pendant lysine and one of the three additional water molecules located in the active site of the variant. The diatomic ligand is bound under oxidising conditions and is removed only after a prolonged period of reduction with H2 and reduced methyl viologen. Once freed of the diatomic ligand, the R479K variant catalyses both H2 oxidation and evolution but with greatly decreased rates compared to the native enzyme. Key kinetic characteristics are revealed by protein film electrochemistry: most importantly, a very low activation energy for H2 oxidation that is not linked to an increased H/D isotope effect. Native electrocatalytic reversibility is retained. The results show that the sluggish kinetics observed for the lysine variant arise most obviously because the advantage of a more favourable low-energy pathway is massively offset by an extremely unfavourable activation entropy. Extensive efforts to establish the identity of the diatomic ligand, the tight binding of which is an unexpected further consequence of replacing the pendant arginine, prove inconclusive.
1. Introduction
Hydrogenases, which catalyse the production and oxidation of molecular hydrogen (H2), are not only essential for many microorganisms but also serve as paradigms for understanding, in detail, the importance of proton-coupled electron transfer (whether concerted or not) in enzyme mechanisms.1 Attached to an electrode, hydrogenases display highly efficient H2 activation,2 and as ‘reversible’ electrocatalysts,1 they can inspire the atomic-level design of novel catalysts for future renewable energy technologies, most obviously in the development of green H2 as a major resource and fuel.3–6 The two membrane-bound group 1 (ref. 7) [NiFe]-hydrogenases of Escherichia coli serve as exemplars for catalyst design since they are highly similar, structurally, yet display subtly diverging catalytic properties. Hydrogenase-1 (‘Hyd-1’) is active only in the H2 oxidation direction at neutral pH,8 but combines unidirectionality with the second desirable property of O2-tolerance, defined as the ability to sustain H2 oxidation activity in the presence of O2.9–11 In contrast, ‘Hyd-2’ displays typical O2 sensitivity, but is capable of efficient H2 production as well as H2 oxidation under standard physiological conditions.12,13The buried active site of [NiFe]-hydrogenases contains a highly conserved inner coordination shell: the Ni atom is coordinated by four strictly conserved cysteine thiolates, two of which form a bridge to a Fe(CO)(CN)2 fragment that appears to remain as a low-spin FeII site during catalysis,14,15 while the other two bind in a terminal manner (Fig. 1). Hydrogenases are usually inactive upon aerobic isolation and characterised by the presence of a hydroxide ligand located in a bridging position between the Ni(III) and Fe(II) atoms (Ni-B, ‘ready’ state) and with additional oxygenation of a cysteine ligand (Ni-A, ‘unready’).2,16 Research into synthetic, structural analogues of hydrogenases3,6,16–18 as well as a Ni-substituted rubredoxin19,20 have demonstrated that the inner coordination shell alone is ineffective at catalysing reversible and bidirectional H2 activation.21,22 ‘Bidirectionality’ refers to the ability of the catalyst to function in either the proton reduction or H2 oxidation direction depending on the applied potential, whereas ‘reversibility’ refers to the ability of the catalyst to be bidirectional with minimal overpotential requirement either side of the equilibrium potential. It follows therefore that a reversible catalyst is always bidirectional, but a bidirectional catalyst may not necessarily display reversibility.23 Looking to the outer coordination shell for further insight, a number of catalytically-important, non-coordinating amino acids have been identified in various [NiFe]-hydrogenases, and two of these appear crucial for activity. One such residue is a glutamate24–26 (E28/E14 in Hyd-1/Hyd-2 respectively), the carboxylate of which lies close (<4 Å) to the Ni-coordinating terminal cysteine C576/C546;25,27 the other is a strictly conserved ‘pendant’ arginine27,28 (R509/R479) that is located in a canopy immediately above the catalytic metals.
It has long been proposed that the H2 molecule is cleaved or formed heterolytically, i.e. involving a transition state of the form H−⋯H+, and studies of reduced catalytic intermediates reveal a hydridic H-atom coordinated in a bridging position between Ni and Fe.2,16,29–31 It has also been proposed that the primary base for abstraction of H+ during the initial heterolytic cleavage is the Ni–S thiolate of C576/546 (Fig. 1)16 and it has been postulated that this proton is then transferred to the nearby glutamate residue (E28/E14).26,32,33 However, recent work has shown that the glutamate in E. coli Hyd-1 is not necessary for the initial deprotonation step of H2 activation;34 furthermore, alteration of this residue does not render the enzyme completely inactive as in other homologues.26,33–35
The importance of the pendant arginine (R509/R479) is not surprising considering its conservation and its close proximity and position above the intermediate hydrido ligand. The exact role of the arginine is disputed in the literature and only recently have experiments been performed that were able to demonstrate a crucial mechanistic relevance across various enzyme counterparts and techniques.27,28,36,37 Studies based on computational analyses37 and kinetic and spectroscopic analyses36 suggest critical roles for the arginine in polarising H2 for proton abstraction by an active site cysteine, controlling substrate/ligand access, conferring conformational rigidity to keep the active site in a catalytically-accessible state, and mediating proton transfer between tautomeric reduced active-site states. In our recent work, based on structural and kinetic data for the arginine-to-lysine substituted variant of Hyd-1 (R509K), we postulated a direct role of the guanidine/guanidinium head group of R509 to act as a primary Lewis base during catalysis and, together with the active site metal (Lewis acid), activate H2 as a Frustrated Lewis Pair (FLP)38,39 – an atomic-level mechanism that is generally accepted for [FeFe]-hydrogenases40,41 and the best functional mimics.3
As with other examples where arginine plays a direct role in catalysis, such a proposal carries the assumption that the high pKa of the guanidinium headgroup is somehow beneficial: either it is significantly lowered from that of free arginine (>12)42 by the local environment or there is direct, transient coupling of deprotonation to other steps involving a strong base, in this case H–H bond formation or cleavage.27,43–52 Proton transfer to an adjacent and strongly basic intermediate, such as a metal hydrido species, could be a favourable step in a sequence.53 For several enzymes, including various flavoenzymes and ascorbate peroxidase, there is a compelling case for direct participation of an arginine guanidinium/guanidine group as a proton relay.43–48,54–57 These examples include recent neutron diffraction studies revealing a deprotonated (guanidine) side chain in an environment otherwise lacking any special features that would stabilise such a state.54 Such direct evidence proves that calculations alone cannot rule out a direct acid–base role for arginine and it is thus safe to say that any general objection to a direct role for arginine in catalysis is untenable.
We recently designed a unique overexpression system for Hyd-2,12,25 greatly facilitating the ability to produce variants analogous to those available for Hyd-1, thus allowing the pendant arginine question to begin to be addressed with a standard, O2-sensitive [NiFe]-hydrogenase that is naturally proficient in H2 production as well as oxidation.12,13 We therefore substituted R479 for lysine in Hyd-2 and carried out a detailed study of the structure and kinetics of the R479K variant to make a ‘like-for-like’ comparison with Hyd-1 R509K. The crystallographic data and protein film electrochemistry results for R479K confirm the vital importance of the pendent arginine for catalytic H2 production as well as oxidation. Additionally, replacing the arginine by lysine allows formation of an inactive oxidised state of the enzyme, in which the vacant coordination site on the Ni is occupied by a tightly-bound diatomic ligand.
2. Methods
2.1 Molecular biology
Cloning steps for the overproduction of Hyd-2 have been described previously.12 The R479K mutation was made on pMAK-hybC plasmid constructs, and following confirmation by DNA sequencing, the codon changes were each transferred to strain HJ001-hyp (Table S1†).27,58,592.2 Enzyme production and purification
All protein was over-produced from E. coli MC4100-derived K-12 strain ‘HJ001-hyp’12,59 or the relevant variant strain transformed with plasmid pOC for over-expression of the hybO gene containing a 3′-hexahistidine tag (Table S1†).12 This system produces enzyme that is not translocated to the membrane.25 Enzymes were purified as previously described.12 Briefly, the catalytic cores of native Hyd-2 and R479K (membrane extrinsic subunits, HybOC) were isolated from the cell cytoplasm by Ni-affinity chromatography. A second purification step was performed: the resulting product was used for steady-state solution kinetics and PFE experiments, and to prepare samples for crystallography, EPR and IR spectroscopy. For clarity, the native over-produced Hyd-2 (otherwise abbreviated Hyd-2-NOP) and variant over-produced R479K enzymes will be referred to as ‘Hyd-2’ and ‘R479K’ respectively throughout this paper.2.3 Protein film electrochemistry
All electrochemical experiments were performed with a gastight, water-jacketed electrochemical cell contained inside an anaerobic (N2) glovebox (MBraun or Belle Technologies, O2 <3 ppm). Precise gas mixtures were created from component gases of the highest purity (BOC; H2 >99.99% purity, O2 99.5% purity, CO 99.97% purity and Ar 99.998% purity) using mass flow controllers (Sierra Instruments). Electrochemical control was achieved using an Autolab potentiostat (PGSTAT128N) controlled by Nova software (EcoChemie). The pyrolytic graphite ‘edge’ (PGE) working electrode (area 0.03 cm2) was rotated at a constant speed (ω > 1000 rpm) to minimise any limitations from H2 mass transport either during its oxidation (as substrate) or production (as inhibitor). A Pt wire (counter) and saturated calomel electrode (SCE) reference completed the circuit, the SCE electrode being housed in a side arm containing 0.1 M NaCl that was electrochemically connected to the main compartment via a Luggin capillary. All potentials quoted have been converted to the Standard Hydrogen Electrode (SHE) scale using the correction ESHE = ESCE + 0.241 at 25 °C.60To prepare each enzyme film, the PGE electrode was abraded with sandpaper (P400 Hookit) and thoroughly rinsed with ultrapure water. Enzyme (1–5 µg) was applied to the working electrode by pipetting directly onto the electrode surface, or immersing the electrode in a dilute enzyme ‘forming’ solution whilst monitoring activity as a function of potential in 100% H2. All enzyme films were prepared in this way unless otherwise stated, i.e., for some experiments with R479K a ‘modified’ electrode system was used, as previously described28,61,62 wherein enzyme is covalently linked to pyrene–butyric acid on multi-walled carbon nanotubes. All enzyme films were extensively ‘activated’ (i.e., the electrode was poised at a reducing potential under H2 until no further increase in activity is observed) to reduce oxidised species formed during aerobic purification. The electrode potential was stepped between −0.66 V (300 s) where activation is faster for readily activated state(s) and −0.40 V (15 s) to monitor the H2 oxidation current. The progress of activation under H2 was monitored across a wider potential range by cyclic voltammetry at a scan rate of 30 mV s−1. Where necessary, the activation period was extended for several hours or overnight.
2.4 Steady-state activity measurements
Prior to steady-state activity measurements, enzyme stocks were reductively activated by purging with 100% H2 (BOC, >99.99% purity) for 12–18 hours either in the absence or presence of electrochemically reduced methyl viologen (1 mM to give >1400-fold excess). The initial velocity of H2 oxidation by R479K was measured as previously described12,28 using 2 mM benzyl viologen as electron acceptor (E = −358 mV, ε = 8.4 mM−1 cm−1 at 604 nm (ref. 63)) in H2-saturated, 100 mM potassium phosphate buffer at pH 6.0. The H2-only-activated enzyme stock was then purged with Ar for at least 12 hours. The H2 production activity was measured using reduced methyl viologen as electron donor (E = −446 mV, ε = 13.9 mM−1 cm−1 at 604 nm (ref. 63)) as previously described12,13 in Ar-saturated 100 mM potassium phosphate buffer at pH 6.0. Only enzyme activated in the absence of methyl viologen was used for H+ reduction activity measurements.2.5 Protein crystallization
Enzymes were crystallized using conditions described elsewhere.12 Purified R479K was concentrated to 5 mg mL−1 (assessed by Bradford assay,64 Sigma) using centrifugal concentrators with a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecular weight cut-off (Vivaspin). The as-isolated enzyme was transferred to an anaerobic glove box for crystallisation without further treatment; reduced enzyme was obtained by exposing the enzyme solution to a flow of 100% H2 for 20 h at 4 °C in an anaerobic (N2) glovebox (Belle Technologies, <0.3 ppm O2); CO-inhibited enzyme was produced by further exposure of reduced enzyme to 100% CO (BOC 100% purity) for 20 minutes. Re-exposure to O2 following extensive reductive activation was performed using 100% O2 (BOC 99.9999% purity). Crystals of each form of R479K were obtained using the sitting drop vapour diffusion technique: 1 µL of protein solution was mixed with an equal volume of de-oxygenated crystallization solution (100 mM Bis-Tris pH 5.5–5.9, 200 mM MgCl2 and 18–22% w/v PEG 3350). Crystallization trays were set up manually under anaerobic conditions (Belle Technologies, <0.3 ppm O2) and incubated at 23 °C (ambient glovebox temperature). Regardless of gas treatments, long rod-shaped crystals appeared after approximately 24 h; these were manually transferred to an anaerobic cryo-protecting solution (100 mM Bis-Tris pH 5.5–5.9, 200 mM MgCl2, 23% w/v PEG 3350 and 15% v/v glycerol) for 30–60 s before flash-cooling in liquid nitrogen.
000 molecular weight cut-off (Vivaspin). The as-isolated enzyme was transferred to an anaerobic glove box for crystallisation without further treatment; reduced enzyme was obtained by exposing the enzyme solution to a flow of 100% H2 for 20 h at 4 °C in an anaerobic (N2) glovebox (Belle Technologies, <0.3 ppm O2); CO-inhibited enzyme was produced by further exposure of reduced enzyme to 100% CO (BOC 100% purity) for 20 minutes. Re-exposure to O2 following extensive reductive activation was performed using 100% O2 (BOC 99.9999% purity). Crystals of each form of R479K were obtained using the sitting drop vapour diffusion technique: 1 µL of protein solution was mixed with an equal volume of de-oxygenated crystallization solution (100 mM Bis-Tris pH 5.5–5.9, 200 mM MgCl2 and 18–22% w/v PEG 3350). Crystallization trays were set up manually under anaerobic conditions (Belle Technologies, <0.3 ppm O2) and incubated at 23 °C (ambient glovebox temperature). Regardless of gas treatments, long rod-shaped crystals appeared after approximately 24 h; these were manually transferred to an anaerobic cryo-protecting solution (100 mM Bis-Tris pH 5.5–5.9, 200 mM MgCl2, 23% w/v PEG 3350 and 15% v/v glycerol) for 30–60 s before flash-cooling in liquid nitrogen.
2.6 X-ray data collection and structure determination
X-ray diffraction data were collected at beamline i04-1, Diamond Light Source, at a wavelength of 0.92 Å using a Pilatus 6M hybrid pixel array detector. All data were collected at 100 K using a helical line scan to minimise photo-reduction. Data reduction was performed using DIALS65 and Aimless.66 Initial phase estimates were generated by molecular replacement within Phaser67 using the Hyd-2 pdb file 6EHQ12 as a search model. The electron-density maps were inspected using COOT,68 residue 479 was manually changed to lysine and crystallographically ordered water molecules assigned. The resulting model was refined using REFMAC5 (ref. 69) followed by additional rounds of manual rebuilding and refinement until no further improvements were possible. Structure analysis was performed using a combination of CastP,70 Caver2 (ref. 71) and Pymol.722.7 EPR spectroscopy
Continuous-wave EPR spectroscopy was carried out on as-isolated R479K samples at concentrations of 90–400 µM in 100 mM MES, 100 mM NaCl, 10% glycerol, pH 6.0. An X-band (9.1–9.9 GHz) Bruker EMX spectrometer equipped with a low-temperature helium flow cryostat (Oxford Instruments) was used to make measurements at 10 K and 80 K. Thin-walled quartz EPR tubes (Fluorochem) were used, high precision (1.6 mm) tubes for the 400 µM sample and standard 4 mm tubes for all other samples. Data were collected at a microwave power of 2.00 mW and a modulation amplitude of 1.0 mT. Spectra were averaged over >12 hours for the 400 µM sample. EPR simulations were performed using the EasySpin 5.2.25 routines for a Matlab scripting environment (The Mathworks, Natick, NJ).732.8 Infrared spectroscopy
Solution-based transmission infrared (IR) experiments were performed on 5–6 µL samples of 1 mM R479K in 20 mM Tris, pH 7.2, 150 mM NaCl sandwiched between two CaF2 windows (31.8 mm diameter, 1.5 mm thickness, Crystan) separated by a 25 µm Teflon spacer (Kromatek Ltd) coated with vacuum grease and enclosed in a commercial IR transmission cell (PIKE). Spectra were measured at room temperature inside an anaerobic, dry glovebox (Glove Box Technology Ltd, <2 ppm of O2, <85 °C dew point) on a Bruker Vertex 80 FTIR spectrometer equipped with a mercury cadmium telluride (MCT) detector cooled with liquid N2. Each spectrum was recorded as an average of 1024 interferograms using OPUS software (Bruker) in the double-sided, forward–backward mode with a resolution of 2 cm−1, an aperture setting of 1 mm and a scan velocity of 20 Hz. Data were processed using home-written routines in MATLAB™ and OPUS software (Bruker). The figures were prepared in Origin graphical software (OriginLabCorp).All samples were stored under liquid nitrogen or at −80 °C in a sealed anaerobic vial until use. Prior to measurements, as-isolated samples were reductively activated: large volume dilute samples (>50 µL, ∼100 µM) were flushed with humidified 100% H2 for 24 hours in an anaerobic glovebox (Belle Technologies, O2 <3 ppm), whereas small-volume, concentrated samples (<10 µL at ∼1 mM) were activated under 4 bar H2 (Büchi Tinyclave pressure vessel) inside an anaerobic glovebox (Glove Box Technology Ltd, <3 ppm O2) for 19 hours. The various gas treatments of samples for identification of the diatomic ligand included an in-line ultra-high capacity O2 scrubber (Restek™ Super-Clean™) and were performed as explained in Results. All gases were obtained from BOC (H2 >99.99% purity, O2 99.5% purity, CO 99.97% purity and Ar 99.998% purity).
In-crystallo microspectroscopic electrochemical IR experiments were performed using single crystals of R479K enzyme under electrochemical control as previously described.74 Prior to any experiments, regardless of the oxidised/reduced state of the samples, the crystals were reductively activated, electrochemically, in a microspectroscopic electrochemical cell at ca. −600 mV until no further changes were observed in the vCO and vCN bands in the IR spectra over a period of approximately 10 min. Typically, this activation period was at least 1 h. Unless otherwise stated, all in-crystallo microspectroscopic electrochemical data were collected using a Vertex 80v FTIR spectrometer with a Hyperion 3000 IR microscope (Bruker) at Diamond Light Source, MIRIAM beam line B22. The high-sensitivity photovoltaic MCT 50 µm pitch detector was cooled using liquid N2. The stage of the IR microscope was contained in an N2-purged Perspex housing to minimise contamination by atmospheric O2 and water vapour. Each spectrum was recorded using OPUS software (Bruker) in the double-sided, forward backward mode as an average of 512 interferograms at 80 kHz scanner velocity and 4 cm−1 resolution using a 6 mm aperture. Electrochemical control was achieved using an Autolab 128N potentiostat and Nova 1.10 software (Metrohm). All subsequent in-crystallo IR and electrochemical data analysis was performed using Origin graphical software (OriginLabCorp).
2.9 QM/MM calculations
QM/MM geometry optimizations were performed using the ASH multiscale modelling program (https://ash.readthedocs.io), version 0.7, developed by R. Bjornsson. ORCA version 5.0 (ref. 75) was used as QM program and the OpenMM library version 7.5 (ref. 76) as MM program, both programs being interfaced to ASH. The CHARMM36 force field77 was used to describe the protein and the TIP3P forcefield78 to describe water, with a simple nonbonding model for the metal clusters (based on Hirshfeld-calculated atom charges and Lennard-Jones parameters for sulfide from CHARMM). An additive QM/MM expression was used, with electrostatic embedding to describe the QM-MM electrostatic interaction, linkatoms used to cap the QM-MM boundary and a charge-shifting scheme as implemented in ASH. The QM-region was described using density functional theory with the r2SCAN functional79–81 and the D4 dispersion82 correction with a ZORA83,84 scalar relativistic Hamiltonian (polarized by the MM point charges). The relativistically recontracted ZORA-def2-TZVP basis set85,86 was used. The resolution of identify approximation87 was used to speed up Coulomb integrals using a decontracted Coulomb-fitting basis set88 (SARC/J keyword in ORCA). Calculated vibrational frequencies were scaled by the factor 0.9781. This scaling factor was derived from a calculation of free CO at the same level of theory compared to the experimental frequency. Precise details of all computational methods are provided as ESI.†3. Results
3.1 Production of R479K
All enzymes were overproduced and the R479K mutation was incorporated into the overexpression system as reported elsewhere.12 Previous investigations revealed that the enzymatic characteristics of native, over-produced Hyd-2 are identical with those of natively (chromosomally) expressed Hyd-2; therefore differences in enzymatic parameters from over-produced variants can be attributed to the point mutation.12 The variant protein was over-produced to similar yields and purity as over-produced native Hyd-2 and both enzymes purified as the HybOC dimer (Fig. S1†).3.2 Steady-state solution activities
The steady-state H2 oxidation and H+ reduction activities were measured for R479K at pH 6.0 (Table 1) and are clearly much lower than observed for Hyd-2.12 However, in view of the difficulty in removing the diatomic ligand bound tightly to as-isolated R479K (see later), it was important to establish the fraction of enzyme that is actually active. For H2 oxidation, R479K that had been activated under H2 in the presence of reduced methyl viologen showed an approximately five-fold increase in rate compared to enzyme that had been activated without methyl viologen. Judging from the data in Table 1 (which reports the rates found in the absence of reduced methyl viologen to allow for direct comparison to other literature) we could therefore be confident that regardless of the method of activation, the H2 oxidation activity of R479K lies between 2-10% of the native enzyme. For proton reduction, the rate of the R479K variant was less than 0.4% that of native Hyd-2, having activated the enzyme in the presence of H2 alone due to the difficulty in removing the excess methyl viologen from the activated sample used for H2 oxidation measurements. Given the activity range achieved for H2 oxidation we applied this five-fold difference to the H+ reduction activities measured to give a range of H+ reduction activity for R479K that is 0.4–2% of Hyd-2.| Parameter | Native Hyd-2 (ref. 12) | Hyd-2 R479K | Native Hyd-1 (ref. 28) | Hyd-1 R509K28 |
|---|---|---|---|---|
| a Measured with benzyl viologen (E = −358 mV (ref. 63)). b Measured with methylene blue (E = 11 mV (ref. 63)). c Measured with methyl viologen (E = −446 mV (ref. 63)). d Measured at −0.175 V. e Measured at 0 V. f Measured at −0.200 V. g Measured at 0 V. h Measured at −0.45 V. i Measured at 20 °C; n.d. not determined. j N.B. all absolute steady-state turnover rates reported here were measured using enzyme activated in the presence of H2 alone, i.e. in the absence of reduced methyl viologen. | ||||
| H2 oxidation steady-state turnover rate (s−1) | 218.0 ± 17.0a | 5.0 ± 1.0a | 257.4 ± 30.4b | 3.3 ± 1.1b |
| H+ reduction steady state turnover rate (s−1) | 7.0 ± 1.0c | 0.03 ± 0.01c | n.d. | n.d. |
 (µM) (µM) |
23.0 ± 7.0d | 214.0 ± 45.0d | 19.8 ± 4.1e | 6.5 ± 1.1e |
| ΔH‡ (kJ mol−1) H2 oxidation | 43.7 ± 2.0f | 23.0 ± 2.6f | 44.7 ± 0.9g | 39.4 ± 1.4g |
| ΔH‡ (kJ mol−1) H+ reduction | 39.1 ± 0.6h | 38.1 ± 0.5h | n.d. | n.d. |
 at −0.660 V (µM) at −0.660 V (µM) |
240.0 ± 10.0 | 1077.0 ± 96.0 | n.d. | n.d. |
| K i CO at −0.175 V (µM) | 3.1 ± 0.6i | 3.8 ± 0.5i | n.d. | n.d. |
3.3 Protein film electrochemical analysis of kinetic parameters
 values (Table 1) at pH 6.0 and 30 °C for Hyd-2 and R479K were determined from currents measured at different potentials in cyclic voltammograms2,28,94 recorded at a scan rate of 5 mV s−1 over a range of H2 concentrations (Fig. S3†). The conditions were returned to 100% H2 every fourth cycle to correct for slow film loss94 during the course of the experiment. Results are shown in Fig. 3 alongside relevant data obtained for E. coli Hyd-1, to allow comparisons to be made. For Hyd-2,
values (Table 1) at pH 6.0 and 30 °C for Hyd-2 and R479K were determined from currents measured at different potentials in cyclic voltammograms2,28,94 recorded at a scan rate of 5 mV s−1 over a range of H2 concentrations (Fig. S3†). The conditions were returned to 100% H2 every fourth cycle to correct for slow film loss94 during the course of the experiment. Results are shown in Fig. 3 alongside relevant data obtained for E. coli Hyd-1, to allow comparisons to be made. For Hyd-2,  (23 ± 7 µM at −0.175 V) does not change significantly with potential: in contrast, R479K shows a much higher
(23 ± 7 µM at −0.175 V) does not change significantly with potential: in contrast, R479K shows a much higher  (214 ± 45 µM at −0.175 V) with a significant potential dependence (Fig. 3A), the
(214 ± 45 µM at −0.175 V) with a significant potential dependence (Fig. 3A), the  value increasing two-fold between −0.35 and −0.20 V. As seen in Fig. 3B, the large increase in value for R479K relative to Hyd-2 is not observed for the corresponding mutation in Hyd-1 (R509K), but is observed for the Hyd-1 D118A variant (D118 corresponds to D103 in Hyd-2). Substantial anaerobic inactivation of R479K (Fig. 2B) impaired precise measurement of
value increasing two-fold between −0.35 and −0.20 V. As seen in Fig. 3B, the large increase in value for R479K relative to Hyd-2 is not observed for the corresponding mutation in Hyd-1 (R509K), but is observed for the Hyd-1 D118A variant (D118 corresponds to D103 in Hyd-2). Substantial anaerobic inactivation of R479K (Fig. 2B) impaired precise measurement of  at potentials more positive than −50 mV, therefore the measured
at potentials more positive than −50 mV, therefore the measured  at higher potentials is likely an underestimate, since even a 100% H2 atmosphere (corresponding to a concentration of approximately 0.76 mM at 20 °C) is below 4 ×
at higher potentials is likely an underestimate, since even a 100% H2 atmosphere (corresponding to a concentration of approximately 0.76 mM at 20 °C) is below 4 ×  . Measurements of
. Measurements of  were also carried out at 20 °C for R479K (Fig. S4†) due to a convex Eyring plot for H2 oxidation (see later).
were also carried out at 20 °C for R479K (Fig. S4†) due to a convex Eyring plot for H2 oxidation (see later).
 | ||
Fig. 3
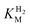 for (A) Hyd-2 and R479K is plotted next to the reported for (A) Hyd-2 and R479K is plotted next to the reported  of (B) Hyd-1, R509K, and D118A.28 of (B) Hyd-1, R509K, and D118A.28 is calculated from Hanes–Wolf or Michaelis Menten plots populated with data from 5 mV s−1 scans at 10 different H2 concentrations (0.3–100%), see ESI.† Films were returned to 100% H2 every fourth scan to account for film loss. Error bars represent the standard deviation of three repeats. Other conditions: 30 °C, ω = 3000 rpm, pH 6.0, total gas flow rate of 1000 sccm. is calculated from Hanes–Wolf or Michaelis Menten plots populated with data from 5 mV s−1 scans at 10 different H2 concentrations (0.3–100%), see ESI.† Films were returned to 100% H2 every fourth scan to account for film loss. Error bars represent the standard deviation of three repeats. Other conditions: 30 °C, ω = 3000 rpm, pH 6.0, total gas flow rate of 1000 sccm. | ||
Comparing the proton reduction currents at different H2 concentrations showed that R479K is less prone to product inhibition than Hyd-2 (Fig. 2 and S3†).13,95 Indeed, competitive inhibition by H2 calculated by poising the enzyme film at −660 mV while altering the H2 concentration (Fig. S5†) showed that R479K experiences a  of approximately 1.1 mM compared to 0.24 mM for native Hyd-2 (Table 1).13
of approximately 1.1 mM compared to 0.24 mM for native Hyd-2 (Table 1).13
 | ||
| Fig. 4 Determining the stability of oxidised inactive states for Hyd-1 and Hyd-2 enzymes. After full inactivation under anaerobic, oxidizing conditions, at high pH, the gas is switched to H2 (see Fig. S6†) buffer exchanged and the potential is slowly (0.1 mV s−1) lowered from the highly oxidizing potential (approx. +0.36 V). Other conditions: 30 °C, ω = 1000 rpm, pH 6.0, 100% H2 at 1000 sccm. The Hyd-2 and R509K data have been smoothed with the Savitzky–Golay method, 41 points per window using Origin software, due to lower signal-to-noise. | ||
 between the native enzyme and R479K (Fig. 3A).13,96 Importantly, replacement of CO by H2 is rapid and reversible in both cases.
between the native enzyme and R479K (Fig. 3A).13,96 Importantly, replacement of CO by H2 is rapid and reversible in both cases.
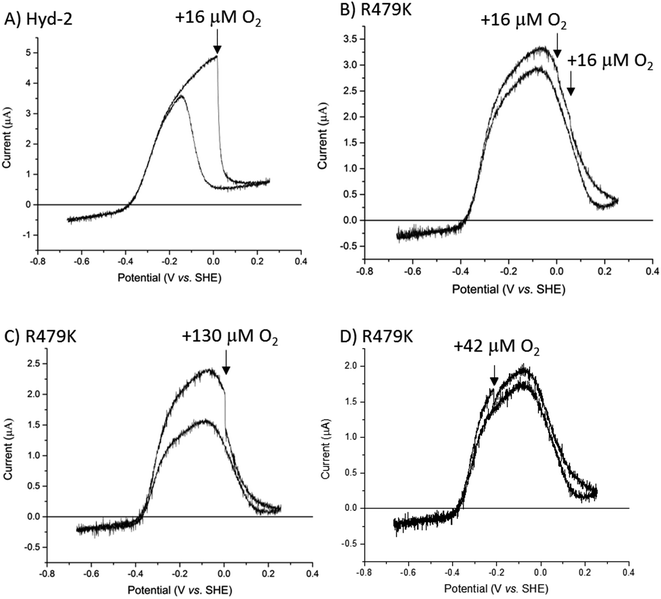
As an inhibitor of [NiFe]-hydrogenases, O2 is much more complicated than CO. The O2 sensitivity of Hyd-2 was measured first by injecting O2-saturated buffer into the electrochemical cell under a constant flow of H2. In this experiment, the transiently invading O2 begins to be removed almost immediately from the cell under the flow of H2.96 The immediate response to the burst of O2 and the duration of the effect of O2 on the current (H2 oxidation rate) depends on (a) the rate that O2 binds to the active site and (b) the rate at which the enzyme can deliver electrons and protons to the active site to ensure its complete reduction to harmless H2O.9–11,97 Normally, the final stage of a successful ‘rescue’ is reduction of the Ni(III)-µ(OH−) product (Ni-B) and transport of the resulting water molecule away from the active site. As we found in earlier studies, Hyd-2 is highly sensitive to O2 attack:13 injection of O2 to give a short-lived burst of just 16 µM O2 almost fully inactivates the enzyme (Fig. 6A) and activity does not recover until the potential is lowered below 0 V during the reverse scan. In contrast, the cyclic voltammetry for R479K under 100% H2 was almost unaffected by two sequential injections of the same amount of O2-saturated buffer (Fig. 6B). Indeed, the voltammetry was only significantly disturbed by O2 at concentrations around 130 µM (Fig. 6C), and exposure to this level of O2 still only lowered the current by approximately half the level seen when delivering 16 µM O2 to R479K. However, R479K does not recover from the O2 attack until the applied potential is very reducing, and the current level is not fully recovered until the following scan (Fig. S8†). Exposure of R479K to O2 (42 µM) at a potential where there is no obvious anaerobic inactivation (−0.215 V) also showed minimal effect on H2 oxidation ability (Fig. 6D).
To examine more closely the apparently ‘robust’ defence that R479K offers against transient exposure to O2, we tested the effects of prolonged exposure to O2 during turnover (H2 oxidation) and non-turnover conditions (Fig. 7) – the non-turnover experiments (argon/O2 in the absence of H2) more closely resembling conditions the enzymes would experience during isolation and purification. After measuring cyclic voltammograms to establish activity, experiments were performed in ‘chronoamperometric mode’ at a constant potential of −0.1 V, at which there is minimal anaerobic oxidative inactivation of both Hyd-2 and the R479K variant (Fig. 7), and minimal interference from background (direct) O2-reduction at the graphite electrode. The reference experiment against which to compare all others was conducted with Hyd-2 (Fig. 7A). Starting first under 100% H2 (approximately 0.8 mM at 20 °C) then 80% H2/20% Ar, an essentially instantaneous and complete extinction of activity was observed when 80% H2/20% O2 was introduced into the cell headspace, equivalent to 0.64 mM H2/0.29 mM O2. When the H2 level was restored to 100%, 57% of the activity was recovered spontaneously, no further increase being observed following periods when a highly reducing potential was applied (by stepping the applied potential to −0.659 V for 1 or 10 minutes and then back to −0.1 V to check for recovery of current). For the R479K variant (Fig. 7C), in the directly comparable experiment to that just described for Hyd-2 (Fig. 7A), the introduction of O2 caused the current to decrease slowly to almost zero over the course of 30 minutes, followed by only a slow partial spontaneous recovery when the atmosphere was restored to 100% H2, reaching 7% after 30 minutes at −0.1 V. Application of more reducing conditions resulted in 44% recovery. In view of the high  value for R479K (Fig. 3A and Table 1) the experiment was repeated, starting with 100% H2, changing to 10% H2/90% Ar (80 µM H2), then 30% H2/70%Ar, before introducing 70% O2 (0.24 mM H2/1 mM O2) (Fig. 7E). Under this lower H2 level, the decrease in activity upon introducing O2 was faster than under 80% H2 (Fig. 7C) but still very slow compared with Hyd-2. No spontaneous recovery occurred when a 100% H2 atmosphere was restored (the slight increase in current is due to removal of O2 and thus removal of direct O2 reduction at the electrode surface13) and more reducing conditions resulted in up to 29% recovery.
value for R479K (Fig. 3A and Table 1) the experiment was repeated, starting with 100% H2, changing to 10% H2/90% Ar (80 µM H2), then 30% H2/70%Ar, before introducing 70% O2 (0.24 mM H2/1 mM O2) (Fig. 7E). Under this lower H2 level, the decrease in activity upon introducing O2 was faster than under 80% H2 (Fig. 7C) but still very slow compared with Hyd-2. No spontaneous recovery occurred when a 100% H2 atmosphere was restored (the slight increase in current is due to removal of O2 and thus removal of direct O2 reduction at the electrode surface13) and more reducing conditions resulted in up to 29% recovery.
In the non-turnover experiments (Fig. 7B, D and F), O2 (in Ar) was introduced to the cell headspace after first establishing activity then flushing all H2 from the system with 100% Ar. In all experiments the headspace was restored to 100% H2 after approximately 30 minutes of exposure to O2. At this point all traces showed a small current increase due to removal of background O2-reduction as O2 is flushed from the system. However, whereas the H2 oxidation activity of Hyd-2 increased gradually and spontaneously to 81% of the initial value after exposure to 20% O2, the R479K variant showed almost negligible recovery (2% when exposed to 70% O2 and 3% when exposed to 20% O2). Only after applying a much more reducing potential was R479K seen to recover significant H2 oxidation current (up to 59% and 43% after exposure to 20% and 70% O2, respectively). In contrast, Hyd-2 undergoes a high degree of recovery after switching to 100% H2 (81% in the experiment shown in Fig. 7B) which does not increase further after more reducing conditions are applied.
Immediate conclusions can be drawn from these experiments: reaction of O2 with Hyd-2 is fast and, regardless of whether or not H2 is also present, yields an inactive state that is readily activated upon reduction; in contrast, the reaction of O2 with R479K is slow, and largely escapes attention in transient exposure experiments monitored by cyclic voltammetry (Fig. 6), but yields an inactive state (or states) that requires much more forcing conditions for reactivation.
The ΔH‡ value obtained for H2 oxidation by Hyd-2 (ref. 12) at −0.2 V (Table 1 and Fig. 8B) agrees well with data measured previously.98 In contrast, the R479K variant showed a convex deviation of the Eyring plot (Fig. 8C) with the slope becoming zero or slightly negative at the most oxidising potential (0 V vs. SHE) and the highest temperatures. Based on data obtained below approx. 25 °C, where the gradient is steepest, a value of ΔH‡ = 23 kJ mol−1 was estimated for H2 oxidation by R479K at −0.20 V. Notwithstanding the likelihood that this value is an underestimate, it still represents a very large decrease (up to 21 kJ mol−1) compared to native Hyd-2 (ΔH‡ = 43.7 kJ mol−1). This result was consistent across the potential range −0.2 to 0 V (Fig. 8A).
The activation enthalpy for H+ reduction was also measured over a 10–45 °C temperature range (Table 1 and Fig. 9). Due to the effect of product (H2) inhibition12,13 data were obtained under 100% Ar at pH 6 by cyclic voltammetry between −0.65 and −0.17 V at 5 mV s−1. The Eyring plot in the H+ reduction potential window was linear across the entire temperature range used for Hyd-2 and R479K (Fig. 9B and C). The ΔH‡ value for R479K at −0.55 V is lower than that of Hyd-2 by approximately 1.5 kJ mol−1 and both enzymes show a slight potential dependence, the activation enthalpy increasing as the potential is lowered (Fig. 9A).
In addition to extracting values for ΔH‡, Eyring plot data can be used to calculate the activation entropy of the reaction (ΔS‡). In PFE, because the enzyme coverage of the electrode used in the temperature dependence experiments is unknown, the intercept of the y axis cannot be used to calculate ΔS‡, instead we rely on the value of ΔH‡ and the difference in steady-state rates measured in solution (Table 1) to estimate the difference in activation entropy for the two enzymes (ΔΔS‡) at a given potential (see ESI†).
Using the steady-state H2 oxidation rates in addition to the value of ΔH‡ calculated at −0.2 V, (to achieve a potential comparable with the benzyl viologen electron acceptor used in the assay, Table 1) the difference in activation entropy between Hyd-2 and R479K, ΔΔS‡, is estimated to be −101 to −87 J K−1 mol−1, which is approximately 50 J K−1 mol−1 more negative than the additional entropic barrier calculated by the same method for Hyd-1 vs. R509K.28 The activation enthalpy for H+ reduction by Hyd-2 is significantly lower in 100% Ar than when measured in 100% H2,98 which probably reflects a contribution from product inhibition. The difference in activation enthalpies for Hyd-2 and R479K for H+ reduction are not as dramatic as for H2 oxidation, but the reaction is still entropy-controlled with a ΔΔS‡ estimated to be −49 to −36 J K−1 mol−1.
3.4 Structures of Hyd-2 R479K
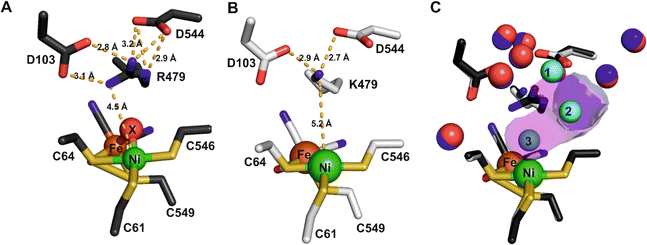
The structure of the aerobically purified ‘as isolated’ R479K variant (pdb accession 6SYO, Table S3†) was solved to a resolution of 1.25 Å (Fig. 10B and 11A). The active site is fully formed with minimal change in the position of sidechains relative to the native enzyme, except residue K479 (Fig. 10C), suggesting any changes in kinetic parameters are a consequence of replacing the guanidinium group with a primary amine. The K479 side chain adopts an atypical conformation by rotation about the Cγ–Cδ bond, allowing Nζ to contribute in the hydrogen bond network between the Oδ1 atom of the D103 and Oδ2 atom of D544. The rotated conformation increases both the distance between the head group of residue 479 and the Ni-atom (5.2 Å) and the diameter of the terminus of the hydrophobic channel immediately adjacent to the active site (2.8 Å) relative to the native enzyme (with values of 4.5 Å and 2.2 Å respectively).12 | ||
| Fig. 11 Electron density (2Fo-Fc and omit) maps and resultant structural models of the active site of R479K after different gas treatment regimes. (A) The as-isolated enzyme showing additional density adjacent to the Ni atom represented as diatomic ligand X–Y. (B) The reduced active site after exposure to an atmosphere of 100% H2 for 20 hours displays near spherical density (in 2Fo-Fc and omit maps) next to the Ni atom, suggesting a water molecule has replaced the diatomic ligand. For comparison, an omit map has been included for the water molecule next to D103, which also shows near spherical density. (C) After exposure of the reduced enzyme to pure oxygen, showing that electron density for diatomic ligand X–Y reappears in the active site. (D) Exposure of the reduced enzyme to CO indicates binding of CO to the Ni atom of the active site. The 2Fo-Fc electron density maps (blue mesh) are contoured at 1.5 r.m.s.d corresponding to an absolute value of 0.6 electrons per Å3. Omit maps calculated in the absence of Ni-coordinating ligand (pink mesh) are contoured at 5 r.m.s.d., an absolute value of 0.66 electrons per Å3. All hydrogen-bonds stabilising the bound ligands are shown as dashed lines and their lengths indicated. This figure is reproduced in the ESI (Fig. S15†) without the omit density to highlight the contiguous 2Fo-Fc density linking the Ni atom and bound ligand. Temperature factors for the components of each active site are shown in Table S5.† | ||
Exposure of the protein sample to H2 flow for 9 hours prior to crystallization resulted in crystals in which the occupancy of the diatomic species had decreased to approximately 40% of that in the as-isolated structure. Increasing the duration of H2 exposure to 20 hours appeared to remove the bound diatomic ligand, which was replaced by electron density assigned to a coordinated water molecule (Fig. 11B, pdb accession 6SZD, Table S3†). In a more aggressive procedure, a solution of R479K at 5 mg mL−1 was diluted 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a 1 mM solution of electrochemically reduced methyl viologen then purged with 100% H2 for 40 hours. Steady-state solution assays showed that R479K activated in this manner displayed approximately 5-fold higher activity, however, inspection of electron density maps suggested this method was no more effective at removing the bound diatomic ligand than simply exposing to H2 for 20 hours. The increase in steady-state rate upon additional electrochemical reduction is likely due to activation of other slow-to-reactivate states that are not crystallographically resolved.
1 with a 1 mM solution of electrochemically reduced methyl viologen then purged with 100% H2 for 40 hours. Steady-state solution assays showed that R479K activated in this manner displayed approximately 5-fold higher activity, however, inspection of electron density maps suggested this method was no more effective at removing the bound diatomic ligand than simply exposing to H2 for 20 hours. The increase in steady-state rate upon additional electrochemical reduction is likely due to activation of other slow-to-reactivate states that are not crystallographically resolved.
The orientation and geometries of the metal atoms and amino acids in the reduced structures were unchanged from those of the as-isolated structure (RMSD 0.04 Å), with the most significant difference being removal of the diatomic species. Re-exposure of the reduced sample to pure O2 for 1 hour resulted in large increases in electron density for the bound diatomic species, the occupancy level returning to approximately 80% (pdb accession 6SYX, Fig. 11C and Table S3†). The X–Y bond length is approximately 1.2 Å. The X-atom is located 1.85 Å from the Ni atom and the Y-atom is positioned so that it can form hydrogen bonds to the side chain Nζ atom of K479 and a crystallographically ordered water molecule. The resulting stable complex has a Ni–X–Y angle of 163°. A structurally identical complex was also formed upon anaerobic oxidation with DCIP, but with the diatomic ligand showing full occupancy, in agreement with IR-spectroscopy analysis (see below).
All R479K variant structures contain two additional water molecules in the active site (labelled 1 and 2 in Fig. 10C) as a result of replacing the bulky guanidinium head group of residue 479 with a smaller amine group. One of the new water molecules is positioned above the Nζ, atom, similar to that observed previously in the as-isolated structure of Hyd-1 R509K (compare H2O ‘1’ in Fig. 10C with Fig. S14†).27 Ordered water molecule ‘2’ (Fig. 10C) is positioned 4.2 Å above the sulphur of a terminal cysteine ligand (C546) proposed to be involved in the mechanism of H2 activation,16,26,32,33,103 and is 3.6 Å away from the Nζ atom of K479: this water molecule is also observed in Hyd-1 R509K, albeit with a lower occupancy (Fig. S14†). Additionally, H2-reduced R479K contains a third water molecule (H2O ‘3’ in Fig. 10C) that appears to be weakly coordinating (∼2.6 Å) the Ni atom alone, where the distance to the Fe atom is too long (4.0 Å) for it to be considered as a bridging ligand.
3.5 Spectroscopic analysis
 | ||
| Fig. 13 Solution-based transmission IR spectra of R479K following various gas treatments. IR spectra are presented from ∼1 mM samples of R479K prepared as (A) as-isolated, (B) H2-reduced, (C) H2-reduced, oxidised with mediators then exposed to CO, (D) H2-reduced then exposed to O2, (E) H2-reduced then oxidised with DCIP under strict anaerobic conditions, and (F) H2-reduced, left overnight and then oxidised with DCIP prepared the previous day to avoid traces of O2. All samples were prepared and the spectra collected at room temperature in an anaerobic glovebox. All spectra are shown on the same scale (bar), and the vCO and vCN bands are highlighted by dashed lines. Typical IR bands for native Hyd-2 and the oxygen sensitive hydrogenase from D. vulgaris Myazaki F are listed in Table S6.† | ||
An experiment was then conducted to examine the reaction of R479K with CO. For Hyd-2, a CO inhibited state, NiS–CO is formed when the active Nia-SI state (vCO 1945 cm−1) reacts with CO; NiS–CO exhibits a vCO for the endogenous carbonyl ligand at 1944 cm−1, and a new band for the exogenous, inhibitory CO on Ni(II) at 2054 cm−1. The solution potential of the reduced R479K sample was raised by addition of a cocktail of redox mediators (final concentration 0.5 mM in each) of anthraquinone 2 sulfonate (−151 mV), indigo carmine (−29 mV), and phenazine methosulfonate (+139 mV).74 For Hyd-2 this condition favours conversion of reduced states to Nia-SI; however for R479K no significant change in the spectrum was observed. The sample was then flushed with 100% humidified CO gas for 1 hour and left under a slight overpressure of 100% CO overnight (ca. 18 hours). Again, no significant changes were observed in the spectra for R479K (Fig. 13C). The fact that NiS–CO is not obviously formed with this variant is unsurprising given that it also does not exhibit a recognisable Nia-SI state.
To explore the reactivity of R479K with O2, an aliquot of the reduced variant was treated with a flow of humidified 100% O2 for 2 hours, and then left under a slight overpressure of O2 for an additional hour. The IR spectrum of the O2-exposed protein looked largely similar to the as-isolated sample, again showing a species with high wavenumber vCO at 2000 cm−1 together with unusually high wavenumber vCN bands (compare Fig. 13A and D). The fact that the ‘high wavenumber species’ appearing at 2000 cm−1 is lost upon H2 reduction, and reformed upon exposure to O2 demonstrate that this species undergoes reversible redox chemistry at the active site.
To ascertain whether the high-wavenumber species forms only in the presence of O2, the variant was exposed to oxidant under anaerobic conditions: an aliquot of H2-activated R479K (∼1 mM) was treated with an excess (2 mM final concentration) of 2,6-dichlorophenolindophenol (DCIP, 2e− donor/acceptor, Em = +317 mV vs. SHE74) prepared in an anaerobic buffer (20 mM Tris, pH 7.2, 150 mM NaCl). The resulting IR spectrum shows full conversion into the high-wavenumber species having bands at vCO 2000 cm−1 and vCN at 2096 and 2112 cm−1 (Fig. 13E). To exclude the possibility of trace O2 contamination, the procedure was repeated using a DCIP solution that was left inside an anaerobic glovebox overnight. As shown in Fig. 13F, the spectrum is reproduced, confirming that formation of the high wavenumber, oxidised species appears independently of any intentional exposure to O2. The fact that no corresponding set of high wavenumber IR bands is observed for the CO-exposed sample, and that there is no additional band which could be attributed to exogenous CO, indicates that it is not a CO-inhibited state. The cyanide region was also explored for additional CN bands in the anaerobically oxidized state and the raw data are shown in Fig. S16A.† Above 2112 cm−1 no bands are detected even in the second derivative spectrum (Fig. S16B†). Furthermore, the spectrum of the DCIP-oxidised sample shows the high wavenumber species as a single, majority form of the active site, eliminating the possibility that this new species arises from binding of CO released from enzyme molecules with degraded active site, as is observed for FeFe hydrogenases. To confirm this point, we recorded IR spectra on single crystals of R479K at two reducing potentials, −700 and −150 mV with and without CO, and saw no evidence for the species with νCO at 2000 cm−1 in these spectra. Additionally, we confirmed that anaerobic oxidation to form the high wavenumber species could be reproduced in crystals to a majority species with νCO at 2000 cm−1 which was insensitive to treatment with CO (Fig. S17 and ESI discussion†). Oxidative and reductive redox titrations performed on single crystals of R479K at pH 6.0 in the potential range −600 to +400 mV showed that the high wavenumber species is reversibly formed at potentials above +150 mV and is the majority (pure) state at +400 mV (Fig. S18 and ESI discussion†).
3.6 Computational analysis
The experimental shift (33–36 cm−1) of the 2076/2063 cm−1 CN “H2-reduced” bands to 2112/2096 cm−1 upon DCIP addition is consistent with oxidation of the cofactor. The simplest computational model considered which explains the experimental data, contains an oxidized Ni(III)–Fe(II) species without any exogenous ligand bound. This model gives 2122/2111 cm−1 for the CN bands, i.e. a predicted shift of 28–38 cm−1 compared to the H2O-bound Ni(II)–Fe(II) model. Analysis of the spin density clearly reveals Ni-based oxidation (with only a small spin population on the Fe ion) which nonetheless results in this shift of the Fe–CN modes. The experimental 2000 cm−1 peak in the experimental DCIP data could be explained by a strong shift (87 cm−1) of the Fe-bound CO band from 1913 cm−1 (“H2-red”) to 2000 cm−1 (“DCIP”) due to some charge-transfer away from the Fe site. The calculated Ni(III)–Fe(II) model without a ligand on Ni, gives a calculated CO mode of 1967 cm−1 compared to the 1914 cm−1 mode in the H2O-bound Ni(II)–Fe(II) model, amounting to a shift of 53 cm−1 compared to the experimental shift of 87 cm−1. While this comparison suggests that the experimental 2000 cm−1 peak is plausibly assigned to a strongly shifted Fe–CO mode upon Ni oxidation, the calculations of the Ni(III)–Fe(II) model without a ligand do not fully capture this effect.
We next considered Ni(III)–Fe(II) models with a terminal X–Y ligand bound to Ni. These models gave overall similar Fe–CN mode energies and Fe–CO modes as models without the ligand, suggesting the Fe–CN/CO modes to be primarily sensitive to the overall redox state and not Ni coordination. Overall, a Ni(III)–Fe(II) model with a relatively innocent ligand emerges as a plausible model for the DCIP-oxidized state.
We also considered the possibility of additional oxidation to give either a Ni(IV)–Fe(II) or a Ni(III)–Fe(III) species which could conceivably be responsible for a shift of an Fe-bound CO peak to 2000 cm−1. A recent structure of the oxygen-tolerant [NiFe] hydrogenase from H. thermoluteolus, reveals a glutamate coordinating to the Ni.106 In this form of the enzyme, the active site which displays a band at 1993 cm−1 was assigned as a Ni(IV)–Fe(II) species,107 although it has since been proposed that an antiferromagnetically coupled Ni(III)–Fe(III) system is more plausible.108 However, calculations starting from the X-ray structures in this work did not give electron structures consistent with either oxidation to Ni(IV) or Fe(III). Instead, all attempts to force the calculation (see ESI†) to give a local Fe(III) oxidation state resulted instead in S-centred oxidation occurring over one or more thiolates, which is highly unlikely to be a stable species.
4. Discussion
The dimensions of the substrate-access channel28 are unaffected by the mutation apart from a widening where it meets the active site, and CO inhibition (hence diffusion) is rapid, suggesting substrate availability at the active site is not affected significantly. However, crystal structures are static representations and do not account for transient flexing of the molecule, which could be altered in R479K. Comparison with the structure of Hyd-1 R509K shows the sidechain of K509 to be highly flexible relative to other canopy residues,27 and in the absence of the X–Y ligand, residue K479 displays similar flexibility, evidenced by the increased temperature factor of the side chain upon reductive activation of the enzyme (Fig. S15B and Table S5†). Ordering of such a mobile residue upon substrate binding would be entropically unfavourable, but ultimately locks the lysine side chain in the observed conformation: the temperature factor is reduced to that of the rest of the canopy. The observed structure could thus represent a kinetic trap.4.1 Consequences of replacing the active site arginine by lysine on catalytic kinetics
Once the inhibitory diatomic ligand has been removed, R479K displays catalytic activity that differs from Hyd-2 in two distinctive ways. First, the activity is much decreased, not only for H2 oxidation, as was originally observed when the equivalent arginine was replaced by lysine in Hyd-1,27 but also for H+ reduction. Although the R479K variant retains bidirectionality and behaves as a reversible electrocatalyst, there are large attenuations of catalytic activity as measured by steady-state solution kinetics, 2–10% for H2 oxidation (1 bar) and 0.4–2% for H+ reduction: values that may be compared alongside the 1% activity measured for H2 oxidation by Hyd-1 R509K. As with Hyd-1 R509K, the most striking observations lie in the temperature dependences of the rates. Unlike absolute turnover frequency, which in PFE always requires a knowledge of the electroactive coverage, the temperature dependence of rate, from which ΔH‡ is derived, is independent of coverage: ΔH‡ is therefore a diagnostic tool. A surprising feature of Hyd-1 R509K was that the low activity of that variant coincided with a significantly lowered activation enthalpy (by over 5 kJ mol−1), uniform over the temperature range 10–45°, which strongly suggested that the mechanism switches to one that is under entropic control (there being a greatly decreased probability of reaching the transition state). This behaviour is repeated for Hyd-2 R479K but with an additional interesting feature. Whereas the ≥98% drop in H+ reduction activity occurs with a uniform 1 kJ mol−1 decrease in ΔH‡ across the range 14–32 °C, which can again be interpreted in terms of a switch to entropic control, H2 oxidation shows a convex temperature dependence, ΔH‡ apparently varying from 23 kJ mol−1 at low temperature (a decrease of 24 kJ mol−1 compared to Hyd-2) to <0 kJ mol−1 above 23 °C. The low and variable value cannot be attributed to a favourable pre-equilibrium, which would have to involve a very stable intermediate, such as the Michaelis complex, the constant for which is actually higher than for Hyd-2.12,13 The absence of a significant kinetic isotope effect means that H-tunnelling, which could also account for a greatly lowered activation enthalpy, is not important in the rate-determining step. Despite the large decrease in rates, electrocatalysis remains reversible, as seen clearly in the voltammograms that show no evidence for additional overpotential requirement (any inflection) in the region where the current cuts the zero axis: this can be interpreted in terms of catalytic proton and electron transfers remaining tightly coupled rather than occurring in separate steps.1,109Convex Eyring plots have long been considered to signify a change in rate-determining step as the temperature is varied; however, attention has turned recently to a richer interpretation. Work by Arcus, Mulholland and co-workers has highlighted and explained the role of heat capacity changes over the course of a reaction, with ΔCp‡ emerging as an important parameter in enzyme kinetics within the framework of macromolecular rate theory (MMRT).110–113 In MMRT, convex Eyring plots will tend to appear as the specific heat of the transition state becomes sufficiently small compared to that for the precursor enzyme–substrate complex. Low ΔC‡p values are associated with a marked decrease in low-frequency vibrational modes, which can even originate in zones remote from the site of catalysis itself, and it is the large size of enzymes (and the consequent high number of such vibrational modes) that amplifies the contribution of ΔC‡p compared to the reactions of small molecules.
4.2 Options available for the tightly bound ligand in the as-isolated or oxidised states
The as-isolated state of R479K has an extremely tightly bound ligand (X–Y) coordinated to the Ni atom (Fig. S27†) that blocks catalytic activity. The electron density maps are of high enough resolution (Table S3†) to allow confident assignment of ligand identity as a terminally-oriented diatomic species composed of light (2p) elements. The IR spectrum of the adduct is dominated by a blue-shifted vCO at 2000 cm−1, evidence for back donation being greatly decreased and implying that the active site is in an oxidised state. The options for X–Y are CN−, CO, N2, NO and O2, which we now deal with in turn.Cyanide is consistent with the requirement for the inactive complex to form under oxidising conditions (for first row transition metals, CN− behaves mainly as a strong σ donor, and should stabilise Ni(III) relative to Ni(II)) and in terms of the bond angle (163°) that is slightly smaller than that observed for CO. Against this option: first, CN− is a strong IR absorber and the new stretching band that would be expected to be observed was not detected (Fig. S16†); second, since the CN− would have to originate from the breakdown of other active sites, a large fraction of the enzyme would be depleted, yet there is no evidence of this in the crystal structures and extensive reduction restored most of the activity. Moreover, redox titrations show that the formation of the high wavenumber species at 2000 cm−1 is reversible.
Carbon monoxide is consistent with the bond angle. However, inhibition by CO follows a completely different kinetic pattern (it being rapid and reversible), no new Ni–CO band was detected, and the inactive enzyme is formed under oxidising conditions that should not be favourable for a Ni–CO adduct. Furthermore, although a diatomic ligand was observed bound to the active site upon treatment with CO, no IR-band at 2000 cm−1 was observed.
Dinitrogen is an inert molecule under all but highly specialised catalytic conditions, and like CO, its coordination to a metal requires reducing conditions favouring an electron-rich site.
Nitric oxide coordination would be consistent with the bond angle, but it is particularly unclear where NO would originate from. In addition, NO is known to attack Fe–S clusters, and any such damage is not observed.
Dioxygen could bind in different forms, O2, O2− (superoxide) or O22− (peroxide) depending on the oxidation state of the Ni, but under oxidising conditions superoxide bound to Ni(III) would be more likely, and consistent with the lack of an EPR signal. However, the Ni–X–Y bond angle is much larger than expected for metal–O–O bonds; moreover, O2 is not required for appearance of the high wavenumber species in IR experiments.
A puzzling predicament thus arises as to the identity of ligand X–Y, as each of the above-listed options has serious drawbacks, yet the answer must lie in one of them. Taking into consideration the formation of the adduct under oxidising conditions, its inhibitory properties and the availability of a source of X–Y, the ligands N2, CO and NO look extremely unlikely, leaving CN− (supported by bond angle) or an O2-derived ligand as the more likely options. The affinity for either ligand would have to be extremely high, to such a degree that the ligand is sequestered from an extremely dilute source. An alternative but even more unlikely scenario is that the ligand is an O–O species derived from two H2O: such an extreme case would require that the O–O species is stabilised to an unprecedented degree (in electrochemical terms, this would amount to an extreme case of underpotential deposition) so that it cannot be released without imposing a different condition (reduction back to H2O). Although such a high degree of stabilisation is indeed the case, we must regard this option as very unlikely.
5. Conclusions
Questions undoubtedly remain, but the conclusions arising from this work may be summarised as follows:(1) For Hyd-2, as with its O2-tolerant counterpart Hyd-1 (ref. 27) the pendent arginine is essential for rapid H2 oxidation. This study now extends this observation to H2 production which is severely suppressed when the arginine is replaced by lysine. The mechanism of hydrogen activation remains unresolved but must include a key role for the pendent guanidinium group in each direction. In view of the fact that R479K incorporates other changes in structure in addition to the R-to-K exchange, it is not possible to attribute the large rate retardation to the replacement alone.
(2) The normal inner coordination sphere of the active site is essentially unchanged when lysine replaces arginine, emphasising the role of the outer sphere and wider active site.
(3) Despite greatly attenuated rates of H2 oxidation and proton reduction, the R479K variant remains a reversible electrocatalyst. Having a pendent guanidinium group is therefore not a factor for reversibility.
(4) The activation parameters can only be explained if the arginine to lysine substitution results in a great difficulty in sampling a productive conformation to achieve the transition state that is otherwise energetically more favourable than the native enzyme. The value of ΔH‡ for H2 oxidation is particularly low and the curvature of the Eyring plot may reflect a significant change in heat capacitance during the reaction.
(5) Replacement of arginine by lysine in the active site canopy results in an unexpected and unprecedented high affinity for a diatomic ligand that is coordinated to the Ni and hydrogen bonded to the lysine amine group. This ligand is bound very tightly in the as-isolated form and in samples subjected to oxidising conditions, whether aerobic or not.
(6) Extensive investigations to determine the identity of the strongly coordinated diatomic ligand remain inconclusive but in view of the fact that the adduct is formed under oxidising conditions the most likely options are a cyanide, possibly originating from active site breakdown, or a dioxygen species having unprecedented stability.
Data availability
Coordinates and structure factors for each X-ray structure reported in this work have been deposited in the PDB with the following accession numbers: as-isolated enzyme (6SYO), hydrogen reduced enzyme (6SZD), oxygen exposed enzyme (6SYX) and CO exposed enzyme (6SZK).Author contributions
RME, SEB, SBC and FAA conceived and designed study. RME, SEB, SBC purified enzyme. RME, SEB, LK and FAA performed and analysed electrochemical and biochemical data. SBC performed X-ray structure determination. SEB and WKM performed EPR measurements. RME, KLW, PRM, PAA and KAV collected and analysed IR spectroscopic data. RB and YP performed DFT calculations. All authors contributed to writing the manuscript.Conflicts of interest
The authors report no conflict of interests.Acknowledgements
This research was supported by the UK Biological and Biotechnological Sciences Research Council Grants BB/I022309 and BB/L009722 to F. A. A. and BB/R018413 to K. A. V. S. E. B. was supported by a Holaday Scholarship awarded by the Dakota Foundation. L. K. was supported by a Gateway Fellowship provided by RUB Research School, PLUS, financed by the excellence initiative [DFG GSC 98/3]. W. K. M. is supported by the UK EPSRC (EP/L011972/1, grant to CAESR, the Centre for Advanced Spin Resonance). The authors would like to thank Diamond Light Source for beam time (proposals mx12346 and sm21651) and the staff of beamlines i04-1 and B22 for assistance during data collection. P. R. M. thanks the University of Oxford for a Glasstone Research Fellowship and Linacre College Oxford for a Junior Research Fellowship.References
- F. A. Armstrong and J. Hirst, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 14049–14051 CrossRef CAS PubMed
.
- F. A. Armstrong, R. M. Evans, S. V. Hexter, B. J. Murphy, M. M. Roessler and P. Wulff, Acc. Chem. Res., 2016, 49, 884–892 CrossRef CAS PubMed
.
- N. P. Boralugodage, R. J. Arachchige, A. Dutta, G. W. Buchko and W. J. Shaw, Catal. Sci. Technol., 2017, 7, 1108–1121 RSC
.
- B. Ginovska-Pangovska, A. Dutta, M. L. Reback, J. C. Linehan and W. J. Shaw, Acc. Chem. Res., 2014, 47, 2621–2630 CrossRef CAS PubMed
.
- J. Schild, B. Reuillard, A. Morozan, P. Chenevier, E. Gravel, E. Doris and V. Artero, J. Am. Chem. Soc., 2021, 143, 18150–18158 CrossRef CAS PubMed
.
- S. E. Smith, J. Y. Yang, D. L. DuBois and R. M. Bullock, Angew. Chem., Int. Ed., 2012, 51, 3152–3155 CrossRef CAS PubMed
.
- C. Greening, A. Biswas, C. R. Carere, C. J. Jackson, M. C. Taylor, M. B. Stott, G. M. Cook and S. E. Morales, ISME J., 2015, 10, 1–17 Search PubMed
.
- B. J. Murphy, F. Sargent and F. A. Armstrong, Energy Environ. Sci., 2014, 7, 1426–1433 RSC
.
- J. A. Cracknell, A. F. Wait, O. Lenz, B. Friedrich and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 20681–20686 CrossRef PubMed
.
- M. J. Lukey, M. M. Roessler, A. Parkin, R. M. Evans, R. A. Davies, O. Lenz, B. Friedrich, F. Sargent and F. A. Armstrong, J. Am. Chem. Soc., 2011, 133, 16881–16892 CrossRef CAS PubMed
.
- R. M. Evans, A. Parkin, M. M. Roessler, B. J. Murphy, H. Adamson, M. J. Lukey, F. Sargent, A. Volbeda, J. C. Fontecilla-Camps and F. A. Armstrong, J. Am. Chem. Soc., 2013, 135, 2694–2707 CrossRef CAS PubMed
.
- S. E. Beaton, R. M. Evans, A. J. Finney, C. M. Lamont, F. A. Armstrong, F. Sargent and S. B. Carr, Biochem. J., 2018, 475, 1353–1370 CrossRef CAS PubMed
.
- M. J. Lukey, A. Parkin, M. M. Roessler, B. J. Murphy, J. Harmer, T. Palmer, F. Sargent and F. A. Armstrong, J. Biol. Chem., 2010, 285, 3928–3938 CrossRef CAS PubMed
.
- F. Dole, A. Fournel, V. Magro, E. C. Hatchikian, P. Bertrand and B. Guigliarelli, Biochemistry, 1997, 36, 7847–7854 CrossRef CAS PubMed
.
- J. E. Huyett, M. Carepo, A. Pamplona, R. Franco, I. Moura, J. J. G. Moura and B. M. Hoffman, J. Am. Chem. Soc., 1997, 119, 9291–9292 CrossRef CAS
.
- W. Lubitz, H. Ogata, O. Rüdiger and E. Reijerse, Chem. Rev., 2014, 114, 4081–4148 CrossRef CAS PubMed
.
- M. Rakowski DuBois and D. L. DuBois, Chem. Soc. Rev., 2009, 38, 62–72 RSC
.
- B. Ginovska-Pangovska, A. Dutta, M. L. Reback, J. C. Linehan and W. J. Shaw, Acc. Chem. Res., 2014, 47, 2621–2630 CrossRef CAS PubMed
.
- J. W. Slater, S. C. Marguet, H. A. Monaco and H. S. Shafaat, J. Am. Chem. Soc., 2018, 140, 10250–10262 CrossRef PubMed
.
- J. W. Slater, S. C. Marguet, M. E. Gray, M. Sotomayor and H. S. Shafaat, ACS Catal., 2019, 9, 8928–8942 CrossRef CAS
.
- S. Ogo, K. Ichikawa, T. Kishima, T. Matsumoto, H. Nakai, K. Kusaka and T. Ohhara, Science, 2013, 339, 682–684 CrossRef CAS PubMed
.
- C. Wombwell and E. Reisner, Chem. - Eur. J., 2015, 21, 8096–8104 CrossRef CAS PubMed
.
- F. A. Armstrong, B. Cheng, R. A. Herold, C. F. Megarity and B. Siritanaratkul, Chem. Rev., 2023, 123, 5421–5458 CrossRef CAS PubMed
.
- S. Dementin, B. Burlat, A. L. De Lacey, A. Pardo, G. Adryanczyk-Perrier, B. Guigliarelli, V. M. Fernandez and M. Rousset, J. Biol. Chem., 2004, 279, 10508–10513 CrossRef CAS PubMed
.
-
R. M. Evans and S. E. Beaton, Enzymes of Energy Technology, in Methods in Enzymology, ed. F. A. Armstrong, 2018, pp. 91–116 Search PubMed
.
- B. L. Greene, G. E. Vansuch, C.-H. Wu, M. W. W. Adams and R. B. Dyer, J. Am. Chem. Soc., 2016, 138, 13013–13021 CrossRef CAS PubMed
.
- R. M. Evans, E. J. Brooke, S. A. M. Wehlin, E. Nomerotskaia, F. Sargent, S. B. Carr, S. E. V. Phillips and F. A. Armstrong, Nat. Chem. Biol., 2016, 12, 46–50 CrossRef CAS PubMed
.
- E. J. Brooke, R. M. Evans, S. T. A. Islam, G. M. Roberts, S. A. M. Wehlin, S. B. Carr, S. E. V Phillips and F. A. Armstrong, Biochemistry, 2017, 56, 132–142 CrossRef CAS PubMed
.
- S. Foerster, M. Van Gastel, M. Brecht and W. Lubitz, J. Biol. Inorg. Chem., 2005, 10, 51–62 CrossRef CAS PubMed
.
- M. Brecht, M. van Gastel, T. Buhrke, B. Friedrich and W. Lubitz, J. Am. Chem. Soc., 2003, 125, 13075–13083 CrossRef CAS PubMed
.
- H. Ogata, T. Krämer, H. Wang, D. Schilter, V. Pelmenschikov, M. van Gastel, F. Neese, T. B. Rauchfuss, L. B. Gee, A. D. Scott, Y. Yoda, Y. Tanaka, W. Lubitz and S. P. Cramer, Nat. Commun., 2015, 6, 7890 CrossRef CAS PubMed
.
- H. Ogata, K. Nishikawa and W. Lubitz, Nature, 2015, 520, 571–574 CrossRef CAS PubMed
.
- S. Dementin, B. Burlat, A. L. De Lacey, A. Pardo, G. Adryanczyk-Perrier, B. Guigliarelli, V. M. Fernandez and M. Rousset, J. Biol. Chem., 2004, 279, 10508–10513 CrossRef CAS PubMed
.
- R. M. Evans, P. A. Ash, S. E. Beaton, E. J. Brooke, K. A. Vincent, S. B. Carr and F. A. Armstrong, J. Am. Chem. Soc., 2018, 140, 10208–10220 CrossRef PubMed
.
- E. Szőri-Dorogházi, G. Maróti, M. Szőri, A. Nyilasi, G. Rákhely and K. L. Kovács, PLoS One, 2012, 7, e34666 CrossRef PubMed
.
- G. E. Vansuch, C.-H. Wu, D. K. Haja, S. A. Blair, B. Chica, M. K. Johnson, M. W. W. Adams and R. B. Dyer, Chem. Sci., 2020, 11, 8572–8581 RSC
.
- A. M. Escorcia and M. Stein, Front. Chem., 2018, 6, 1–11 CrossRef PubMed
.
- D. W. Stephan and G. Erker, Angew. Chem., Int. Ed., 2010, 49, 46–76 CrossRef CAS PubMed
.
- J. A. Birrell, P. Rodríguez-Maciá, E. J. Reijerse, M. A. Martini and W. Lubitz, Coord. Chem. Rev., 2021, 449, 214191 CrossRef CAS
.
- G. Berggren, A. Adamska, C. Lambertz, T. R. Simmons, J. Esselborn, M. Atta, S. Gambarelli, J. M. Mouesca, E. Reijerse, W. Lubitz, T. Happe, V. Artero and M. Fontecave, Nature, 2013, 499, 66–69 CrossRef CAS PubMed
.
- J. Esselborn, N. Muraki, K. Klein, V. Engelbrecht, N. Metzler-Nolte, U.-P. Apfel, E. Hofmann, G. Kurisu and T. Happe, Chem. Sci., 2016, 7, 959–968 RSC
.
-
D. Voet and J. G. Voet, Biochemistry, John Wiley & Sons, Inc., New York., 4th edn, 2011 Search PubMed
.
- K. L. Pankhurst, C. G. Mowat, E. L. Rothery, J. M. Hudson, A. K. Jones, C. S. Miles, M. D. Walkinshaw, F. A. Armstrong, G. A. Reid and S. K. Chapman, J. Biol. Chem., 2006, 281, 20589–20597 CrossRef CAS PubMed
.
- M. K. Doherty, S. L. Pealing, C. S. Miles, R. Moysey, P. Taylor, M. D. Walkinshaw, G. A. Reid and S. K. Chapman, Biochemistry, 2000, 39, 10695–10701 CrossRef CAS PubMed
.
- Y. V. Guillén Schlippe and L. Hedstrom, Arch. Biochem. Biophys., 2005, 433, 266–278 CrossRef PubMed
.
- G. Tedeschi, S. Ronchi, T. Simonic, C. Treu, A. Mattevi and A. Negri, Biochemistry, 2001, 40, 4738–4744 CrossRef CAS PubMed
.
- J. H. Hong, P. Dilbeck, R. J. Debus and R. L. Burnap, Biochemistry, 2007, 46, 11987–11997 CrossRef PubMed
.
- M. W. Van Der Kamp, F. Perruccio and A. J. Mulholland, Chem. Commun., 2008, 1874–1876 RSC
.
- H. Kwon, J. Basran, J. M. Devos, R. Suardíaz, M. W. van der Kamp, A. J. Mulholland, T. E. Schrader, A. Ostermann, M. P. Blakeley, P. C. E. Moody and E. L. Raven, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 6484–6490 CrossRef CAS PubMed
.
- C. A. Fitch, G. Platzer, M. Okon, B. E. Garcia-Moreno and L. P. McIntosh, Protein Sci., 2015, 24, 752–761 CrossRef CAS PubMed
.
- M. Lučić, D. A. Svistunenko, M. T. Wilson, A. K. Chaplin, B. Davy, A. Ebrahim, D. Axford, T. Tosha, H. Sugimoto, S. Owada, F. S. N. Dworkowski, I. Tews, R. L. Owen, M. A. Hough and J. A. R. Worrall, Angew. Chem., Int. Ed., 2020, 59, 21656–21662 CrossRef PubMed
.
- M. Lučić, M. T. Wilson, D. A. Svistunenko, R. L. Owen, M. A. Hough and J. A. R. Worrall, J. Biol. Inorg. Chem., 2021, 26, 743–761 CrossRef PubMed
.
- R. H. Morris, J. Am. Chem. Soc., 2014, 136, 1948–1959 CrossRef CAS PubMed
.
- H. Kwon, J. Basran, J. M. Devos, R. Suardíaz, M. W. van der Kamp, A. J. Mulholland, T. E. Schrader, A. Ostermann, M. P. Blakeley, P. C. E. Moody and E. L. Raven, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 6484–6490 CrossRef CAS PubMed
.
- C. A. Fitch, G. Platzer, M. Okon, B. E. Garcia-Moreno and L. P. McIntosh, Protein Sci., 2015, 24, 752–761 CrossRef CAS PubMed
.
- M. Lučić, D. A. Svistunenko, M. T. Wilson, A. K. Chaplin, B. Davy, A. Ebrahim, D. Axford, T. Tosha, H. Sugimoto, S. Owada, F. S. N. Dworkowski, I. Tews, R. L. Owen, M. A. Hough and J. A. R. Worrall, Angew. Chem., Int. Ed., 2020, 59, 21656–21662 CrossRef PubMed
.
- M. Lučić, M. T. Wilson, D. A. Svistunenko, R. L. Owen, M. A. Hough and J. A. R. Worrall, J. Biol. Inorg. Chem., 2021, 26, 743–761 CrossRef PubMed
.
- C. M. Hamilton, M. Aldea, B. K. Washburn, P. Babitzke and S. R. Kushner, J. Bacteriol., 1989, 171, 4617–4622 CrossRef CAS PubMed
.
- C. M. Lamont and F. Sargent, Arch. Microbiol., 2016, 199, 495–503 CrossRef PubMed
.
-
A. J. Bard and L. R. Faulkner, Electrochemical methods, Wiley, New york, 2001 Search PubMed
.
- O. Rüdiger, J. M. Abad, E. C. Hatchikian, V. M. Fernandez and A. L. De Lacey, J. Am. Chem. Soc., 2005, 127, 16008–16009 CrossRef PubMed
.
- S. Krishnan and F. A. Armstrong, Chem. Sci., 2012, 3, 1015–1023 RSC
.
-
R. Cammack, V. M. Fernandez, E. Claude Hatchikian and J. J. L. H. D. Peck, in Methods in Enzymology, Academic Press, 1994, vol. 243, pp. 43–68 Search PubMed
.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed
.
-
D. G. Waterman, G. Winter, J. M. Parkhurst, L. Fuentes-Montero, J. Hattne, A. Brewster, N. K. Sauter and G. Evans, CCP4 Newsletter on Protein Crystallography, 2013, vol. 49, pp. 16–19 Search PubMed
.
- P. R. Evans and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2013, 69, 1204–1214 CrossRef CAS PubMed
.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed
.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed
.
- G. N. Murshudov, P. Skubak, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed
.
- W. Tian, C. Chen, X. Lei, J. Zhao and J. Liang, Nucleic Acids Res., 2018, 46, W363–W367 CrossRef CAS PubMed
.
- A. Jurcik, D. Bednar, J. Byska, S. M. Marques, K. Furmanova, L. Daniel, P. Kokkonen, J. Brezovsky, O. Strnad, J. Stourac, A. Pavelka, M. Manak, J. Damborsky and B. Kozlikova, Bioinformatics, 2018, 34, 3586–3588 CrossRef CAS PubMed
.
-
W. L DeLano, {CCP4} Newsletter On Protein Crystallography, 2002, 40 Search PubMed
.
- S. Stoll and A. Schweiger, J. Magn. Reson., 2006, 178, 42–55 CrossRef CAS PubMed
.
- P. A. Ash, S. E. T. Kendall-Price, R. M. Evans, S. B. Carr, A. Brasnett, S. Morra, R. Hidalgo, A. J. Healy, G. Cinque, M. D. Frogley, F. A. Armstrong and K. A. Vincent, Chem. Sci., 2021, 12, 12959–12970 RSC
.
- F. Neese, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2022, 1–15 Search PubMed
.
- P. Eastman, J. Swails, J. D. Chodera, R. T. McGibbon, Y. Zhao, K. A. Beauchamp, L. P. Wang, A. C. Simmonett, M. P. Harrigan, C. D. Stern, R. P. Wiewiora, B. R. Brooks and V. S. Pande, PLoS Comput. Biol., 2017, 13, 1–17 CrossRef PubMed
.
- R. B. Best, X. Zhu, J. Shim, P. E. M. Lopes, J. Mittal, M. Feig and A. D. MacKerell, J. Chem. Theory Comput., 2012, 8, 3257–3273 CrossRef CAS PubMed
.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS
.
- J. Sun, A. Ruzsinszky and J. Perdew, Phys. Rev. Lett., 2015, 115, 1–6 Search PubMed
.
- J. W. Furness, A. D. Kaplan, J. Ning, J. P. Perdew and J. Sun, J. Phys. Chem. Lett., 2020, 11, 8208–8215 CrossRef CAS PubMed
.
- S. Ehlert, U. Huniar, J. Ning, J. W. Furness, J. Sun, A. D. Kaplan, J. P. Perdew and J. G. Brandenburg, J. Chem. Phys., 2021, 154, 061101 CrossRef CAS PubMed
.
- E. Caldeweyher, C. Bannwarth and S. Grimme, J. Chem. Phys., 2017, 147, 034112 CrossRef PubMed
.
- C. Van Wüllen, J. Chem. Phys., 1998, 109, 392–399 CrossRef
.
- E. Van Lenthe, R. Van Leeuwen, E. J. Baerends and J. G. Snijders, Int. J. Quantum Chem., 1996, 57, 281–293 CrossRef CAS
.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC
.
- D. A. Pantazis, X. Y. Chen, C. R. Landis and F. Neese, J. Chem. Theory Comput., 2008, 4, 908–919 CrossRef CAS PubMed
.
- F. Neese, J. Comput. Chem., 2003, 24, 1740–1747 CrossRef CAS PubMed
.
- F. Weigend, Phys. Chem. Chem. Phys., 2006, 8, 1057–1065 RSC
.
- V. Fourmond, E. S. Wiedner, W. J. Shaw and C. Léger, J. Am. Chem. Soc., 2019, 141, 11269–11285 CrossRef CAS PubMed
.
- R. M. Evans, B. Siritanaratkul, C. F. Megarity, K. Pandey, T. F. Esterle, S. Badiani and F. A. Armstrong, Chem. Soc. Rev., 2019, 49, 2039–2052 RSC
.
- K. A. Vincent, A. Parkin and F. A. Armstrong, Chem. Rev., 2007, 107, 4366–4413 CrossRef CAS PubMed
.
- H. R. Pershad, J. L. C. Duff, H. A. Heering, E. C. Duin, S. P. J. Albracht and F. A. Armstrong, Biochemistry, 1999, 38, 8992–8999 CrossRef CAS PubMed
.
- R. M. Evans, B. Siritanaratkul, C. F. Megarity, K. Pandey, T. F. Esterle, S. Badiani and F. A. Armstrong, Chem. Soc. Rev., 2019, 49, 2039–2052 RSC
.
-
R. M. Evans and F. A. Armstrong, in Metalloproteins - Methods and Protocols, ed. J. C. Fontecilla-Camps and Y. Nicolet, Humana Press, New York, 2014, pp. 73–94 Search PubMed
.
- F. A. Armstrong, N. A. Belsey, J. A. Cracknell, G. Goldet, A. Parkin, E. Reisner, K. A. Vincent and A. F. Wait, Chem. Soc. Rev., 2009, 38, 36–51 RSC
.
- C. Leger, S. Dementin, P. Bertrand, M. Rousset, B. Guigliarelli, C. Léger, S. Dementin, P. Bertrand, M. Rousset and B. Guigliarelli, J. Am. Chem. Soc., 2004, 126, 12162–12172 CrossRef CAS PubMed
.
- T. Goris, A. F. Wait, M. Saggu, J. Fritsch, N. Heidary, M. Stein, I. Zebger, F. Lendzian, F. A. Armstrong, B. Friedrich and O. Lenz, Nat. Chem. Biol., 2011, 7, 310–318 CrossRef CAS PubMed
.
- S. V. Hexter, F. Grey, T. Happe, V. Climent and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11516–11521 CrossRef CAS PubMed
.
- J. Dragelj, C. Karafoulidi-Retsou, S. Katz, O. Lenz, I. Zebger, G. Caserta, S. Sacquin-Mora and M. A. Mroginski, Front. Microbiol., 2023, 13, 1073315 CrossRef PubMed
.
- A. Volbeda, L. Martin, C. Cavazza, M. Matho, B. W. Faber, W. Roseboom, S. P. J. Albracht, E. Garcin, M. Rousset and J. C. Fontecilla-Camps, J. Biol. Inorg. Chem., 2005, 10, 239–249 CrossRef CAS PubMed
.
- A. Volbeda, L. Martin, E. Barbier, O. Gutiérrez-Sanz, A. L. De Lacey, P.-P. Liebgott, S. Dementin, M. Rousset and J. C. Fontecilla-Camps, JBIC, J. Biol. Inorg. Chem., 2015, 20, 11–22 CrossRef CAS PubMed
.
- Y. Ilina, C. Lorent, S. Katz, J.-H. Jeoung, S. Shima, M. Horch, I. Zebger and H. Dobbek, Angew. Chem., 2019, 18883–18887 CrossRef
.
- H. Tai, K. Nishikawa, Y. Higuchi, Z.-W. Mao and S. Hirota, Angew. Chem., 2019, 131, 13419–13424 CrossRef
.
- M. E. Pandelia, H. Ogata, L. J. Currell, M. Flores and W. Lubitz, Biochim. Biophys. Acta, Bioenerg., 2010, 1797, 304–313 CrossRef CAS PubMed
.
- H. Ogata, Y. Mizoguchi, N. Mizuno, K. Miki, S. Adachi, N. Yasuoka, T. Yagi, O. Yamauchi, S. Hirota and Y. Higuchi, J. Am. Chem. Soc., 2002, 124, 11628–11635 CrossRef CAS PubMed
.
- Y. Shomura, M. Taketa, H. Nakashima, H. Tai, H. Nakagawa, Y. Ikeda, M. Ishii, Y. Igarashi, H. Nishihara, K. S. Yoon, S. Ogo, S. Hirota and Y. Higuchi, Science, 2017, 357, 928–932 CrossRef CAS PubMed
.
- C. J. Kulka-Peschke, A. C. Schulz, C. Lorent, Y. Rippers, S. Wahlefeld, J. Preissler, C. Schulz, C. Wiemann, C. C. M. Bernitzky, C. Karafoulidi-Retsou, S. L. D. Wrathall, B. Procacci, H. Matsuura, G. M. Greetham, C. Teutloff, L. Lauterbach, Y. Higuchi, M. Ishii, N. T. Hunt, O. Lenz, I. Zebger and M. Horch, J. Am. Chem. Soc., 2022, 144, 17022–17032 CrossRef CAS PubMed
.
- R. Kumar and M. Stein, J. Am. Chem. Soc., 2023, 145, 10954–10959 CrossRef CAS PubMed
.
- O. Lampret, J. Duan, E. Hofmann, M. Winkler, F. A. Armstrong and T. Happe, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 20520–20529 CrossRef CAS PubMed
.
- V. L. Arcus, M. W. van der Kamp, C. R. Pudney and A. J. Mulholland, Curr. Opin. Struct. Biol., 2020, 65, 96–101 CrossRef CAS PubMed
.
- H. A. Bunzel, J. L. R. Anderson, D. Hilvert, V. L. Arcus, M. W. van der Kamp and A. J. Mulholland, Nat. Chem., 2021, 13, 1017–1022 CrossRef CAS PubMed
.
- V. L. Arcus and A. J. Mulholland, Annu. Rev. Biophys., 2020, 49, 163–182 CrossRef CAS PubMed
.
- H. A. Bunzel, H. Kries, L. Marchetti, C. Zeymer, P. R. E. Mittl, A. J. Mulholland and D. Hilvert, J. Am. Chem. Soc., 2019, 141, 11745–11748 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc05641k |
| This journal is © The Royal Society of Chemistry 2023 |