Open Access Article
Open Access ArticleHeterologous expression of the cryptic mdk gene cluster and structural revision of maduralactomycin A†
Jan W. Schwitalla ab,
Ngoc-Thao-Hien Lec,
Soohyun Umd,
Felix Schalk
ab,
Ngoc-Thao-Hien Lec,
Soohyun Umd,
Felix Schalk a,
Mark Brönstrup
a,
Mark Brönstrup e,
Martin Baunachf and
Christine Beemelmanns
e,
Martin Baunachf and
Christine Beemelmanns *abg
*abg
aChemical Biology of Microbe-Host Interactions, Hans-Knöll Institute (HKI), Beutenbergstraße 11a, 07745 Jena, Germany
bHelmholtz Institute for Pharmaceutical Research Saarland (HIPS), Helmholtz Centre for Infection Research (HZI), Campus E8, 66123, Saarbrücken, Germany. E-mail: christine.beemelmanns@helmholtz-hips.de
cDepartment of Pharmaceutical Sciences, Natural Products and Food Research and Analysis (NatuRA), University of Antwerp, Universiteitsplein 1, B-2610 Antwerp, Belgium
dCollege of Pharmacy, Yonsei Institute of Pharmaceutical Sciences, Yonsei University, Incheon, 21983, South Korea
eDepartment of Chemical Biology, Helmholtz Centre for Infection Research, Inhoffenstrasse 7, D-38124 Braunschweig, Germany
fInstitute of Pharmaceutical Biology, University of Bonn, Nussallee 6, 53115 Bonn, Germany
gSaarland University, 66123, Saarbrücken, Germany
First published on 22nd November 2023
Abstract
After conducting an in silico analysis of the cryptic mdk cluster region and performing transcriptomic studies, an integrative Streptomyces BAC Vector containing the mdk gene sequence was constructed. The heterologous expression of the mdk cluster in Streptomyces albus J1074 resulted in the production of the angucyclic product, seongomycin, which allowed for the assesment of its antibacterial, antiproliferative, and antiviral activities. Heterologous production was further confirmed by targeted knock-out experiments involving key regulators of the biosynthetic pathways. We were further able to revise the core structure of maduralactomycin A, using a computational approach.
1. Introduction
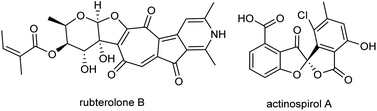
In recent years, actinobacteria associated with insects have garnered significant attention from natural product chemists worldwide.1 This is owing to their frequently remarkable biosynthetic capacity, and their role in host defense mechanisms through the secretion of bioactive compounds.2,3 By combining ecology-based natural product discovery techniques with analytical dereplication strategies, scientists have unveiled a multitude of structurally distinct and protective metabolites.4 As part of our ecology-guided natural product discovery efforts, the bacterial symbiont of fungus-growing termites, named Actinomadura rubteroloni (Actinomadura sp. RB29),5,6 attracted our attention due to its antagonistic effect against parasitic co-isolated fungi of Macrotermes natalensis. MS-guided chemical analyses led to the isolation of antifungal lanthipeptide rubrominins, the polyketide-derived rubterolones,7–10 and the halogenated angucyclic maduralactomycins and spirocyclic actinospirols (Fig. 1).11 In an initial genome-mining endeavour, a potential type II polyketide synthase (PKS) gene cluster region, designated as mdk and spanning approximately 36 kbp, was identified. Subsequent RT-PCR analysis revealed that transcription of the mdk biosynthetic gene cluster (BGC) occurred exclusively when A. rubteroloni was cultivated from spores in modified ISP5 medium; otherwise, it remained dormant.In this study, we tested if the proposed mdk BGC is indeed responsible for the production of angucyclic natural products by heterologous expression. Findings of this study enabled us to isolate the heterologously produced angucyclic product seongomycin and stimulated research on its biological origin. Results of our biosynthetic studies also stimulated the application of computational chemical shift prediction tools, and results of these studies allowed us to revise the planar structure of maduralactomycin A.
2. Experimental
2.1 Materials and methods
Cloning of the mdk gene cluster into the heterologous host was performed based on a conjugation protocol described by Kieser et al., 15 which was modified for tri-parental conjugation. Briefly, an overnight pre-culture of E. coli carrying the mdk containing pDualP vector and E. coli HB101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) prK2013 (carrying the helper plasmid for conjugational transfer) was inoculated and grown to an OD of approx. 0.4 at 30 °C. These cells were washed two times with LB media and cooled to 0 °C on ice. Next, SPIRO BGC containing E. coli were mixed with E. coli HB101
prK2013 (carrying the helper plasmid for conjugational transfer) was inoculated and grown to an OD of approx. 0.4 at 30 °C. These cells were washed two times with LB media and cooled to 0 °C on ice. Next, SPIRO BGC containing E. coli were mixed with E. coli HB101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) prK2013 in a ratio of 1
prK2013 in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Heat shocked spores from S. albus J1074 were added in a ratio of 1
1. Heat shocked spores from S. albus J1074 were added in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, inverted and the Streptomyces/E. coli mixture was plated on MS media with 10 mM MgCl2. After 16–20 h of growth at 30 °C, plates were overlaid first with 1 mL of an antibiotic solution containing spectinomycin (5 mg mL−1) and nalidixic acid (0.5 mg mL−1). For selection of strains carrying the modified plasmid, plates were also overlaid with 1 mL of an antibiotic solution containing apramycin (1.25 mg mL−1) and nalidixic acid (0.5 mg mL−1). After one week of growth exconjugants were selected and grown on MS-agar (20 g per L agar, 20 g per L mannitol, 20 g per L soy flour, supplemented with apramycin (50 μg mL−1) and nalidixic acid (25 μg mL−1)). Positive clones were identified and verified by gene amplification and sequencing (Table S3 and Fig. S5–S8†).
1, inverted and the Streptomyces/E. coli mixture was plated on MS media with 10 mM MgCl2. After 16–20 h of growth at 30 °C, plates were overlaid first with 1 mL of an antibiotic solution containing spectinomycin (5 mg mL−1) and nalidixic acid (0.5 mg mL−1). For selection of strains carrying the modified plasmid, plates were also overlaid with 1 mL of an antibiotic solution containing apramycin (1.25 mg mL−1) and nalidixic acid (0.5 mg mL−1). After one week of growth exconjugants were selected and grown on MS-agar (20 g per L agar, 20 g per L mannitol, 20 g per L soy flour, supplemented with apramycin (50 μg mL−1) and nalidixic acid (25 μg mL−1)). Positive clones were identified and verified by gene amplification and sequencing (Table S3 and Fig. S5–S8†).
LC-ESI-HRMS based metabolomics was performed on a Dionex Ultimate3000 system equipped with a Luna Omega C18 column (100 × 2.1 mm, particle size 1.6 μm, pore diameter 100 Å, Phenomenex) coupled with a Q-Exactive Plus mass spectrometer (Thermo Scientific) equipped with an electrospray ion (HESI) source. The column oven temperature was set to 40 °C; scan range of full MS was set to m/z 180 to 2000 with a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and AGC target 3 × 106 and maximum IT 100 ms under positive mode with centroid data type. MS2 analysis was performed as top10 experiments (positive mode) with a resolution of 17
000 and AGC target 3 × 106 and maximum IT 100 ms under positive mode with centroid data type. MS2 analysis was performed as top10 experiments (positive mode) with a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 and AGC target 1 × 105 and maximum IT 50 ms and a normalized collision energy (NCE) of 28. The spray voltage (+) was set to 4000 volt, and (−) was set to 3300 volt. The capillary temperature (±) was set to 340 °C and probe heater temperature (±) was set to 200 °C. The sheath gas flow (±) was set to 35 L min−1 and Aux gas flow (±) to 5 L min−1. Max spray current (+) and (−) was set to 100 volt. S-Lens RF level was set to 50.
500 and AGC target 1 × 105 and maximum IT 50 ms and a normalized collision energy (NCE) of 28. The spray voltage (+) was set to 4000 volt, and (−) was set to 3300 volt. The capillary temperature (±) was set to 340 °C and probe heater temperature (±) was set to 200 °C. The sheath gas flow (±) was set to 35 L min−1 and Aux gas flow (±) to 5 L min−1. Max spray current (+) and (−) was set to 100 volt. S-Lens RF level was set to 50.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded with 0.1 mL culture medium per well of the 96-well microplates. To test the antiproliferative effect of natural products, the cells were incubated for 72 hours in plates prepared with control and different dilutions of test substances. The GI50 values were defined as being where the inhibition of proliferation is 50% compared to untreated control (Tables S23–S25 and Fig. S35–S36†).
000 cells were seeded with 0.1 mL culture medium per well of the 96-well microplates. To test the antiproliferative effect of natural products, the cells were incubated for 72 hours in plates prepared with control and different dilutions of test substances. The GI50 values were defined as being where the inhibition of proliferation is 50% compared to untreated control (Tables S23–S25 and Fig. S35–S36†).3. Results and discussion
3.1 Gene cluster analysis
We first reanalysed the gene cluster region mdk, which contains about 21 genes including the core PKS-encoding genes (mdkA, mdkB, mdkC, mdkD, mdkE, mdkI), five genes coding for oxidative enzymes (mdkG, mdkOR, mdkO1, mdkO2, mdkO3), and one halogenase (mdkH) (Fig. 2 and S1†), which were flanked by at least three regulatory genes (mdkR, mdkR1, mdkR2).11 While mdkR was annotated as TetR/AcrR-type regulator and is presumably involved in the regulation of antibiotic resistance,21,22 mdkR1 is likely a member of the MerR family of regulators, known for similar functions. On the other hand, mdkR2 belongs to the OmpR family of response regulators, featuring a conserved receiver domain and a DNA binding motif. Prior PCR-based studies indicated that two regulators, mdkR1 and mdkR2,23 control the gene expression of the cluster.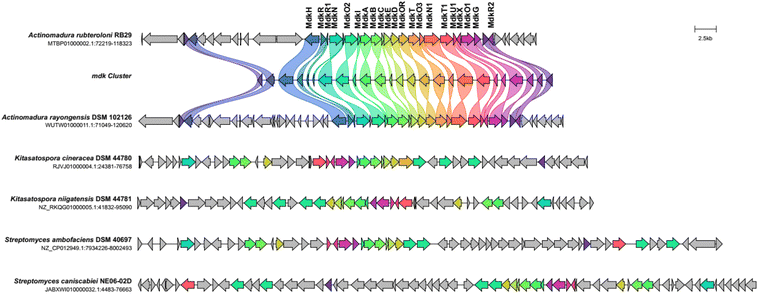 | ||
| Fig. 2 Cblaster analysis using the mdk gene cluster sequence as query sequence. Five dene clusters with similar co-located gene arrangement and identity (threshold of 30% identity) are depicted and homologous gene sequences are shown in the same color code (Fig. S1†). | ||
A functional and evolutionary analysis of the mdk cluster sequence by a combination of cblaster13 and clinker14 tool, showed that the BGC was nearly identically organized to a cluster encoded within the genome of the closely related strain A. rayongensis DSM 102126.5 In contrast, the alp cluster in Streptomyces ambofaciens,23 which encodes the biosynthesis of the structurally very closely related angucycline kinamycin, contained only a smaller core set of similar genes, and an only partial similar gene organization (Fig. 2 and S1†). Gene clusters coding for the biosynthesis of structurally related angucycline natural products, such as jadomycin (jad),24,25 landomycin (lan),26 and simocyclinone (sim) respectively,27 exhibited only moderate overall similarities and different gene organizations (Fig. S4†).
Intriguingly, all analysed angucycline clusters encoded a similar domain organization of the encoded PKS, several monooxygenases and a set of regulators and transporters. However, only mdk encoded the halogenase mdkH, which was otherwise absent in related clusters. A phylogenetic analysis of the chain length factor (MdkB) and the keto synthase (MdkA) of mdk using the Natural product domain seeker 228 revealed that both share a close phylogenetic relationship to the ketosynthase of fluostatin biosynthesis (fls/fluo)29 and the chain length factor of azicemycin biosynthesis (azicB),30 respectively, both of which belong to the compound family angucyclines (Fig. S2–S4†).
We also noted that only one other, distantly related cluster encoded two (mdkR and mdkR1) of the three mdk regulators. Several of the mdk-related BGCs, in particular those of Streptomyces origin, lacked sequences related to mdkR and mdkR1,31 and harboured instead an ORF coding for an AfsA homologue, a protein essential for the biosynthesis of the auto regulatory γ-butyrolactones. Homologous sequences of mdkR2 were only found in every distantly related cluster, such as chaI, which encodes a positive regulator for angucycline production in Streptomyces chattanoogensis.32 Overall, the differences in the encoded regulatory elements pinpointed towards a species-specific transcriptional regulation of the pathways causing us to hypothesize that the biosynthesized metabolites could serve as species-specific signals fulfilling external or internal signalling functions.
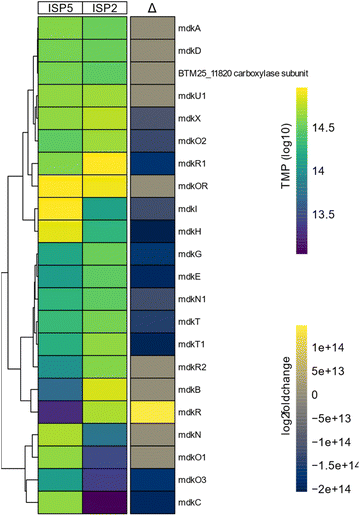
To test more broadly the effect of the inducing cultivation conditions, we performed a differential expression analysis of the mdk gene cluster. For this, A. rubteroloni was first cultivated for seven days under “producing” (ISP5*) and “non-producing” (ISP2) conditions (Tables S1 and S2†), and RNA was isolated from vegetative mycelium.11 Differential expression analysis uncovered that transcript levels of mdkC, mdkE, mdkI (PKS) as well as mdkG (oxidase), mdkO2 (FAD-depended monooxygenase), and mdkH (halogenase) were higher under producing conditions compared to the levels detected for non-producing conditions (Fig. 3).
Transcripts related to precursor supply and regeneration of cofactors (mdkN (acyl-CoA carboxylase), mdkO3 (NADH-dependent reductase))33 were likewise differentially expressed. Furthermore, transcript levels of genes encoding two putative transporter proteins (mdkT and mdkT1) and the elusive mdkX protein (37% similarity to jadX, up to 60% to other jadX-like genes) were increased.34
3.2 Heterologous expression
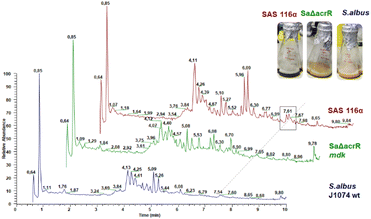
To verify that the mdk cluster is indeed responsible for the biosynthesis of angucycline natural products, the mdk region (see Fig. 2) was obtained from genomic DNA using a CRISPR-Cas based excision system, and then cloned into an integrative Streptomyces BAC (pDualp) vector. The pDualp vector carried two inducible promoters, PnitA35 and Potr,36 which allowed induction of transcription in both reading frames in the presence of either ε-caprolactam or oxytetracycline. The vector containing the mdk gene cluster was then transferred into Streptomyces albus J1074, Streptomyces coelicolor M1146 and Streptomyces lividans TK24 as heterologous hosts by intergeneric conjugation, which was verified via PCR (Fig. S5–S8†). Subsequently, the cluster was heterologously expressed in all three strains (S. albus J1074/SAS116 α, S. coelicolor M1146/1.8PI; S. lividans TK24/1.32PISL) and the induced metabolome analyzed. The metabolome of cultures supernatants retrieved from unmodified strains S. albus J1074, S. coelicolor M1146 and S. lividans TK24 served as control samples. As the presence of the inducer oxytetracycline caused reduced biomass formation of S. albus J1074 and the modified strain SAS116α, both strains were also cultivated in DNPM broth containing only ε-caprolactam. For metabolomic analysis, supernatants of induced cultures were concentrated using a standardized solid-phase extraction (SPE) procedure and subsequently analyzed by liquid-chromatography coupled with tandem mass spectrometry (LC-HRMS/MS). Comparative analysis of culture extracts showed that modified strains (J1074, 1.8PI, 1.32PISL) exhibited an upregulation of known host-specific metabolites in all experiments.Only strain S. albus J1074/SAS116 α37 was found to produce one additional ε-caprolactam-induced metabolite compared to the controls, which correlated to a distinct UV peak at 7.61 min (λmax at 285 and 511 nm, [M + H]+ = 454.0947) and thus was sought to be likely responsible for the greenish color of the culture (Fig. 4). We also performed an extensive targeted metabolomic survey of m/z features that correlated to [M + H]+ values of maduralactomycins/actinospirols and derivatives thereof; however, neither the exact m/z values nor related molecular ion peaks were detected (Fig. S9–S19†).
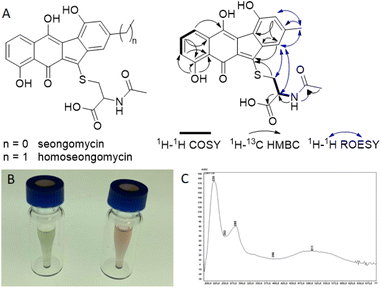
To test if regulators MdkR and MdkR1 are involved in the transcriptional regulation of the mdk pathway, we employed a knock-out strategy using the λ red system, in which a resistance marker replaces the respective gene via homologous recombination in E. coli.38 Using this technique, mdkR and mdkR1 were replaced with a spectinomycin resistance cassette yielding the plasmid ΔacrR. The edited plasmid was then transferred to the heterologous host S. albus J1074/SAS116 α via conjugation yielding strain SaΔacrR (Fig. S5–S8†). Subsequent heterologous expression experiments were performed using the mutated S. albus J1074/SaΔacrR, strain SAS116α carrying the unedited plasmid, and the wild type S. albus J1074 (control). Metabolites of culture supernatants were enriched by SPE extraction and subjected to LC-MS/MS-analysis. Manual targeted searches and network-based analysis of MS-datasets for mdk-related angucyclic metabolite products (Fig. S15–S19 and Tables S6–S8†) revealed several shared secondary metabolite related clusters, including the host-specific natural product family surugamides.39 Only one molecular ion cluster was unique to S. albus J1074/SAS116 α, which contained three connected nodes with related molecular ions features (m/z 454.095, 452.079, 468.074), and were otherwise absent in culture extracts of the deletion mutant strain SaΔacrR and S. albus J1074. To elucidate the structure of the induced metabolite, strain SAS 116α was cultivated and metabolites of the culture supernatant enriched using our standard C18-based SPE protocol. The resulting 10% and 30% MeOH/H2O eluents were subjected to a UV-Vis- and MS-guided protocol using semi-preparative HPLC, yielding 9.24 mg of the major product (m/z 454.095, approx. production titer: 23 mg L−1) together with minor amounts of an analog (m/z = 468.074, m/z = 14 Da) (Fig. S20 and S21†). During the work-up, we also noted a pH-depended UV-absorption related to the produced compound, which was visible in a reversible color shift of the product in solution and solid state. Comparative 1D and 2D NMR analysis of the major isolated compound ([M + H]+ = 454.0947) uncovered matching 1H and 13C chemical shifts (d6-DMSO + 1% d1-TFA) with the previously reported angucyclic metabolite seongomycin (Fig. 5, S22–S32 and Table S9†) and those obtained by computational NMR analysis (Table S10†). The minor metabolite was identified as homoseongomycin based on matching HRMS/MS data and comparison of chemical shift values (Fig. S22 and S23).11,40
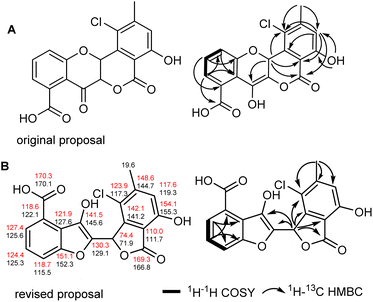
3.3 Structural revision of maduralactomycin A
The formation of seongomycin, which shows a different regiochemical orientation of the substituents in ring A compared to maduralactomycin A (Fig. 6A), stimulated us to revisit the analytical datasets of the previously isolated maduralactomycin A and the proposed biosynthetic pathway, which remained ambiguous despite extensive 13C-labeling and computation efforts (Table S11†).Using of a combination of geometry optimization of determined conformers based on B3LYP/6-31+G(d,p) level of theory in the gas-phase, we re-calculated the 13C and 1H chemical shifts values of twenty-six regioisomers (Fig. S33 and Tables S12–S17†). Shielding tensors were then computed at mPW1PW91/6-311+G(2d,p) level of theory with polarizable continuum model (PCM).41 Extensive comparative NMR analysis allowed us to identify two chemical structures, which showed the best matching chemical shift pattern and correlations of which only one fitted the observed 13C-labling pattern, (Fig. S34 and Tables S19–S21†). Hence, we propose here a revised structure of maduralactomycin A (Fig. 6B), which now is composed of two benzofuranone-like moieties interconnected by C–C bond and carries expected actinospirol-like substitution pattern (methylation, chlorination).
3.4 Biosynthetic reconsiderations
The successful expression of the mdk cluster in S. albus J1074 as heterologous host confirmed the angucyclic nature of the mdk cluster products. The angucyclic product seongomycin contains a ring system akin to the modified structure of maduralactomycin A, featuring a contracted 6,5,5,6 ring arrangement. However, a notable distinction lies in the linkage of the two five-membered ring systems, attributed to the presence of an extra bridging carbon atom (presumably C-14). Further structural disparities encompass the changed position of the methyl and hydroxyl groups within the A-ring and the absence of the halogenation pattern. Consequently, we postulate that seongomycin is likely a by- or shuntproduct of the heterologously expressed mdk pathway. To trace the origin of seongomycin, we then re-analyzed the biosynthetic proposal (Fig. 7). The initial step of the biosynthesis includes a typical type II polyketide pathway, in which the synthase utilizes an acetyl-CoA starter unit and malonyl-CoA extender units to generate a highly reactive acyl-carrier protein (ACP)-bound poly-β-ketone chain (I, Fig. 7). The next biosynthetic step in the biosynthesis involves the didomain polyketide cyclase MdkD, which should initiate the cyclization cascade of poly-β-ketone chain to yield the aromatized intermediate II.42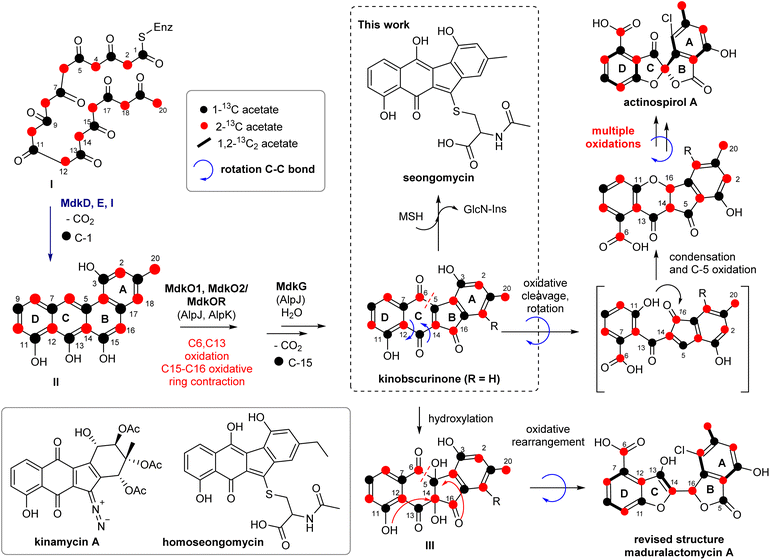 | ||
| Fig. 7 Reconsideration of putative key transformations involved in the biosynthesis of (A) seongomycin and (B) structurally revised maduralactomycin A based on prior 13C-labeling and gene cluster analysis (Fig. S33 and Tables S18–S20†). | ||
The sequence of oxygenation, oxidative ring cleavage and ring contraction, which is accompanied by the loss of C-1 and C-15, is likely catalyzed by the ring opening oxygenase MdkG (AlpJ)43 MdkO1, MdkO2 and MdkOR should yield the precursor kinobscurinone as previously reported. Chlorination is proposed to be catalyzed by MdkH at an early stage of the biosynthesis as only one position in both natural products was regioselectively halogenated. As neither chlorination of seongomycin nor putative biosynthetic precursors were observed within this study, we hypothesize that MdkH is either highly substrate specific or dysfunctional due to the heterologous expression in S. albus J1074 SAS 116α.
The formation of seongomycin, however, also highlights the limitation of S. albus as heterologous host as the AcCys S-conjugate likely represents a mycothiol-based detoxification product of kinobscurinone. The formation of seongomycin can be reasoned as follows: (1) nucleophilic addition of mycothiol (MSH), a protective agent found in Gram-positive Streptomycetes that safeguards cells against toxic and reactive electrophiles, to the intermediate kinobscurinone; (2) subsequent water elimination, double bond isomerization, and a reduction; and (3) cleavage of the MSH-conjugate by a specific amidase, resulting in the release of GlcN-Ins (1-D-myo-inosytyl 2-amido-2-deoxy-α-D-glucopyranoside) and the AcCys S-adduct.44,45 Hence, it can be inferred that when aiming for the heterologous production of natural products formed from reactive intermediates susceptible to 1,4-Michael additions or acetal formation, heterologous hosts known for generating elevated quantities of MSH should be avoided.
3.5 Bioactivities
We also evaluated the pharmacological profile of seongomycin more in detail and performed antibacterial, antifungal, antiviral and cytological tests (Tables S22–S25 and Fig. S34–S37†). However, neither antimicrobial activity nor an antiproliferative effect on the HeLa cell lines was detectable. Although a recent antiviral assay of homoseongomycin, a close derivative of seongomycin, revealed activity of this compound against certain species of alpha viruses,46 seongomycin showed neither antiviral effects against the coronaviruses (SARS-CoV-2, HCoV-229E), Respiratory Syncytial Virus (RSV) nor the human pathogenic alpha virus Chikungunya-Virus (CHIKV).4. Conclusions
In this study, we successfully expressed the mdk cluster in S. albus J1074/SAS116 α, leading to the biosynthesis of the angucyclic product seongomycin. Notably, heterologous production was effectively abolished upon the knockout of two pivotal regulators within the pathways. Additionally, we accomplished a revision of the core structure of maduralactomycin through a computational approach combined with 2D NMR analysis. The work presented herein establishes the chemical and molecular biological groundwork for systematic investigations of the mdk pathway across diverse host systems. It furthermore enables pathway manipulation to identify key biosynthetic intermediates and unravel the nature of the distinctive oxidative biosynthetic rearrangement steps.Author contributions
JWS contributed to conceptualization, data curation, writing – original draft, review and editing. HL, SU, FS contributed by formal analysis, investigation, validation and visualization as well as formal analysis. MBrö, MBau, CB contributed with conceptualization, formal analysis, funding acquisition, supervision, validation, visualization, writing – original draft, review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was also funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Project-ID 239748522 – CRC 1127. This work was also supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT; grant number 2021R1I1A1A0104960613). The work was funded by a grant from the COVID-19 Research Network of the State of Lower Saxony (COFONI) (14-76403-184). We thank Mr Dahse (HKI), Chen-You Ting (HZI), Frederick Mbui (HZI), Ursula Bilitewski (HZI), Benedikt Hellwinkel (Twincore), Natalie Koehler (Twincore), and Thomas Pietschmann (Twincore) for initial biological assays.Notes and references
- E. B. Van Arnam, C. R. Currie and J. Clardy, Chem. Soc. Rev., 2018, 47, 1638–1651 RSC.
- M. Miethke, M. Pieroni, T. Weber, M. Brönstrup, P. Hammann, L. Halby, P. B. Arimondo, P. Glaser, B. Aigle, H. B. Bode, R. Moreira, Y. Li, A. Luzhetskyy, M. H. Medema, J. L. Pernodet, M. Stadler, J. R. Tormo, O. Genilloud, A. W. Truman, K. J. Weissman, E. Takano, S. Sabatini, E. Stegmann, H. Brötz-Oesterhelt, W. Wohlleben, M. Seemann, M. Empting, A. K. H. Hirsch, B. Loretz, C. M. Lehr, A. Titz, J. Herrmann, T. Jaeger, S. Alt, T. Hesterkamp, M. Winterhalter, A. Schiefer, K. Pfarr, A. Hoerauf, H. Graz, M. Graz, M. Lindvall, S. Ramurthy, A. Karlén, M. van Dongen, H. Petkovic, A. Keller, F. Peyrane, S. Donadio, L. Fraisse, L. J. V. Piddock, I. H. Gilbert, H. E. Moser and R. Müller, Nat. Rev. Chem, 2021, 5, 726–749 CrossRef CAS PubMed.
- D. A. van Bergeijk, B. R. Terlouw, M. H. Medema and G. P. van Wezel, Nat. Rev. Microbiol., 2020, 18, 546–558 CrossRef CAS PubMed.
- M. J. Smanski, D. C. Schlatter and L. L. Kinkel, J. Ind. Microbiol. Biotechnol., 2016, 43, 115–128 CrossRef CAS PubMed.
- R. Benndorf, K. Martin, M. Küfner, Z. W. de Beer, J. Vollmers, A.-K. Kaster and C. Beemelmanns, Int. J. Syst. Evol. Microbiol., 2020, 70, 5255–5262 CrossRef CAS PubMed.
- R. Benndorf, H. Guo, E. Sommerwerk, C. Weigel, M. Garcia-Altares, K. Martin, H. Hu, M. Kuefner, Z. W. de Beer, M. Poulsen and C. Beemelmanns, Antibiotics, 2018, 7, E83 CrossRef PubMed.
- H. Guo, R. Benndorf, D. Leichnitz, J. K. Klassen, J. Vollmers, H. Görls, M. Steinacker, C. Weigel, H. M. Dahse, A. K. Kaster, Z. W. de Beer, M. Poulsen and C. Beemelmanns, Chem. – Eur. J., 2017, 23, 9338–9345 CrossRef CAS PubMed.
- H. Guo, R. Benndorf, S. König, D. Leichnitz, C. Weigel, G. Peschel, P. Berthel, M. Kaiser, C. Steinbeck, O. Werz, M. Poulsen and C. Beemelmanns, Chem. – Eur. J., 2018, 24, 11319–11324 CrossRef CAS PubMed.
- Y. Yan, J. Yang, Z. Yu, M. Yu, Y. Ma, L. Wang, C. Su, J. Luo, G. P. Horsman and S. Huang, Nat. Commun., 2016, 7, 13083 CrossRef CAS PubMed.
- X. Cai, Y.-M. Shi, N. Pöhlmann, O. Revermann, I. Bahner, S. J. Pidot, F. Wesche, H. Lackner, C. Büchel, M. Kaiser, C. Richter, H. Schwalbe, T. P. Stinear, A. Zeeck and H. B. Bode, Angew. Chem., Int. Ed., 2017, 56, 4945 CrossRef CAS PubMed.
- H. Guo, J. W. Schwitalla, R. Benndorf, M. Baunach, C. Steinbeck, H. Görls, Z. W. de Beer, L. Regestein and C. Beemelmanns, Org. Lett., 2020, 22, 2634–2638 CrossRef CAS PubMed.
- K. Blin, S. Shaw, A. M. Kloosterman, Z. Charlop-Powers, G. P. van Weezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, W29–W35 CrossRef CAS PubMed.
- C. L. M. Gilchrist, T. J. Booth, B. van Wersch, L. van Grieken, M. H. Medema and Y.-H. Chooi, Bioinf. Adv., 2021, 1, vbab016 Search PubMed.
- C. L. M. Gilchrist and Y.-H. Chooi, Bioinformatics, 2021, 37, 2473–2475 CrossRef CAS PubMed.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Practical Streptomyces genetics, John Innes Foundation Norwich, 2000, vol. 291 Search PubMed.
- B. Gust, T. Kieser and K. Chater, PCR targeting system in Streptomyces coelicolor A3 (2), John Innes Centre, 2002, vol. 3, 2, pp. 1–39 Search PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W. T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C. C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C. C. Liaw, Y. L. Yang, H. U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. P. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P. M. Allard, P. Phapale, L. F. Nothias, T. Alexandrov, M. Litaudon, J. L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D. T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- D. Petras, V. V. Phelan, D. Acharya, A. E. Allen, A. T. Aron, N. Bandeira, B. P. Bowen, D. Belle-Oudry, S. Boecker, D. A. Jr Cummings, J. M. Deutsch, E. Fahy, N. Garg, R. Gregor, J. Handelsman, M. Navarro-Hoyos, A. K. Jarmusch, S. A. Jarmusch, K. Louie, K. N. Maloney, M. T. Marty, M. M. Meijler, I. Mizrahi, R. L. Neve, T. R. Northen, C. Molina-Santiago, M. Panitchpakdi, B. Pullman, A. W. Puri, R. Schmid, S. Subramaniam, M. Thukral, F. Vasquez-Castro, P. C. Dorrestein and M. Wang, Nat. Methods, 2022, 19, 134–136 CrossRef CAS PubMed.
- N. T. H. Le, T. Vermeyen, R. Aerts, W. A. Herrebout, L. Pieters and E. Tuenter, Molecules, 2023, 28, 214 CrossRef CAS PubMed.
- N. T. H. Le, E. Van Roy, E. Dendooven, L. Peeters, M. Theunis, K. Foubert, L. Pieters and E. Tuenter, Phytochemistry, 2021, 190, 112863 CrossRef PubMed.
- I. Ostash, B. Ostash, A. Luzhetskyy, A. Bechthold, S. Walker and V. Fedorenko, FEMS Microbiol. Lett., 2008, 285, 195–202 CrossRef CAS PubMed.
- L. Cuthbertson and J. R. Nodwell, Microbiol. Mol. Biol. Rev., 2013, 77, 440–475 CrossRef CAS PubMed.
- R. Bunet, L. Song, M. V. Mendes, C. Corre, L. Hotel, N. Rouhier, X. Framboisier, P. Leblond, G. L. Challis and B. Aigle, J. Bacteriol., 2011, 193, 1142–1153 CrossRef CAS PubMed.
- K. Kulowski, E. W. Pienkowski, L. Han, K. Q. Yang, L. C. Vining and C. R. Hutchinson, J. Am. Chem. Soc., 1999, 121, 1786–1794 CrossRef CAS.
- Y. H. Chen, C. C. Wang, L. Greenwell, U. Rix, D. Hoffmeister, L. C. Vining, J. Rohr and K. Q. Yang, J. Biol. Chem., 2005, 280, 22508–22514 CrossRef CAS PubMed.
- P. Patrikainen, P. Kallio, K. Fan, K. D. Klika, K. A. Shaaban, P. Mäntsälä, J. Rohr, K. Yang, J. Niemi and M. Metsä-Ketelä, Chem. Biol., 2012, 19, 647–655 CrossRef CAS PubMed.
- A. Trefzer, S. Pelzer, J. Schimana, S. Stockert, C. Bihlmaier, H. P. Fiedler, K. Welzel, A. Vente and A. Bechthold, Antimicrob. Agents Chemother., 2002, 46, 1174–1182 CrossRef CAS PubMed.
- L. J. Klau, S. Podell, K. E. Creamer, A. M. Demko, H. W. Singh, E. E. Allen, B. S. Moore, N. Ziemert, A. C. Letzel and P. R. Jensen, J. Biol. Chem., 2022, 298, 102480 CrossRef CAS PubMed.
- Z. Feng, J. H. Kim and S. F. Brady, J. Am. Chem. Soc., 2010, 132, 11902–11903 CrossRef CAS PubMed.
- Y. Ogasawara and H.-W. Liu, J. Am. Chem. Soc., 2009, 131, 18066–18068 CrossRef CAS PubMed.
- J. y. Kato, N. Funa, H. Watanabe, Y. Ohnishi and S. Horinouchi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2378–2383 CrossRef CAS PubMed.
- Z. Zhou, Q. Xu, Q. Bu, Y. Guo, S. Liu, Y. Liu, Y. Du and Y. Li, ChemBioChem, 2015, 16, 496–502 CrossRef CAS PubMed.
- V. Blanc, D. Lagneaux, P. Didier, P. Gil, P. Lacroix and J. Crouzet, J. Bacteriol., 1995, 177, 5206–5214 CrossRef CAS PubMed.
- A. W. Robertson, S. M. Forget, C. F. Martinez-Farina, N. E. McCormick, R. T. Syvitski and D. L. Jakeman, J. Am. Chem. Soc., 2016, 138, 2200–2208 CrossRef CAS PubMed.
- M. Matsumoto, Y. Hashimoto, Y. Saitoh, T. Kumano and M. Kobayashi, Biosci., Biotechnol., Biochem., 2016, 80, 1230–1237 CrossRef CAS PubMed.
- W. Wang, T. Yang, Y. Li, S. Li, S. Yin, K. Styles, C. Corre and K. Yang, ACS Synth. Biol., 2016, 5, 765–773 CrossRef CAS PubMed.
- M. Myronovskyi and A. Luzhetskyy, Nat. Prod. Rep., 2019, 36, 1281–1294 RSC.
- B. Gust, G. L. Challis, K. Fowler, T. Kieser and K. F. Chater, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 1541–1546 CrossRef CAS PubMed.
- F. Xu, B. Nazari, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2017, 139, 9203–9212 CrossRef CAS PubMed.
- C. M. Woo, S. L. Gholap and S. B. Herzon, J. Nat. Prod., 2013, 76, 1238–1241 CrossRef CAS PubMed.
- H. T. N. Le, E. Van Roy, E. Dendooven, L. Peeters, M. Theunis, K. Foubert, L. Pieters and E. Tuenter, Phytochemistry, 2021, 190, 112863 CrossRef CAS PubMed.
- S.-C. Tsai, Annu. Rev. Biochem., 2018, 87, 503–531 CrossRef CAS PubMed.
- S. Lösgen, Ö. Schlörke, K. Meindl, R. Herbst-Irmer and A. Zeeck, Eur. J. Org Chem., 2007, 13, 2191–2196 CrossRef.
- G. Newton and R. Fahey, Arch. Microbiol., 2002, 178, 388–394 CrossRef CAS PubMed.
- V. K. Jothivasan and C. J. Hamilton, Nat. Prod. Rep., 2008, 25, 1091–1117 RSC.
- S. C. Lin, C. W. Lehman, A. K. Stewart, L. Panny, N. Bracci, J. L. C. Wright, M. Paige, W. K. Strangman and K. Kehn-Hall, Antiviral Res., 2021, 191, 105087 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The datasets supporting this article have been uploaded as part of the ESI and contains details for chemical procedures, 1D and 2D NMR of described compounds, as well as HRMS data and bioassay data. See DOI: https://doi.org/10.1039/d3ra05931f |
| This journal is © The Royal Society of Chemistry 2023 |