Open Access Article
Open Access ArticleCopper nanoparticles embedded fungal chitosan as a rational and sustainable bionanozyme with robust laccase activity for catalytic oxidation of phenolic pollutants†
Efrata Getachew Mekonnena,
Kassie Nigus Shitawb,
Bing-Joe Hwangbc,
Yitayal Admassu Workiedf,
Ebrahim M. Abda*ae and
Menbere Leul Mekonnen *df
*df
aBiotechnology Department, Addis Ababa Science, and Technology University, P. O. Box 1647, Addis Ababa, Ethiopia. E-mail: ebrahim.mama@aastu.edu.et
bDepartment of Chemical Engineering, National Taiwan University of Science and Technology, Taipei 106, Taiwan
cNational Synchrotron Radiation Research Center, Hsinchu, Taiwan
dIndustrial Chemistry Department, Addis Ababa Science and Technology University, P. O. Box 1647, Addis Ababa, Ethiopia. E-mail: menbere.leul@aastu.edu.et
eBioprocess and Biotechnology Center of Excellence, Addis Ababa Science and Technology University, P. O. Box 1647, Addis Ababa, Ethiopia
fNanotechnology Center of Excellence, Addis Ababa Science and Technology University, P. O. Box 1647, Addis Ababa, Ethiopia
First published on 1st November 2023
Abstract
Despite their potential for oxidation of persistent environmental pollutants, the development of rational and sustainable laccase nanozymes with efficient catalytic performance remains a challenge. Herein, fungal-produced chitosan–copper (CsCu) is proposed as a rational and sustainable bionanozyme with intrinsic laccase activity. The CsCu nanozyme was prepared by in situ reduction of copper on chitosan extracted from Irpex sp. isolate AWK2 a native fungus, from traditional fermented foods, yielding a low molecular weight chitosan with a 70% degree of deacetylation. Characterizations of the nanozyme using SEM-EDX, XRD, and XPS confirmed the presence of a multi-oxidation state copper on the chitosan matrix which is consistent with the composition of natural laccase. The laccase memetic activity was investigated using 2,4-DP as a substrate which oxidized to form a reddish-pink color with 4-AP (λmax = 510 nm). The CsCu nanozyme showed 38% higher laccase activity than the pristine Cu NPs at pH 9, indicating enhanced activity in the presence of chitosan structure. Further, CsCu showed significant stability in harsh conditions and exhibited a lower Km (0.26 mM) which is competitive with that reported for natural laccase. Notably, the nanozyme converted 92% of different phenolic substrates in 5 h, signifying a robust performance for environmental remediation purposes.
1 Introduction
Phenolic compounds are persistent environmental pollutants that cause a public health concern. These compounds emerge from industrial and municipal sources as well as from the decomposition of organic matter and often end up in the aquatic system exerting a toxic effect.1 Hence, it's of high significance to find suitable remediation for such persistent environmental pollutants. Although laccase enzymes have potential in the remediation of phenolic compounds, their intrinsic limitations such as low stability, high purification cost, etc. have greatly hindered their application in real industrial settings.2,3Laccase is a multi-copper oxidase capable of oxidizing a wide range of phenolic substrates. In a typical laccase-catalyzed reaction, the substrates are oxidized at the T1 Cu site, and electrons are transferred to the T2/T3 trinuclear Cu cluster where molecular oxygen is converted into the water through a cysteine–histidine (Cys–His) pathways.4 Hence, the reaction product being water qualifies laccase as a green catalyst. However, laccase shares the intrinsic limitations associated with natural enzymes and hence could not be effectively used under extreme operational conditions for environmental remediation purposes. In this regard, artificial enzymes such as nanozymes could be considered as suitable alternatives.4,5 For instance, various nanomaterials such as copper oxide,6 cerium oxide,7 etc. have shown laccase mimicking properties for degradation of different phenolic compounds.
Nanozymes are enzyme-mimetic nanomaterials that emerged as robust catalysts that could be alternatives for natural enzymes in various application areas. The progress in nanotechnology allows synthetic protocols to be tailored so that nanomaterials can be easily designed to mimic the required enzyme structures at a lower cost. This could also avoid the lengthy purification steps required to produce natural enzymes.8,9 After the first report on the peroxidase mimetic ability of Fe3O4 nanoparticles by Gao and co-workers in 2007, the utility of nanozymes has progressed much in terms of both type and mechanistic understanding.10,11 Among these, copper and other transition metal-based nanozymes have been reported as laccase mimics for sensing and environmental remediation purposes.4,7,12 However, most of these laccase nanozymes often lack either rational design or sustainability which in turn affects their intrinsic enzyme activity. Hence the development of sustainable and robust nanozymes mimicking both the structure and function of natural enzymes could be beneficial.13
Given that the active site of laccase lies on copper–cysteine–histidine, various laccase nanozymes are reported by coordinating copper with oligonucleotides and dipeptides.5,14–16 However, such protein/amino acid structures are expensive and would not serve at extreme operational conditions for environmental remediation purposes. Recently, copper–tannic acid has been presented as a more tolerant laccase nanozymes for phenolic compound oxidation.17 Hence, it's critical to look for more robust and cheaper rational designs of nanozymes mimicking the natural laccase with its activity as well as structure.18
In this study, a rational design of laccase bionanozyme is demonstrated by using fungal-produced chitosan as a scaffold for the copper active center to form Chitosan–Copper (CsCu) laccase nanozyme. The hydroxyl and amino groups in chitosan could endow the nanozyme similar reductive microenvironment for Cu2+/Cu+ as the amino acids in laccase. Such a microenvironment enhances the electron transfer similar to the cysteine–histidine pathway in the natural laccase.12,14 Chitosan is a biodegradable and abundant biopolymer which is an important factor for sustainable industrial-scale production of nanozymes. Current extraction of chitosan depends on marine sources which often suffer from the seasonal nature of the feedstock and non-uniform chitosan product.19 To counter this, chitosan is extracted from a fungus isolated from traditional fermented food. This demonstrates cheaper and sustainable feedstock for chitosan and laccase nanozymes construction. The prepared nanozyme was optimized for its laccase memetic activity and showed promising degradation of various phenolic compound substrates.
2 Experimental
2.1 Materials
All chemicals and reagents used were analytical grades. 2,4-Dichlophenol, 4-aminoantipyrine, copper chloride, hydroquinone, phenol, and chloroform were purchased from Alpha Chemika, India. Hydrazine Isopropanol, magnesium sulfate heptahydrate, ammonium sulfate, and calcium chloride, were obtained from Loba Chemie, India. Ethanol, sodium carbonate, carbonic acid, Britton's buffer, and trisbuffer were obtained from Research-LAB Fine Chemical Industries India. Boric acid and dipotassium phosphate were purchased from HiMedia Laboratories India. Sodium hydroxide was purchased at Central Drug House India. Acetic acid, lactophenol blue, acetone, peptone, glucose, yeast extract, potato dextrose broth, and potato dextrose agar were purchased from Sisco Research Laboratories, India.2.2 Isolation of fungi
The isolation of native fungal species from Kocho, a traditional fermented food, was done following standard procedures.20 Briefly, about 250 grams of Kocho were collected from a farmer's homestead plot in Aleta Wendo, located in the Sidama region, Ethiopia. The Kocho had been buried in the earthen pits and covered with layers of leaf sheets for three months before sampling. Then, ten grams of the Kocho sample was homogenized in 90 mL of sterile saline water. An aliquot of serially diluted samples was inoculated onto Petri plates containing potato dextrose agar (PDA) medium and incubated at 28 °C for 2 to 5 days. The obtained fungal isolates were then re-streaked on the same media to obtain a pure culture with a uniform morphological appearance in size, shape, spore color, and mycelium color.2.3 Screening for chitosan-producing fungi
Fungal isolates were refreshed on PDA agar plates at 28 °C for 8 days. Once the culture reached the late exponential phase, which can be determined by spore germination, a spore suspension was obtained by adding sterile distilled water and then gently scraping the surface of the agar plates with a spatula. Subsequently, spore suspensions of each isolate were plated on fresh PDA and allowed to germinate at 28 °C. After the incubation period, plugs from fungal cultures were taken using sterile pipette tips and inoculated in 100 mL of the flask containing potato dextrose broth (PDB) and incubated at 28 °C. Fungal biomass was harvested by filtration from late exponential phase cultures, washed using sterile distilled water, and freeze-dried to extract chitosan.212.4 Chitosan production in submerged fermentation
Spore suspension that was obtained from pre-cultures was observed under a light microscope with a hematocytometer, and its concentration was adjusted to 108 spores per mL. Afterward, the spore suspension was inoculated into 2000 mL Erlenmeyer flasks containing 1500 mL of the sterile production media made up of glucose (2%), peptone (1%), yeast extract (0.1%), (NH4)2SO4 (0.5%), K2HPO4 (0.1%), NaCl (0.1%), CaCl2·H2O (0.01%), and MgSO4·7H2O (0.05%). Fungal biomass was harvested from late exponential-phase cultures to extract chitosan. Additional methods for extracting and characterizing chitosan from fungi are available in the ESI.†2.5 DNA extraction, PCR amplification and sequencing
Fungal DNA was extracted by following the procedures described elsewhere22 with a slight modification. Briefly, 200 mg of fresh fungal mycelium was dissolved in 800 μL of extraction buffer (0.1 M Tris–HCl pH 8, 10 mM EDTA pH 8, 2.5 M NaCl, 3.5% CTAB, and 150 μL of 20 mg mL−1 proteinase K). The mixture was vortexed at high speed for five minutes before incubation for 30 minutes in a water bath at 65 °C. It was then centrifuged at room temperature for 10 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The supernatant was mixed with an equal volume of phenol–chloroform–isoamyl alcohol (25
000 rpm. The supernatant was mixed with an equal volume of phenol–chloroform–isoamyl alcohol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and centrifuged at room temperature for 10 minutes at 10
1) and centrifuged at room temperature for 10 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The supernatant was collected once more, and an equal volume of chloroform–iso-amyl-alcohol (24
000 rpm. The supernatant was collected once more, and an equal volume of chloroform–iso-amyl-alcohol (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added before centrifugation. The supernatant was collected again, and an equivalent amount of ice-cold isopropanol was added, and further incubated at 20 °C for 2 hours. The suspension was centrifuged at 13
1) was added before centrifugation. The supernatant was collected again, and an equivalent amount of ice-cold isopropanol was added, and further incubated at 20 °C for 2 hours. The suspension was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes to pellet the DNA. Finally, the DNA pellet was washed in 800 mL of 70% ethanol, air-dried, and dissolved in 50 μL of TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA). The quality of the extracted DNA was assessed after electrophoresis on 0.8% agarose gels.
000 rpm for 15 minutes to pellet the DNA. Finally, the DNA pellet was washed in 800 mL of 70% ethanol, air-dried, and dissolved in 50 μL of TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA). The quality of the extracted DNA was assessed after electrophoresis on 0.8% agarose gels.
Polymerase Chain Reaction (PCR) amplifications of the fungal internal transcribed spacer region of the ribosomal RNA (ITS) were performed using a thermal cycler (Model: DW-B960) by employing two sets of previously published primers (synthesized by Eurofins, India): ITS1 5′-TCCGTAGGTGAACCTGCGG-3′ and ITS4 5′-TCCTCCGCTTATTGATATGC3′. The PCR reaction was prepared in 50 μL reaction mixtures, which contained 25 μL PCR mix, 2.5 μL DNA sample, 2.25 μL of each primer, and 17.5 μL of ddH2O. The PCR reaction was set as follows: 3 minutes at 94 °C pre-denaturation, 35 cycles of 10 seconds at 94 °C denaturation, 10 seconds of annealing at 50 °C, 10 seconds at 72 °C extension, and 120 seconds at 72 °C for a final extension. The PCR products were evaluated by DNA electrophoresis and visualized and documented using a UV light gel documentation device. PCR primers were used to sequence the purified PCR fragments under the recommended conditions. Taxonomic identification of isolated fungi was carried out by a homology search of the sequencing data using the BLASTn program against the NCBI GenBank database.
2.6 Synthesis of chitosan–copper (CsCu) nanozyme
Chitosan was produced by the deacetylation of chitin obtained from the mycelia of fungus isolated from Kocho samples. The synthesis of chitosan–copper nanocomposite (CsCu) was carried out according to some previously reported methods with slight modifications.23 Briefly, 14 mL of 1% chitosan (w/v) solution in acetic acid (2%, v/v) was added to 100 mL of CuCl2 solution (0.015 M) under stirring for 30 minutes. The pH of the solution was then adjusted to 9 using an ammonia solution. This was followed by the dropwise addition of 500 μL of hydrazine as a reducing agent. The resulting reddish-brown solution was filtered, washed, and vacuum-dried overnight at 60 °C. Similarly, optimization of the nanozyme composition was performed by varying the copper-to-chitosan mass ratio.2.7 Characterization of the nanozyme
The morphology of as-synthesized nanozyme was characterized by a field emission scanning electron microscope (FESEM JSM-6500F). All absorbance measurements were conducted using a double-beam UV-vis spectrophotometer (Jasco770). Functional group identification was done by FTIR spectrometer (Nicolet Evolution-300). The crystallinity and phase composition of the prepared nanozyme were checked by an X-ray powder diffractometer (D/MAX-2500 Bruker) with, CuKα source (λ = 1.5406 Å), isolated with a Ni foil filter. The elemental states in the nanocomposite were investigated by X-ray photoelectron spectrometer using XPS, VG ESCA Scientific Theta Probe with a monochromatic Al Kα X-ray source of 1486.6 eV.2.8 Catalytic activity assay
The preliminary laccase-like activity of CsCu nanozymes was investigated by using 2,4-D as the substrate and 4-AP as the chromogenic agent which forms a reddish-pink adduct with the oxidized phenolic compound. This colored reaction product absorbs at 510 nm.5,12 In a typical test, 700 μL buffer solution was first added to a test tube followed by the addition of 100 μL of 2,4-DP (1 mg mL−1), 4-AP (1 mg mL−1) and 1 mg CsCu respectively. After incubating for 30 minutes, the absorbance of the supernatant was measured at 510 nm. Controlled experiments containing 2,4-DP and 4-AP in the absence of nanozyme and only 2,4-DP with nanozyme were tested in parallel. Unless mentioned otherwise, this protocol was used as a standard assay for the entire work.Further to this, the unit activity of the nanozyme was analyzed by varying the amount of nanozyme at fixed substrate concentration. Typically, 2,4-DP (1 mg mL−1, 100 μL) and 4-AP (1 mg mL−1, 100 μL) were run for 1 h and the initial rates at different nanozyme concentrations were used to calculate the nanozyme activity as shown in eqn (1),
 | (1) |
2.9 Steady-state kinetic analysis
Kinetic studies were performed by measuring the initial rates at different concentrations of 2,4-DP (10–100 μg mL−1) and at a fixed nanozyme concentration. For all tests, the concentration of 4-AP was in excess (0.15 mg mL−1). The initial rate was then determined from a time course measurement of absorbance at 510 nm. Kinetic parameters such as Km and Vmax were then determined using the Lineweaver–Burk plot (eqn (2)) which is the linear form of the Michaelis–Menten (eqn (3))
 | (2) |
 | (3) |
2.10 Effect of different solution parameters
The effect of different parameters viz pH, amount of nanozyme, ionic strength, on the catalytic activity of the CsCu nanozyme was evaluated using one factor at a time approach. Accordingly, the mass of the nanozyme required for catalyzing the chromogenic reaction was optimized by conducting the reactions at various amounts of CsCu (1–2.5 mg) at constant pH and substrate concentrations. The effect of pH on the laccase mimetic activity was studied by varying the solution pH from 1–10 using Britons–Robinson (BR) buffer. Further to this, the effect of buffer species on the laccase activity was investigated by using different buffer components (borates, carbonates, and BR) at the optimum pH obtained in the previous experiment. Similarly, the effect of ionic strength on the activity of CsCu nanozymes was studied by adding different concentrations of NaCl (0–500 mM).2.11 Stability and recyclability of CsCu nanozyme
The thermal stability of the CsCu nanozyme was evaluated by conducting the laccase activity test at different temperatures ranging from 25–100 °C using a water bath system. Further, the storage stability of CsCu nanozymes was tested by dispersing 1.5 mg of CsCu nanozyme in BR buffer. An aliquot of this nanozyme dispersion was taken for a similar chromogenic reaction at different time intervals. The recyclability of CsCu was also examined by measuring the catalytic activity for three consecutive reaction cycles. Briefly, 100 μL of 1 mg mL−1 both 2,4-DP and 4-AP were added to 700 μL of borate buffer (pH = 9). Next 1.5 mg of CsCu was added to the reaction mixture and incubated for 30 min. The reaction mixture was then centrifuged (at 6000 rpm for 5 minutes) and was washed with distilled water. The cleaned nanozyme was then used for the subsequent cycle of the catalytic assay. The efficiency of the nanozyme at each cycle was compared by measuring the absorbance at 510 nm.2.12 Phenolic compounds oxidation test
The phenolic compound oxidation efficiency of the prepared CsCu nanozyme was studied by using three phenolic compound substrates namely phenol, 2,4-DP, and hydroquinone. Briefly, 1 mg mL−1 of each phenolic compound was subjected to catalysis by 1.5 mg of nanozyme in 700 μL of buffer (pH 9) in the presence of 4-AP. While the reaction was allowed to proceed the absorbance of the resulting solution was measured at different time intervals. The phenolic degradation efficiency of the nanozyme was also investigated using real wastewater taken from the Akaki River in found Addis Ababa, Ethiopia. Briefly, a known amount of 2,4-DP was spiked into a semi-treated wastewater sample (0.16% v/v). Next, the spiked sample was similarly added to the nanozyme/4-AP system. The absorbance of the resulting colored solution was measured at different times to calculate the % oxidation efficiency using Beer's Law (eqn (4))
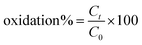 | (4) |
3 Results and discussion
3.1 Isolation and screening of chitosan-producing fungi
Nine morphologically distinct fungal isolates were obtained from an aliquot of serially diluted Kocho samples inoculated onto Petri plates containing PDA media (Fig. S1†). The fungal isolates could rapidly develop mycelia within a week of cultivation. Chitosan yield from fungal isolates labeled AWK8, AWK9, AWK12, and AWK15 in the PDB medium ranged from 4 mg L−1 to 14 mg L−1, which was relatively low and insignificant compared to their biomass (Table S1†). The other five isolates AWK1, AWK2, AWK3, AWK6, and AWK7 could produce chitosan in the range of 21 mg L−1 to 61.2 mg L−1. Further, their chitosan yields were improved when isolates were cultured in a minimal salt medium under the same fermentation conditions. AWK2 produced the highest amounts of mycelial chitosan (135.58 mg L−1), followed by AWK7 (114.76 mg L−1), and AWK1 produced the least (63.16 mg L−1) (Table S1†).3.2 Molecular identification of chitosan-producing fungi
To molecularly identify three of the top chitosan-producing fungi (AWK1, AWK2, and AWK7), a region that carries much of the diversity information was PCR-amplified by employing the universal ITS primers. Thus, sequence analysis of the ITS rDNA showed that fungal isolates belong to the genera Irpex, Diaporthe, and Schizophyllum. Furthermore, AWK2 was over 99% similar to GenBank Irpex sp. isolate FS16, with BLASTn query coverage of 93–100%. The other isolate, AWK1, had a 99.69% similarity with the Schizophyllum commune strain C77P. Also, isolate AWK7 revealed the highest (97.67%) sequence similarities with Diaporthe sp. isolate F255. Consequently, the chitosan-producing fungi were identified as Irpex sp. isolate AWK2, Schizophyllum commune strain AWK1 and Diaporthe sp. isolate AWK7 (Table S2†).3.3 Extraction and characterization of chitosan from Irpex sp. isolate AWK2
Chitosan was successfully extracted by deacetylation of chitin obtained from the mycelia of Irpex sp. isolate AWK2. The degree of deacetylation (DDA) as determined from the FTIR spectrum was 70%. The viscosimetric average molecular weight of the extracted chitosan was 7.94 × 103 Da. The results are comparable with low molecular weight commercial chitosan from marine sources. Given the intrinsic advantage of the feedstock, fungi could be a viable option for chitosan production.The morphology of the extracted chitosan from the fungus was characterized by SEM. As can be seen in the SEM micrograph (Fig. 1a), the extracted chitosan appeared as layered flakes with microfibril structures. Fig. 1b shows the FTIR spectrum of extracted chitosan. As seen in the spectrum, the broad peak at 3269 cm−1 corresponds to the OH stretching while the NH stretching at 3443 cm−1 is superimposed on the OH absorption. The sharp medium peak at 2925 cm−1 is due to C–H stretching. The sharp medium peaks at 1633, 1547 and 1368 cm−1 correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of residual amide, the NH bending, and C–N stretching respectively. The sharp intense peak at 1022 cm−1 belongs to the C–O stretching.24,25
O stretching of residual amide, the NH bending, and C–N stretching respectively. The sharp intense peak at 1022 cm−1 belongs to the C–O stretching.24,25
3.4 Synthesis and characterization of CsCu nanozymes
CsCu nanozyme was successfully synthesized using a facile wet method involving in situ reduction of copper onto the chitosan matrix. The morphologies of the CsCu nanocomposite were investigated by SEM. As seen from the SEM micrographs (Fig. 2a), the copper nanoparticles are successfully anchored on the chitosan flake structures. Further, the EDS spectrum (Fig. S2†) also confirms the presence of Cu, N, O, and C which are all expected elemental compositions from the precursors.The phase composition and crystallinity of the prepared nanozyme were studied using powdered XRD. The XRD pattern (Fig. 2b) of CsCu shows diffraction at 2θ ca 14.7° and 22.8° from crystalline chitosan flakes.26 The peaks at 2θ 29.63°, 32.72°, 58.2° and 74.3° belongs to diffractions from (110), (111), (220) and (311) planes of Cu2O (JCPDS nO. 65–3288). While the peaks at 43.6° and 50.7° are characteristics diffractions peaks from (111) and (200) planes of face-centered cubic (fcc) Cu respectively (JCPDS 04-0836). Also, the peak at 2θ 46.7° represents diffraction from (112) planes of CuO (JCPDS no. 04-0836).27,28
Further, XPS was used to elucidate the surface chemical states of the nanozyme. The wide scan XPS spectrum (Fig. 3a) shows peaks corresponding to Cu 2p, N 1s, O 1s, and C 1s where the C, N, and O are from the chitosan. The high-resolution spectrum of Cu 2p (Fig. 3b) shows peaks at 931.3 and 951.06 eV corresponding to the Cu 2p3/2 and Cu 2p1/2 of a Cu0/Cu+ moiety respectively. While the higher binding energy peaks at 933.0 and 952.6 eV correspond to Cu2+ states accounting for 22.78% of the total copper. The shake-up satellite peak at 942.5 eV is also a characteristic of d9 configuration which confirms the presence of Cu2+.12,29 The results indicate that the nanozyme has the multi-oxidation forms of copper, consistent with the T1/T2/T3 centers of the natural laccase enzyme.30,31 Deconvolution of the high-resolution spectrum N 1s spectrum (Fig. 3c) resulted in two peaks at 399.6 and 398.4 eV corresponding to copper-coordinated and free amino groups, respectively. This shows that the copper atoms are involved in coordination with the amino group of chitosan. In addition, deconvolution of the narrow scan spectrum of O 1s (Fig. 3d) revealed three peaks at binding energies of 531.2, 529.7, and 523.2 eV corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, Cu–O, and C–O bonds, respectively. This further corroborates the interaction of copper with the chitosan matrix.32
O, Cu–O, and C–O bonds, respectively. This further corroborates the interaction of copper with the chitosan matrix.32
 | ||
| Fig. 3 (a) Wide scan XPS spectrum of CsCu; high-resolution XPS spectra of (b) Cu 2p; (c) N 1s; (d) O 1s. | ||
3.5 Laccase memetic activity of CsCu nanozyme
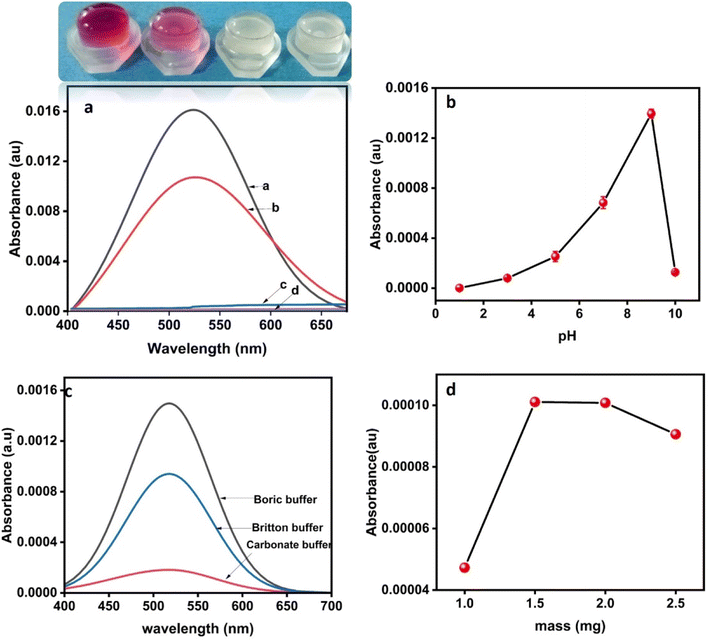
A preliminary test on the laccase memetic activity of the CsCu nanozyme was done by using 2,4-DP as a model substrate and 4-AP as a chromogenic agent in a standard buffer (pH 7). As seen in Fig. 4a and the inset photograph, in the presence of CsCu the reaction mixture turned reddish-pink with absorption maxima at 510 nm. This indicates the catalytic oxidation of 2,4-DP into a quinone intermediate which further reacted with 4-AP to form a colored adduct.12 The role of the CsCu nanozyme was further evidenced by the absence of a colored product when the two substrates are allowed to react in the absence of the nanozyme (Fig. 4b). Interestingly, chitosan has no any laccase activity, however, CsCu owes 38% higher catalytic activity than pristine copper NPs (Fig. 4b–d), This evidences the role of the chitosan scaffold to disperse the copper nanoparticles which otherwise is prone to aggregation. In addition, the amino and hydroxyl group of chitosan could enhance the electron transfer, similar to the amino acid microenvironment in natural laccase, thereby facilitating the oxidation of 2,4-DP.14Similar to the laccase enzyme, the mechanism of phenolic compound oxidation by CsCu can be proposed in three steps.33 The catalytic reaction starts with the binding of the substrate with nanozyme where it is oxidized, resulting in the reduction of Cu+ into Cu0. Subsequently, the transfer of electrons between the copper species will be enhanced by the amino and hydroxyl groups of the chitosan which is similar to the amino acid microenvironment in laccase. Finally, the reduction of molecular oxygen in water could take place by regenerating the nanozyme.12,33,34
The catalytic activity of CsCu at different pH values was measured to determine the optimal pH. As seen in Fig. 4b, the catalytic activity of the CsCu nanozyme increased gradually and reaches its maximum at pH 9. In general, the laccase memetic activity of the nanozyme is inhibited in lower pH regions. This could be due to the dissolution of the metals as well as the chitosan in an acidic medium. Further, as shown in Fig. 4c, the laccase memetic activity at the optimum pH depends on the buffer species. Accordingly, the CsCu nanozyme has outstanding catalytic efficiency in borate buffer. This could be ascribed to improved binding of substrates facilitated by the electrostatic repulsion between CsCu and Borates.35
Similarly, the effect of the dose of the nanozymes was studied at different doses of CsCu and fixed substrate concentration (1 mg L−1, 100 μL). As shown in Fig. 4d, the activity of the nanozymes increases with increasing CsCu mass, peaking at 1.5 mg, followed by a decrease in activity with a further increase in mass. This mass of nanozymes was then selected as optimum for further experiments. The specific unit activity (SA) of the nanozyme was then analyzed based on initial rates calculated from the different masses used above. Accordingly, the specific activity of nanozymes determined from the slope of a plot of b vs. mass of nanozymes (Fig. S2†) was found to be 0.01 U mg−1.
3.6 Enzyme kinetics of CsCu nanozyme
Steady-state kinetic studies were done at various concentrations of 2,4-DP at a fixed concentration of CsCu nanozyme. As seen in Fig. 5a and b, the rate of catalytic oxidation increased with substrate concentration and followed a typical Michaelis–Menten model. The kinetics parameters were then computed from the Lineweaver–Burk plot. Accordingly, the Km and Vmax were calculated to be 0.26 mM and 5.14 μM min−1, respectively. The lower Km often indicates an enzyme's higher affinity towards its substrate. Table 1 shows a survey of kinetic parameters of recently reported natural laccase enzymes and different nanozymes. As seen in Table 1, CsCu nanozyme shows comparable and even superior Km. This could be attributed to the rational design of our nanozyme which boosted the ease of substrate binding.| Catalyst | Substrate | Km/(mM) | Vmax/(μM min−1) | References |
|---|---|---|---|---|
| a NB: Cys:cysteine; GNF: Guanine copper phosphate; BSA: bovine serum albumin; CH: cysteine–histidine; CMC: Carboxymethylcellulose. | ||||
| Cu-Cys | 2,4-DP | 0.14 | 1.44 | 36 |
| BSA-Cu | Guaiacol | 0.159 | 4 | 37 |
| GNF | Catechol | 1.84 | 5.4 | 38 |
| CH-Cu | 2,4-DP | 0.42 | 7.3 | 16 |
| CMC-Pt | 2,4-DP | 0.22 | — | 39 |
| Laccase | 2,4-DP | 0.24 | 4.3 | 7 |
| Laccase | 2,4-DP | 0.40 | 3.5 | 40 |
| CsCu | 2,4-DP | 0.26 | 5.14 | This work |
3.7 Catalytic stability and recyclability of CsCu nanozyme
The stability of nanozymes is critical as it is one of the factors which rivals natural enzymes. Hence the thermal stability of CsCu nanozyme was examined after 30 minutes of incubation at different temperatures (25 to 100 °C). As shown in Fig. 6a, the catalytic activity of the CsCu nanozyme gradually increased reaching its maxima at 50 °C. Further, the nanozyme maintained 73.98% of its original activity at 70 °C indicating the capacity of the nanozyme to withstand such high operational temperature. Meanwhile, the nanozyme activity decreases after 50 °C which could be due to the decomposition of the CsCu structure. However, natural laccase could not effectively function at much higher temperatures due to the instability of the protein scaffold. For instance, in a study conducted by Lou and colleagues7 as well as Liu and coworkers,14 the laccase enzyme lost half of its activity at 50 °C. Further, in a recent study12 natural laccase enzyme and CACu nanozyme retained 25% and 13% of their activity at 70 °C and 100 °C, respectively. Hence compared with these nanozymes and natural laccase, the rationally designed CsCu nanozyme in this study showed a robust performance. Further, as seen in Fig. 6b, the CsCu nanozyme retained 53% of its catalytic activity after being stored in a buffer for 25 days. Recent studies showed laccase enzyme can maintain only 12% of its activity after 12 days of storage.41 This demonstrates enhanced temporal stability of the prepared CsCu nanozyme. | ||
| Fig. 6 (a) Thermal stability; (b) temporal stability; (c) recyclability; (d) effect of salinity on the CsCu nanozyme activity. | ||
The reusability of nanozymes is also a critical factor for an economically feasible application. Hence the recyclability of CsCu nanozyme was studied by recovering the nanozyme through centrifuging and washing after each cycle. As shown in Fig. 6c, the CsCu nanozyme maintained 37% of its activity after the third cycle. The decrease in activity after repeated cycles could be due to the leaching-out of copper from the polymer matrix which in turn degrades the nanozyme performance.6
The degradation of phenolic compounds in real wastewater is one of the applications for laccase enzymes. However, the laccase instability due to extreme operational conditions such as high salinity limits its application.6,12 Therefore, the stability of the CsCu nanozyme in saline conditions was assessed under different NaCl concentrations. As shown in Fig. 6d, the catalytic activity gradually increased with NaCl concentrations with 98.3% higher activity at 500 mM NaCl. Further, as seen in Fig. S3,† the CsCu showed improved activity in the presence of different salts irrespective of the ionic species. This could be attributed to the increased surface charges which could enhance the catalytic activity.12 The result compliments the lower activity of natural laccase enzyme in a saline environment due to the salting-out effect.42 Added with abundant chitosan used for its preparation CsCu represents a robust and sustainable nanozyme suitable for practical applications.
3.8 Catalytic oxidation of phenolic compounds
The application of the CsCu nanozyme for the catalytic oxidation of different phenolic substrates was tested in the presence of the chromogenic agent 4-AP. The % oxidation was determined from the absorbance measured at different time intervals using eqn (4). As seen in Fig. 7a, the nanozyme converts more than 80% 2,4-DP and phenol, and 60% hydroquinone (HQ) in one hour and over 92% of the three substrates in 5 h. The conversion efficiency of 2,4-DP and phenol at three and six hours is not statistically significant, while the difference for HQ is statistically significant at p < 0.05. This suggests that 3 h could be enough to effectively oxidize phenol and 2,4-DP, while HQ needs more time to oxidize completely. This could be attributed to the difference in the molecular structure of the substrates. Hydroquinone with two hydroxyl groups could take longer to completely oxidize than 2,4-DP which contains only one hydroxyl group. As seen in Table 2, in this study higher oxidation percentage (92%) is obtained even in a shorter time. This could be ascribed to the chitosan matrix which facilitates substrate binding and faster electron transfer. For instance, the laccase nanozyme constructed from a dipeptide coordinated to copper (Cys-Cu-Asp) required over 10 h to convert more than 90% of phenolic compounds which is double than required by the CsCu nanozyme.12 This result suggests that the prepared nanozyme is effective in the catalytic degradation of diverse phenolic pollutants.| Catalyst | Substrates | Percent of degradation | Total time used (h) | Ref. |
|---|---|---|---|---|
| Fe-nano zeolite/activated carbon | Phenol | 62.3 | 4 | 43 |
| Fe3O4-NH2-PEI-laccase | Phenol | 70 | 10 | 3 |
| Cu-1-methyl imidazole | 2,4-DP | 91.8 | 10 | 44 |
| Fe-N-C-single atom | Phenol | 83 | 10 | 45 |
| CA-Cu | 2,4-DP | 90 | 10 | 12 |
| Cu-adenosine monophosphate | Phenol | 65 | 5 | 46 |
| Ce(IV) MOF | 2,4-DP | 42.88 | 12 | 7 |
| BPA | 76–86 | |||
| CsCu | Hydroquinone | 92 | 5 | This work |
| 2,4-DP | 92 | |||
| Phenol |
Further, to study the feasibility of the nanozyme activity in real wastewater treatment similar assay was conducted using phenolic compounds spiked wastewater sample (0.16% v/v 2,4-DP in wastewater). As shown in Fig. 7b, 62.5% 2,4-DP was oxidized in 2 h. However, unlike the synthetic phenolic compounds, the spiked sample percent conversion declined after 2 h. This could be ascribed to the presence of interfering ions in the wastewater matrix which occupies catalytic sites of the nanozyme.
4 Conclusions
In summary, fungal chitosan–copper nanocomposite (CsCu) is demonstrated as a rational and sustainable laccase mimetic bionanozyme. Low molecular weight chitosan (7.94 × 103 Da) with a 70% average degree of deacetylation was obtained from fungal isolate identified as Irpex sp. isolates AWK2. Chitosan due to its polycationic nature boosted the laccase mimetic activity of copper nanoparticles by enhancing the electron transfer in the catalytic reactions. Further morphological and structural characterizations showed three species of copper distributed on the chitosan matrix which is consistent with the composition of natural laccase. The prepared nanozyme showed improved laccase memetic activity for the oxidation of different phenolic compounds both in synthetic and real wastewater obeying the Michaelis–Menten model. The calculated Km value is lower when compared with previously reported nanozymes as well as natural laccase indicating excellent binding affinity. The nanozyme retained its room temperature activity at higher temperatures up to 70 °C and under saline conditions. Given the sustainable feedstock used for the production, the rationally prepared CsCu bionanozyme represents a robust and sustainable nanozyme with promising utility in environmental remediation.Data availability
The data that support the findings of this study are available on request from the corresponding author.Author contributions
EGM: investigation, material preparation, writing original draft, software; KNS: investigation, material characterization; BJH: investigation, resources; YAW: supervision; EMA: conceptualization, supervision, methodology, editing; MLM: conceptualization, funding acquisition, resources, methodology, writing – review & editing.Conflicts of interest
The authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors are grateful to Addis Ababa Science and Technology University for the financial support under the University's interdisciplinary internal research grant number IG 07/2022.References
- W. A. William, A. M. Messai and P. G. Penny, in Phenolic Compounds, ed. S.-H. Marcos, P.-T. Mariana and G.-M. Maria del Rosario, IntechOpen, Rijeka, 2017, ch. 17, DOI:10.5772/66927.
- Z. Chen, W.-D. Oh and P.-S. Yap, Chemosphere, 2022, 307, 135824 CrossRef CAS PubMed.
- T.-T. Xia, M. Feng, C.-L. Liu, C.-Z. Liu and C. Guo, Eng. Life Sci., 2021, 21, 374–381 CrossRef CAS PubMed.
- L. Lei, X. Yang, Y. Song, H. Huang and Y. Li, New J. Chem., 2022, 46, 3541–3550 RSC.
- T. D. Tran, P. T. Nguyen, T. N. Le and M. I. Kim, Biosens. Bioelectron., 2021, 182, 113187 CrossRef CAS PubMed.
- T. Maity, S. Jain, M. Solra, S. Barman and S. Rana, ACS Sustain. Chem. Eng., 2022, 10, 1398–1407 CrossRef CAS.
- S. Liang, X.-L. Wu, J. Xiong, X. Yuan, S.-L. Liu, M.-H. Zong and W.-Y. Lou, Chem. Eng. J., 2022, 450, 138220 CrossRef CAS.
- B. Das, J. L. Franco, N. Logan, P. Balasubramanian, M. I. Kim and C. Cao, Nanomicro Lett., 2021, 13, 193 CAS.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- X. Xu, J. Wang, R. Huang, W. Qi, R. Su and Z. He, Catal. Sci. Technol., 2021, 11, 3402–3410 RSC.
- A. Robert and B. Meunier, ACS Nano, 2022, 5(2), 6956–6959 CrossRef PubMed.
- H. Liang, F. Lin, Z. Zhang, B. Liu, S. Jiang, Q. Yuan and J. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 1352–1360 CrossRef CAS PubMed.
- S. Shams, W. Ahmad, A. H. Memon, Y. Wei, Q. Yuan and H. Liang, RSC Adv., 2019, 9, 40845–40854 RSC.
- J. Wang, R. Huang, W. Qi, R. Su, B. P. Binks and Z. He, Appl. Catal., B, 2019, 254, 452–462 CrossRef CAS.
- H. Ma, N. Zheng, Y. Chen and L. Jiang, Colloids Surf., A, 2021, 613 Search PubMed.
- Y. Cai, J. Zhou, J. Huang, W. Zhou, Y. Wan, M. A. Cohen Stuart and J. Wang, J. Colloid Interface Sci., 2023, 645, 458–465 CrossRef CAS PubMed.
- I. Aranaz, A. R. Alcántara, M. C. Civera, C. Arias, B. Elorza, A. Heras Caballero and N. Acosta, Polymers, 2021, 13, 3256 CrossRef CAS PubMed.
- M. F. Decarlini, M. L. Aimar, A. M. Vázquez, S. Vero, L. I. Rossi and P. Yang, Biocatal. Agric. Biotechnol., 2017, 12, 275–285 CrossRef.
- P. Pochanavanich and W. Suntornsuk, Lett. Appl. Microbiol., 2002, 35, 17–21 CrossRef CAS PubMed.
- I. Gontia-Mishra, N. A. Tripathi and S. Tiwari, Indian J. Biotechnol., 2014, 13, 536–539 CAS.
- T. Jayaramudu, K. Varaprasad, R. D. Pyarasani, K. K. Reddy, K. D. Kumar, A. Akbari-Fakhrabadi, R. V. Mangalaraja and J. Amalraj, Int. J. Biol. Macromol., 2019, 128, 499–508 CrossRef CAS PubMed.
- R. Varma and S. Vasudevan, ACS Omega, 2020, 5, 20224–20230 CrossRef CAS PubMed.
- J. Cheng, H. Zhu, J. Huang, J. Zhao, B. Yan, S. Ma, H. Zhang and D. Fan, Food Sci. Nutr., 2020, 8, 1987–1994 CrossRef CAS PubMed.
- S. A. Mathew and S. Arumainathan, ACS Omega, 2022, 7, 18732–18744 CrossRef CAS PubMed.
- L. Chen, Y. Zhang, P. Zhu, F. Zhou, W. Zeng, D. D. Lu, R. Sun and C. Wong, Sci. Rep., 2015, 5, 9672 CrossRef PubMed.
- D. Mardiansyah, T. Badloe, K. Triyana, M. Q. Mehmood, N. Raeis-Hosseini, Y. Lee, H. Sabarman, K. Kim and J. Rho, Sci. Rep., 2018, 8, 10639 CrossRef PubMed.
- C.-K. Wu, M. Yin, S. O'Brien and J. T. Koberstein, Chem. Mater., 2006, 18, 6054–6058 CrossRef CAS.
- L. Arregui, M. Ayala, X. Gómez-Gil, G. Gutiérrez-Soto, C. E. Hernández-Luna, M. Herrera de los Santos, L. Levin, A. Rojo-Domínguez, D. Romero-Martínez, M. C. N. Saparrat, M. A. Trujillo-Roldán and N. A. Valdez-Cruz, Microb. Cell Factories, 2019, 18, 200 CrossRef CAS PubMed.
- M. Loi, O. Glazunova, T. Fedorova, A. F. Logrieco and G. Mulè, J. Fungi, 2021, 7, 1048 CrossRef CAS PubMed.
- NIST X-ray Photoelectron spectroscopy Database 2.0, https://srdata.nist.gov/xps/Default.aspx, accessed June 01 2023.
- J. R. Jeon, P. Baldrian, K. Murugesan and Y. S. Chang, Microb. Biotechnol., 2012, 5, 318–332 CrossRef PubMed.
- J. Yoon, B. D. Liboiron, R. Sarangi, K. O. Hodgson, B. Hedman and E. I. Solomon, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13609–13614 CrossRef CAS PubMed.
- S. Tian, C. Zhang, M. Yu, Y. Li, L. Fan and X. Li, Anal. Chim. Acta, 2022, 1204, 339725 CrossRef CAS PubMed.
- M. Guan, M. Wang, W. Qi, R. Su and Z. He, Front. Chem. Sci. Eng., 2020, 15, 310–318 CrossRef.
- S. Rashtbari and G. Dehghan, J. Hazard. Mater., 2021, 406, 124340 CrossRef CAS PubMed.
- T. D. Tran, P. T. Nguyen, T. N. Le and M. I. Kim, Biosens. Bioelectron., 2021, 182, 113187 CrossRef CAS PubMed.
- L. Yang, X.-Y. Guo, Q.-H. Zheng, Y. Zhang, L. Yao, Q.-X. Xu, J.-C. Chen, S.-B. He and W. Chen, Sens. Actuators, B, 2023, 393, 134165 CrossRef CAS.
- Y. Wang, C. He, W. Li, J. Zhang and Y. Fu, Catal. Lett., 2017, 147, 2144–2152 CrossRef CAS.
- P. Makam, S. S. R. K. C. Yamijala, V. S. Bhadram, L. J. W. Shimon, B. M. Wong and E. Gazit, Nat. Commun., 2022, 13, 1505 CrossRef CAS PubMed.
- M. L. Toledo, M. M. Pereira, M. G. Freire, J. P. A. Silva, J. A. P. Coutinho and A. P. M. Tavares, ACS Sustain. Chem. Eng., 2019, 7, 11806–11814 CrossRef CAS.
- N. L. M. Tri, P. Q. Thang, L. Van Tan, P. T. Huong, J. Kim, N. M. Viet, N. M. Phuong and T. M. Al Tahtamouni, J. Water Process. Eng., 2020, 33, 101070 CrossRef.
- Y. Lei, B. He, S. Huang, X. Chen and J. Sun, Molecules, 2022, 27, 4712 CrossRef CAS PubMed.
- Y. Yin, L. Shi, W. Li, X. Li, H. Wu, Z. Ao, W. Tian, S. Liu, S. Wang and H. Sun, Environ. Sci. Technol., 2019, 53, 11391–11400 CrossRef CAS PubMed.
- H. Huang, L. Lei, J. Bai, L. Zhang, D. Song, J. Zhao, J. Li and Y. Li, Chin. J. Chem. Eng., 2021, 29, 167–175 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06619c |
| This journal is © The Royal Society of Chemistry 2023 |