Open Access Article
Open Access ArticleCOF-based artificial probiotic for modulation of gut microbiota and immune microenvironment in inflammatory bowel disease†
Qingqing
Deng
ac,
Lu
Zhang
a,
Xuemeng
Liu
ac,
Lihua
Kang
*b,
Jiadai
Yi
ac,
Jinsong
Ren
 *ac and
Xiaogang
Qu
*ac and
Xiaogang
Qu
 *ac
*ac
aState Key Laboratory of Rare Earth Resources Utilization and Laboratory of Chemical Biology, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, P. R. China. E-mail: kanglh@jlu.edu.cn; jren@ciac.ac.cn; xqu@ciac.ac.cn
bCancer Center, First Affiliated Hospital, Jilin University, Changchun, Jilin 130061, P. R China
cUniversity of Science and Technology of China, Hefei, Anhui 230029, China
First published on 27th December 2022
Abstract
Conventional strategies for treating inflammatory bowel disease merely relieve inflammation and excessive immune response, but fail to solve the underlying causes of IBD, such as disrupted gut microbiota and intestinal barrier. Recently, natural probiotics have shown tremendous potential for the treatment of IBD. However, probiotics are not recommended for IBD patients, as they may cause bacteremia or sepsis. Herein, for the first time, we constructed artificial probiotics (Aprobiotics) based on artificial enzyme-dispersed covalent organic frameworks (COFs) as the “organelle” and a yeast shell as the membrane of the Aprobiotics to manage IBD. The COF-based artificial probiotics, with the function of natural probiotics, could markedly relieve IBD by modulating the gut microbiota, suppressing intestinal inflammation, protecting the intestinal epithelial cells, and regulating immunity. This nature-inspired approach may aid in the design of more artificial systems for the treatment of various incurable diseases, such as multidrug-resistant bacterial infection, cancer, and others.
Introduction
Inflammatory bowel disease (IBD) is a complex chronic inflammation of the gastrointestinal (GI) tract, leading to anemia, malnutrition, and even death. The pathological manifestations of IBD involve excessive inflammatory response, imbalance of immune responses, especially dysbiosis of gut microbiota, and disrupted intestinal barrier functions, which seriously affect gastrointestinal (GI) tract function.1–4 Clinical intervention treatments mainly concentrate on using immunosuppressive drugs, amino salicylates, and corticosteroids to relieve the symptoms of IBD by inhibiting inflammation and regulating disordered immune responses.5–9 However, these conventional drugs can cause systemic toxic side effects and result in serious complications, such as microbial infection, colorectal tumor, and autoimmune disease. To overcome the issues, great efforts have been devoted to developing targeted or stimuli-responsive delivery systems, which could specifically accumulate in the inflamed colon.10–14 Nevertheless, all these palliative approaches merely relieve inflammation and excessive immune response, and fail to solve the underlying causes of IBD, such as disrupted gut microbiota and intestinal barrier function.15 Therefore, there is an urgent need to seek a new strategy to regulate the gut microbiota, suppress intestinal inflammation, repair the intestinal barrier and modulate the immune response for the effective management of IBD.Probiotics are living microorganisms that exert health benefits beyond the metabolic effect of its nutritional components.16 Probiotic bacteria have been used as promising and novel agents for the treatment of various gastrointestinal inflammatory disorders based on the following mechanisms: (i) modulating gut microbiota through the consumption of free oxygen and the release of some bioactive molecules, (ii) decreasing the secretion of inflammatory cytokine, (iii) regulating the immune system to suppress intestinal inflammation, and (iv) protecting the intestinal epithelial barrier function and repairing mucosal damage.17–19 However, for complex cases such as inflammatory bowel disease, a combination of multiple probiotics with different functions is usually required to obtain an ideal therapeutic effect. Moreover, probiotics are not recommended for IBD patients, as they may cause bacteremia or even sepsis.16,20
As an emerging class of porous crystalline materials, covalent organic frameworks (COFs) with well-defined porosity, tunable composition, ultrahigh surface area and excellent biocompatibility have been developed as salient materials for separation, catalysis, sensing, drug delivery, and other biomedical applications.21–27 Particularly, Schiff base-linked COFs have attracted great attention because of their interesting physical and chemical properties.28 Furthermore, it is worth noting that due to the highly active imine (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) bond, the Schiff base and its derivatives possess excellent antioxidant activity.29–32 By taking advantage of these unique features, we speculated that a Schiff base-linked COF could be an ideal platform for the treatment of IBD.
N) bond, the Schiff base and its derivatives possess excellent antioxidant activity.29–32 By taking advantage of these unique features, we speculated that a Schiff base-linked COF could be an ideal platform for the treatment of IBD.
Herein, for the first time, we designed and constructed a COF-based artificial probiotic to modulate gut microbiota, suppress intestinal inflammation, regulate the immune microenvironment, and protect the intestinal epithelial cells for the treatment of IBD. As illustrated in Fig. 1, the artificial probiotics were created by encapsulating the COFs dispersed with ultrasmall gold nanoparticles in the yeast shell (denoted as YS, composed primarily of β-glucan). In this design, COFs with uniform pore sizes were utilized as “sponge” to scavenge the free radicals. Owing to the well-defined porosity, ultrahigh surface area and excellent biocompatibility, the COFs were utilized as scaffolds to disperse and protect the artificial enzyme. The COFs dispersed with artificial enzymes served as “organelle,” and the yeast β-glucan served as the membrane of the artificial probiotic. This artificial probiotic can mimic the function of natural probiotics for the treatment of IBD by (i) modulating the gut microbiota by “secreting” gluconic acid and depleting oxygen,18,33,34 (ii) suppressing the inflammatory response, (iii) protecting the intestinal epithelial cells,35,36 and (iv) switching the polarization of macrophages from the classically pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype to regulate the imbalance in the immune microenvironment and inhibit excessive immune response.37–39 Taken together, the COF-based artificial probiotics provide an effective intervention strategy to facilitate recovery from IBD. This nature-inspired approach may aid the design of more artificial systems for the treatment of various incurable diseases such as multidrug-resistant bacterial infection, cancer, and others.
Results and discussion
To verify our hypothesis, the artificial probiotics (denoted as Aprobiotics) were created by encapsulating porous covalent organic frameworks (COFs) supported by ultrasmall gold nanoparticles in yeast β-glucan. The detailed synthesis process is provided in ESI.† Briefly, COFs were prepared by a facile solution-phase synthesis method at room temperature.40–42 Then, the COF@Au was obtained by in situ reduction method.43,44 Finally, COF@Au nanoparticles were encapsulated in the empty YS by electrostatic force-driven self-deposition to construct the artificial probiotic.45,46The morphology and size of the COF, COF@Au, and Aprobiotics were characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). After reduction in situ, the ultrasmall Au nanoparticles were distributed throughout the COFs. In the COF@Au nanosystem, the size and morphology of COFs were well maintained (∼120 nm). SEM and TEM images indicated that the obtained ellipsoid YS products were enwrapped COF@Au nanoparticles (Fig. 2a and S1–S5†). Meanwhile, X-ray photon spectroscopy (XPS) further confirmed the distribution of C, O, N, and Au elements in the same particle. Moreover, the XPS result demonstrated that Au 4f peaks were observed at 83.7 and 87.5 eV, which were not found in the COF (Fig. 2d and S6†). The presence of Au nanoparticles and COF was further verified by the X-ray powder diffraction (XRD) pattern (Fig. 2b).47 The zeta potential changed from −15.1 mV to −18.4 mV, which demonstrated that the gold nanoparticles were successfully dispersed in the COF. Then, due to the COF@Au (−18.4 mV) encapsulation by PEI-YS (7.6 mV), the zeta potential reached −8.7 mV (Fig. 2c). All results proved the successful preparation of the artificial probiotic.
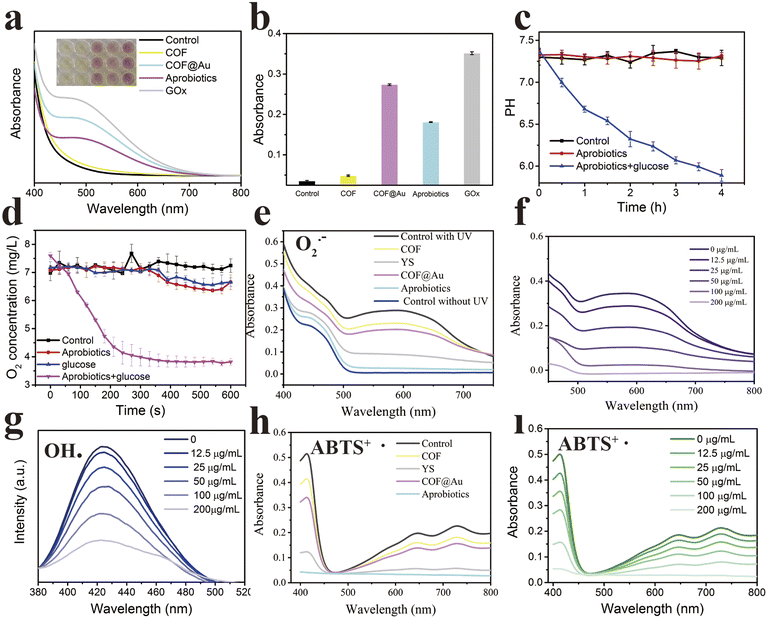
After confirming successful preparation of the artificial probiotic, the bioactivities of the Aprobiotics were first explored in vitro, including glucose oxidase-like activity and antioxidant activity. Due to the excellent glucose oxidase-like activity, the artificial probiotics may increase gut microbiota richness and diversity. First, the glucose oxidase-like catalytic ability was carefully studied. The successful oxidation of glucose into gluconic acid was tested using colorimetric assays. As shown in Fig. 3a–c, activity is attributed to the immobilization of ultrasmall gold nanoparticles, leading to the production of gluconic acid and the decrease of pH. Aside from the oxidation of glucose, the Aprobiotics can deplete oxygen, decreasing O2 concentration from 7.60 to 3.82 mg L−1 in 600 s (Fig. 3d).48,49 These results strongly demonstrated that the Aprobiotics could “secrete” gluconic acid and deplete oxygen for the regulation of gut microbiota.
Recently, numerous biological membranes have been developed and applied in functional hybrids.50–52 The membrane of Aprobiotics is primarily composed of β-glucan, which is a polysaccharide derived from the yeast cell wall. The triple-helix conformation of β-glucan as free-radical quencher may reveal excellent anti-oxidation activity. The anti-oxidation activity was studied by the ability to remove the superoxide anion radical (O2˙−), hydroxyl radical (OH˙), and 2, 2′-azinobis (3-ethylbenzothiazoline 6-sulfonate) (ABTS+˙).53–55 The O2˙−, as a highly oxidizing ROS, was chosen to investigate the anti-oxidation ability of COF, COF@Au, and the Aprobiotics.
As shown in Fig. 3e and f, the Aprobiotic membrane (β-glucan) showed extremely high ROS scavenging effects when incubated with O2˙−. The scavenging efficiency of Aprobiotics reached 90%, which exhibited that the Aprobiotics possess outstanding antioxidant capacity in a concentration-dependent manner. The hydroxyl radical (OH˙) is a typical type of ROS in the inflammatory response, which was selected to study the antioxidant activity of the Aprobiotics. The Aprobiotics showed the highly sensitive and concentration-dependent scavenging of OH˙ (Fig. 3g and S12†). Furthermore, ABTS+˙ was employed to further assess the antioxidant activity of the Aprobiotics. As shown in Fig. 3h and i, more than 80% of the ABTS+˙ was scavenged by the Aprobiotics (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg mL−1). The outstanding antioxidant activity of the Aprobiotics was attributed to the triple helix structure of the membrane. We proved that this artificial probiotic readily removed all types of representative ROS to reduce the level of oxidative stress.
μg mL−1). The outstanding antioxidant activity of the Aprobiotics was attributed to the triple helix structure of the membrane. We proved that this artificial probiotic readily removed all types of representative ROS to reduce the level of oxidative stress.
To further prove the in vitro antioxidant ability of the Aprobiotics, cell-based studies were performed. Firstly, the cellular uptake of Aprobiotics was investigated using RAW 264.7 and CT26 cells (Fig. S16 and S17†). The results showed that the Rhodamine B–labeled Aprobiotics could be taken up effectively after 4 h. The cytotoxicity of the Aprobiotics was also assessed by methyl thiazolyl tetrazolium (MTT) assay (Fig. S18 and 19†), which demonstrated that the Aprobiotics had negligible cytotoxicity. These results demonstrated that the Aprobiotics have potential for further biological applications.
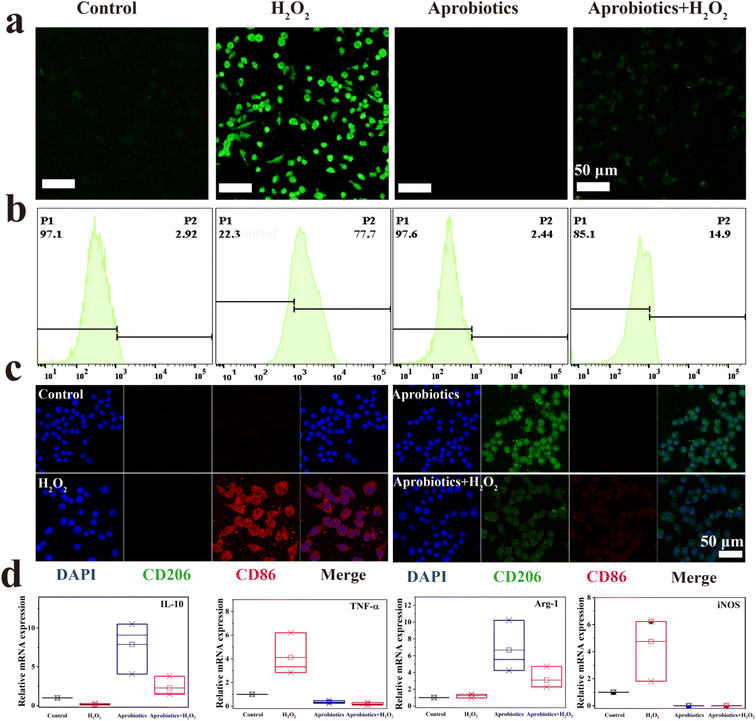
Hydrogen peroxide (H2O2) is a critical mediator in the inflammatory response process, leading to oxidative stress and inflammation.56,57 The antioxidant activity of the Aprobiotics encouraged us to study the cytoprotection against ROS-induced damage. Fig. S20 and S21† showed that H2O2 caused a sharp decrease of cell viability. The cell viability of Raw 264.7 and CT26 was reduced to 14.25% and 21.49%, respectively. However, after adding Aprobiotics (200 μg mL−1), cell viability was maintained at 98.66% and 90.03%, respectively. To prove the cytoprotective effect of Aprobiotics, 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) was employed as a ROS probe to study intracellular ROS. Bright-green fluorescence was found after treatment with H2O2 (Fig. 4a and S22†). However, the cell incubated with both Aprobiotics and H2O2 showed very weak fluorescence, similar to the control group. Moreover, the flow cytometry analysis further evidenced the effective antioxidant property of the Aprobiotics (Fig. 4b). All results exhibited that the Aprobiotics have the potential to relieve oxidative damage and protect the intestinal epithelial cells.
Previous studies have found that the symptoms of IBD can be alleviated by reprogramming the pro-inflammatory M1-phenotype macrophages, which could inhibit the excessive inflammatory response.58 Therefore, the immunomodulatory effect of Aprobiotics was explored by examining the phenotype of macrophages. M1-phenotype macrophages are recognized by surface markers CD86, inducible nitric oxide synthase (iNOS), and pro-inflammatory cytokine, such as tumor necrosis factor-α (TNF-α), which is related to the pro-inflammatory microenvironment resulting in the excessive inflammatory response in IBD. In contrast, M2-phenotype macrophages secrete high levels of surface marker CD206 and arginase-1 (Arg-1) and generate anti-inflammatory mediators (IL-10), which results in anti-inflammation, cell proliferation, and tissue repair.59,60 Compared with the control group, H2O2-treated cells exhibited the M1 phenotype with greater production of CD86 and less production of CD206 (Fig. 4c). When macrophages were exposed to additional Aprobiotics, the generation of CD206 increased compared with the control group. However, the cells incubated with both Aprobiotics and H2O2 had levels of CD206 and CD86 similar to the control group. In addition, qPCR assay was utilized for the quantitative exploration of the level of related cytokines. The Aprobiotic-treated group demonstrated decreased expression level of M1-related RNA (iNOS and TNF-a), and the expression of M2-related RNA (Arg-1 and IL-10) significantly increased, indicating the production of anti-inflammatory cytokines induced by Aprobiotics (Fig. 4d). All these outcomes demonstrated that the Aprobiotics could effectively regulate the transition of macrophages from M1 phenotype to M2 phenotype, inhibiting the excessive inflammatory response.
Before exploring the treatment effect of Aprobiotics in vivo, we studied their biosafety. Firstly, the in vivo toxicity of Aprobiotics was evaluated by monitoring body weight and the hematological parameters of mice. No evident differences from the control group on body weight and blood indexes were exhibited after treatment with Aprobiotics (Fig. S23 and 24†). Hematoxylin and eosin (H&E) staining assay of major organs also confirmed the excellent biocompatibility of the Aprobiotics (Fig. S25†). These results demonstrated that there was no obvious toxicity from Aprobiotics in vivo.
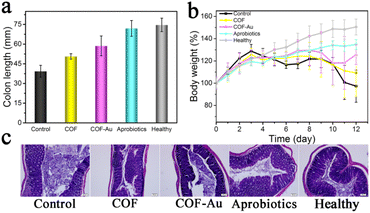
Inspired by the excellent anti-inflammation activity and good biosafety of Aprobiotics, their therapeutic effect was further explored in vivo using DSS-induced ulcerative colitis (UC) models. The successful construction of UC was indicated by the decrease of body weight (Fig. 5b). Afterward, COF, COF@Au, or Aprobiotics were orally administered for 7 days. The therapeutic effect of Aprobiotics was assessed by measuring the colon length (Fig. 5a) and body weight (Fig. 5b). Compared with the control group, the mice treated with Aprobiotics exhibited longer colon length (Fig. 5a). Moreover, the colon tissues were taken out for H&E staining assay (Fig. 5c). Compared with the healthy colon, inflammatory signs of colitis (inflammatory cell infiltration, ulceration, and goblet cell damage) were obviously observed in the ulcerative colitis mice (Fig. 5c).
However, after treatment with Aprobiotics, the excessive inflammatory response and disrupted intestinal barrier showed significant improvement in histopathologic features, similar to healthy colon tissue. These results indicated that Aprobiotics had outstanding therapeutic effects in DSS-induced UC models.
The disruption of gut microbiota is the underlying cause of IBD, but conventional strategies fail to modulate the imbalanced gut microbiota. Previous studies have demonstrated that natural probiotics can modulate gut microbiota through the consumption of free oxygen and the release of some bioactive molecules for the treatment of IBD. Therefore, we investigated whether Aprobiotic treatment could regulate the gut microbiota in IBD. Mice faecal samples were analyzed by 16S ribosomal RNA gene sequencing in the V3–V4 regions, which displayed that Aprobiotic treatment significantly increased the gut bacterial richness of IBD mice (Fig. 6a). The principal component analysis revealed that IBD mice treated with Aprobiotics possessed similar gut microbiota profiles (Fig. 6b and c). We further studied the phylum level of the gut microbiome, which showed that Aprobiotic treatment significantly increased the relative abundance of Clostridiales (responsible for inducing Treg cells), and Lactobacillus (beneficial to IBD animal models) (Fig. 6d–f). All of these are critical factors to relieve the symptom of IBDs.15,61 These results suggest that the benefits of Aprobiotics are partially attributed to their modulation of the gut microbiome.
The cell-based study has confirmed that Aprobiotics could effectively regulate the transition of macrophages from M1 phenotype to M2 phenotype, inhibiting the excessive inflammatory response. Therefore, we examined whether the Aprobiotics could modulate the imbalanced immune microenvironment in vivo. Colons tissues were taken out for immunohistochemistry assay. Compared with the healthy colon, high levels of TNF-α and IL-6 revealed that the pro-inflammation M1 macrophages significantly increased in the control group (Fig. 6g). By contrast, after treatment with Aprobiotics, the proportion of M1 macrophages decreased significantly. Overall, these results demonstrated that the excessive immune response in IBD was reversed by the Aprobiotics, and that a new anti-inflammatory microenvironment was re-established for the treatment of IBD.
Conclusions
In summary, we successfully constructed COF-based artificial probiotics by encapsulating the porous covalent organic frameworks (COFs) dispersed with ultrasmall gold nanoparticles in a yeast shell for the effective treatment of IBD. This artificial probiotic can treat DSS-induced IBD via four biological functions: (i) regulating gut microbiota, (ii) suppressing the inflammatory response, (iii) protecting the intestinal epithelial cells, and (iv) reprogramming the macrophage to regulate the imbalanced immune microenvironment. Taken together, the artificial probiotics provide a promising strategy for the treatment of IBD by modulating the gut microbiota, suppressing intestinal inflammation, protecting the intestinal epithelial cells, and regulating immunity. This nature-inspired approach may aid the design of more artificial systems for the treatment of other incurable diseases such as multidrug-resistant bacterial infection, cancer, and others.Data availability
Data are available in the ESI† online.Author contributions
Q. Deng, L. Kang, J. Ren and X. Qu conceived the project. Q. Deng and X. Liu designed the experiments; Q. Deng, X. Liu, and J. Yi performed the experiments. Q. Deng, L. Zhang, and X. Liu analyzed the data. Q. Deng, L. Zhang, X. Liu, and J. Ren wrote the paper. All authors approved the final version.Ethical statement
All animal experiments were carried out in accordance with the NIH guidelines for the care and use of laboratory animals. The Jilin University Animal Care and Use Committee has approved the experiments. Six-week-old Kunming mice (20 g) were purchased from the Laboratory Animal Center of Jilin University (Changchun, China). The animal handling procedures were in accordance with the guidelines of the Animal Ethics Committee of Jilin University for animal experiments.Conflicts of interest
All authors declare no competing interests.Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFF1200701, 2019YFA0709202) and the National Nature Science Foundation of China (21820102009, 91856205 and 22237006).Notes and references
- J. L. Round and S. K. Mazmanian, Nat. Rev. Immunol., 2009, 9, 313–323 CrossRef CAS PubMed.
- J. Halfvarson, C. J. Brislawn, R. Lamendella, Y. Vázquez-Baeza, W. A. Walters, L. M. Bramer, M. D'Amato, F. Bonfiglio, D. McDonald, A. Gonzalez, E. E. McClure, M. F. Dunklebarger, R. Knight and J. K. Jansson, Nat. Microbiol., 2017, 2, 17004 CrossRef CAS PubMed.
- S. Citi, Science, 2018, 359, 1097–1098 CrossRef CAS PubMed.
- R. Caruso, B. C. Lo and G. Nunez, Nat. Rev. Immunol., 2020, 20, 411–426 CrossRef CAS PubMed.
- L. Steidler, W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers and E. Remaut, Science, 2000, 289, 1352–1355 CrossRef CAS PubMed.
- B. Tirosh, N. Khatib, Y. Barenholz, A. Nissan and A. Rubinstein, Mol. Pharm., 2009, 6, 1083–1091 CrossRef CAS PubMed.
- S. Zhang, J. Ermann, M. D. Succi, A. Zhou, M. J. Hamilton, B. Cao, J. R. Korzenik, J. N. Glickman, P. K. Vemula, L. H. Glimcher, G. Traverso, R. Langer and J. M. Karp, Sci. Transl. Med., 2015, 7, 300ra128 Search PubMed.
- Q. Zhang, H. Tao, Y. Lin, Y. Hu, H. An, D. Zhang, S. Feng, H. Hu, R. Wang, X. Li and J. Zhang, Biomaterials, 2016, 105, 206–221 CrossRef CAS PubMed.
- Y. Tian, J. Xu, Y. Li, R. Zhao, S. Du, C. Lv, W. Wu, R. Liu, X. Sheng, Y. Song, X. Bi, G. Li, M. Li, X. Wu, P. Lou, H. You, W. Cui, J. Sun, J. Shuai, F. Ren, B. Zhang, M. Guo, X. Hou, K. Wu, L. Xue, H. Zhang, M. V. Plikus, Y. Cong, C. J. Lengner, Z. Liu and Z. Yu, Gastroenterology, 2019, 156, 2281–2296.e2286 CrossRef CAS PubMed.
- S. Bertoni, Z. Liu, A. Correia, J. P. Martins, A. Rahikkala, F. Fontana, M. Kemell, D. Liu, B. Albertini, N. Passerini, W. Li and H. A. Santos, Adv. Funct. Mater., 2018, 28, 1806175 CrossRef.
- N. Dammes, M. Goldsmith, S. Ramishetti, J. L. J. Dearling, N. Veiga, A. B. Packard and D. Peer, Nat. Nanotechnol., 2021, 11, 1030–1038 CrossRef PubMed.
- G. Chen, F. Wang, M. Nie, H. Zhang, H. Zhang and Y. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2112704118 CrossRef CAS PubMed.
- B. M. Scott, C. Gutierrez-Vazquez, L. M. Sanmarco, J. A. da Silva Pereira, Z. Li, A. Plasencia, P. Hewson, L. M. Cox, M. O'Brien, S. K. Chen, P. M. Moraes-Vieira, B. S. W. Chang, S. G. Peisajovich and F. J. Quintana, Nat. Med., 2021, 27, 1212–1222 CrossRef CAS PubMed.
- D. S. Wilson, G. Dalmasso, L. Wang, S. V. Sitaraman, D. Merlin and N. Murthy, Nat. Mater., 2020, 9, 923–928 CrossRef PubMed.
- Y. Lee, K. Sugihara, M. G. Gillilland, 3rd, S. Jon, N. Kamada and J. J. Moon, Nat. Mater., 2020, 19, 118–126 CrossRef CAS PubMed.
- J. Suez, N. Zmora, E. Segal and E. Elinav, Nat. Med., 2019, 25, 716–729 CrossRef CAS PubMed.
- T. Kuhbacher, S. J. Ott, U. Helwig, T. Mimura, F. Rizzello, B. Kleessen, P. Gionchetti, M. Blaut, M. Campieri, U. R. Folsch, M. A. Kamm and S. Schreiber, Gut, 2006, 55, 833–841 CrossRef CAS PubMed.
- A. M. O'Hara and F. Shanahan, Sci. World J., 2007, 7, 31–46 CrossRef PubMed.
- J. Zhou, M. Li, Q. Chen, X. Li, L. Chen, Z. Dong, W. Zhu, Y. Yang, Z. Liu and Q. Chen, Nat. Commun., 2022, 13, 3432 CrossRef CAS PubMed.
- T. Kuhn, M. Koch and G. Fuhrmann, Small, 2020, 16, 2003158 CrossRef CAS PubMed.
- C. S. Diercks and O. M. Yaghi, Science, 2017, 355, eaal1585 CrossRef PubMed.
- M. Li, S. Qiao, Y. Zheng, Y. H. Andaloussi, X. Li, Z. Zhang, A. Li, P. Cheng, S. Ma and Y. Chen, J. Am. Chem. Soc., 2020, 142, 6675–6681 CrossRef CAS PubMed.
- Q. Sun, C. W. Fu, B. Aguila, J. Perman, S. Wang, H. Y. Huang, F. S. Xiao and S. Ma, J. Am. Chem. Soc., 2018, 140, 984–992 CrossRef CAS PubMed.
- N. Huang, P. Wang and D. Jiang, Nat. Rev. Mater., 2016, 1, 16068 CrossRef CAS.
- H. Wang, Z. Zeng, P. Xu, L. Li, G. Zeng, R. Xiao, Z. Tang, D. Huang, L. Tang, C. Lai, D. Jiang, Y. Liu, H. Yi, L. Qin, S. Ye, X. Ren and W. Tang, Chem. Soc. Rev., 2019, 48, 488–516 RSC.
- T. Zhang, G. Zhang and L. Chen, Acc. Chem. Res., 2022, 55, 795–808 CrossRef CAS PubMed.
- Q. Fang, J. Wang, S. Gu, R. B. Kaspar, Z. Zhuang, J. Zheng, H. Guo, S. Qiu and Y. Yan, J. Am. Chem. Soc., 2015, 137, 8352–8355 CrossRef CAS PubMed.
- X. H. Han, K. Gong, X. Huang, J. W. Yang, X. Feng, J. Xie and B. Wang, Angew. Chem., Int. Ed. Engl., 2022, 61, e202202912 CAS.
- S. M. Sondhi, S. Arya, R. Rani, N. Kumar and P. Roy, Med. Chem. Res., 2011, 21, 3620–3628 CrossRef.
- K. P. Rakesh, H. M. Manukumar and D. C. Gowda, Bioorg. Med. Chem. Lett., 2015, 25, 1072–1077 CrossRef CAS PubMed.
- S. Yu, Y. Wang, S. Wang, J. Zhu and S. Liu, Ind. Eng. Chem. Res., 2019, 59, 1031–1037 CrossRef.
- A. GÜMÜŞ, V. Okumu and S. GÜMÜŞ, Turk. J. Chem., 2020, 44, 1200–1215 CrossRef PubMed.
- W. Luo, C. Zhu, S. Su, D. Li, Y. He, Q. Huang and C. Fan, ACS Nano, 2010, 4, 7451–7458 CrossRef CAS PubMed.
- M. Zhao, C. Chen, Z. Yuan, W. Li, M. Zhang, N. Cui, Y. Duan, X. Zhang and P. Zhang, Exp. Ther. Med., 2021, 22, 1312 CrossRef CAS PubMed.
- G. Şener, H. Toklu, F. Ercan and G. Erkanlı, Int. Immunopharmacol., 2005, 5, 1387–1396 CrossRef PubMed.
- U. Bacha, M. Nasir, S. Iqbal and A. A. Anjum, BioMed Res. Int., 2017, 2017, 1–14 CrossRef PubMed.
- K. H. Lee, M. Park, K. Y. Ji, H. Y. Lee, J. H. Jang, I. J. Yoon, S. S. Oh, S. M. Kim, Y. H. Jeong, C. H. Yun, M. K. Kim, I. Y. Lee, H. R. Choi, K. S. Ko and H. S. Kang, Immunobiology, 2014, 219, 802–812 CrossRef CAS PubMed.
- Y. Nie, Q. Lin and F. Luo, Int. J. Mol. Sci., 2017, 18, 1372 CrossRef PubMed.
- H. Yuan, P. Lan, Y. He, C. Li and X. Ma, Molecules, 2019, 25, 57 CrossRef PubMed.
- C. Hu, L. Cai, S. Liu and M. Pang, Chem. Commun, 2019, 55, 9164–9167 RSC.
- Y. Shi, S. Liu, Z. Zhang, Y. Liu and M. Pang, Chem. Commun, 2019, 55, 14315–14318 RSC.
- F. Benyettou, G. Das, A. R. Nair, T. Prakasam, D. B. Shinde, S. K. Sharma, J. Whelan, Y. Lalatonne, H. Traboulsi, R. Pasricha, O. Abdullah, R. Jagannathan, Z. Lai, L. Motte, F. Gándara, K. C. Sadler and A. Trabolsi, J. Am. Chem. Soc., 2020, 142, 18782–18794 CrossRef CAS PubMed.
- S.-Y. Ding, J. Gao, Q. Wang, Y. Zhang, W.-G. Song, C.-Y. Su and W. Wang, J. Am. Chem. Soc., 2011, 133, 19816–19822 CrossRef CAS PubMed.
- Y. Lin, L. Wu, Y. Huang, J. Ren and X. Qu, Chem. Sci., 2015, 6, 1272–1276 RSC.
- E. R. Soto and G. R. Ostroff, Bioconjugate Chem., 2008, 19, 840–848 CrossRef CAS PubMed.
- X. Zhou, X. Zhang, S. Han, Y. Dou, M. Liu, L. Zhang, J. Guo, Q. Shi, G. Gong, R. Wang, J. Hu, X. Li and J. Zhang, Nano Lett., 2017, 17, 1056–1064 CrossRef CAS PubMed.
- Y. Tao, E. Ju, J. Ren and X. Qu, Adv. Mater., 2015, 27, 1097–1104 CrossRef CAS PubMed.
- M. C. Ortega-Liebana, J. Bonet-Aleta, J. L. Hueso and J. Santamaria, Catalysts, 2020, 10, 333 CrossRef CAS.
- L. Zhang, Z. Wang, Y. Zhang, F. Cao, K. Dong, J. Ren and X. Qu, ACS Nano, 2018, 12, 10201–10211 CrossRef CAS PubMed.
- Q. Zhang, D. Dehaini, Y. Zhang, J. Zhou, X. Chen, L. Zhang, R. H. Fang, W. Gao and L. Zhang, Nat. Nanotechnol., 2018, 13, 1182–1190 CrossRef CAS PubMed.
- K. Zhang, R. Deng, Y. Sun, L. Zhang and J. Li, Chem. Sci., 2017, 8, 7098–7105 RSC.
- L. Sun, Y. Gao, Y. Wang, Q. Wei, J. Shi, N. Chen, D. Li and C. Fan, Chem. Sci., 2018, 9, 5967–5975 RSC.
- K. Korschelt, R. Ragg, C. S. Metzger, M. Kluenker, M. Oster, B. Barton, M. Panthofer, D. Strand, U. Kolb, M. Mondeshki, S. Strand, J. Brieger, M. Nawaz Tahir and W. Tremel, Nanoscale, 2017, 9, 3952–3960 RSC.
- N. Singh, M. A. Savanur, S. Srivastava, P. D'Silva and G. Mugesh, Angew. Chem., Int. Ed. Engl., 2017, 56, 14267–14271 CrossRef CAS PubMed.
- T. Liu, B. Xiao, F. Xiang, J. Tan, Z. Chen, X. Zhang, C. Wu, Z. Mao, G. Luo, X. Chen and J. Deng, Nat. Commun., 2020, 11, 2788 CrossRef CAS PubMed.
- Q. Wang, C. Cheng, S. Zhao, Q. Liu, Y. Zhang, W. Liu, X. Zhao, H. Zhang, J. Pu, S. Zhang, H. Zhang, Y. Du and H. Wei, Angew. Chem., Int. Ed. Engl., 2022, 61, e202201101 CAS.
- Y. Liu, Y. Cheng, H. Zhang, M. Zhou, Y. Yu, S. Lin, B. Jiang, X. Zhao, L. Miao, C.-W. Wei, Q. Liu, Y.-W. Lin, Y. Du, C. J. Butch and H. Wei, Sci. Adv., 2020, 6, eabb2695 CrossRef CAS PubMed.
- B.-C. Lee, J. Y. Lee, J. Kim, J. M. Yoo, I. Kang, J.-J. Kim, N. Shin, D. J. Kim, S. W. Choi, D. Kim, B. H. Hong and K.-S. Kang, Sci. Adv., 2020, 6, eaaz2630 CrossRef CAS PubMed.
- J. H. Song, T. Yoon, S. M. Lee, C. H. Mun, D. Kim, J. Han, J. W. Kim, Y. Bae, T. Kim, Y. N. Park, M. H. Cho, Y. B. Park and K. H. Yoo, Adv. Funct. Mater., 2021, 32, 2107433 CrossRef.
- S. Zhao, Y. Li, Q. Liu, S. Li, Y. Cheng, C. Cheng, Z. Sun, Y. Du, C. J. Butch and H. Wei, Adv. Funct. Mater., 2020, 30, 2004692 CrossRef CAS.
- D. W. Zheng, R. Q. Li, J. X. An, T. Q. Xie, Z. Y. Han, R. Xu, Y. Fang and X. Z. Zhang, Adv. Mater., 2020, 32, 2004529 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc04984h |
| This journal is © The Royal Society of Chemistry 2023 |