Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The influence of chirality on the behavior of oligonucleotides inside cells: revealing the potent cytotoxicity of G-rich L-RNA†
Chen-Hsu
Yu
and
Jonathan T.
Sczepanski
 *
*
Department of Chemistry, Texas A&M University, College Station, Texas 77843, USA. E-mail: jon.sczepanski@chem.tamu.edu
First published on 30th December 2022
Abstract
Due to their intrinsic nuclease resistance, mirror image L-oligonucleotides are being increasingly employed in the development of biomedical research tools and therapeutics. Yet, the influence of chirality on the behavior of oligonucleotides in living systems, and specifically, the extent to which L-oligonucleotides interact with endogenous biomacromolecules and the resulting consequences remain unknown. In this study, we characterized the intracellular behavior of L-oligonucleotides for the first time, revealing important chirality-dependent effects on oligonucleotide cytotoxicity. We show that exogenously delivered L-oligonucleotides have the potential to be highly cytotoxic, which is dependent on backbone chemistry, sequence, and structure. Notably, for the sequences tested, we found that single-stranded G-rich L-RNAs are more cytotoxic than their D-DNA/RNA counterparts, exhibiting low nanomolar EC50 values. Importantly, RNA-seq analysis of differentially expressed genes suggests that G-rich L-RNAs stimulate an innate immune response and pro-inflammatory cytokine production. These data not only challenge the general perception that mirror image L-oligonucleotides are nontoxic and nonimmunogenic, but also reveal previously unrecognized therapeutic opportunities. Moreover, by establishing sequence/structure toxicity relationships, this work will guide how future L-oligonucleotide-based biotechnologies are designed and applied.
Introduction
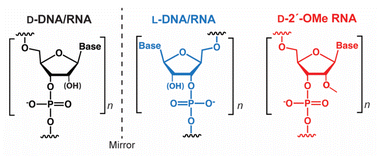
Native nucleic acids, D-DNA and D-RNA, are chiral molecules as a result of their D-(deoxy)ribose sugars. As such, they have enantiomers, referred to as mirror image L-DNA and L-RNA, which consist of L-(deoxy)ribose sugars (Fig. 1). Although L-oligonucleotides no longer exist in nature, they can be prepared synthetically in the laboratory, where they have been studied since the 1970s.1 From a biotechnology perspective, L-oligonucleotides have several advantageous properties relative to their native counterparts.1 Most notably, L-oligonucleotides are highly resistant to degradation by cellular nucleases, providing them with superior stability in harsh biological environments.2,3L-Oligonucleotides are also expected to avoid potentially toxic off-target interactions with endogenous nucleic acids because D- and L-oligonucleotides are incapable of forming contiguous WC base pairs with each other.2,4 Furthermore, as enantiomers, D- and L-oligonucleotides have the same physical properties in terms of duplex thermostability and hybridization kinetics.4–6 Thus, well-established design principles can be directly applied to L-oligonucleotides without further optimization, representing a key advantage over other chemically modified oligonucleotides.7,8 These favorable characteristics have fuelled the recent development of many promising L-oligonucleotide-based biotechnologies, including L-aptamers,9–12 microarrays,2 molecular sensors,13 live cell imaging probes,14–16 and drug delivery.17,18Given the stereospecific nature of biomolecular interactions, the behavior of D- and L-oligonucleotides within living systems is expected to be different. For example, the inability of D- and L-oligonucleotides to form contiguous WC base pairs with each other implies that L-oligonucleotides will interact far less with endogenous nucleic acids relative to their native counterparts.2,4 However, L-oligonucleotides are still susceptible to non-specific protein interactions, which are likely to be distinct from their native counterparts. Nevertheless, the influence of chirality on the behavior of oligonucleotides in living systems and, specifically, the extent to which L-oligonucleotides interact with endogenous biomacromolecules has not been studied carefully. Given the large number of proteins that have been shown to bind nucleic acids in a nonspecific or “promiscuous” manner, it is reasonable to predict that interactions across the mirror are abundant, potentially leading to undesired effects. Indeed, we recently showed that L-RNA binds tightly to the polycomb repressive complex 2 (PRC2) and prevents it from interacting with its intended targets.19 If L-oligonucleotides are to be routinely employed in living cells and organisms, then it is imperative that we understand how they interact with these environments and their potential consequences. Such studies may also reveal whether chirality as a design parameter can be harnessed to induce and/or modulate a desired biological response, opening the door to new applications. In this study, we conducted the first detailed characterization of how L-oligonucleotides interact with biological systems at the cellular level, revealing important chirality-dependent behaviors of L-oligonucleotides. Notably, we show that G-rich L-oligonucleotides have potent cytotoxicity and a dramatic impact on gene expression, suggesting they make extensive interactions with endogenous proteins.
Results and discussion
G-rich L-RNA is cytotoxic to human cells
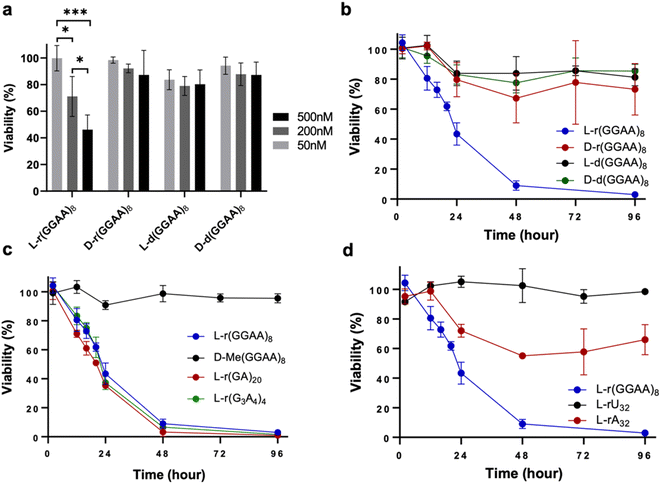
We previously reported that guanine (G)-rich L-RNAs, but not other sequences, bound tightly to PRC2 in vitro, suggesting that G-rich sequences have a propensity for interacting with native proteins. Therefore, we chose the model G-rich sequence (GGAA)8 as a starting point for our studies. We prepared both D and L stereoisomers of (GGAA)8 DNA and RNA (D/L-d(GGAA)8 and D/L-r(GGAA)8, respectively) (Table S1†). As an initial test for disruption of cellular pathways, we transfected HeLa cells with the different (GGAA)8 variants and measured cell viability after 24 hours using an CCK-8 assay. Unless stated otherwise, all oligonucleotides were transfected into cells using Lipofectamine 3000 (see ESI†). Of the four (GGAA)8 variants tested, only L-r(GGAA)8 exhibited a strong concentration-dependent decrease in cell viability after 24 hours, suggesting that the L-RNA strand was uniquely cytotoxic upon transfection (Fig. 2a). We confirmed these results by conducting a time-dependent toxicity assay over the course of 96 hours (Fig. 2b). Notably, while cells treated with D-r(GGAA)8 showed no cytotoxic effects during this timeframe, L-r(GGAA)8 killed nearly all cells after just 48 hours. The half-maximal effective concentration (EC50) of L-r(GGAA)8 was determined to be 58 ± 26 nM (Table 1 and Fig. S1†), which is ∼5-fold lower than D-r(GGAA)8. We extracted the various (GGAA)8 oligonucleotides from cells after 24 hours and analysed their length by gel electrophoresis. The results revealed that comparable levels of full-length material for all four versions of (GGAA)8 were present inside cells after 24 hours (Fig. S2†). The persistence of D-r(GGAA)8 and D-d(GGAA)8 is likely due to the Cy5 dye positioned at the 3′-end, which has been shown to greatly increase the half-life of oligonucleotides in serum-supplemented media and inside cells by protecting against 3′-exonuclease degradation.20 Importantly, the presence of full-length D-d(GGAA)8 and D-r(GGAA)8 inside cells after 24 hours indicated that their much lower cytotoxicity relative to L-r(GGAA)8 cannot be attributed to nuclease degradation. Unsurprisingly,21 the 2′-O-methyl (2′-OMe) RNA version of D-r(GGAA)8, D-Me(GGAA)8, showed no visible signs of cytotoxicity in HeLa cells (Fig. 2c and Table 1), highlighting the divergent effects of 2′-OMe and L-RNA modifications on cytotoxicity. Two other G-rich L-RNAs, L-r(GA)20 and L-r(G3A4)4, showed a similar toxicity profile as L-r(GGAA)8 (Fig. 2c) and even lower EC50 values (Table 1 and Fig. S1†), indicating that potent cytotoxicity is a common characteristic of G-rich L-RNA. Consistent with the (GGAA)8 series, the D-RNA versions of these RNAs, D-r(GA)20 and D-r(G3A4)4, were at least 30-fold less cytotoxic than their L-RNA counterparts (Table 1 and Fig. S1†). In addition, we found that all three G-rich L-RNAs L-r(GGAA)8, L-r(GA)20 and L-r(G3A4)4 were cytotoxic to MCF7 cells (Fig. S3†), demonstrating that the observed cytotoxicity is not specific to HeLa cells.| Sequence | EC50 (nM) |
|---|---|
| L-r(GGAA)8 | 58 ± 26 |
| D-r(GGAA)8 | 279 ± 68 |
| L-d(GGAA)8 | 2056 ± 854 |
| D-d(GGAA)8 | 634 ± 253 |
| D-Me(GGAA)8 | 1974 ± 687 |
| L-r(GC/GC) | >3000 |
| L-r(G3A4)4 | 12 ± 3 |
| D-r(G3A4)4 | 339 ± 122 |
| L-r(GAAA)10 | 42 ± 14 |
| L-rA32 | 191 ± 42 |
| L-r(GA)20 | 28 ± 12 |
| D-r(GA)20 | >3000 |
| L-r(GA)15 | 34 ± 15 |
| L-r(GA)10 | 55 ± 24 |
| L-r(GA)5 | 413 ± 197 |
| L-r(GU)20 | 133 ± 60 |
To determine whether the observed cytotoxicity of L-RNA was selective for G-rich sequences, we also examined L-poly[A] (L-rA32) and L-poly[U] (L-rU32) (Fig. 2d). HeLa cells treated with L-rU32 were mostly viable (>90%) after 96 hours, whereas cells treated with L-rA32 were ∼60% viable after the same time. The EC50 for L-rA32 was calculated to be 191 ± 42 nM, indicating that it was still much less cytotoxic than those containing both G and A (EC50 ∼ 10–50 nM) (Table 1 and Fig. S1†). Nevertheless, the observed cytotoxicity of L-rA32 suggested that there may be an effect from purine-rich sequences in addition to G-rich sequences on the cytotoxicity of L-RNA. To test this, we prepared an L-RNA containing only G and U, L-r(GU)20, and found that it was ∼4-fold less toxic than the analogous sequence containing only G and A, L-r(GA)20 (Table 1 and Fig. S1†). Thus, in addition to guanine, the overall purine content appears to contribute the cytotoxicity of G-rich L-RNA. One possible explanation is that the greater hydrophobicity of A relative to the other nucleobases22 results in increased cytotoxic L-RNA–protein interactions.23 Additional studies are needed to explore this idea further. Finally, we attempted to address whether cytotoxicity was proportional to the percent G in a sequence by comparing L-r(GAAA)10 (25% G) and L-r(GGGA)10 (75% G) to L-r(GGAA)8 and L-r(GA)20, both of which contain 50% G. Unfortunately, L-r(GGGA)10 formed aggregates under almost all conditions tested, which precluded cellular delivery and analysis. The EC50 of L-r(GAAA)10 was found to be within error of both L-r(GGAA)8 and L-r(GA)20, despite having half the number of Gs relative to As (Table 1 and Fig. S1†). Thus, while we cannot comment on sequences with >50% G, these data suggest that for G/A-rich L-RNA sequence with 25–50% G, the cytotoxicity is not proportional to the percent G.
Overall, these results demonstrate that G-rich L-RNAs have the potential to be highly cytotoxic to human cells, especially in the presence of adenine (i.e., G/A-rich sequences). Cytotoxic and antiproliferative activities have previously been reported for G-rich D-oligonucleotides.24–28 These results now indicate that such properties extend across the chiral mirror. These results also show that the stereochemistry of an oligonucleotide can influence its cytotoxicity and, in the case of G-rich sequences, we find that L-RNA is considerably more cytotoxic than its D-counterparts for those sequences tested herein. To the best of our knowledge, the first time any cytotoxicity has been assigned to L-oligonucleotides.
Cytotoxicity of G-rich L-RNAs is dependent on structure and length
The cytotoxicity of oligonucleotides has been shown to be dependent on secondary structure and folding. For example, a positive correlation between G-quadruplex (G4) formation and cytotoxicity has been reported for G-rich D-DNA.28 In the case of G-rich L-RNA, potent cytotoxicity was observed for both G4-forming sequences (e.g., L-r(GGAA)8) and those that cannot fold into G4s (e.g., L-r(GA)20) (Fig. 2 and Table 1),19 suggesting that the cytotoxic effects are mostly independent of the potential for G4 formation. In support of this notion, we obtained circular dichroism spectra of two cytotoxic sequences L-r(GGAA)8 and L-r(GA)20 under conditions used during transfection (25 mM Tris pH 7.4, 1 mM EDTA, and 50 mM KCl in Opti-MEM), as well as under intracellular K+ concentrations (140 mM KCl) (Fig. S4†). These data confirmed that L-r(GGAA)8, but not L-r(GA)20, form folded G4 structures under both sets of conditions. Thus, while we can't completely rule out the involvement of G4 structures, they do not appear to be the major determinant of L-RNA toxicity.In addition to G4s, single-stranded (ss) and double-stranded (ds) oligonucleotides can also influence cytotoxic interactions.29 To test this, we prepared a G-rich L-RNA, L-r(GC/GC), that contained the same number of Gs as L-(GGAA)8 but was designed to fold into a hairpin such that the majority of G residues where base paired. Cells treated with L-r(GC/GC) hairpin (200 nM) showed no cytotoxic effects after 96 hours (Fig. 3). The EC50 for L-r(GC/GC) hairpin was calculated to be >3 μM (Table 1 and Fig. S1†). Thus, we conclude that the cytotoxicity of G-rich L-RNA is dependent on the RNA being single-stranded.
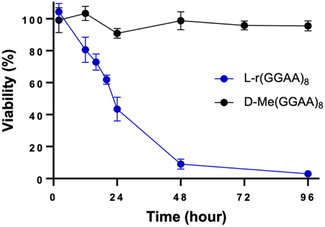 | ||
| Fig. 3 Time-dependent viability assay (CCK-8) of HeLa cells treated with 200 nM of L-r(GC/GC) hairpin. L-r(GGAA)8 is shown for reference. Data are mean ± S.D. (n = 3 biological replicates). | ||
In addition to structure, we also examined the influence of length on cytotoxicity of L-RNA. We prepared a series of incrementally shorter versions of L-(GA)20 (Table S1†). As shown in Table 1, L-r(GA)20, L-r(GA)15, L-r(GA)10, which contain 20, 15, and 10 Gs, respectively, were equally cytotoxic to HeLa cells (EC50 ∼ 30–50 nM) following transfection. However, when the length was further reduced to contain only five Gs (L-r(GA)5), the cytotoxicity was reduced dramatically (EC50 = 413 ± 197 nM). This suggests that a minimum of ten closely spaced Gs is sufficient for L-RNA cytotoxicity.
Cellular uptake and intracellular distribution of L-oligonucleotides
We next examined cellular uptake and localization of L-oligonucleotides with the goal determined whether these factors were correlated with cytotoxicity. We first measured the uptake of the various 3′-Cy5-labeled oligonucleotides (200 nM) into HeLa cells by flow cytometry two hours after transfection (Fig. 4a). In general, the RNA series of oligonucleotides were uptaken by HeLa cells better than the DNA series (D/L-d(GGAA)8) when transfected by Lipofectamine 3000. However, the difference in uptake between DNA and RNA series was only about 2–3-fold, whereas the EC50 values for the D/L-d(GGAA)8 were at least 10-fold higher than any of the cytotoxic L-RNA strands. Among the RNA series, little correlation was found between uptake and cytotoxicity. All D-RNA and L-RNA oligonucleotides tested, as well as D-Me(GGAA)8, had similar uptake into HeLa cells, despite L-r(GGAA)8 being substantially more cytotoxic than the others (Fig. 4a and Table 1). Overall, while we can't completely rule out a role for uptake, these data suggest that the potent cytotoxicity of G-rich L-RNA relative to the other oligonucleotides tested herein is not due to increased cellular uptake.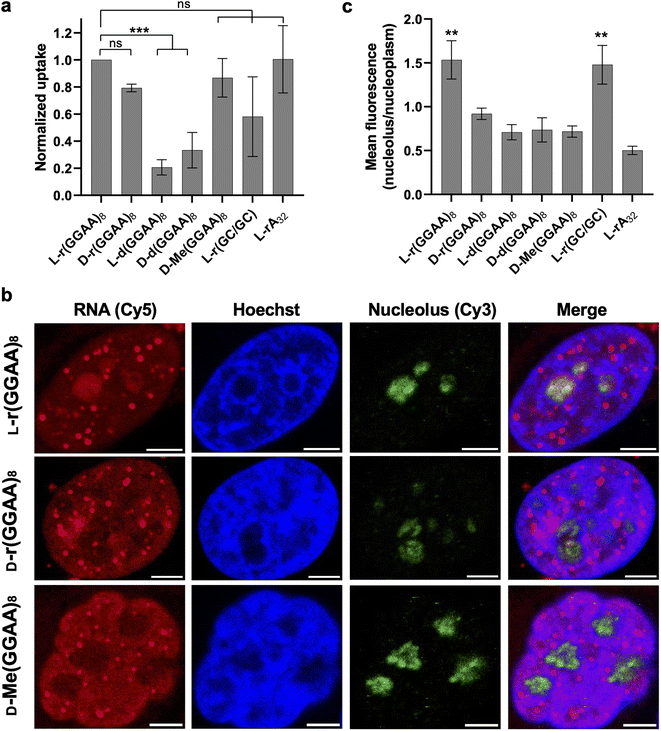 | ||
| Fig. 4 Uptake and localization of L-oligonucleotides into HeLa cells. (a) Cellular uptake of the various oligonucleotides studied in this work. HeLa cells were transfected with 200 nM oligonucleotide and uptake was measured 2 hours later by flow cytometry. Data are mean ± S.D. (n = 3 biological replicates). ***P < 0.001. (b) Representative fluorescent confocal microscopy images of HeLa cells transfected with 200 nM of the indicated oligonucleotides. See also Fig. S5.† Nucleolus was stained with an anti-fibrillarin antibody and Cy3-labeled secondary antibody. All scale bars: 5 μm. (c) Nucleolar enrichment of G-rich L-RNA. Data is the mean fluorescent intensity within the nucleolus divided by the mean fluorescent intensity within the nucleoplasm. Values greater than one indicate nucleolar enrichment (n = 4 cells). **P < 0.01. | ||
Fluorescent confocal microscopy was then used to determine the subcellular localization of the different 3′-Cy5-labeled oligonucleotides post transfection (Fig. 4b and S5†). After 2 hours all oligonucleotides primarily localized into the cell nucleus, consistent with earlier reports showing that synthetic oligonucleotides transfected with lipid-based reagents rapidly accumulate in the nucleus.21,30,31 However, there were interesting differences in the subnuclear localization of these strands. L-r(GGAA)8, D-r(GGAA)8, and D-Me(GGAA)8 formed discrete nuclear foci that were not present in cells treated with other oligonucleotides (Fig. 4b and S5†). Similar structures have been widely observed upon transfection of backbone-modified antisense oligonucleotides (ASOs) into cells, and have been attributed to non-specific protein interactions, and in particular, with nuclear paraspeckle-associated proteins (e.g., P54nrb).21,23,29,32 The lack of nuclear foci for D/L-d(GGAA)8 and L-r(CG/GC) hairpin are consistent with this model, as DNA and double-stranded oligonucleotides do not interact with paraspeckle proteins.29 While future studies are needed to determine the exact nature and composition of the foci formed by these RNAs, these observations suggest that L-r(GGAA)8 can seed the formation of protein-rich nuclear structures when transfected into cells. Another major difference among the oligonucleotides examined was nucleolar localization. Whereas most of the strands were excluded from the nucleolus, L-r(GGAA)8 was significantly enriched in this compartment (Fig. 4c and S5†). L-r(GGAA)8 was also the only oligonucleotide to both form nuclear foci and accumulate in the nucleolus, suggesting that the combination of these activities underly its potent cytotoxic effects. This notion is consistent with previous studies showing that ASO-mediated sequestration of nuclear proteins, including paraspeckle proteins, into the nucleolus induces cellular toxicity through nucleolar stress.21,29
Together, these data show that G-rich L-RNA form nuclear foci and accumulate in the nucleolus when transfected into HeLa cells, behaviors that are highly indicative of cytotoxic protein interactions. These data also demonstrate that in addition to sequence and structure, the subcellular localization and trafficking of oligonucleotides is dependent on its chirality.
G-rich L-RNAs induce apoptosis
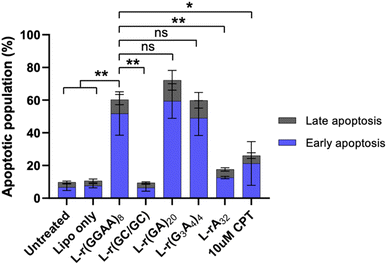
To determine the type of cell death related to cytotoxic G-rich L-RNA, HeLa cells were stained with annexin V and propidium iodide (PI) 24 hours post transfection and fluorescence was measured by flow cytometry. AnnexinV detects the externalization of phosphatidylserine on early apoptotic cells, while PI is used to detect necrotic or late apoptotic cell due to the loss of membrane integrity.33 Very few (<10%) apoptotic or necrotic cells were detected in untreated and Lipofectamine-only treated cells after 24 hours (Fig. 5 and S6†). However, treatment with cytotoxic G-rich L-RNAs resulted in ∼50% apoptotic cells (annexin V positive and PI negative) and 10–20% late apoptotic cells (annexin V and PI positive) following 24 hour exposure. The extent of apoptotic cells for G-rich L-RNAs exceeded that of camptothecin (CPT), a common positive control for apoptosis. None of the toxic L-RNAs induced significant necrosis (annexin V negative and PI positive) (Fig. S6†). As expected, the non-toxic L-RNA hairpin L-r(GC/GC) did not trigger apoptosis or necrosis. L-rA32 also induced far less apoptosis than any of G-rich L-RNAs, consistent with its lower overall cytotoxicity. Together, these results suggest that toxic G-rich L-RNA induce apoptotic cell death.Differential gene expression induced by G-rich L-RNA indicate activation of innate immunity
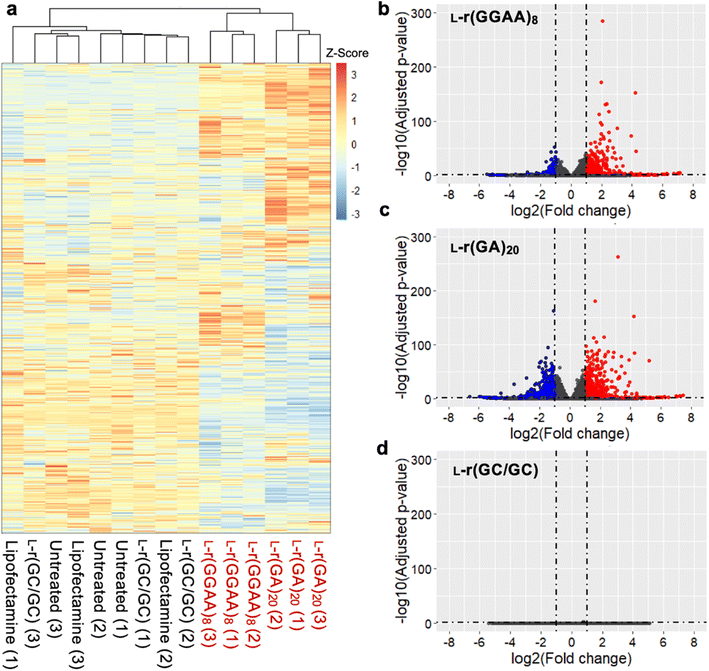
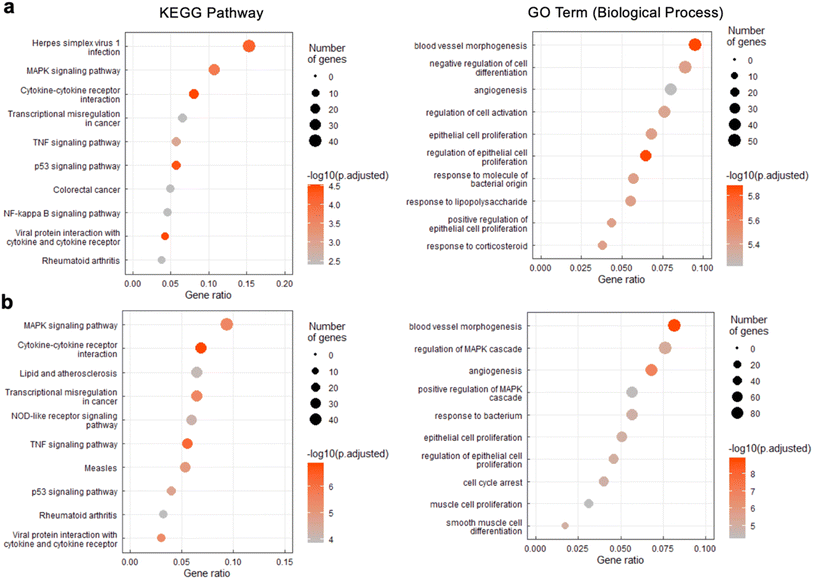
To better understand the source of cytotoxicity for G-rich L-RNAs, and to reveal the extent of their biological interactions, we performed RNA-seq analysis of total RNA isolated from HeLa cells 12 hours after treatment with 200 nM L-r(GGAA)8 and L-r(GA)20. Cells treated with Lipofectamine only and non-toxic L-r(GC/GC) served as controls for these experiments. As shown in Fig. 6a, gene expression differed significantly in cells treated with cytotoxic L-r(GGAA)8 and L-r(GA)20 compared to the controls, which were clustered together. Interestingly, gene expression profiles for L-r(GGAA)8 and L-r(GA)20 were also clustered separately, indicating that they interact differently with cells. While L-r(GGAA)8 is capable of folding into a G-quadruplex structure, L-r(GA)20 is not (Fig. S4†), representing a potential source for these differences. Treatment with L-r(GGAA)8 caused significant upregulation of 507 genes (log2(ratio) ≥ 1, P-value < 0.05) and downregulation of 267 genes (log2(ratio) ≤ −1, P-value < 0.05) relative to the Lipofectamine only control (Fig. 6b and ESI File 1†). In cells treated with L-r(GA)20, 689 genes were upregulated and 1015 genes were downregulated relative to the Lipofectamine only control (Fig. 6c and ESI File 1†). The expression level of several significantly dysregulated genes was confirmed by qRT-PCR, which showed good agreement with the RNA-seq data (Fig. S7†). No genes were differentially expressed in cells treated with non-toxic L-r(GC/GC). Overall, these data reveal that G-rich L-RNA induce dramatic perturbations in gene expression levels, suggesting extensive engagement with cellular proteins.To interpret the potential biological consequence of changes in the transcriptome caused by L-r(GGAA)8 and L-r(GA)20, over-representation analyses were performed to identify biological pathways and processes enriched in differentially expressed genes. Although there were some differences, KEGG pathway analysis revealed a common set of enriched pathways for L-r(GGAA)8 and L-r(GA)20, including mitogen-activated protein kinase (MAPK) signaling pathway, cytokine–cytokine receptor interaction, (tumour necrosis factor) TNF signaling pathway, p53 signaling pathway, and transcriptional misregulation (Fig. 7). Collectively, these pathways strongly suggest that L-r(GGAA)8 and L-r(GA)20 stimulate an innate immune response, possibly via engagement of pattern-recognition receptors such as Toll-like receptors (TLRs).34–36 Consistently, several key pro-inflammatory cytokines central to the innate immune response, including TNF, interleukin (IL)-1, IL-6, IL-12, and CXCL8, were among the most highly overexpressed genes (Table S2†). Moreover, the gene ontology (GO) biological process analyses identified blood vessel morphogenesis, angiogenesis, and cell proliferation among the top 10 enriched terms for both cytotoxic L-RNAs (Fig. 7), which is consistent with cytokine overexpression, and in particular, the very high level (>100-fold) of the angiogenic chemokine CXCL1 (Table S2†).37–39 A number of pro-apoptotic genes associated with inflammatory cytokine production and downstream p53 activation were also upregulated in response to L-r(GGAA)8 and L-r(GA)20 (Table S2†).40,41 In support of TLR receptor involvement, siRNA-mediated knockdown of TLR3 and/or TLR7 resulted in a ∼70% reduction in TNF expression following G-rich L-RNA treatment as compared to WT cells (Fig. S8†). While TLR engagement and immune system-related cytotoxicity has been documented for synthetic oligonucleotides,42–45 including those with unnatural sugars,46 strong immunostimulatory actives by G-rich L-RNA was surprising given the complete stereochemical inversion of the sugar backbone, as well as prior reports concluding that L-oligonucleotides have very low immunogenic potential.9,47 These prior studies, however, did not examine G-rich L-RNAs, highlighting the importance of sequence activity relationship studies. We acknowledge that immune system-related cytotoxicity may not be the only cause of apoptosis induced by G-rich L-RNA. For example, accumulation of gapmer ASOs in the nucleoli, which we also observed for L-r(GGAA)8, was shown to induce p53 activation and apoptotic cell death.21 Further experiments beyond the scope of this work will be required to fully identify the pathways involved in L-RNA toxicity, as well as the specific receptors (e.g., TLRs) responsible for these signalling outcomes.
Conclusions
In conclusion, we characterized the intracellular behavior of L-oligonucleotides for the first time, revealing that the chirality of an oligonucleotide can have a profound effect on its behavior in living systems. Most notably, we found that single-stranded G-rich L-RNAs were highly cytotoxic to human cells, whereas D-RNA versions of the same sequence showed mild or no cytotoxicity. While the number of sequences examined herein is likely too limited to generalize that G-rich L-RNAs are more cytotoxic that their D-counterparts, our results highlight the profound influence of stereochemistry on the behavior of oligonucleotides inside cells. Furthermore, we showed that cytotoxic L-RNA induce dramatic perturbations in gene expression levels. Given that L-oligonucleotides are incapable of forming contiguous WC base pairing interactions with native DNA and RNA, we propose that these effects are the results of L-RNA–protein interactions, such has been observed previously for cytotoxic D-oligonucleotides. While future proteomics studies will be needed for confirmation and to identify key L-RNA–protein interactions responsible for the observed cytotoxic effects, an analysis of differentially expressed genes suggests that G-rich L-RNA stimulate an innate immune response and pro-inflammatory cytokine production. Thus, these data challenge the general perception that mirror image L-oligonucleotides are nontoxic and nonimmunogenic. Moreover, this work demonstrates that the antiproliferative properties of G-rich D-oligonucleotides (e.g., AS1411), which are currently being pursued as anticancer agents, extends across the chiral mirror to G-rich L-RNA.23–27,48Due to their favorable properties, such as nuclease resistance, L-ON are being increasingly employed in biomedical applications. However, our results emphasize that caution should be taken when employing L-oligonucleotides in cells and organisms. Moving forward, it will be important to better characterize off-target interactions underlying these cytotoxic effects and to establish detailed structure toxicity relationships. Such information could be used to better predict toxicity from sequence and potentially allow for toxic motifs to be eliminated at the design stage, which provided the key motivation for this work. Towards this goal, this study offers some preliminary design considerations for minimizing the potential toxic effects of L-oligonucleotides. For example, based on the sequences tested herein, L-DNA appears to be less cytotoxic than L-RNA, making it a better choice for most biomedical applications. However, if L-RNA must be used, long (>10 nt), single-stranded regions rich in G residues should be avoided, especially in the context of G/A-rich sequences. Furthermore, complementary interactions (i.e., hybridization) appear to mitigate the cytotoxic effects of G-rich L-RNA, as evident by L-r(GC/GC) hairpin. This suggests that the cytotoxicity and potential antiproliferative activities of L-RNA could be regulated through programmable base pairing interactions, opening the door to more-targeted anticancer therapies. Finally, while most applications of L-oligonucleotides may seek to evade an immune response (e.g., Spiegelmers), L-oligonucleotides could also be highly useful as immunostimulants (e.g., vaccine adjuvants), especially considering their lack of off-target hybridization and superior nuclease resistance. Thus, further evaluation of the immunostimulatory effects of L-RNA, as well as the specific pathways and receptors involved, is highly warranted.
Overall, this work represents an important step in understanding the intracellular behavior of L-oligonucleotides and the establishment of an L-oligonucleotide “interactome”, both of which are expected to have a broad impact on how future L-oligonucleotide-based technologies are designed and applied. Our results also reveal previously unrecognized therapeutic opportunities for L-oligonucleotides. More broadly speaking, this work should motivate similar investigations into other modified oligonucleotides, such as XNAs, that are also perceived to be highly bioorthogonal, which may reveal unforeseen biological effects.
Data availability
Raw RNA-seq read and processed data are available in the NCBI GEO repository under the accession number GSE205338. The complete list of differentially expressed genes for L-r(GGAA)8 and L-r(GA)20 can be accessed in the ESI File 1 document.† All other data generated during all experiments is available from the author upon reasonable request.Author contributions
Chen-Hsu Yu: methodology, investigation, formal analysis, visualization, writing – review & editing. Jonathan Sczepanski: conceptualization, writing – original draft, supervision, project administration, funding acquisition.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors thank Professor Jerome Menet for his thoughtful discussion regarding our RNA-seq data. This work was supported by the National Institute of General Medical Sciences (R35GM124974) of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was provided by the Welch Foundation (A-1909-20190330). We thank the Texas A&M Microscopy and Imaging Center (RRID: SCR_022128) for acquisition of the confocal microscopy images. Portions of this research were conducted with the advanced computing resources provided by Texas A&M High Performance Research Computing.References
- B. E. Young, N. Kundu and J. T. Sczepanski, Mirror-Image Oligonucleotides: History and Emerging Applications, Chem.–Eur. J., 2019, 25(34), 7981–7990 CrossRef PubMed.
- N. C. Hauser, R. Martinez, A. Jacob, S. Rupp, J. D. Hoheisel and S. Matysiak, Utilising the left-helical conformation of L-DNA for analysing different marker types on a single universal microarray platform, Nucleic Acids Res., 2006, 34(18), 5101–5111 CrossRef PubMed.
- H. Urata, E. Ogura, K. Shinohara, Y. Ueda and M. Akagi, Synthesis and properties of mirror-image DNA, Nucleic Acids Res., 1992, 20(13), 3325–3332 CrossRef PubMed.
- K. Hoehlig, L. Bethge and S. Klussmann, Stereospecificity of Oligonucleotide Interactions Revisited: No Evidence for Heterochiral Hybridization and Ribozyme/DNAzyme Activity, PLoS One, 2015, 10(2), e0115328, DOI:10.1371/journal.pone.0115328.
- H. Urata, K. Shinohara, E. Ogura, Y. Ueda and M. Akagi, Mirror-image DNA, J. Am. Chem. Soc., 1991, 113(21), 8174–8175 CrossRef CAS.
- M. Szabat, D. Gudanis, W. Kotkowiak, Z. Gdaniec, R. Kierzek and A. Pasternak, Thermodynamic features of structural motifs formed by β-L-RNA, PLoS One, 2016, 11, e0149478, DOI:10.1371/journal.pone.0149478.
- A. M. Kabza, B. E. Young and J. T. Sczepanski, Heterochiral DNA Strand-Displacement Circuits, J. Am. Chem. Soc., 2017, 139(49), 17715–17718 CrossRef CAS.
- T. L. Mallette, M. N. Stojanovic, D. Stefanovic and M. R. Lakin, Robust Heterochiral Strand Displacement Using Leakless Translators, ACS Synth. Biol., 2020, 9(7), 1907–1910 CrossRef CAS PubMed.
- A. Vater and S. Klussmann, Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics, Drug Discovery Today, 2015, 20, 147–155 CrossRef CAS.
- S. Dey and J. T. Sczepanski, In vitro selection of l-DNA aptamers that bind a structured d-RNA molecule, Nucleic Acids Res., 2020, 48(4), 1669–1680 CrossRef CAS.
- C.-Y. Chan and C. K. Kwok, Specific Binding of a D-RNA G-Quadruplex Structure with an L-RNA Aptamer, Angew. Chem., Int. Ed., 2020, 59(13), 5293–5297 CrossRef CAS PubMed.
- J. Chen, M. Chen and T. F. Zhu, Directed evolution and selection of biostable l-DNA aptamers with a mirror-image DNA polymerase, Nat. Biotechnol., 2022, 40, 1601–1609 CrossRef CAS.
- T. A. Feagin, D. P. V. Olsen, Z. C. Headman and J. M. Heemstra, High-Throughput Enantiopurity Analysis Using Enantiomeric DNA-Based Sensors, J. Am. Chem. Soc., 2015, 137(12), 4198–4206 CrossRef CAS.
- W. Zhong and J. T. Sczepanski, A Mirror Image Fluorogenic Aptamer Sensor for Live-Cell Imaging of MicroRNAs, ACS Sens., 2019, 4(3), 566–570 CrossRef CAS.
- G. Ke, C. Wang, Y. Ge, N. Zheng, Z. Zhu and C. J. Yang, l-DNA Molecular Beacon: A Safe, Stable, and Accurate Intracellular Nano-thermometer for Temperature Sensing in Living Cells, J. Am. Chem. Soc., 2012, 134(46), 18908–18911 CrossRef CAS PubMed.
- L. Cui, R. Peng, T. Fu, X. Zhang, C. Wu, H. Chen, H. Liang, C. J. Yang and W. Tan, Biostable L-DNAzyme for sensing of metal ions in biological systems, Anal. Chem., 2016, 88, 1850–1855 CrossRef.
- H. B. D. Thai, K.-R. Kim, K. T. Hong, T. Voitsitskyi, J.-S. Lee, C. Mao and D.-R. Ahn, Kidney-Targeted Cytosolic Delivery of siRNA Using a Small-Sized Mirror DNA Tetrahedron for Enhanced Potency, ACS Cent. Sci., 2020, 6(12), 2250–2258 CrossRef PubMed.
- A. Y. Lee, K.-R. Kim, J. H. Yu and D.-R. Ahn, L-DNA linear duplex: An efficient drug delivery carrier with a simple structure, J. Ind. Eng. Chem., 2019, 74, 187–192 CrossRef.
- C. E. Deckard and J. T. Sczepanski, Polycomb repressive complex 2 binds RNA irrespective of stereochemistry, Chem. Commun., 2018, 54(85), 12061–12064 RSC.
- A. Lacroix, E. Vengut-Climent, D. de Rochambeau and H. F. Sleiman, Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale, ACS Cent. Sci., 2019, 5(5), 882–891 CrossRef PubMed.
- W. Shen, C. L. De Hoyos, M. T. Migawa, T. A. Vickers, H. Sun, A. Low, T. A. Bell, M. Rahdar, S. Mukhopadhyay, C. E. Hart, M. Bell, S. Riney, S. F. Murray, S. Greenlee, R. M. Crooke, X.-h. Liang, P. P. Seth and S. T. Crooke, Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index, Nat. Biotechnol., 2019, 37(6), 640–650 CrossRef PubMed.
- P. Shih, L. G. Pedersen, P. R. Gibbs and R. Wolfenden, Hydrophobicities of the nucleic acid bases: distribution coefficients from water to cyclohexane, J. Mol. Biol., 1998, 280(3), 421–430 CrossRef CAS.
- W. Shen, C. L. De Hoyos, H. Sun, T. A. Vickers, X.-h. Liang and S. T. Crooke, Acute hepatotoxicity of 2′ fluoro-modified 5–10–5 gapmer phosphorothioate oligonucleotides in mice correlates with intracellular protein binding and the loss of DBHS proteins, Nucleic Acids Res., 2018, 46(5), 2204–2217 CrossRef CAS PubMed.
- P. J. Bates, D. A. Laber, D. M. Miller, S. D. Thomas and J. O. Trent, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer, Exp. Mol. Pathol., 2009, 86(3), 151–164 CrossRef CAS PubMed.
- X. Xu, F. Hamhouyia, S. D. Thomas, T. J. Burke, A. C. Girvan, W. G. McGregor, J. O. Trent, D. M. Miller and P. J. Bates, Inhibition of DNA Replication and Induction of S Phase Cell Cycle Arrest by G-rich Oligonucleotides, J. Biol. Chem., 2001, 276(46), 43221–43230 CrossRef CAS PubMed.
- H. Qi, C.-P. Lin, X. Fu, L. M. Wood, A. A. Liu, Y.-C. Tsai, Y. Chen, C. M. Barbieri, D. S. Pilch and L. F. Liu, G-Quadruplexes Induce Apoptosis in Tumor Cells, Cancer Res., 2006, 66(24), 11808–11816 CrossRef CAS.
- C. R. Ireson and L. R. Kelland, Discovery and development of anticancer aptamers, Mol. Cancer Ther., 2006, 5(12), 2957–2962 CrossRef CAS.
- E. W. Choi, L. V. Nayak and P. J. Bates, Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides, Nucleic Acids Res., 2010, 38(5), 1623–1635 CrossRef CAS PubMed.
- L. L. Flynn, R. Li, I. L. Pitout, M. T. Aung-Htut, L. M. Larcher, J. A. L. Cooper, K. L. Greer, A. Hubbard, L. Griffiths, C. S. Bond, S. D. Wilton, A. H. Fox and S. Fletcher, Single Stranded Fully Modified-Phosphorothioate Oligonucleotides can Induce Structured Nuclear Inclusions, Alter Nuclear Protein Localization and Disturb the Transcriptome In Vitro, Front. Genet., 2022, 13, 791416 CrossRef CAS PubMed.
- D. Castanotto, M. Lin, C. Kowolik, L. Wang, X.-Q. Ren, H. S. Soifer, T. Koch, B. R. Hansen, H. Oerum, B. Armstrong, Z. Wang, P. Bauer, J. Rossi and C. A. Stein, A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells, Nucleic Acids Res., 2015, 43(19), 9350–9361 CrossRef CAS.
- C. F. Bennett, M. Y. Chiang, H. Chan, J. E. Shoemaker and C. K. Mirabelli, Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides, Mol. Pharmacol., 1992, 41(6), 1023–1033 CAS.
- S. T. Crooke, T. A. Vickers and X.-h. Liang, Phosphorothioate modified oligonucleotide–protein interactions, Nucleic Acids Res., 2020, 48(10), 5235–5253 CrossRef.
- L. C. Crowley, B. J. Marfell, A. P. Scott and N. J. Waterhouse, Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry, Cold Spring Harb. Protoc., 2016 DOI:10.1101/pdb.prot087288.
- J.-H. Shi and S.-C. Sun, Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor κB and Mitogen-Activated Protein Kinase Pathways, Front. Immunol., 2018, 9, 1849 CrossRef.
- T. Kawasaki and T. Kawai, Toll-Like Receptor Signaling Pathways, Front. Immunol., 2014, 5, 461 Search PubMed.
- T. Kawai and S. Akira, TLR signaling, Cell Death Differ., 2006, 13(5), 816–825 CrossRef CAS.
- T. Liu, L. Zhang, D. Joo and S.-C. Sun, NF-κB signaling in inflammation, Signal Transduction Targeted Ther., 2017, 2(1), 17023 CrossRef.
- R. M. Morris, T. O. Mortimer and K. L. O'Neill, Cytokines: Can Cancer Get the Message?, Cancers, 2022, 14(9), 2178 CrossRef CAS PubMed.
- S. Singh, A. Sadanandam and R. K. Singh, Chemokines in tumor angiogenesis and metastasis, Cancer Metastasis Rev., 2007, 26(3), 453–467 CrossRef PubMed.
- J. Yue and J. M. López, Understanding MAPK Signaling Pathways in Apoptosis, Int. J. Mol. Sci., 2020, 21(7), 2346 CrossRef PubMed.
- J. D. Webster and D. Vucic, The Balance of TNF Mediated Pathways Regulates Inflammatory Cell Death Signaling in Healthy and Diseased Tissues, Front. Cell Dev. Biol., 2020, 8, 365 CrossRef PubMed.
- A. S. Alharbi, A. J. Garcin, K. A. Lennox, S. Pradeloux, C. Wong, S. Straub, R. Valentin, G. Pépin, H.-M. Li, M. F. Nold, C. A. Nold-Petry, M. A. Behlke and M. P. Gantier, Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists, Nucleic Acids Res., 2020, 48(13), 7052–7065 Search PubMed.
- A. Bishani and E. L. Chernolovskaya, Activation of Innate Immunity by Therapeutic Nucleic Acids, Int. J. Mol. Sci., 2021, 22(24), 13360 CrossRef PubMed.
- T. O. Kabilova, M. I. Meschaninova, A. G. Venyaminova, V. P. Nikolin, M. A. Zenkova, V. V. Vlassov and E. L. Chernolovskaya, Short Double-Stranded RNA with Immunostimulatory Activity: Sequence Dependence, Nucleic Acid Ther., 2012, 22(3), 196–204 CrossRef PubMed.
- K. S. Frazier, Antisense Oligonucleotide Therapies: The Promise and the Challenges from a Toxicologic Pathologist's Perspective, Toxicol. Pathol., 2014, 43(1), 78–89 CrossRef PubMed.
- M. J. Lange, D. H. Burke and J. C. Chaput, Activation of Innate Immune Responses by a CpG Oligonucleotide Sequence Composed Entirely of Threose Nucleic Acid, Nucleic Acid Ther., 2018, 29(1), 51–59 CrossRef PubMed.
- B. Wlotzka, S. Leva, B. Eschgfaller, J. Burmeister, F. Kleinjung, C. Kaduk, P. Muhn, H. Hess-Stumpp and S. Klussmann, In vivo properties of an anti-GnRH spiegelmer: an example of an oligonucleotide-based therapeutic substance class, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 8898–8902 CrossRef CAS.
- R. Yazdian-Robati, P. Bayat, F. Oroojalian, M. Zargari, M. Ramezani, S. M. Taghdisi and K. Abnous, Therapeutic applications of AS1411 aptamer, an update review, Int. J. Biol. Macromol., 2020, 155, 1420–1431 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, materials and methods. ESI-MS spectra for all oligonucleotides used in this study. ESI Fig. S1–S23 and Tables S1–S2, including the sequences of all oligonucleotides used in this study. See DOI: https://doi.org/10.1039/d2sc05511b |
| This journal is © The Royal Society of Chemistry 2023 |