 Open Access Article
Open Access ArticleEvolving approaches in glioma treatment: harnessing the potential of copper metabolism modulation
Riccardo Cazzoli†
a,
Agata Zamborlin†bc,
Maria Laura Erminic,
Antonietta Salernoa,
Manuela Curciod,
Fiore Pasquale Nicolettad,
Francesca Iemmad,
Orazio Vittorio‡
af,
Valerio Voliani‡ ce and
Giuseppe Cirillo‡
ce and
Giuseppe Cirillo‡ *d
*d
aChildren's Cancer Institute Australia, Lowy Cancer Research Centre, University of New South Wales, Sydney, NSW, Australia
bNEST-Scuola Normale Superiore, Piazza San Silvestro, 12 – 56127 Pisa, Italy
cCenter for Nanotechnology Innovation, Istituto Italiano di Tecnologia, Piazza San Silvestro, 12 – 56127 Pisa, Italy
dDepartment of Pharmacy Health and Nutritional Science, University of Calabria, 87036 Rende, Italy. E-mail: giuseppe.cirillo@unical.it; Tel: +39 0984493208
eDepartment of Pharmacy, School of Medical and Pharmaceutical Sciences, University of Genoa, Viale Cembrano, 4 – 16148 Genoa, Italy
fSchool of Biomedical Sciences, University of New South Wales, Sydney, NSW, Australia
First published on 21st November 2023
Abstract
The key properties and high versatility of metal nanoparticles have shed new perspectives on cancer therapy, with copper nanoparticles gaining great interest because of the ability to couple the intrinsic properties of metal nanoparticles with the biological activities of copper ions in cancer cells. Copper, indeed, is a cofactor involved in different metabolic pathways of many physiological and pathological processes. Literature data report on the use of copper in preclinical protocols for cancer treatment based on chemo-, photothermal-, or copper chelating-therapies. Copper nanoparticles exhibit anticancer activity via multiple routes, mainly involving the targeting of mitochondria, the modulation of oxidative stress, the induction of apoptosis and autophagy, and the modulation of immune response. Moreover, compared to other metal nanoparticles (e.g. gold, silver, palladium, and platinum), copper nanoparticles are rapidly cleared from organs with low systemic toxicity and benefit from the copper's low cost and wide availability. Within this review, we aim to explore the impact of copper in cancer research, focusing on glioma, the most common primary brain tumour. Glioma accounts for about 80% of all malignant brain tumours and shows a poor prognosis with the five-year survival rate being less than 5%. After introducing the glioma pathogenesis and the limitation of current therapeutic strategies, we will discuss the potential impact of copper therapy and present the key results of the most relevant literature to establish a reliable foundation for future development of copper-based approaches.
1. Introduction
Copper is an essential micronutrient involved in fundamental processes that are conserved throughout all forms of life. It plays a role in enzyme catalysis, redox reactions, mitochondrial respiration, and free radical scavenging.1 Copper also contributes to many biological processes such as embryogenesis, growth, and metabolism.2The regulation of the intra-cellular copper quantities is thus fundamental for the orchestration of cell physiology, especially because copper is potentially toxic if found free in the cytosol rather than bound to metallothioneins. Although its redox activity is essential for enzymatic reactions, elevated copper levels may damage lipids, proteins, DNA, and other biomolecules by ultimately inducing cell apoptosis.3 Moreover, copper may interfere with other metal-driven activities. It can impact the correct functioning of proteins containing iron–sulfur clusters4 or displace zinc (or other metals) from metalloproteins,5 resulting in inhibition of their activity. This occurs because copper is a critical component of different metalloenzymes as the matrix metalloproteinase (MMP-9), which is essential to the metastatic process.
Imbalances in copper homeostasis have been implicated in the pathogenesis of neurodegenerative disorders that are associated to toxic effects caused by oxidative stress, such as Wilson's disease (WD),6 Menkes disease,7 Alzheimer's disease (AD),8 Parkinson's disease and amyotrophic lateral sclerosis,9 as well as idiopathic pulmonary fibrosis,10 rheumatoid arthritis and diabetes mellitus.11 Furthermore, there is increasing evidence that copper-dependent cell proliferation, known as cuproplasia,12 plays a fundamental role in the development and/or progression of different types of tumors,13–16 such as breast, thyroid, cervical, ovarian, lung, pancreatic, prostate, gastric, oral and bladder cancers.17–25
Voli et al. (2020) demonstrated the role of copper in modulating PD-L1 expression and contributing to cancer immune evasion by inhibiting EGFR and AKT phosphorylation in neuroblastoma (NB) and glioblastoma (GBM) cells, suggesting that tumors that exhibit increased intra-tumoral copper distribution could be targeted by selectively reducing their levels of copper.26 An excess of copper may be one of the factors triggering cancer stem-like cells to initiate the tumor and induce its progression. It is well-known, indeed, that copper is involved in the epithelial-mesenchymal (EMT) and the mesenchymal-to-epithelial (MET) transitions within the tumor microenvironment (TME).1 When this occurs, cell migration, invasion and tumor growth will be promoted through hypoxia-related genes. Under hypoxic conditions, the hypoxia-inducing factor (HIF) genes are deeply involved in the survival of cancer cells by maintaining immature, stem-like tumor cells. Knockdown of these genes would reduce the expression of angiogenesis-associated vascular endothelial growth factor (VEGF), leading to partial sympathetic neural differentiation of tumor stem cells.
In this review, the recent researches investigating the impact of copper in glioma treatment will be discussed, highlighting the possibility to use either copper overload or depletion as a therapeutic strategy. Copper nanoparticles (Cu NPs) will be explored as the approach to provide tumors with excess Cu allowing the production of reactive oxygen species (ROS), while the use of copper chelators will be discussed as the strategy to reduce the Cu concentration. Moreover, the synthetic routes available for the fabrication of highly engineered Cu NPs, as well as the therapeutic outcome of both approaches will be presented, to open the discussion about future developments in the field and the possible effective translation to pre-clinical and clinical studies.
2. Copper nanoparticles for cancer treatment
Currently, the most relevant therapeutic strategies for cancer treatment are based on surgery, chemotherapy, and radiotherapy.27 Conventional chemotherapy is based on the use of cytotoxic agents mainly acting by blocking DNA synthesis and/or cell replication.28 Each approach suffers from some severe limitations, mainly due to the uncomplete removal of cancer cells (surgery),29 or side effects on healthy cells and tissues (chemo- and radio-therapy).30,31 Thus, current trends in oncology focus on the design and development of efficient cancer nanomedicines,32,33 defined as miniature-sized products with ideal properties for interaction with living tissues, including (but not limited to) reduced toxicity, high surface-area-to-volume ratio, and high chemical versatility.34 Several nanoplatforms are currently in development, including polymer nanoparticles and micelles, liposomes and lipid nanoparticles, inorganic and metal nanoparticles.35 Each material shows advantages and disadvantages arising from their intrinsic properties, allowing or denying specific applications.36Metal NPs, including noble (e.g. gold, silver, platinum, and palladium) and non-noble (e.g. iron, zinc, titanium, cerium, nickel, copper, magnesium, barium, calcium, and bismuth) metal-based NPs have recently gained significant interest for multipurpose biomedical applications, including cancer treatment.37–39
Copper nanoparticles (NPs) possess significant advantages both in terms of synthesis and application within the realm of metal NPs.40 Notably, the production of copper NPs is more cost-effective than that of other noble metal NPs, owing to the abundant natural availability of the metal.41 This abundance facilitates efficient large-scale production and straightforward storage and handling of highly engineered nanosystems.42 The distinctive properties of copper NPs, including their crystallinity, surface strain, and the prevalence of defect sites, contribute to their rapid dissolution.43 This characteristic leads to noteworthy biological effects, as copper plays a substantial role in various biological functions. Furthermore, copper NPs exhibit versatility in undergoing a wide array of reactions due to their accessible oxidation states (0, I, II, and III).44 This chemical adaptability not only optimizes the synthetic procedures, but also allows tailored functionalization, thereby enabling precise modulation of biological responses at the tissue and cellular levels.45
Over the last decades, the interest over copper NPs is increasing due to their features and associated potential applications in nano-biomedicine,46 including the possibility to induce copper-dependent apoptosis (cuproptosis) of cancer cells via oxidative stress.8 Although their potential applications, in vivo studies should better elucidate the toxicity of copper nanoparticles on normal cells since they are not yet fully understood.47–49
Within the present review, we will focus on the application of copper NPs in glioma treatment since the hypoxic environment contributes to the development of therapeutic resistance and invasiveness into normal brain tissues,50 thus increasing the need for a more effective therapeutic regimen. In the following paragraphs, the different types of copper NPs tested for glioma treatment will be discussed, highlighting the synthetic procedure and the main biomedical outcome (Table 1).
| NPs | Synthetic route | Mean diameter | Main outcome | Ref. | |
|---|---|---|---|---|---|
| D | H | ||||
| a CPNDs: copper peroxide nanodots; Cu HARS: copper high-aspect ratio structure; Cu USNPs: copper ultrasmall NPs D: dry; DSF: disulfiram; DOX: doxorubicin; H: hydrated; HM: hollow mesoporous SiO2; IDX: indoximod, NPs: nanoparticles; PTX: paclitaxel; TMZ: temozolomide; UCNs: polymer-templated Cu NPs. | |||||
| Cu | High-voltage discharge | 2.1 | Cell membrane depletion nucleus budding | 51 | |
| CuO | Flame spray pyrolysis | 209 | ROS generation | 52 | |
| CuO–Fe | |||||
| CuO | Alkaline precipitation | 141 | ROS generation | 53 | |
| CuO | Alkaline precipitation | 136 | ROS generation | 54 | |
| CuO | Alcothermal precipitation | 30–60 | Apoptosis induction | 55 | |
| UCNs | Alkaline precipitation | 20–30 | TMZ stabilization | 56 | |
| Au–CuO | Hydrothermal method (Cnici benedicti extract) | 13 | Cell cycle blocked at G2-M phase | 57 | |
| ZnO–CuO | 28 | ||||
| CuS | Hydrothermal method | 50 | Photothermal ablation | 58 | |
| Cu2S | Hot injection colloidal approach | 15–20 | Photothermal ablation | 59 | |
| Synergistic DOX delivery | |||||
| CuS | Alkaline hydrothermal method | 86 | Photothermal ablation | 60 | |
| Synergistic DOX delivery | |||||
| HM-CuS | Reduction + ion exchange | 172 | Photothermal ablation | 61 | |
| Synergistic DSF delivery | |||||
| HM-CuS | Reduction + ion exchange | 4 | Synergistic TMZ delivery | 62 | |
| Chemodynamic therapy | |||||
| Photothermal ablation | |||||
| Starvation therapy | |||||
| HM-CuS | Reduction + ion exchange | 190 | Photothermal ablation | 63 | |
| Macrophage repolarization | |||||
| CdSe/Cu2S | Hydrothermal method | 13–17 | Photothermal ablation | 64 | |
| CPNDs | Alkaline precipitation | 80 | Synergistic PTX delivery (prodrug) | 65 | |
| Chemodynamic therapy | |||||
| Photodynamic therapy | |||||
| Au@Cu2−xSe | Reductive coating | 21 | Inhibition of autophagy flux | 66 | |
| Cu2−xSe | Reduction method | 3 | Photodynamic therapy | 67 | |
| Cu2−xSe | Reduction method | 3 | Synergistic DSF delivery | 68 | |
| Cu2−xSe | Reduction method | 3 | Synergistic IDX delivery | 69 | |
| Immunotherapy | |||||
| CuHARS | Redox self-assembly | 20 | Cell tracking | 70 | |
| CuHARS | Redox self-assembly | 20 | Nitrogen oxide production | 71 | |
| CuHARS | Redox self-assembly | 20 | Apoptosis | 72 | |
3. Synthesis of copper nanoparticles for glioma treatment
The preparation of copper NPs can be accomplished with various methods including physical, wet-chemical, and green synthesis.As for gold NPs preparation, the wet methods result widely used for their straightforwardness, while physical methods usually require specific equipment. It is worth reporting that several papers describe the synthesis of copper NPs by employing microorganisms, among which plants, algae, bacteria, and fungi, which spontaneously synthesize copper nanoparticles.73
3.1. Copper nanoparticles by top-down approaches
Physical syntheses of NPs are techniques which use mechanical or thermodynamic processes. In general, when compared to wet methods, the physical processes often involve large and expensive set-up requiring high energy. On the other hand, the particle distribution can reach an interesting uniformity and the contamination of chemicals can be removed.74 For example, the solvated metal atom dispersion method allows to avoid purification processes and byproducts formation.75 This method generates metal atoms through vaporization of the bulk material, subsequently condensed into clusters with a solvent at low temperature (around 70 K). Reaching the room temperature, NPs are separated from the solvent by evaporation. For copper, the nature of the solvent, together with the reaction conditions, have been proved to be crucial for determining the nanoparticle size.76The voltage discharge method belongs to the top-down methodologies and it is often referred as a simple and cheap physical method for nanoparticle synthesis.77 Briefly, the bulk material of two electrodes is eroded by an increase of temperature associated with a current flow.78 Vodyanoy et al. used two metal electrodes immersed in water and connected to a high-voltage generator.51 Applying an AC voltage (15 kV), an electric discharge was generated between the electrodes for 1 hour. Depending on the distance between the electrodes, the generated plasma produced a satisfactory dispersion of the metal. After 12 h sedimentation and 2 h centrifugation, 2 nm NPs were recovered from the supernatant. Size and distribution were measured with atomic force microscopy, revealing a quite narrow distribution with a standard deviation of 0.6 nm and a polydispersity index of 0.082. A 15% of the atoms were oxidized and the material was investigated on rat brain glioma cancer cells RG2.
The ablation of copper can also be accomplished by employing a pulsed laser in a liquid environment. Tilaki et al. investigated the influence of the medium on the size and shape of the obtained NPs. They used a Nd:YAG laser in water and acetone.79 TEM images of the resulting particles showed an average diameter of 30 and 3 nm in water and acetone, respectively, with a more regular shape in the second medium. Colloidal copper in acetone had a longer shelf life compared to the particles in water (10 months vs. two weeks). Furthermore, the degree of oxidation of copper NPs in water was higher due to the oxygen dissolved into the medium. The presence of copper oxide is a significant information since it can crucially influence the bio-toxicity of the particles. CuO NPs, indeed, may improve the production of ROS, which can alter the metabolism of cells.40 Iron-doped CuO NPs were used by Joshi et al. on C6 glioma cell line.52 The synthesis was accomplished through flame spray pyrolysis, a physical method where a solution containing precursors (usually metal salts) is sprayed into a flame. The formed droplets, after the evaporation of the solvent and pyrolysis of the precursor, result in the production of the metal oxide NPs. Combustion process, aerosols technology and materials of precursors determine the characteristics of the particles in terms of morphology, homogeneity, and size.80 In particular, CuO-NPs and 10% iron-doped CuO-NP (CuO-Fe-NPs) ultrafine powders were produced using copper naphthenate and iron naphthenate precursor solutions in xylene, a methane/oxygen flame and a glass fiber to collect the NPs.52 Flow rate, flame precombustion characteristics and precursor concentration were finely tuned to obtain spherical 50 nm particles, stable in water after functionalization with dimercaptosuccinate (ζ-potentials of −40.3 and −37.2 mV).
3.2. Copper nanoparticles by bottom-up approaches
Wet-chemical synthesis procedures are referred to as bottom-up approaches, and they allow a better tuning of size, shape, and chemical composition of the final metal NPs compared to the top-down approaches.81 NPs have to be purified from the chemical and biological unreacted precursors, which could limit the biocompatibility of the NPs, especially for the non-green procedures. The persistence of toxic reactants, indeed, may affect both the biological investigation and the potential applications. Reduction of the metal ions in solution is one of the preferred methods to produce metal NPs due to the versatility of the process. Stabilizing agents of low or high molecular weight can be employed to protect the metal NPs from aggregation and environmental stress factors.82 It is worth to remember that the conjugation of stabilizing agents on the surface of metal NPs can modify the biological interactions of the nanostructures.Copper NPs (CuNPs) and copper-oxide NPs (CuO NPs) for the treatment of gliomas can be prepared by reduction of copper salts. Joshi et al. synthetized CuO NPs of around 141 nm of hydrodynamic diameter by a wet chemical method.53 Following the procedure proposed by Bulcke et al., and by Kobayashi et al., Cu(NO)3 was reduced in an alkaline solution (NaOH) to induce the NPs formation at 75 °C.54,83 Dimercaptosuccinic acid (DMSA) was used to coat CuO NPs, and a bovine serum albumin (BSA) coating was used to improve the stability of the resulting CuO NPs. The protein coating increased the hydrodynamic diameter to 175 nm, with an increase of the zeta potential from −42.5 mV to −14.1 mV for CuO NPs. Precipitation by addition of high salt buffer allowed their purification. The copper content was quantified photometrically using bathocuproine disulfonate by absorption at 405 nm against a calibration curve of CuCl2, and atomic absorption spectroscopy was used for validation.53 Joshi et al. suggested that the toxic effects observed in C6 glioma cells was due to the release of copper ions from the NPs. Kukia et al. produced Cu NPs of 30 and 60 nm (as per scanning electron microscopy – SEM analysis) through an alcothermal method.55 CuSO4 was used as a Cu2+ source, and sodium borohydride (NaBH4) reduced the ions. The reduction of Cu2+ is followed by the change of the colloid solution color from blue to green, and eventually to brown. Polyvinylpyrrolidone K30 (PVP) was selected as stabilizing agent, and further stabilization was provided by ascorbic acid added at 60 °C to avoid Cu oxidation to CuO.84 Cu NPs were purified through precipitation and washing in ethanol. These Cu NPs exerted toxicity to a glial rat cancer cell line (B92) by stimulating apoptosis, especially for the smaller NPs at higher concentrations.
Wang et al., proposed a biomimetic polymer-templated Cu NPs (UCNs) to stabilize a temozolomide intermediate56 (Fig. 1).
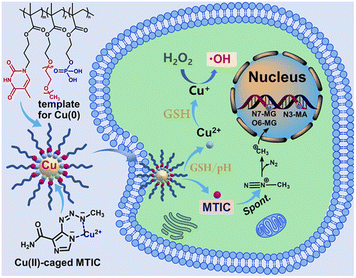 | ||
| Fig. 1 Schematic representation of UCNs for TMZ stabilization. Reproduced with permission from ref. 56. Copyright© 2021, American Chemical Society. | ||
Temozolomide is the first-line treatment for glioblastoma, and it was designed as a prodrug: temozolomide crosses the blood–brain barrier and converts spontaneously into the active 5-(3-methyl-1-triazeno)imizadole-4-carboxamide (MTIC) molecule.85 This prolongs the short half-life of MTIC, but the conversion kinetics is slow and at the same time MTIC can be released extracellularly. UCNs were designed as delivery system of the active form MTIC. First, Cu2+ ions from CuSO4 were complexed by a DNA-mimetic polymer, and then reduced to Cu using hydroxylamine chloridrate in an alkaline environment (pH 11.5). After stirring for 2 hours, UCNs were purified through dialysis using 10 kDa MWCO membrane against pH 9.6. Copper nanoclusters stabilized by these biomimetic polymers were 20–30 nm (TEM). The reduction of Cu was not complete, as an autoxidation took place.
Moreover, the polymer avoided Cu escape into the solution. Copper was quantified spectrophotometrically by detection of the complex between Cu2+ and diethyldithiocarbamate at 450 nm. The persistence of Cu2+ is crucial as only this ion can complex MTIC.86 Finally, temozolomide was added to UCNs and the drug was spontaneously hydrolyzed to MTIC, which in turn complexed Cu2+. These delivery systems were tested on temozolomide-resistant and -sensitive glioblastoma cells (T98G and U-87MH, respectively). Interestingly, authors conjugated the effect of CuNPs to the therapeutic effect of MTIC paving the way for the reduction of temozolomide resistance burden.
New trends in the bottom-up approach for the synthesis of CuNPs and CuO NPs involve the use of green reducing agents. For example, Elemike et al. used the Alchornea cordifolia aqueous leaf extract to reduce CuSO4 and obtain a dark green colloidal suspension of 16 nm Cu2O/CuO NPs tested against HeLa cancer cells (cervical adenocarcinoma).87 Wringhtia titoria extract was used by Rajagopal et al. to reduce CuSO4 to obtain 15 nm CuNPs applied to breast cancer cell line (MCF-7).88 Sankar et al. discovered the capability of Ficus religiosa leaf extract to reduce CuSO4 and produce CuNPs with a hydrodynamic diameter of 577 nm.89 These NPs had anticancer activity and were tested by Kalaiarasi et al. in A549 lung cancer cells, in which they stimulated apoptosis and anticancer activity via inhibition of the histone deacetylase.90 As far as glioma is concerned, Cnici benedicti water extract were used for the green synthesis of bimetallic nanoparticles such as Au–CuO and CuO–ZnO NPs, obtaining higher efficiency in the presence of Au rather than ZnO (Fig. 2).57
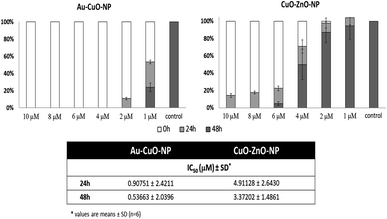 | ||
| Fig. 2 Cytotoxicity of biosynthesized Au–CuO and CuO–ZnO nanoparticles on C6 cells. Reproduced with permission from ref. 57. Copyright© 2018 Production and hosting by Elsevier B.V. on behalf of King Saud University. | ||
3.3. Copper sulfide nanoparticles
Over the past few years, considerable attention has been paid to biomedical applications of copper sulfide nanostructures (CuxSy) as multimodal imaging, due to the paramagnetic behavior and to the possibility to vary the stoichiometries in order to tune the absorption above 900 nm.91 In this spectral region, almost no interference of biological tissues occurs, thus it is possible to significantly reduce background noise and enhance the spatial imaging resolution,92,93 including magnetic resonance imaging (MIR), upconversion luminescence imaging, and photoacoustic imaging.94,95Moreover, copper sulfide NPs are recognized as an effective alternative to Au NPs for photothermal therapy, which originates from a d–d electronic transition,96 whereas surface plasmon resonance is involved in the case of Au NPs.97 Interestingly, the acidic environment of tumor tissues induces the Cu2+ release from CuS NPs, further promoting the ROS production via Fenton-like reaction with H2O2.98 By combining MIR and PTT properties of CuS, theranostic platforms for image-guided ablation therapy can be designed, with possibility to target glioma cells by conjugation with PEGylated c(RGDfK) peptide (Fig. 3).58
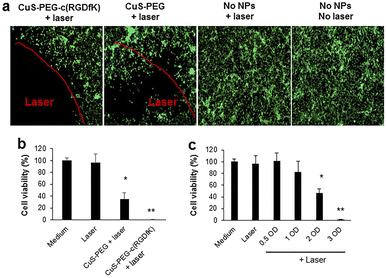 | ||
| Fig. 3 (a) Photothermal ablation of U87 cells mediated by CuS-PEG-c(RGDfK) or CuS-PEG NPs. (b) U87 viability upon treatment with CuS-PEG-c(RGDfK) or CuS-PEG NPs. (c) U87 viability upon treatment with increasing concentrations of CuS-PEG-c(RGDfK) NPs. Reproduced with Permission from ref. 58. Copyright© 2018, American Chemical Society. | ||
Poulose et al. reported on the anticancer efficiency of Cu2S NPs synthesized by hot injection colloidal approach involving the use of Cu and S precursors in the absence of any cytotoxic ligands. NPs were coated with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[folate(polyethylene glycol)-2000] (DSPE-PEG-Fol) for targeting the glioma cancer cells and taking advantages from a targeted NIR-responsive photothermal ablation in synergism with the chemotherapeutic action of a loaded cytotoxic agent such as DOX.59 A similar chemo- and photothermal co-therapy was obtained by loading DOX into the mesoporous SiO2 coated CuS NPs.60 In details, CuS NPs were capped with cetyltrimethylammonium bromide to confer the positive charge suitable for the growth of the mesoporous silica shell.
In a different approach, Lan et al. synthesized hollow mesoporous CuS NPs (HM-CuS NPs) as disulfiram delivery vehicle.61 At first, Cu2O NPs were prepared by reduction of CuCl2 with hydrazine, and then subjected to ion exchange in the presence of Na2S. Then, disulfiram was loaded by exploiting its ability to chelate copper, and a targeting effect was obtained by coating with transferrin. The final nanosystem was found to selectively kill glioma cells both in vitro and in vivo as a result of CuS acid-responsive dissolution and copper ions release characteristics (Fig. 4).
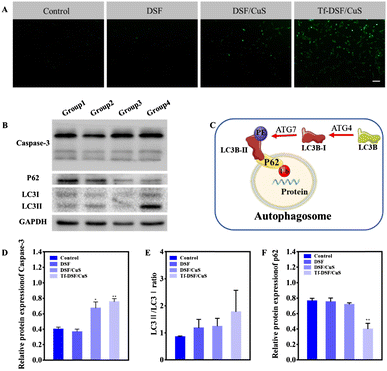 | ||
| Fig. 4 ROS generation autophagy induction by Tf-DSF/CuS. (A) H2DCFDA (green) staining indicates ROS generation. Scale bar = 100 μm. (B) Western blot analysis of apoptosis-related protein (caspase-3) and autophagy markers (LC3I/II, P62). (C) Schematic process of autophagy. Densitometry of caspase-3 (D), LCII/LC3I (E), and p62 (F) in western blots. Reproduced with permission from ref. 61. Copyright© 2021 Elsevier B.V. | ||
Hollow mesoporous copper sulfide NPs (HM-CuS NPs) were used for a chemotherapy (CT)/starvation therapy (ST)/chemodynamic therapy (CDT)/photothermal therapy (PTT) of glioblastoma.62 A complex HM-CuS NPs nanosystem was prepared by loading with temozolomide as a chemotherapeutic, while hyaluronic acid was used as a coating material for inducing targeting efficiency and preventing drug from premature leakage. Glucose oxidase (GOx) was also inserted on the NPs shell to reduce the intracellular glucose level thus inducing ST, and to generate H2O2 (acting as a substrate of Cu based Fenton-like reaction) upon production of gluconic acid further promoting the copper ions release due to pH reduction. NPs surface was further modified with lactoferrin (Lf) to enhance the blood–brain barrier penetration (Fig. 5).
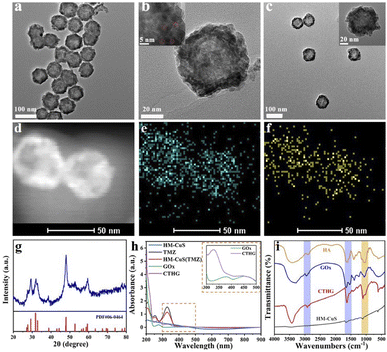 | ||
| Fig. 5 (a) SEM and (b) TEM image of HM-CuS NPs. (c) TEM images of CTHG-Lf NPs. (d) STEM image and (e) and (f) elemental mapping of Cu and S of CTHG-Lf NPs. (g) XRD of CTHG-Lf NPs. (h) UV-vis-NIR absorption spectra of TMZ, HM-CuS, HM-CuS(TMZ), GOx and CTHG NPs. (i) FT-IR of HA, GOx, HM-CuS NPs and CTHG NPs. Reproduced with permission from ref. 62. Copyright© 2021 Elsevier B.V. | ||
The surface of HM-CuS NPs was modified with Lauric acid and a PEGylated peptide acting as a gatekeeper and pH-responsive element enabling translocation within cancer cells, respectively, to fabricate a carrier for the ISRIB stress granules inhibitor.63 The activity of the nanosystem was related to a combined PTT and remodeling of the immunosuppressive microenvironment of brain metastases.
CuS was finally used as a component of quantum dots platforms (QDs). Mohamed et al. developed a strategy to encapsulate Cu2S within CdSe QDs by hydrothermal treatment in the presence of Jatropha curcas oil as a capping coordinating and stabilizing reagent.64 The ultimate aim was to combine the property of Cd as X-ray contrast agent with the Cu2S induced PTT, and the system was further engineered by conjugation with PEG and folic acid as biocompatible and targeting elements, respectively.
3.4. Other copper nanoparticles
A different class of copper NPs was proposed by Lin et al., who prepared copper peroxide nanodots (CPNDs) as chemodynamic therapy agent boosting the production of ROS within the cancer cells.99 Authors exploited the well-known ability of metal peroxides to act as a H2O2 source to promote ROS production via Fenton-like reactions.100 They synthesized 16.3 nm CPNDs by the reaction of CuCl2, H2O2, and sodium hydroxide in an aqueous solution containing polyvinyl pyrrolidone at room temperature for 30 min. The CPNDs were effectively internalized within cancer cells by enhanced permeability and retention effect, and then decomposed in the acidic endo/lysosomal compartments, with the release of large amount of H2O2 (Fig. 6).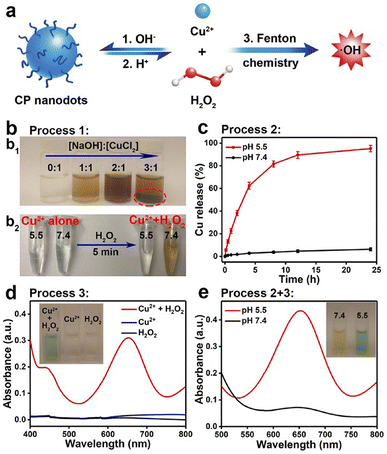 | ||
| Fig. 6 (a) Schematic representation of formation and dissociation of CP nanodots for ˙OH production. (b1) Photograph of CP materials obtained in the presence of PVP at different molar ratios of NaOH to CuCl2. (b2) Photograph of CuCl2 solutions with different pH values before and after the addition of H2O2. (c) Cumulative Cu release from CP nanodots in different pH conditions. (d) UV-vis spectra and photographs (inset) of TMB aqueous solution treated with H2O2, Cu2+, or Cu2+ plus H2O2 for 30 min. (e) Colorimetric detection of ˙OH generated by CP nanodots at different pH values. Reproduced with permission from ref. 99. Copyright© 2019 American Chemical Society. | ||
A further development of this approach consisted in embedding CPNDs within a hydroxypropyl chitin hydrogel together with RGD-peptide-modified paclitaxel prodrug nanoparticles for a combined photodynamic/chemodynamic/chemotherapy treatment of postoperative glioblastoma, reaching a consistent extent of the survival time of postoperative glioma mice.65
To overcome the radioresistance of glioblastoma, Xu et al. proposed core–shell copper selenide coated gold NPs (Au@Cu2−xSe NPs) that inhibited DNA repair mechanisms and protective autophagy on U-87MG cancer cells.66 Inner AuNPs of 13 nm were produced following Turkevich method, while the protective layer was produced by reduction of SeO2 and CuSO4 with ascorbic acid.101 This step changed the color of the colloid suspension to dark green. Dithiol PEG (5 kDa) was used to modify the surface of the NPs.
The purification was performed via dialysis with a molecular weight cut-off (MWCO) of 30 kDa. The final size was around 21 nm, and it was assessed by TEM. Copper was quantified by using Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES).
Cyclodextrin (CD)-modified Cu2−xSe NPs were proposed for a photodynamic chemotherapy (Doxorubicin as cytotoxic drug), by taking advantages of the ability of Near Infrared Radiation (NIR) irradiated NPs to produce a huge amount of ROS by means of both electron transfer and energy transfer mechanisms.67 Spherical NPs (3.3 nm as per TEM analysis) were synthesized by adding CuCl2 to a selenium precursor obtained by reduction of Se powder with NaBH4 and removing the excess copper by ultracentrifugation. Afterward, NPs were modified with CD and HS-PEG-SH to allow a pH-controlled release of the cytotoxic agent after delivery into malignant glioblastoma by opening the blood–brain-barrier with the assistance of focused ultrasound. The same material was tested as a vehicle for boosting the anticancer performance of orally administrated disulfiram after loading with hypoxia-inducible factor-1α (HIF-1α) inhibitor and coating with tumor cell membrane.68 Noteworthly, the same research group explored the possibility to use such cell membrane-coated NPs in an immunotherapy protocol by selectively targeting the tumor-associated macrophages instead of glioma cells. Authors were able to re-activate the immune responses through remodeling the tumor immunosuppressive microenvironment by loading an inhibitor of indoleamine-2,3-dioxygenease (indoximod) and an inhibitor for reducing the expression of PD-L1 (JQ1).69
Along with copper nanospheres, materials with different morphologies have been proposed for glioma applications, as the biocomposite of copper and cysteine suggested by Kelly et al.102,103 These copper structures (CuHARS) include copper and L-cysteine and demonstrated high stability with no aggregation in liquid media. L-Cys was dissolved in a NaOH solution, and CuSO4 was added as a source of copper ions. The excess of copper was removed by a short HCl treatment, and the product was purified by precipitation and washings with water.70,71 CuHARS sizes ranged from about 20 nm in diameter and hundreds of nm in length. Karan et al. integrated CuHARS in cellulose discs to test them on CRL2303 glioma cells.70 CuHARS were slowly but completely biodegraded in biological media, probably because of copper complexation to enzymes and proteins. The entrapment in cellulose scaffold prolongs the persistence of CuHARS and allows the interaction with cells to exert their action. Karekar et al. tested the toxicity of CuHARS on CRL2303 glioma and SH-EP1 neuroblastoma cells.72 CuHARS were more toxic than the silver analogue to these cell lines. The same group evaluated the immunomodulatory potential of CuHARS on CRL2303 glioma cells, in which they increased nitrogen oxide production while reducing the viability of glioma cells and avoiding harming healthy cells (brain microvascular endothelial cells, BMVECs).71
Interesting anticancer applications have been demonstrated for ultrasmall-in-nanoarchitectures (NAs) that comprise copper ultrasmall NPs (USNPs) of <2 nm.104 NAs are hybrid nanomaterials in which ultrasmall metal NPs are entrapped in polymeric aggregates and protected by a silica shell.105,106 The final size is around 100–150 nm, with a silica shell of approximately 20 nm. NAs are promising tools in anticancer therapy because they do not accumulate in the body nor harm the organs, moreover NAs can combine different therapeutic and diagnostic approaches in a single system.107–113 Cu USNPs are prepared by reduction of glutathione-complexed copper ions from CuSO4 using NaBH4. Cu content in the whole NAs was quantified by Inductively Coupled Plasma Mass Spectrometry (ICP-MS).104 CuNAs favored the recovery from burnt skin in in vivo models after a single topical application, with a significant anti-inflammatory activity and without systemic toxicity.104 On the other side, CuNAs also showed to slow down the metastatic cascade in pancreatic chorioallantoic membrane (CAM) cancer models of BxPC-3.
4. Copper chelation therapy as a therapeutic strategy against glioma
The notion that copper is a key limiting factor for tumor progression has encouraged the development of copper-specific chelators as therapies to inhibit this process.10 Copper chelators are small molecules that bind to copper ions and reduce their availability in the body. These compounds mimic the regulatory chaperones that manage cellular copper influx and efflux across cell membranes in physiological conditions. They can directly induce apoptosis through the generation of ROS and the inhibition of cytochrome C oxidase, which would decrease the level of ATP produced. These phenomena are directly linked to the role played by copper in essential cellular processes such as energy production. Additionally, copper chelators can prevent the recruitment of bone marrow-derived endothelial progenitor cells (EPC). This results in the inhibition of angiogenesis and tumor progression by affecting signaling pathways such as PI3K/AKT and MAPK/ERK, and/or via the regulation of epigenetic changes, also during late-stages.7Copper chelating agents such as tetrathiomolybdate (TM), D-penicillamine (DPA) and triethylenetetramine dihydrochloride (trientine/TETA), alone or in combination therapies,114 are already employed for the treatment of Wilson's disease.115,116 Clinical trials revealed that these therapies are generally well tolerated, since selectively targeting cancer and exerting little toxicity on normal cells.117,118 Based on their efficacy, these compounds and other similar drugs are currently being investigated for their adaptability to cancer therapeutics.7 As a matter of fact, TM has been shown to inhibit tumor growth and angiogenesis in animal models of breast cancer by reducing the expression of the highly-angiogenic factor Lysil oxidase (LOX) and the metalloproteinases MMP-2 and MMP-9, which are involved in the generation of metastases.119,120 This compound is also currently used in phase I or II clinical trials for the treatment of BRAF V600E mutated tumors,121–123 head and neck carcinoma,124 mesothelioma118 and pancreatic duct adenocarcinoma.125 Similarly, D-penicillamine treatment reduces metastatic melanoma cells.126 Furthermore, it was able to delay progression of glioblastoma by inhibits LOX activity and reducing VEGF expression.127 Despite reducing excessive copper levels in the system by secretion through the urine, it was also demonstrated that DPA chelation leads to the generation of hydrogen peroxide (H2O2) and other ROS, resulting in copper-dependent cytotoxic effects.128 Consistently, even though DPA was involved in different clinical trials for the treatment of Wilson's129 and Alzheimer's disease,8 as well as glioblastoma,15 DPA chelation of copper in brain (where copper concentration is usually higher) did not ameliorate patients' neurological symptoms, due to ROS-dependent toxicity.130 However, around 60% of patients showed an improvement after 4 years of therapy.131 The combination of DPA with inhibitors of hydroperoxide metabolism is thus more common than the treatment involving the single agent. Alternatively, despite having a reduced effect compared to DPA, TETA shows an improved safety profile. This drug was originally introduced for the treatment of DPA-intolerant patients with Wilson's disease132 and its efficacy to hinder tumor growth in vivo, with a molecular mechanism involving inhibition of IL-8 production, was first demonstrated in hepatocellular carcinomas.133 In addition, it was shown that TETA exerts an anti-tumor effect not only via the regulation of copper transporters' flux, but also by the interaction with multiple anticancer targets, which results in the reduction of oxidative stress and polyamine metabolism, an important energy source for cell's growth.134 Remarkably, it was demonstrated how TETA can cross the blood–brain-barrier (BBB) and target the central nervous system (CNS) by surface modified liposomes, which enable a higher-dose drug delivery and successfully delays neurodegeneration,135 resulting in the most promising copper-chelating agents to target brain tumors.
A similar molecular mechanism is exerted by the FDA-approved tetraethylenepentamine (TEPA), an analogue of trientine. Recently, it was reported how TEPA can increase immune cell infiltration in the TME, downregulating STAT3, EGFR, AKT, and GSK3b phosphorylation, which inhibits the transcription of PD-L1 and thus reduces tumor growth in NB and GBM mouse models, improving survival.26 Furthermore, another study showed that TEPA downregulates EMT-associated cancer invasion in vitro by directing reducing the levels of TGF-β and downstream signaling pathways in three tumor types: human triple-negative breast cancer (TNBC), diffuse intrinsic pontine glioma (DIPG) and NB. In the same study, it was also shown how metastases are suppressed in vivo, even if small doses of TEPA were used, making it stand out against other copper chelating agents, or other therapies directly targeting TGF- β, as an attractive non-toxic alternative.136
The number of ongoing clinical trials for copper chelators as single agents is much bigger than the number of trials concerning combination therapies. A deeper investigation of the molecular mechanisms acting in synergy during treatment would boost consideration of these novel agents as an effective therapeutic strategy against cancer.
5. Conclusions and perspectives
The involvement of copper in the development and/or progression of different types of tumors suggests new therapeutic opportunities, with three main routes evoked for explain the biological mechanisms to kill cancer cells by using copper, namely (i) the use of Cu NPs as ROS inducers; (ii) Cu chelation by suitable chemical species, and (iii) induction of Cu overload by local release of copper in the tumor area followed by ROS production and cuproptosis. Each strategy shows great potentiality allowing hypothesizing effective translation to the pre-clinical and clinical practice, although some key issues should be addressed. Cu NPs, indeed, may lead to a moderate risk of off-target toxicity; copper chelators are characterized by safer profile and much less toxicity but often they need to be used in combination with other therapeutic(s) for an effective cancer treatment; while fewer groups tested Cu overload as a therapeutic regimen.In this review, we mainly discussed the recent development in using Cu NPs, since they showed the key advantage to allow customizing the particles physico-chemical properties, loading Cu chelators and drugs on the same structure, including ROS inducers and conventional chemotherapeutics. Open questions and possible improvements are in the area of specificity, immune system clearance of the particles, ADMET, and off target toxicity. Solving these challenges will contribute to enhance the translation of copper overload/depletion in oncology, even if additional investigations are required to improve the rational design of nanoparticle architectures to tailor the features for the desired therapeutic activity.
We expect that in the future many clinical trials will start to test combinational therapies more than single agent ones, Cu-related treatments are often wide range and multitargeting, i.e. TEPA downregulation of kinases and mitochondrial activity. These wide spectra therapeutics are suitable for combination with highly specific drugs, such as EGFR inhibitors for example, resulting in a stronger and more articulated inhibition of tumor growth. A multitarget therapy combined with a highly specific one can tackle the cancer cells ability to grow resistance upregulating other pathways, i.e. mTOR upregulation as resistance to MAPKs inhibition.
Moreover, the design of nanoparticle from an industrial perspective is urgently required to hypothesize an effective clinical translation of Cu NPs. Although these issues still need to be solved, we hope that this review can open a discussion among the scientific community and show the potential applicability of Cu NPs for the treatment of brain cancer.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge funding from the National Health and Medical Research Council Ideas Grants (grant number GNT2012567).Notes and references
- J. Lopez, D. Ramchandani and L. Vahdat, Met. Ions Life Sci., 2019, 19, 303–330 CAS.
- A. Parmar, G. Pascali, F. Voli, L. Lerra, E. Yee, A. Ahmed-Cox, K. Kimpton, G. Cirillo, A. Arthur, D. Zahra, G. Rahardjo, G. J. Liu, N. Lengkeek, F. Saletta, A. Charil, M. Kavallaris and O. Vittorio, Theranostics, 2018, 8, 5645–5659 CrossRef CAS.
- F. Michniewicz, F. Saletta, J. R. C. Rouaen, R. V. Hewavisenti, D. Mercatelli, G. Cirillo, F. M. Giorgi, T. Trahair, D. Ziegler and O. Vittorio, Chemmedchem, 2021, 16, 2315–2329 CrossRef CAS.
- L. Macomber and J. A. Imlay, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8344–8349 CrossRef CAS.
- O. Wise and O. Coskuner, J. Comput. Chem., 2014, 35, 1278–1289 CrossRef CAS.
- G. J. Brewer, Proc. Soc. Exp. Biol. Med., 2000, 223, 39–46 CAS.
- Z. Tumer and L. B. Moller, Eur. J. Hum. Genet., 2010, 18, 511–518 Search PubMed.
- R. Squitti, P. M. Rossini, E. Cassetta, F. Moffa, P. Pasqualetti, M. Cortesi, A. Colloca, L. Rossi and A. Finazzi-Agro, Eur. J. Clin. Invest., 2002, 32, 51–59 CrossRef CAS.
- R. Giampietro, F. Spinelli, M. Contino and N. A. Colabufo, Mol. Pharmaceutics, 2018, 15, 808–820 CrossRef CAS.
- R. Janssen, B. de Brouwer, J. H. von der Thusen and E. F. M. Wouters, Med. Hypotheses, 2018, 120, 49–54 CrossRef CAS.
- J. Lowe, R. Taveira-da-Silva and E. Hilario-Souza, IUBMB Life, 2017, 69, 255–262 CrossRef CAS.
- V. Oliveri, Front. Mol. Biosci., 2022, 9, 841814 CrossRef CAS.
- C. J. Taylor, J. B. Qiao, N. C. Colon, C. Schlegel, E. Josifi and D. H. Chung, Surgery, 2011, 150, 162–168 CrossRef.
- D. Denoyer, S. Masaldan, S. La Fontaine and M. A. Cater, Metallomics, 2015, 7, 1459–1476 CrossRef CAS.
- S. Brem, S. A. Grossman, K. A. Carson, P. New, S. Phuphanich, J. B. Alavi, T. Mikkelsen, J. D. Fisher and N. A. B. T. Therapy, Neuro-Oncology, 2005, 7, 246–253 CrossRef CAS.
- D. Yoshida, Y. Ikeda and S. Nakazawa, Neurosurgery, 1995, 37, 287–292 CrossRef CAS.
- S. Basu, M. K. Singh, T. B. Singh, S. K. Bhartiya, S. P. Singh and V. K. Shukla, World J. Surg., 2013, 37, 2641–2646 CrossRef.
- X. Ding, M. Jiang, H. Y. Jing, W. Sheng, X. W. Wang, J. Q. Han and L. H. Wang, Environ. Sci. Pollut. Res., 2015, 22, 7930–7935 CrossRef CAS.
- V. Pavithra, T. G. Sathisha, K. Kasturp, D. S. Mallika, S. J. Amos and S. Ragunatha, J. Clin. Diagn. Res., 2015, 9, Bc25–Bc27 CAS.
- A. K. Baltaci, T. K. Dundar, F. Aksoy and R. Mogulkoc, Biol. Trace Elem. Res., 2017, 175, 57–64 CrossRef CAS.
- M. Stepien, M. Jenab, H. Freisling, N. P. Becker, M. Czuban, A. Tjonneland, A. Olsen, K. Overvad, M. C. Boutron-Ruault, F. R. Mancini, I. Savoye, V. Katzke, T. Kuhn, H. Boeing, K. Iqbal, A. Trichopoulou, C. Bamia, P. Orfanos, D. Palli, S. Sieri, R. Tumino, A. Naccarati, S. Panico, H. B. Bueno-de-Mesquita, P. H. Peeters, E. Weiderpass, S. Merino, P. Jakszyn, M. J. Sanchez, M. Dorronsoro, J. M. Huerta, A. Barricarte, S. Boden, B. van Guelpen, N. Wareham, K. T. Khaw, K. E. Bradbury, A. J. Cross, L. Schomburg and D. J. Hughes, Carcinogenesis, 2017, 38, 699–707 CrossRef CAS.
- X. P. Zhang and Q. Yang, J. Int. Med. Res., 2018, 46, 4863–4873 CrossRef CAS.
- F. Chen, J. Wang, J. F. Chen, L. J. Yan, Z. J. Hu, J. F. Wu, X. D. Bao, L. K. Lin, R. Wang, L. Cai, L. S. Lin, Y. Qiu, F. Q. Liu and B. C. He, Oral Dis., 2019, 25, 80–86 CrossRef.
- L. Aubert, N. Nandagopal, Z. Steinhart, G. Lavoie, S. Nourreddine, J. Berman, M. K. Saba-El-Leil, D. Papadopoli, S. C. Lin, T. Hart, G. Macleod, I. Topisirovic, L. Gaboury, C. J. Fahrni, D. Schramek, S. Meloche, S. Angers and P. P. Roux, Nat. Commun., 2020, 11, 3701 CrossRef CAS.
- S. A. K. Saleh, H. M. Adly, A. A. Abdelkhaliq and A. M. Nassir, Curr. Neurol., 2020, 14, 44–49 CAS.
- F. Voli, E. Valli, L. Lerra, K. Kimpton, F. Saletta, F. M. Giorgi, D. Mercatelli, J. R. C. Rouaen, S. Shen, J. E. Murray, A. Ahmed-Cox, G. Cirillo, C. Mayoh, P. A. Beavis, M. Haber, J. A. Trapani, M. Kavallaris and O. Vittorio, Cancer Res., 2020, 80, 4129–4144 CrossRef CAS.
- S. R. Knight, C. A. Shaw, R. Pius, T. M. Drake, L. Norman, A. O. Ademuyiwa, A. O. Adisa, M. L. Aguilera-Arevalo, S. W. Al-Saqqa, I. S. Al-Slaibi, A. Bhangu, B. M. Biccard, P. Brocklehurst, A. Costas-Chavarri, K. M. Chu, A. J. Dare, M. Elhadi, C. J. Fairfield, J. E. Fitzgerald, D. N. Ghosh, J. Glasbey, M. I. V. Henegouwen, J. C. A. Ingabire, T. P. Kingham, M. C. M. Lapitan, I. Lawani, B. Lieske, R. J. Lilford, J. Martin, K. A. Mclean, R. L. Moore, D. Morton, D. Nepogodiev, F. Ntirenganya, F. Pata, T. D. Pinkney, A. U. Qureshi, A. Ramos-De la Medina, A. M. Riad, H. K. Salem, J. Simoes, R. T. Spence, N. J. Smart, S. Tabiri, H. S. Thomas, T. G. Weiser, M. A. West, J. Whitaker, E. M. Harrison, G. Collaborative and G. S. W. Grp, Lancet, 2021, 397, 387–397 CrossRef.
- D. T. Debela, S. G. Y. Muzazu, K. D. Heraro, M. T. Ndalama, B. W. Mesele, D. C. Haile, S. K. Kitui and T. Manyazewal, SAGE Open Med., 2021, 9, 20503121211034366 Search PubMed.
- S. Tohme, R. L. Simmons and A. Tsung, Cancer Res., 2017, 77, 1548–1552 CrossRef CAS.
- S. Zimmermann, R. Dziadziuszko and S. Peters, Cancer Treat. Rev., 2014, 40, 716–722 CrossRef CAS.
- K. Wang and J. E. Tepper, Ca-Cancer J. Clin., 2021, 71, 437–454 CrossRef.
- S. N. Bhatia, X. Chen, M. A. Dobrovolskaia and T. Lammers, Nat. Rev. Cancer, 2022, 22, 550–556 CrossRef CAS.
- M. Rasool, A. Malik, S. Waquar, M. Arooj, S. Zahid, M. Asif, S. Shaheen, A. Hussain, H. Ullah and S. H. Gan, Bioengineered, 2022, 13, 759–773 CrossRef CAS.
- M. J. Nirmala, U. Kizhuveetil, A. Johnson, G. Balaji, R. Nagarajan and V. Muthuvijayan, RSC Adv., 2023, 13, 8606–8629 RSC.
- X. J. Gao, N. Ran, X. Dong, B. F. Zuo, R. Yang, Q. B. Zhou, H. M. Moulton, Y. Q. Seow and H. F. Yin, Sci. Transl. Med., 2018, 10, eaat0195 CrossRef.
- D. Mundekkad and W. L. C. Cho, Int. J. Mol. Sci., 2022, 23, 1685 CrossRef CAS.
- J. J. Xu, W. C. Zhang, Y. W. Guo, X. Y. Chen and Y. N. Zhang, Drug Delivery, 2022, 29, 664–678 CrossRef CAS.
- B. Klebowski, J. Depciuch, M. Parlinska-Wojtan and J. Baran, Int. J. Mol. Sci., 2018, 19, 4031 CrossRef.
- R. Khursheed, K. Dua, S. Vishwas, M. Gulati, N. K. Jha, G. M. Aldhafeeri, F. G. Alanazi, B. H. Goh, G. Gupta, K. R. Paudel, P. M. Hansbro, D. K. Chellappan and S. K. Singh, Biomed. Pharmacother., 2022, 150, 112951 CrossRef CAS.
- M. L. Ermini and V. Voliani, ACS Nano, 2021, 15, 6008–6029 CrossRef CAS.
- A. Fathollahi, S. J. Coupe, A. H. El-Sheikh and E. O. Nnadi, J. Environ. Manage., 2021, 282, 111950 CrossRef CAS.
- M. C. Crisan, M. Teodora and M. Lucian, Appl. Sci., 2022, 12, 141 CrossRef CAS.
- M. P. Nikolova and M. S. Chavali, Biomimetics, 2020, 5, 27 CrossRef CAS.
- M. B. Gawande, A. Goswami, F. X. Felpin, T. Asefa, X. X. Huang, R. Silva, X. X. Zou, R. Zboril and R. S. Varma, Chem. Rev., 2016, 116, 3722–3811 CrossRef CAS.
- A. Semisch, J. Ohle, B. Witt and A. Hartwig, Part. Fibre Toxicol., 2014, 11, 10 CrossRef.
- F. Shen, Y. Fang, Y. Wu, M. Zhou, J. Shen and X. Fan, J. Nanobiotechnol., 2023, 21, 20 CrossRef.
- R. Aishajiang, Z. S. Liu, T. J. Wang, L. Zhou and D. Yu, Molecules, 2023, 28, 2303 CrossRef CAS.
- N. Asif, R. Ahmad, S. Fatima, S. Shehzadi, T. Siddiqui, A. Zaki and T. Fatma, Sci. Rep., 2023, 13, 6246 CrossRef CAS.
- F. Y. Shen, Y. Fang, Y. J. Wu, M. Zhou, J. F. Shen and X. Q. Fan, J. Nanobiotechnol., 2023, 21, 20 CrossRef.
- I. Paw, R. C. Carpenter, K. Watabe, W. Debinski and H. W. Lo, Cancer Lett., 2015, 362, 1–7 CrossRef CAS.
- V. Vodyanoy, Y. Daniels, O. Pustovyy, W. A. MacCrehan, S. Muramoto and G. Stan, Int. J. Nanomed., 2016, 11, 1567–1576 CrossRef CAS.
- A. Joshi, H. Naatz, K. Faber, S. Pokhrel and R. Dringen, Neurochem. Res., 2020, 45, 809–824 CrossRef CAS.
- A. Joshi, W. Rastedt, K. Faber, A. G. Schultz, F. Bulcke and R. Dringen, Neurochem. Res., 2016, 41, 3004–3019 CrossRef CAS.
- F. Bulcke, K. Thiel and R. Dringen, Nanotoxicology, 2014, 8, 775–785 CAS.
- N. R. Kukia, A. Abbasi and S. M. A. Froushani, Dhaka Univ. J. Pharm. Sci., 2018, 17, 105–111 CrossRef CAS.
- X. Wang, A. D. Hu, K. Du and F. D. Feng, ACS Appl. Bio Mater., 2021, 4, 8004–8012 CrossRef CAS.
- R. Dobrucka, M. Kaczmarek, M. Lagiedo, A. Kielan and J. Dlugaszewska, Saudi Pharm. J., 2019, 27, 373–383 CrossRef.
- L. Cui, C. Xiong, M. Zhou, S. Shi, D. S. L. Chow and C. Li, Bioconjugate Chem., 2018, 29, 4062–4071 CrossRef CAS.
- A. C. Poulose, S. Veeranarayanan, M. S. Mohamed, Y. Nagaoka, R. R. Aburto, T. Mitcham, P. M. Ajayan, R. R. Bouchard, Y. Sakamoto, Y. Yoshida, T. Maekawa and D. S. Kumar, Nanoscale, 2015, 7, 8378–8388 RSC.
- S. W. Peng, Y. Y. He, M. Er, Y. Z. Sheng, Y. Q. Gu and H. Y. Chen, Biomater. Sci., 2017, 5, 475–484 RSC.
- Q. H. Lan, C. C. Du, R. J. Yu, J. Y. Zhai, Y. N. Shi, L. F. Kou, J. Xiao, C. T. Lu, Y. Z. Zhao and Q. Yao, Int. J. Pharm., 2021, 607, 120978 CrossRef CAS.
- Y. Cao, L. H. Jin, S. Zhang, Z. J. Lv, N. Yin, H. Zhang, T. Q. Zhang, Y. H. Wang, Y. Chen, X. R. Liu and G. Zhao, Eur. J. Pharm. Sci., 2023, 180, 106319 CrossRef CAS.
- F. Tong, H. Hu, Y. Xu, Y. Zhou, R. Xie, T. Lei, Y. Du, W. Yang, S. He, Y. Huang, T. Gong and H. Gao, Acta Pharm. Sin. B, 2022, 13, 3471–3488 CrossRef.
- M. Sheikh Mohamed, A. C. Poulose, S. Veeranarayanan, R. Romero Aburto, T. Mitcham, Y. Suzuki, Y. Sakamoto, P. M. Ajayan, R. R. Bouchard, Y. Yoshida, T. Maekawa and D. Sakthi Kumar, Nanoscale, 2016, 8, 7876–7888 RSC.
- X. Cao, S. N. Li, W. L. Chen, H. D. Lu, L. Ye, Z. Y. Min, S. B. Sun, C. H. Teng, H. Y. Yin, Q. Zhang, W. C. He, X. Z. Wang, W. Lv, L. Y. Lv and H. L. Xin, ACS Appl. Mater. Interfaces, 2022, 14, 27623–27633 CrossRef CAS.
- Q. Xu, H. Zhang, H. H. Liu, Y. B. Han, W. B. Qiu and Z. Li, Biomaterials, 2022, 280, 121287 CrossRef CAS.
- H. Zhang, T. T. Wang, H. H. Liu, F. Ren, W. B. Qiu, Q. Sun, F. Yan, H. R. Zheng, Z. Li and M. Y. Gao, Nanoscale, 2019, 11, 7600–7608 RSC.
- S. Yang, Y. B. Han, B. L. Bao, C. H. Hu and Z. Li, Composites, Part B, 2022, 243, 110117 CrossRef CAS.
- T. T. Wang, H. Zhang, W. B. Qiu, Y. B. Han, H. H. Liu and Z. Li, Bioact. Mater., 2022, 16, 418–432 CAS.
- A. Karan, M. Darder, U. Kansakar, Z. Norcross and M. A. DeCoster, Int. J. Environ. Res. Public Health, 2018, 15, 844 CrossRef.
- N. Prajapati, A. Karan, E. Khezerlou and M. A. DeCoster, Front. Chem., 2021, 8, 629835 CrossRef.
- N. Karekar, A. Karan, E. Khezerlou, N. Prajapati, C. D. Pernici, T. A. Murray and M. A. DeCoster, Nanomater., 2019, 9, 1282 CrossRef CAS.
- S. C. Mali, A. Dhaka, S. Sharma and R. Trivedi, Inorg. Chem. Commun., 2023, 149, 110448 CrossRef.
- C. Dhand, N. Dwivedi, X. J. Loh, A. N. J. Ying, N. K. Verma, R. W. Beuerman, R. Lakshminarayanan and S. Ramakrishna, RSC Adv., 2015, 5, 105003–105037 RSC.
- S. Sathiyavimal, S. Vasantharaj, D. Bharathi, M. Saravanan, E. Manikandan, S. S. Kumar and A. Pugazhendhi, J. Photochem. Photobiol., B, 2018, 188, 126–134 CrossRef CAS.
- A. A. Ponce and K. J. Klabunde, J. Mol. Catal. A: Chem., 2005, 225, 1–6 CrossRef CAS.
- J. Jablonska, K. Jankowski, M. Tomasik, D. Cykalewicz, P. Uznanski, S. Caluch, M. Szybowicz, J. Zakrzewska and P. Mazurek, SN Appl. Sci., 2021, 3, 244 CrossRef CAS.
- M. Tokushige, T. Nishikiori and Y. Itoc, Russ. J. Electrochem., 2010, 46, 619–626 CrossRef CAS.
- R. M. Tilaki, A. I. Zad and S. M. Mahdavi, Appl. Phys. A: Mater. Sci. Process., 2007, 88, 415–419 CrossRef CAS.
- W. Y. Teoh, R. Amal and L. Madler, Nanoscale, 2010, 2, 1324–1347 RSC.
- G. Habibullah, J. Viktorova and T. Ruml, Nanoscale Res. Lett., 2021, 16, 47 CrossRef CAS.
- R. Javed, M. Zia, S. Naz, S. O. Aisida, N. Ul Ain and Q. Ao, J. Nanobiotechnol., 2020, 18, 172 CrossRef.
- Y. Kobayashi, T. Maeda, K. Watanabe, K. Ihara, Y. Yasuda and T. Morita, J. Nanopart. Res., 2011, 13, 5365–5372 CrossRef CAS.
- T. M. D. Dang, T. T. T. Le, E. Fribourg-Blanc and M. C. Dang, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2011, 2, 015009 Search PubMed.
- H. Strobel, T. Baisch, R. Fitzel, K. Schilberg, M. D. Siegelin, G. Karpel-Massler, K. M. Debatin and M. A. Westhoff, Biomedicines, 2019, 7, 69 CrossRef CAS.
- X. Li, F. L. Shao, J. Sun, K. Du, Y. Sun and F. D. Feng, ACS Appl. Mater. Interfaces, 2019, 11, 41935–41945 CrossRef CAS.
- E. E. Elemike, D. C. Onwudiwe and M. Singh, J. Inorg. Organomet. Polym., 2020, 30, 400–409 CrossRef CAS.
- G. Rajagopal, A. Nivetha, M. Sundar, T. Panneerselvam, S. Murugesan, P. Parasuraman, S. Kumar, S. Ilango and S. Kunjiappan, Heliyon, 2021, 7, e07360 CrossRef CAS.
- R. Sankar, R. Maheswari, S. Karthik, K. S. Shivashangari and V. Ravikumar, Mater. Sci. Eng., C, 2014, 44, 234–239 CrossRef CAS.
- A. Kalaiarasi, R. Sankar, C. Anusha, K. Saravanan, K. Aarthy, S. Karthic, T. L. Mathuram and V. Ravikumar, Biotechnol. Lett., 2018, 40, 249–256 CrossRef CAS.
- N. ul Ain, J. A. Nasir, Z. Khan, I. S. Butler and Z. Rehman, RSC Adv., 2022, 12, 7550–7567 RSC.
- S. X. Shi, X. F. Wen, T. T. Li, X. X. Wen, Q. Z. Cao, X. L. Liu, Y. Y. Liu, M. D. Pagel and C. Li, ACS Appl. Bio Mater., 2019, 2, 3203–3211 CrossRef CAS.
- S. M. H. AL-Jawad, A. A. Taha and A. M. Redha, J. Sol-Gel Sci. Technol., 2019, 91, 310–323 CrossRef CAS.
- J. X. Ge, L. Chen, B. X. Huang, Y. Gao, D. D. Zhou, Y. Zhou, C. Chen, L. Wen, Q. Li, J. F. Zeng, Z. Y. Zhong and M. Y. Gao, ACS Appl. Mater. Interfaces, 2022, 14, 8838–8846 CrossRef CAS.
- D. Y. Gao, Z. H. Sheng, Y. B. Liu, D. H. Hu, J. Zhang, X. J. Zhang, H. R. Zheng and Z. Yuan, Adv. Healthcare Mater., 2017, 6, 1601094 CrossRef.
- Y. Xie, L. Carbone, C. Nobile, V. Grillo, S. D'Agostino, F. Della Sala, C. Giannini, D. Altamura, C. Oelsner, C. Kryschi and P. D. Cozzoli, ACS Nano, 2013, 7, 7352–7369 CrossRef CAS.
- S. S. Gambhir, Nat. Rev. Cancer, 2002, 2, 683–693 CrossRef CAS.
- H. X. Tang, C. G. Liu, J. T. Zhang, X. Zheng, D. Y. Yang, R. K. Kankala, S. B. Wang and A. Z. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 47289–47298 CrossRef CAS.
- L. S. Lin, T. Huang, J. B. Song, X. Y. Ou, Z. T. Wang, H. Z. Deng, R. Tian, Y. J. Liu, J. F. Wang, Y. Liu, G. C. Yu, Z. J. Zhou, S. Wang, G. Niu, H. H. Yang and X. Y. Chen, J. Am. Chem. Soc., 2019, 141, 9937–9945 CrossRef CAS.
- C. C. Huang, W. T. Chia, M. F. Chung, K. J. Lin, C. W. Hsiao, C. Jin, W. H. Lim, C. C. Chen and H. W. Sung, J. Am. Chem. Soc., 2016, 138, 5222–5225 CrossRef CAS.
- J. Turkevich, P. C. Stevenson and J. Hillier, J. Phys. Chem., 1953, 57, 670–673 CrossRef CAS.
- K. C. Kelly, J. R. Wasserman, S. Deodhar, J. Huckaby and M. A. DeCoster, J. Visualized Exp., 2015, 101, 52901 Search PubMed.
- S. Deodhar, J. Huckaby, M. Delahoussaye and M. A. DeCoster, IOP Conf. Ser.: Mater. Sci. Eng., 2014, 64, 012014 CAS.
- M. L. Ermini, M. Summa, A. Zamborlin, V. Frusca, A. K. Mapanao, E. Mugnaioli, R. Bertorelli and V. Voliani, Nanoscale Adv., 2023, 5, 1212–1219 RSC.
- A. K. Mapanao, P. Sarogni, M. Santi, M. Menicagli, A. Gonnelli, A. Zamborlin, M. L. Ermini and V. Voliani, Biomater. Sci., 2022, 10, 6135–6145 RSC.
- D. Cassano, J. David, S. Luin and V. Voliani, Sci. Rep., 2017, 7, 43795 CrossRef CAS.
- D. Cassano, A. K. Mapanao, M. Summa, Y. Vlamidis, G. Giannone, M. Santi, E. Guzzolino, L. Pitto, L. Poliseno, R. Bertorelli and V. Voliani, ACS Appl. Bio Mater., 2019, 2, 4464–4470 CrossRef CAS.
- A. K. Mapanao, G. Giannone, M. Summa, M. L. Ermini, A. Zamborlin, M. Santi, D. Cassano, R. Bertorelli and V. Voliani, Nanoscale Adv., 2020, 2, 3815–3820 RSC.
- A. Zamborlin, M. L. Ermini, M. Summa, G. Giannone, V. Frusca, A. K. Mapanao, D. Debellis, R. Bertorelli and V. Voliani, Nano Lett., 2022, 22, 5269–5276 CrossRef CAS.
- D. Cassano, M. Summa, S. Pocoví-Martínez, K. Mapanao, R. Bertorelli and V. Voliani, Part. Part. Syst. Charact., 2019, 36, 1800464 CrossRef.
- M. Santi, V. Frusca, M. L. Ermini, A. K. Mapanao, P. Sarogni, A. Gonnelli, N. Giannini, A. Zamborlin, L. Biancalana, F. Marchetti and V. Voliani, J. Mater. Chem. B, 2023, 11, 325–334 RSC.
- M. Santi, A. K. Mapanao, D. Cassano, Y. Vlamidis, V. Cappello and V. Voliani, Cancers, 2020, 12, 1063 CrossRef CAS.
- C. Avigo, D. Cassano, C. Kusmic, V. Voliani and L. Menichetti, J. Phys. Chem. C, 2017, 121, 6955–6961 CrossRef CAS.
- I. Mohr and K. H. Weiss, Ann. Transl. Med., 2019, 7, S69 CrossRef CAS.
- S. Baldari, G. Di Rocco and G. Toietta, Int. J. Mol. Sci., 2020, 21, 1069 CrossRef CAS.
- J. Lu, Mol. Cancer Ther., 2010, 9, 2458–2467 CrossRef CAS.
- A. Gupte and R. J. Mumper, Cancer Treat. Rev., 2009, 35, 32–46 CrossRef CAS.
- A. Crowe, C. Jackaman, K. M. Beddoes, B. Ricciardo and D. J. Nelson, PLoS One, 2013, 8, e73684 CrossRef CAS.
- Y. L. Liu, C. L. Bager, N. Willumsen, D. Ramchandani, N. Kornhauser, L. Ling, M. Cobham, E. Andreopoulou, T. Cigler, A. Moore, D. LaPolla, V. Fitzpatrick, M. Ward, J. D. Warren, C. Fischbach, V. Mittal and L. T. Vahdat, npj Breast Cancer, 2021, 7, 108 CrossRef CAS.
- Q. Pan, C. G. Kleer, K. L. van Golen, J. Irani, K. M. Bottema, C. Bias, M. De Carvalho, E. A. Mesri, D. M. Robins, R. D. Dick, G. J. Brewer and S. D. Merajver, Cancer Res., 2002, 62, 4854–4859 CAS.
- S. Sammons, D. Brady, L. Vahdat and A. K. S. Salama, Melanoma Manag., 2016, 3, 207–216 CrossRef.
- S. Baldari, G. Di Rocco, M. C. Heffern, T. A. Su, C. J. Chang and G. Toietta, Cancers, 2019, 11, 659 CrossRef CAS.
- M. M. Xu, M. Casio, D. E. Range, J. A. Sosa and C. M. Counter, Clin. Cancer Res., 2018, 24, 4271–4281 CrossRef CAS.
- C. Cox, S. D. Merajver, S. Yoo, R. D. Dick, G. J. Brewer, J. S. J. Lee and T. N. Teknos, Arch. Otolaryngol., 2003, 129, 781–785 CrossRef.
- Z. Yu, R. T. Zhou, Y. C. Zhao, Y. Pan, H. Liang, J. S. Zhang, S. Tai, L. Jin and C. B. Teng, Cell Proliferation, 2019, 52, e12568 CrossRef.
- S. X. Qiao, C. M. Cabello, S. D. Lamore, J. L. Lesson and G. T. Wondrak, Apoptosis, 2012, 17, 1079–1094 CrossRef CAS.
- T. Mammoto, A. Jiang, E. Jiang, D. Panigrahy, M. W. Kieran and A. Mammoto, Am. J. Pathol., 2013, 183, 1293–1305 CrossRef CAS.
- C. C. Winterbourn and D. Metodiewa, Free Radical Biol. Med., 1999, 27, 322–328 CrossRef CAS.
- J. M. Walshe, Am. J. Med., 1956, 21, 487–495 CrossRef CAS.
- G. J. Brewer, C. A. Terry, A. M. Aisen and G. M. Hill, Arch. Neurol., 1987, 44, 490–493 CrossRef CAS.
- K. H. Weiss, F. Thurik, D. N. Gotthardt, M. Schafer, U. Teufel, F. Wiegand, U. Merle, D. Ferenci-Foerster, A. Maieron, R. Stauber, H. Zoller, H. H. Schmidt, U. Reuner, H. Hefter, J. M. Trocello, R. H. J. Houwen, P. Ferenci, W. Stremmel and E. Consortium, Clin. Gastroenterol. Hepatol., 2013, 11, 1028–1035 CrossRef CAS.
- J. M. Walshe, Lancet, 1982, 319, 643–647 CrossRef.
- M. Moriguchi, T. Nakajima, H. Kimura, T. Watanabe, H. Takashima, Y. Mitsumoto, T. Katagishi, T. Okanoue and K. Kagawa, Int. J. Cancer, 2002, 102, 445–452 CrossRef CAS.
- M. T. Hyvonen, S. Ucal, M. Pasanen, S. Peraniemi, J. Weisell, M. Khomutov, A. R. Khomutov, J. Vepsalainen, L. Alhonen and T. A. Keinanen, Biochem. J., 2016, 473, 1433–1441 CrossRef.
- R. Tremmel, P. Uhl, F. Helm, D. Wupperfeld, M. Sauter, W. Mier, W. Stremmel, G. Hofhaus and G. Fricker, Int. Res. J. Pharm., 2016, 512, 87–95 CAS.
- E. M. Poursani, D. Mercatelli, P. Raninga, J. L. Bell, F. Saletta, F. V. Kohane, D. P. Neumann, Y. Zheng, J. R. C. Rouaen, T. R. Jue, F. T. Michniewicz, P. Schadel, E. Kasiou, M. Tsoli, G. Cirillo, S. Waters, T. Shai-Hee, R. Cazzoli, M. Brettle, I. Slapetova, M. Kasherman, R. Whan, F. Souza-Fonseca-Guimaraes, L. Vahdat, D. Ziegler, J. G. Lock, F. M. Giorgi, K. Khanna and O. Vittorio, Cell Biosci., 2023, 13, 132 CrossRef CAS.
Footnotes |
| † Equal contribution. |
| ‡ Joint last authors. |
| This journal is © The Royal Society of Chemistry 2023 |
