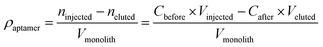Open Access Article
Open Access ArticleOnline highly selective recognition of domoic acid by an aptamer@MOFs affinity monolithic column coupled with HPLC for shellfish safety monitoring†
Fang Songa,
Zhexiang Zhangb,
Xuerong Xu*a and
Xucong Lin *b
*b
aCollege of Economics and Management, Fujian Agriculture and Forestry University, Fuzhou 350001, China. E-mail: xxrfafu@163.com
bEngineering Technology Research Center on Reagent and Instrument for Rapid Detection of Product Quality and Food Safety in Fujian Province, Fuzhou University, Fuzhou, 350108, China. E-mail: linxucong@aliyun.com
First published on 20th October 2023
Abstract
Enabling cost-effective safety monitoring of shellfish is an important measure for the healthy development of the coastal marine economy. Herein, a new aptamer@metal–organic framework (MOF)-functionalized affinity monolithic column was proposed and applied in selective in-tube solid-phase microextraction (IT-SPME) coupled with HPLC for the accurate recognition of domoic acid (DA) in shellfish. Using a surface engineering strategy, ZIF-8 MOF was grown in situ inside the poly(epoxy-MA-co-POSS-MA) hybrid monolith. A high BET surface area and abundant metal reactive sites of the MOF framework were obtained for anchoring massive aptamers with terminal-modified phosphate groups. Various characterizations, such as SEM, elemental mapping, XRD, and BET, were performed, and the affinity performance was also studied. The presence of a massive amount of aptamers with a super coverage density of 3140 μmol L−1 bound on ZIF-8 MOF activated a high-performance bionic-affinity interface, and perfect specificity was exhibited with little interference of tissue matrixes, thus assuring the highly selective capture of DA from the complex matrixes. Under the optimal conditions, DA toxins in shellfish were detected with the limit of detection (LOD) of 7.0 ng mL−1 (equivalent to 14.0 μg kg−1), representing a 5–28 fold enhancement in detection sensitivity over traditional SPE or MIP adsorbents reported previously. The recoveries of fortified mussel and clam samples were achieved as 91.8 ± 1.2%–94.1 ± 1.9% (n = 3) and 91.2 ± 1.1%–94.5 ± 3.6% (n = 3), respectively. This work sheds light on a cost-effective method for online selective IT-SPME and the accurate monitoring of DA toxins using an aptamer@MOF-mediated affinity monolith system coupled with the inexpensive HPLC-UV technique.
1 Introduction
Over the last decade, harmful algal blooms (HABs), including toxic microalgae, have been reported and have increased remarkably. Domoic acid (DA), a typical marine neurotoxin,1,2 can accumulate in bivalve tissues and cause serious amnesic shellfish poisoning (ASP) in human beings.3 Widespread DA toxic incidents have occurred globally. The maximum residue limit (MRL) of DA in bivalve mollusks is 20 mg kg−1 DA as stipulated by the European Union Regulation (EC) No 853/2004.4 The daily inspection and monitoring of DA has great significance in strengthening the risk control of shellfish.Currently, HPLC-UV detection with automatic sampling units has been developed and widely used for quantifying the DA toxin.5 Due to the low content of DA in natural shellfish with complicated matrixes, it is hard to directly achieve the precise quantitation of DA by common HPLC-UV methods.6,7 Pretreatment is necessary, and therefore, developing an efficient recognition strategy that can be online integrated with HPLC for the automatic analysis of DA with good selectivity and sensitivity would be significant. Some common methods such as the traditional liquid–liquid extraction (LLE)8,9 and solid-phase extraction (SPE)10 have been reported for the DA extraction. As the most popular extraction techniques, SPE phases, such as ion-exchange resins,11 amorphous titania,12 molecularly imprinted polymer (MIP),13,14 or C18 reversed-phase polymer,15 have been demonstrated for the purification of DA in shellfish. Magnetic solid-phase extraction (MSPE) was also developed for the isolation of DA using UiO-66 modified magnetite@silica microspheres as the sorbents.16 Though the traditional SPE and MSPE techniques are currently widely adopted and exhibit effective extraction performance, there still are problems to be resolved, such as high solvent consumption, high particle dispersity, and unsatisfactory selectivity.6,8,17 Further, these methods are usually used in an offline manual way and hardly online coupled with LC for automatic analysis,6,18 resulting in a number of issues, such as non-automated operation, multiple steps, analyte loss, long-time operation periods, and low efficiency. Improvements that have attached attention including attempts to automate the techniques, since SPE and MSPE processes generally require offline procedures (loading, washing, elution) and do not seem to be particularly able to cope with batch samples. In particular, the selectivity in traditional SPE and MSPE methods toward the target DA toxin is limited, and certain analogues could be co-adsorbed, resulting in a non-negligible unspecific adsorption and obvious background interference.
As for the other issues, in-tube solid-phase microextraction (IT-SPME), as an emerging extraction protocol, has been proposed and even applied as a powerful and reliable tool for online sample pretreatment, which integrates SPME with HPLC for automatic analysis.19,20 This can effectively reduce most the problems that occur in the offline mode, while the system also exhibits other merits, such as options for miniaturization, automation, and green characteristics.21 Using a functional adsorbent, selective IT-SPME methods have been proposed as an ideal online sample preparation technique, opening up effective access to the automatic identification of a target analyte.22 Recently, using a MIP monolith, a specific recognition strategy of domoic acid was explored to accomplish selective IT-SPME.23 Various advantages, such as skilled preparation, physicochemical robustness, and reusability for DA recognition, were achieved; however, it also suffers from some issues, such as the inevitable template bleeding, tedious elution process, high capital expenditure, and low conversion efficiency. Consequently, there is still a requirement to further develop a cost-effective and specific means for the online recognition of DA in shellfish.
Aptamers, as inexpensive nucleic acid molecules with a flexible structure, and good biocompatibility, which can fold into 3D shapes and bind to specific targets, possess high stability and selectivity to target molecules and have been developed for the special analysis of target analytes.23,24 As a new bionic material, aptamer-based affinity monolithic columns with various unique merits, such as high tolerance, fast mass transfer, good affinity performance, and convenient coupling with instruments,25 have been widely designed as a promising approach with a “molecular recognition” mechanism for selective IT-SPME for the online specific recognition of various analytes.26 Indeed, various methacrylate-based organic polymer monoliths and polyhedral oligomeric silsesquioxane (POSS)-based hybrid affinity monoliths modified with aptamers have been reported and used for the IT-SMPE of trace contaminants (e.g., mycotoxins, BPA, MC-LR).27–29 Using gold nanoparticles (AuNPs) as a medium, aptamer@AuNPs-functionalized monoliths with a high Brunauer–Emmett–Teller (BET) surface area and large content of aptamers have also been designed for perfect specific performance.30–32 The selective IT-SPME of the massive amount of aptamers on the affinity monolith offers an attractive opportunity for the online selective capture and precise measurement of various analytes.33 This sheds light on a promising approach for the online accurate recognition of DA. So far, aptamer-based affinity materials have received much attention, while online selective IT-SPME for the accurate analysis of DA based on affinity monoliths is still absent.
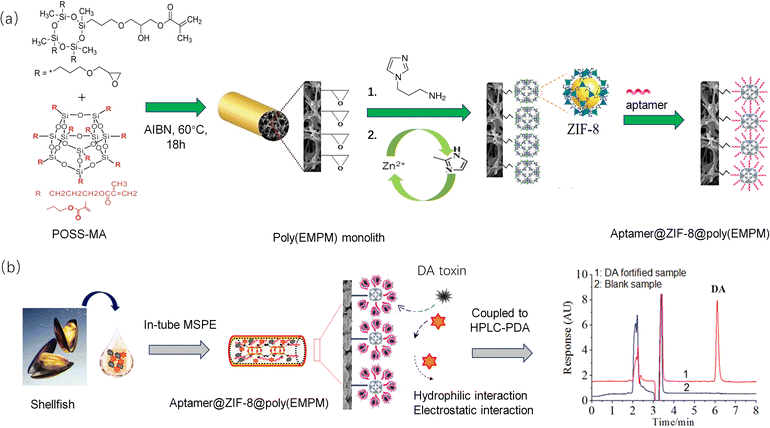
Inspired by the above facts, a novel aptamer@metal–organic framework (aptamer@MOF)-functionalized monolith with massive aptamers is proposed and was adopted for selective IT-SPME for the online specific recognition of DA (Fig. 1). Using monomers such as Epoxy-MA and POSS-MA, multiple epoxy groups could be introduced on the stereostructure, and thus the resultant poly(epoxy-MA-co-POSS-MA) was designed as a matrix possessing abundant epoxy groups for effectively tethering aminoimidazole molecules and subsequently growing ZIF-8-MOFs. Meanwhile, ZIF-8 MOFs possessing perfect surface chemistry and sufficient Zn2+ ions were employed for chelation with phosphate to immobilize the aptamers. Attributed to a large surface area and abundant Zn2+ active sites, massive amounts of aptamers could be efficiently anchored at the MOF sites in the monolith. The high coverage density of aptamers modified in the affinity column could enhance the binding capacity or specific recognition ability. Thus, a mass of affinity interaction sites were engineered, which enabled the online specific bio-recognition of DA. Herein, various characterizations, such as SEM, elemental mapping, XRD, and BET, were performed, and the performance of the aptamer@MOF-modified affinity monolith was verified. The as-developed method was applied for the online recognition of DA in shellfish samples. As result, it sheds light on a promising method for selective IT-SPME for the online identification of DA toxins utilizing a facile HPLC method.
2 Experimental
2.1 Chemicals and materials
Methacryl trisepoxy cyclosiloxane (Epoxy-MA), zinc nitrate hexahydrate (99%), γ-methacryloxypropyltrimethoxylsilane (γ-MAPS, 98%), POSS-methacryl substituted (POSS-MA, 98%), 2-methylimidazole (98%), 2,2′-azobis(2-methylpropionitrile) (AIBN, 99%), and 1-(3-aminopropyl)imidazole (APIM, 99%), were provided by Sigma-Aldrich (USA). 1-Propanol, 1,4-butanediol, polyethylene glycol (PEG, average Mn 400), tryptophan (Trp, 98%), and tris(hydroxymethyl)aminomethane (Tris, 99.8%) were purchased from Aladdin (Shanghai, China). Domoic acid (DA, 98%) was purchased from Acros Organics (USA). Based on ref. 34, oligonucleotide aptamer targeting DA (5′-P-C6-ATAGG AGTCA CGACG ACCAG AAAAA TAATT TAAAT TTTCT ACCCA ATGCT TTTCG CATAA TATGT GCGTC TACCT CTTGA-3′, denoted as Apt), and control oligonucleotide (5′-P-C6-AAAAA ACCCC CCTTT TTTGG GGGGT TTTTT AAAAA AAAAA AACCC CCCTT TTTTT TGGGG GGTTT TTTTT AAAAA A-3′, denoted as control ssDNA), were synthesized by Sangon Biotech. Co. (Shanghai, China). A binding buffer solution (BB), and Tris–EDTA buffer solution (TE) were prepared. The pretreatment of the aptamer and control ssDNA was performed according to ref. 30–32.2.2 Preparation of the aptamer@ZIF-8 affinity monolith
Fig. 1 shows the synthesis of the aptamer@ZIF-8@poly(EMPM) monolith. Before the polymerization, the capillary was washed with HCl and NaOH solutions in order, and γ-MAPS was used for introducing alkenyl functional groups on the capillary for further immobilization of the polymer.A homogeneous prepolymer mixture consisting of Epoxy-MA (35.00 mg), POSS-MA (35.00 mg), 1-propanol (49.40 mg), PEG-400 (80.60 mg), and AIBN (2.00 mg) was made with vortex oscillation for 3 min and then debubbled under ultrasonic treatment for 15 min. The solution was injected into the pretreated capillary to a 10 cm length with a syringe, which was then sealed at both ends and thermostatically heated at 60 °C in the water bath for reaction for 18 h. The poly(epoxy-MA-co-POSS-MA) monolith (poly(EMPM) matrix monolith) was finally obtained and rinsed with methanol.
Next, 200 μL of 1-(3-aminopropyl)imidazole solution (1 mol L−1) was pumped into the above MP parent monolith, and then the epoxy-ring-opening reaction proceeded in a water bath at 65 °C for 12 h. After rinsing with methanol (10 MPa), 20 μL of Zn(NO3)2 methanol solution (20 mmol L−1) was pumped into the imidazole-modified monolith and held there for 30 min to achieve the modification with Zn2+ ions. Then, 20 μL of dimethylimidazole methanol solution (20 mmol L−1) was pumped into the imidazole-Zn2+-modified monolith and held there for 30 min for the in situ growth of ZIF-8 MOF. The process of zinc nitrate and dimethylimidazole modification was repeated until the self-assembly of the ZIF-8 in the monolithic column was achieved. The obtained monolith was rinsed with methanol.
Finally, 20 μL of aptamer solution (100 μM) was pumped into the ZIF-8-modified monolith using Tris–HCl buffer solution (pH 7.50) as the mobile phase at 0.05 mL min−1. The Apt@ZIF-8 affinity monolith was finally achieved. Additionally, a control monolithic column was prepared with control ssDNA following the same procedure.
2.3 Online recognition with the Apt@ZIF-8 monolith
As shown in Fig. S1,† the analysis of DA was performed using the Apt@ZIF-8@ poly(EMPM) monolith coupled with HPLC-UV (LC-20A, Japan). Four steps, namely sample loading, washing, elution, and detection, were adopted in order. Briefly, 20 μL of sample solution was injected into the sample loop, and pumped into the Apt@ZIF-8 monolith at 0.05 mL min−1 and 250 psi. Then 20 μL of the binding buffer solution (BB) was pumped in to rinse the Apt@ZIF-8 monolith. Next, 20 μL of EB solution was applied to release DA from the Apt@ZIF-8 monolith. Finally, the elution was measured using HPLC-UV to detect DA.2.4 Aptamer coverage density and dynamic binding capacity
The aptamer coverage density on the affinity monolith was measured and calculated according to the following formula:27where, ρaptamer is the aptamer coverage density on the ZIF-8-modified monolith, Cbefore and Cafter are the concentrations of the aptamer solutions before and after the procedure of aptamer binding in the monolithic column, respectively, Vinjected and Veluted are the volumes of washing solution and eluent, respectively, and Vmonolith is the volume for the monolithic column.
The dynamic binding capacity of DA was calculated via the equation shown below. Here, 60 ng mL−1 of DA solution was used to evaluate the aptamer binding capacity, and 20 μL aliquots of the eluent were collected for DA analysis to draw the breakthrough curve.
| Qmax = C(VR − V0) |
2.5 Specificity and cross-reactivity analysis
The specific recognition of DA by the matrix monolith, ssDNA control monolith, and Apt@ZIF-8-modified monolith was assessed. Meanwhile, tryptophan with a similar molecular structure was used for evaluation of the cross-reactivity.2.6 Sample preparation
Natural samples of clams and mussels were purchased from local markets and confirmed negative for DA residue by LC-MS (in ESI†). The muscle tissue of the shellfish samples was taken out, rinsed with water, and a certain amount of DA was added. The concentrations of DA spiked in the fortified samples were 30, 100, and 200 μg kg−1, respectively. The fortified samples were first homogenized, and then 1.0 g of the homogenate of shellfish muscle was weighed and put into a 10 mL centrifuge tube. Next, 2 mL of the extracting solution (vethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) vwater = 1
vwater = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added and mixed under ultrasound for 3 min. The extraction solution was centrifuged at RCF 11125×g for 10 min and the supernatant was filtered through a 0.22 μm membrane.
1) was added and mixed under ultrasound for 3 min. The extraction solution was centrifuged at RCF 11125×g for 10 min and the supernatant was filtered through a 0.22 μm membrane.
3 Results and discussions
3.1 Fabrication of hybrid affinity monolith
Fig. 1 shows the preparation of the aptamer@ZIF-8-MOFs-modified affinity monolith with the post-column in situ MOF growth strategy. Using thermal-initiated polymerization, the poly(epoxy-MA-co-POSS-MA) monolithic column (noted as poly(EMPM) matrix monolith) was synthesized and adopted to load abundant epoxy groups for tethering aminoimidazole molecules. After the coordination reactions of Zn2+ with the imidazole groups, the typical ZIF-8-MOFs were built on the monolith, and enabled an efficient loading of aptamers for the affinity performance.For a suitable monolithic column, the optimal recipe of poly(EMPM) was evaluated and their performance assessed. As can be seen from Fig. S2,† with the content of monomers increasing, the polymer phase inside the capillary remained homogeneous, while the distribution of polymerization clusters became compact. The permeability of the monolithic column gradually decreased from 12.19 × 10−14 m2 to 0 (Table 1, a–c). As the proportion of crosslinking agent POSS-MA decreased from 22.5% to 12.5%, the crosslinking degree of the polymerization in the monolithic column decreased (Table 1, d–g). The sizes of the polymerization clusters became significantly smaller (in Fig. S3†), while the porosity in the polymerization phase became more obvious, with increasing permeability from 1.27 × 10−14 m2 to 14.63 × 10−14 m2. The as-prepared monolithic column with a proper permeability (6.87 × 10−14 m2) was the optimal and selected as the matrix for the consequent in situ growth of the MOF.
| Column designation | Monomer-to-solvent ratio | Epoxy-MAa (%, w/w) | POSS-MAa (%, w/w) | Propanolb (%, w/w) | PEG-400 (%, w/w)b | Cycle of MOF growthc (n) | Permeability Kd (×10−14 m2) |
|---|---|---|---|---|---|---|---|
| a The percentages of POSS-MA, and Epoxy-MA in the monomer mixtures, respectively.b The percentages of two porogenic reagents in porogenic solvents, respectively. Porogenic reagents were composed of propanol and PEG-400.c The n refers to the number of in situ growth cycles of ZIF-8 MOF in the column.d The permeability was measured using methanol. The viscosity of methanol was 0.544. | |||||||
| a | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 75 |
12.5 | 12.5 | 28.5 | 46.5 | — | 12.19 |
| b | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
17.5 | 17.5 | 24.7 | 40.3 | — | 2.08 |
| c | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 55 |
22.5 | 22.5 | 20.9 | 34.1 | — | 0 |
| d | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
12.5 | 22.5 | 24.7 | 40.3 | — | 0 |
| e | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
15.0 | 20.0 | 24.7 | 40.3 | — | 1.27 |
| f | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | — | 6.87 |
| g | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
22.5 | 12.5 | 24.7 | 40.3 | — | 14.63 |
| h | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | 2 | 5.93 |
| i | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | 4 | 5.21 |
| j | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | 6 | 4.69 |
| k | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | 8 | 4.26 |
| l | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 65 |
20.0 | 15.0 | 24.7 | 40.3 | 10 | 2.98 |
The optimization of the MOF self-assembly process of the affinity monolithic columns was further demonstrated. As shown in Table 1, h–l, the permeability of the MOF-modified monolithic column decreased from 6.87 × 10−14 m2 to 2.98 × 10−14 m2 with the increase in the number of MOF self-assembly cycles. The response of DA significantly increased after online affinity recognition with the aptamer@MOF-coated monolithic column and reached a maximum at 93.5% with the cycle number of MOF self-assemblies of 8 (in Fig. S4†). Based on the optimization, the aptamer@MOF-modified hybrid-silica monolithic column was achieved and was found to be favourable as an ideal functional medium for the further online specific identification.
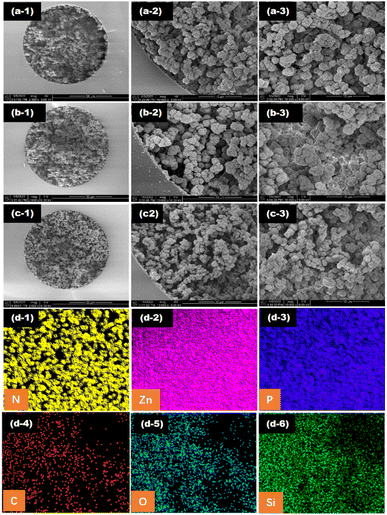
3.2 Characterization of the affinity monolith
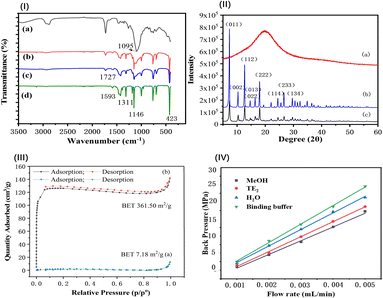
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– at 1146, 1311, and 1593 cm−1 for imidazole and 423 cm−1 for Zn–N in MOF (Fig. 3(I-d)) were distinct and appeared in both the MOF-modified monolith and aptamer@MOF-modified monolith (Fig. 3(I-b and c)).
N– at 1146, 1311, and 1593 cm−1 for imidazole and 423 cm−1 for Zn–N in MOF (Fig. 3(I-d)) were distinct and appeared in both the MOF-modified monolith and aptamer@MOF-modified monolith (Fig. 3(I-b and c)).
In Fig. 3(I-c), it can be seen that the infrared absorption of Zn–N at 423 cm−1 was decreased in comparison to that in Fig. 3(I-b), indicating that the binding of phosphate in the aptamer with Zn2+ occurred and the aptamers were bound on the ZIF-modified monolith. These results indicate that ZIF-8 MOF was successfully synthesized on the monolithic columns. With the modification of the aptamer, the strength of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– and Zn–N peaks were weakened in the aptamer@MOF-modified monolith (Fig. 3(I-c)).
N– and Zn–N peaks were weakened in the aptamer@MOF-modified monolith (Fig. 3(I-c)).
The distinct X-ray diffraction (XRD) peaks illustrated the good crystalline morphology in both the ZIF-8 MOFs and aptamer@ZIF-8 modified monoliths (Fig. 3(II-b and c)). A broadened peak band was observed in the poly(epoxy-MA-co-POSS-MA) monolith, suggesting the polymer phase was amorphous (Fig. 3(II-a)). Diffraction peaks were observed mainly distributed at 7.3°, 10.36°, 12.72°, 18.04°, and 24.52° in the aptamer@ZIF-8 modified monolith and were ascribed to the (011), (002), (112), (222), and (233) crystalline planes, respectively. The XRD pattern of the ZIF-8-MOF@polymer monolith was closely consistent with that of the ZIF-8 MOF, indicating that the in situ growth of ZIF-8 MOF had been successfully achieved without damaging its skeleton integrity and crystallinity.
In Fig. S5,† a high thermogravimetric stability of ZIF-8 MOF and poly(EMPM) monolith could be observed before 300 °C, which was consistent with the literature.37 A slight weight loss of less than 4.1% was exhibited before 120 °C, due to the moisture volatilization from air-drying the aptamer@ZIF@poly(EMPM) monolith. Obvious weight losses occurred at 120–335 °C and 335–500 °C, which were mainly caused by the dissociation of the aptamers and ZIF-8@poly(EMPM). The thermal stability of aptamer@ZIF@poly(EMPM) could be maintained at a room temperature of 25 °C to allows its use for in-tube SPME.
In Fig. 3(III), it can be seen that a high BET specific surface for ZIF-8-modified monolith was achieved at 361.50 m2 g−1, which was rather higher than that of the poly(epoxy-MA-co-POSS-MA) monolith (7.18 m2 g−1) or silsesquioxane-based hybrid monoliths (1.1–59.3 m2 g−1).38 This phenomenon indicated that ZIF-8 MOF composed of imidazole groups and zinc ions were anchored on the surface of the monolithic column. The high BET nature and surface ionization offered by the ZIF-8 MOFs were favourable for providing a support matrix and enough reactive sites for a better coverage density of aptamers with negatively charged 5′-terminal phosphate groups.
In Fig. 3(IV), the mechanical stability of the aptamer@MOF modified hybrid monolith was also evaluated under different mobile phases. Linear correlation coefficients between the back pressure and the flow velocity were obtained with the coefficient of determination (R2) above 0.9970. The as-prepared aptamer@MOF-modified monolith illustrated a good mechanical stability in various mobile phases, including methanol, Tris–HCl buffer, water, and the TE elution solution.
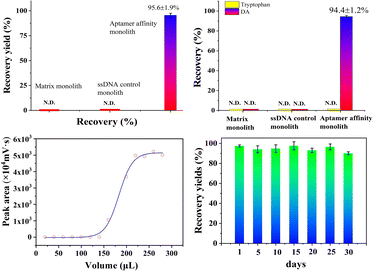
To further evaluate the selectivity of the as-prepared aptamer@MOF monolith, the cross-reactivity was investigated by using tryptophan as the classical structural analogue. For the ssDNA control monolith, the analytes were almost in the outflow while no signals were seen in the eluent (Fig. S7a1 and a2†), suggesting that there was no effective retention of DA and tryptophan. Using the ZIF-8 MOF-modified monolith, DA and tryptophan were effectively adsorbed while no analytes could be effectively eluted, with no effective signals observed in the outflow and eluent when using the ZIF-8 MOF-modified monolith (Fig. S7b1 and b2†). When using the aptamer@MOF affinity monolith, an excellent recovery value of DA (up to 94.4%) and negligible detection of tryptophan (nearly 0% of tryptophan) were achieved (Fig. S7c1 and c2†), which indicated that the DA target could be selectively recognized by the aptamer@MOF affinity monolith even with the co-existence of tryptophan. The selectivity of the aptamer-based monolith towards DA toxin was verified showing it was capable of the specific recognition.
Besides, the lifetime of the aptamer@MOF@poly(EMPM) monolith was also estimated (in Fig. 4d). The recovery yields were in a satisfactory stable state and could be maintained at a high level of more than 90% after being consecutively used over 30 days, indicating that the aptamer@MOF@poly(EMPM) monolith was rather stable and serviceable.
3.3 Method validation
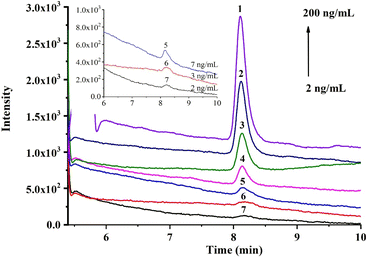
Under the optimal conditions, the online recognition and quantitative analysis of DA were studied. The response of DA was enhanced with the increase in concentration (Fig. 5), and the linear calibration curve (in Fig. S8†) was obtained in the range of 20–200 ng mL−1 with a good coefficient of determination (R2) of 0.9968 (in Table S3†). Based on the selective capture of the affinity monolith, the limit of detection (LOD, S/N = 3) and limit of quantification (LOQ, S/N = 10) were achieved as 7 and 20 ng mL−1 for DA solution, equivalent to 14 μg kg−1 and 40 μg kg−1 for shellfish samples, respectively, which fall in the general DA content range for most natural clams and mussels. Moreover, the repeatability of this method was also investigated (in Table S4†). Satisfactory RSD results regarding the recovery yields of DA, such as the intra-day, inter-day, and batch-to-batch RSD (n = 3), were achieved at 1.5%, 1.6% and 3.3%, respectively. Thus, good repeatability in the selective recognition performance could be fulfilled using this aptamer@MOF-functionalized affinity monolith.3.4 Sample analysis
To verify the applicability of the aptamer@MOF affinity monolith, the practical analysis of OA in clam and mussel samples was investigated. Attributed to the superior selective capture performance of the affinity monolith, trace DA in natural shellfish samples were successfully detected. As listed in Table 2, for three concentration levels of DA of 30, 100, and 200 μg kg−1 spiked in clam and mussel samples, values of 27.74 ± 0.71–28.36 ± 1.08, 91.78 ± 1.17–91.86 ± 1.57, and 188.09 ± 3.81–182.43 ± 2.15 μg kg−1 were obtained, respectively. Acceptable recoveries of 91.8 ± 1.2%–94.1 ± 1.9% (n = 3) and 91.2 ± 1.1%–94.5 ± 3.6% (n = 3) were achieved, respectively. The results demonstrated that trace DA in shellfishes could be effectively identified via the online recognition of this affinity monolith. To further assess the accuracy of this method, the classical SPE-LC-MS method with commercial Sep-pak SAX cartridges was adopted. In Table 2, it can be seen that the relative error between the aptamer@MOF monolith-HPLC-UV and SPE-LC-MS was no more than 5%. The results obtained by these two methods were consistent, as demonstrated by the performances of the as-prepared aptamer-based affinity monolith, such as the accuracy, reliability, and practicability, which were also acceptable for sensitive monitoring purposes.| Sample | DA spiked in samples (ng g−1) | Found by this method (mean ± SD, n = 3) (ng g−1) | Average recoveries with this method (mean ± SD, n = 3) (%) | Found by LC-MS (mean ± SD, n = 3) (ng g−1) | Average recoveries in LC-MS (mean ± SD, n = 3) (%) |
|---|---|---|---|---|---|
| Clam | 0 | 0 | N/A | 0 | N/A |
| 30 | 27.74 ± 0.71 | 92.5 ± 2.4 | 26.71 ± 1.04 | 89.0 ± 3.9 | |
| 100 | 91.78 ± 1.17 | 91.8 ± 1.2 | 88.63 ± 3.02 | 88.6 ± 3.4 | |
| 200 | 188.09 ± 3.81 | 94.1 ± 1.9 | 186.75 ± 1.13 | 93.1 ± 0.6 | |
| Mussel | 0 | 0 | N/A | 0 | N/A |
| 30 | 28.36 ± 1.08 | 94.5 ± 3.6 | 26.13 ± 0.98 | 87.1 ± 3.8 | |
| 100 | 91.86 ± 1.57 | 91.9 ± 1.6 | 88.85 ± 1.06 | 88.9 ± 1.2 | |
| 200 | 182.43 ± 2.15 | 91.2 ± 1.1 | 183.87 ± 3.39 | 91.9 ± 1.8 |
In Table 3, a comparison of this method with other HPLC-UV techniques for DA analysis reported previously is presented. Using ion-pairing/acidifying agent and reversed-phase ionic liquid dispersive liquid–liquid microextraction (RP-IL-DLLME), a highly sensitive LOD of DA was achieved, but required a time-consuming large sample volumes sample pre-concentration, with the gradient elution or the tedious LLE operation needed to be performed off line. Using SPE with SAX cartridges, MIP monolith, and magnetic MIP SPE approaches, the traditional ion-exchange interaction or MIP spatial selective recognition towards DA was adopted. Due to the non-specific nature of ion exchange and the difficulty for eluting trace target molecules from MIP materials, the detection sensitivity in these reports was in the 30–200 ng mL−1 range. In this work, massive aptamers were anchored on the MOF in the monolith for providing highly efficient bionic-affinity interactions and molecular sieving interactions, resulting in a better adsorption performance of DA, with an LOD of 7.0 ng mL−1, displaying a 5–28-fold magnitude enhancement in the detection sensitivity compared to the common SPE or MIP adsorbents reported previously. This work exhibited an online bionic recognition process of DA in shellfish with good selectivity and sensitivity, with an enhanced quantitative capacity for the analysis of DA in shellfish.
| Pretreatment strategy | Absorbent | Coupled to HPLC | Extraction selectivity | LOD (ng mL−1) | Recoveries (%) | Ref. |
|---|---|---|---|---|---|---|
| a A large sample volume sample pre-concentration with the gradient elution.b DLLME: dispersive liquid–liquid microextraction, RP-IL: reversed-phase ionic liquid. N.: not. N.A.: Not available from the literature.c Recovery of DA from the certified mussel tissue reference material (MUS-1B). | ||||||
| Ion pair | Ion-pairing/acidifying agent | Online | No | 0.04a | 97.7 ± 3.0–104.7 ± 1.5 | 36 |
| DLLME | RP-ILb | Off line | No | 0.03 | N.A. | 6 |
| SPE | SAX cartridges | Off line | No | 30 | 90 ± 3c | 38 |
| SPME | Fe3O4@MIP | Off line | Yes | 200 | 87.6 ± 7.0–88.3 ± 6.2 | 13 |
| IT-SPME | MIP monolith | Online | Yes | 76 | 89.3 ± 3.0–91.3 ± 3.4 | 23 |
| IT-SPME | Aptamer@MOF monolith | Online | Yes | 7 | 91.9 ± 1.6–94.5 ± 3.6 | This work |
4 Conclusions
In this study, a novel aptamer@ZIF-8-MOFs-functionalized affinity monolithic column was prepared and enabled online selective biomimetic recognition for the accurate quantitation of DA toxin in shellfish coupled with HPLC. Based on the MOF surface engineering, ZIF-8 MOF was in situ grown inside a poly(epoxy-MA-co-POSS-MA) hybrid monolith, and high BET and abundant metal reactive sites were provided, allowing effectively anchoring massive numbers of aptamers with a super coverage density of more than 3000 μmol L−1 and forming a reactive bionic-affinity interface for the specific recognition of DA. Under the optimal conditions, a perfect specificity of DA was exhibited with little interference of the tissue matrixes, as evidenced by the best recognition capacity of DA from the complex matrixes with a high efficiency. Efficient adsorption and desorption from the aptamer@ZIF-8-MOF monolithic column was achieved and the sensitive detection was achieved, with an LOD for DA of 7.0 ng mL−1 (equivalent to 14.0 μg kg−1), representing a 5–28-fold enhancement in detection sensitivity compared to the traditional SPE or nanoparticle adsorbents reported previously. Applied to natural shellfish samples analysis, good results for DA, such as recovery and RSD in the fortified samples of 91.8 ± 1.2%–94.1 ± 1.9% (n = 3) and 91.2 ± 1.1%–94.5 ± 3.6% (n = 3), respectively, were obtained, which were consistent with that of the classical SPE-LC-MS. This study sheds light on a facile and powerful approach for bionic capturing and accurately analyzing DA toxin in shellfish with high performance by using an aptamer@MOF-mediated affinity monolith online coupled with general HPLC-UV. This study might be helpful for providing cost-effective technology for public health and food safety management in coastal areas.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was funded by Scientific Research and Develop Project of Fujian Province (2021Y0043, 2020R1013001).Notes and references
- H. Aboualaalaa, M. L'Bachir El kbiach, B. Rijal Leblad, F. Hervèd, A. Hormat-Allah, L. Baudy, I. Ennaskhi, I. Hammi, M. Ibghi, H. Elmortaji, E. Abadie, J. Luc Rolland, Z. Amzil and M. Laabir, Toxicon, 2022, 219, 106916 CrossRef CAS PubMed.
- Z. Wang, F. Wang, C. Wang, C. Xie, T. Tang, J. Chen, S. Ji, S. Zhang, Y. Zhang and T. Jiang, Harmful Algae, 2023, 127, 102438 CrossRef CAS PubMed.
- J. Maduníc, K. Hercog, M. Geríc, A. Domijan, B. Zegură and G. Gajski, Toxicology, 2022, 470, 153157 CrossRef PubMed.
- A. F. Saeed, S. A. Awan, S. Ling, R. Wang and S. Wang, Algal Res., 2017, 24, 97–110 CrossRef.
- H. Jin, L. Lian, H. Zhou, S. Yan and W. Song, Chemosphere, 2018, 209, 328–337 CrossRef CAS PubMed.
- Q. Wang, L. Liang, J. Sun and J. Zhou, Heliyon, 2022, 8(8), e10152 CrossRef CAS PubMed.
- L. M. Botana, P. Hess, R. Munday, A. Nathalie, S. L. Degrasse, M. Feeley, T. Suzuki, M. V. D. Berg, V. Fattori and E. G. Gamarro, Trends Food Sci. Technol., 2017, 59, 15–24 CrossRef CAS.
- A. B. Kanu, J. Chromatogr. A, 2021, 1654, 462444 CrossRef CAS PubMed.
- C. I. Silvestre, J. L. Santos, J. L. Lima and E. A. Zagatto, Anal. Chim. Acta, 2009, 652, 54–65 CrossRef CAS PubMed.
- J. Płotka-Wasylka, N. Szczepanská, M. de la Guardia and J. Namieśnik, Trends Anal. Chem., 2016, 27, 23–43 CrossRef.
- E. V. Piletska, F. N. Villoslada, I. Chianella, A. Bossi, K. Karim, M. J. Whitcombe, S. A. Piletsky, G. J. Doucette and J. S. Ramsdell, Anal. Chim. Acta, 2008, 610, 35–43 CrossRef CAS PubMed.
- I. O. M. Chan, V. W. H. Tsang, K. K. Chu, S. K. Leung, M. H. W. Lam, T. C. Lau, P. K. S. Lam and R. S. S. Wu, Anal. Chim. Acta, 2007, 583, 111–117 CrossRef CAS PubMed.
- Z. Lin, S. Chen, L. Li, A. Peng and Z. Huang, Heliyon, 2020, 6, e05287 CrossRef CAS PubMed.
- M. Jiang, J. Tang, N. Zhou, J. Liu, F. Tao, F. Wang and C. Li, Talanta, 2022, 236, 122885 CrossRef CAS PubMed.
- Z. Wang, K. L. King, J. S. Ramsdell and G. J. Doucette, J. Chromatogr. A, 2007, 1163, 169–176 CrossRef CAS PubMed.
- W. Zhang, Z. Yan, J. Gao, P. Tong, W. Liu and L. Zhang, J. Chromatogr. A, 2015, 1400, 10–18 CrossRef CAS PubMed.
- P. Hess, S. Morris, L. A. Stobo, N. A. Brown, J. D. G. Mcevoy, G. Kennedy, P. B. Young, D. Slattery, E. McGovern, T. McMahon and S. Gallacher, Trends Anal. Chem., 2005, 24(4), 358–367 CrossRef CAS.
- Z. Lin, D. Wang, A. Peng, Z. Huang and Y. Lin, J. Sep. Sci., 2016, 39, 3254–3259 CrossRef CAS PubMed.
- H. Kataoka, J. Chromatogr. A, 2021, 1636, 461787 CrossRef CAS PubMed.
- Y. Moliner-Martinez, R. Herráez-Hernández, J. Verdú-Andrés, C. Molins-Legua and P. Campíns-Falcó, TrAC, Trends Anal. Chem., 2015, 71, 205–213 CrossRef CAS.
- J. Płotka-Wasylka, N. Szczepànska, M. de la Guardia and J. Namiésnik, TrAC, Trends Anal. Chem., 2015, 73, 19–38 CrossRef.
- (a) I. D. Souza, L. P. Melo, I. C. S. F. Jardim, J. C. S. Monteiro, A. M. S. Nakano and M. E. C. Queiroz, Anal. Chim. Acta, 2016, 932, 49–59 CrossRef CAS PubMed; (b) Y. Vitta, Y. Moliner-Martínez, P. Campíns-Falcó and A. Fernández Cuervo, Talanta, 2010, 82(3), 952–956 CrossRef CAS PubMed; (c) R. MirzajaniZ and R. F. Kardani, Microchem. J., 2017, 130, 93–101 CrossRef.
- F. Yang, R. Wang, G. Na, Q. Yan, Z. Lin and Z. Zhang, Anal. Bioanal. Chem., 2018, 410, 1845–1854 CrossRef CAS PubMed.
- (a) A. D. Keefe, S. Pai and A. Ellington, Aptamers as therapeutics, Nat. Rev. Drug Discovery, 2010, 9, 537–550 CrossRef CAS PubMed; (b) Q. Xie, Z. Xu, G. Huang, C. Lin and X. Lin, J. Chromatogr. A, 2023, 1688, 463728 CrossRef CAS PubMed.
- (a) Q. Zhang, Y. Yang, C. Zhang, Y. Zheng and X. Wang, Food Control, 2020, 119, 107461 CrossRef; (b) J. Xu, J. Chi, C. Lin, X. Lin and Z. Xie, J. Chromatogr. A, 2020, 1620, 461026 CrossRef CAS PubMed; (c) T. Zhao, M. Zhang, Q. Peng, X. Lin and Z. Xie, Anal. Chim. Acta, 2021, 1185, 339077 CrossRef CAS PubMed.
- Y. Ma, L. Hao, X. Lin, X. Liu, X. Qiu, X. Zhang and X. Hu, J. Chromatogr. A, 2020, 1611, 460617 CrossRef CAS PubMed.
- Y. Chen, X. Ding, D. Zhu, X. Lin and Z. Xie, Talanta, 2019, 200, 193–202 CrossRef CAS PubMed.
- T. Zhao, S. Tong, S. Zhou, C. Lin, X. Lin and Z. Xie, Analyst, 2021, 146(18), 5732–5739 RSC.
- W. Ma, S. Wan, C. Lin, X. Lin and Z. Xie, Talanta, 2020, 219, 121275 CrossRef CAS PubMed.
- J. Xu, J. Chi, C. Lin, X. Lin and Z. Xie, J. Chromatogr. A, 2020, 1620, 461026 CrossRef CAS PubMed.
- J. Chi, M. Chen, L. Deng, X. Lin and Z. Xie, Analyst, 2018, 143(21), 5210–5217 RSC.
- L. Zhao, Y. Sun, L. Mao, H. Lian and D. Sheng, Talanta, 2021, 225, 121993 CrossRef CAS PubMed.
- Y. Lv, Y. Shang, L. Li, Y. Zhang and Q. Ma, Analyst, 2023, 148(8), 1815–1823 RSC.
- L. Zhao, H. Guo, H. Chen, B. Zou, C. Yang, X. Zhang, Y. Gao, M. Sun and L. Wang, Bioengineering, 2022, 9(11), 684 CrossRef CAS PubMed.
- G. Zheng, H. Wu, H. Che, X. Li, Z. Zhang, J. Peng, M. Guo and Z. Tan, Toxins, 2022, 14, 862 CrossRef CAS PubMed.
- L. L. Mafra Jr, C. Léger, S. S. Bates and M. A. Quilliam, J. Chromatogr. A, 2009, 1216(32), 6003–6011 CrossRef PubMed.
- (a) U. P. N. Tran, K. K. A. Le and N. T. S. Phan, ACS Catal., 2011, 1(2), 120–127 CrossRef CAS; (b) G. Kibar, J. Inorg Organomet. Polym. Mater., 2023, 33, 831–840 CrossRef CAS.
- P. Hess, S. Gallacher, L. A. Bates and N. Brown, J. AOAC Int., 2001, 84(5), 1657–1667 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra05901d |
| This journal is © The Royal Society of Chemistry 2023 |