Open Access Article
Open Access ArticleBacteria-derived topologies of Cu2O nanozymes exert a variable antibacterial effect†
Ashish Kumar Shukla *a,
Vinod Moryaa and
Bhaskar Datta
*a,
Vinod Moryaa and
Bhaskar Datta *ab
*ab
aDepartment of Biological Engineering, Indian Institute of Technology Gandhinagar, Gandhinagar 382055, India. E-mail: ashish.shukla@iitgn.ac.in
bDepartment of Chemistry, Indian Institute of Technology Gandhinagar, Gandhinagar 382055, India. E-mail: bdatta@iitgn.ac.in
First published on 2nd October 2023
Abstract
The ability of bacteria to facilitate fabrication of nanomaterials has been adapted towards bacterial sensing applications. In this work, we fabricate spherical, cubic and truncated octahedron topologies of Cu2O nanoparticles via E. coli-facilitated redox reaction in an electrochemical setup. The Cu2O nanoparticles exhibit cytochrome c oxidase-like activity with the spherical topology displaying higher catalytic rate compared to the other geometries. The topology-dependent catalytic behavior of Cu2O nanoparticles has not been reported previously. The Cu2O nanozymes also display E. coli killing activity in a topology-correlated manner. The E. coli mediated redox reaction in an electrochemical setup is being reported for the first time for synthesis of different topologies of Cu2O which also exert a variable antibacterial effect.
Introduction
Nanozymes have emerged as a unique class of nanomaterials due to their biocompatibility, affordability, and distinctive catalytic properties.1–4 These synthetic nanomaterials imitate the activity of enzymes and find applications in therapeutics,5,6 biosensors,7–10 and for environmental remediation.11–13 The fabrication of nanozymes14,15 has been reported previously through various methods,16–19 including biosynthesis,20 electrochemical synthesis,21 and wet chemical methods.14,22 The rich biochemical and electrochemical capabilities of bacteria have been leveraged for fabrication of distinct morphologies of nanoparticles.23,24 The chemical and physical properties associated with specific bacterial strains can affect the size and shape of nanoparticles being produced and which would suit specific applications.17 The bacteria-mediated synthesis of nanomaterials is purported to render unique properties that are difficult to achieve with conventional fabrication techniques. Bacteria-facilitated synthesis of metal nanomaterials involves reduction of metal ions by characteristic reducing agents associated with the bacterial strain.25 Previously, bacteria-mediated nanoparticle growth has been reported using various reducing agents such as enzymes (nitrate reductase26 and hydrogenase27) and NADH- and NADH dependent enzyme cofactors.28 These agents and cofactors provide the necessary reducing power, enabling the transformation of metal ions into nanoparticles. Understanding the diverse range of reducing agents and cofactors involved in bacteria-mediated nanoparticle growth is crucial for harnessing the full potential of this biosynthesis approach. Specifically, cupric reductase NDH-2 is associated with the surface of E. coli and has been utilized for the reduction of Cu2+ to Cu+.29 In this process, bacteria play a pivotal role in reducing metal ions to their metallic forms, leading to the formation of nanoparticles with unique properties. The ability of bacteria to drive topological diversity in nanomaterials is worthy of deeper scrutiny. Different morphologies of nanomaterials can significantly affect their chemical reactivity due to inherent differences in arrangement of atoms, bonding, and surface energy.30 While nanoparticles with highly reactive surfaces are desirable, they are challenging to produce. Surfaces with high reactivity usually possess high surface energy, leading to rapid shrinkage during nanoparticle growth. The bacteria-derived biosynthesis of nanozymes continues to be explored in the context of different metal oxides and composites. While such synthesis relies on the ability of bacteria to catalyse formation of nanomaterials of uniform size and shape, the process is complex with multiple parameters that need to be optimized.In this work, we synthesized spherical, cubic and truncated octahedron of cupric oxide (Cu2O) nanozyme via a bacteria-mediated redox reaction in an electrochemical setup. The Cu2O nanozyme displayed cytochrome c oxidase (CcO)-like activity with topology playing an unambiguous role in the catalytic activity of the nanozyme. Further, the Cu2O nanozymes demonstrated E. coli killing in a topology-dependant manner. While the bacterial-mediated synthesis of nanomaterials and antibacterial properties of nanomaterials have been studied previously, we have not found any reports that converge both activities. The bacteria-facilitated synthesis of different topologies of Cu2O nanoparticles and the differences in their cognate antibacterial activities suggests a distinctive and hitherto less explored combination of sensing and remediation.
Results and discussion
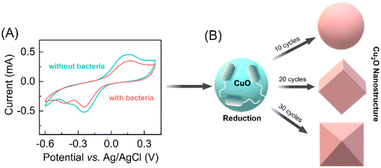
The synthesis of distinct morphologies of the Cu2O NPs were carried out by a bacteria-mediated redox reaction in an electrochemical setup. Previously, a variety of the electrochemical methods have been deployed for the synthesis of the different metal nanoparticles using three-electrode electrochemical setups.31–33 In our experiments we also used three-electrode system for cyclic voltammetry study which include a CuO thin film on FTO substrate as a working electrode (WE), platinum (Pt) as a counter, and Ag/AgCl as a reference electrode. The CuO thin film was synthesized by modification of our previously reported method.29 Characterization was performed by a combination of FESEM and XRD (see ESI†). In order to assess the bacteria-mediated redox processes at the CuO thin film, cyclic voltammetry was performed in the absence and presence of bacteria (1 × 104 CFU mL−1 DH-Alpha), in 30 mM KNO3 electrolyte solution. The electrode was swept cathodically to reduce the CuO nanoparticles to Cu2O nanoparticles, prospectively facilitated in the presence of bacteria. Based on the reductive changes being observed in the absence of bacteria, CuO nanoparticles present on the electrode surface are undergoing a two-electron reduction (Cu(II) to Cu(0)). In contrast, the presence of bacteria appears to facilitate a one-electron reduction of CuO nanoparticles (Cu(II) to Cu(I)) (Fig. 1A). The presence of cupric reductase NDH-2 on E. coli surface has been previously suggested to promote direct one-electron transfer from Cu(II) to Cu(I).29 Specifically, when bacteria come in contact with a CuO electrode, it triggers a redox reaction (E. coli + Cu2+ + e− → E. coli + Cu+) at electrode surface. This reaction involves the reduction of copper oxide nanoparticles (CuO NPs) to cupric oxide nanoparticles (Cu2O NPs), facilitated by cupric reductase NDH-2 enzyme present on the surface of the bacteria. Notably, a prominent oxidative peak is observed in the absence of bacteria that corresponds to the stripping of Cu(0) formed during the direct two-electron reduction.34 The oxidative peak is significantly lower intensity in presence of bacteria, suggesting the reduction of Cu(II) to Cu(I) and concomitant formation of Cu2O nanostructures on the surface of the WE. After the redox-reaction the surface of the FTO electrode clearly shows a brick-colored deposition following the reduction of CuO NPs to Cu2O NPs in the presence of bacteria (see ESI†).Based on our initial observations pertaining to the bacteria-facilitated synthesis of Cu2O nanostructures, we hypothesized that provision of different reaction times at the electrode surface could influence the topology of Cu2O nanostructures. In previous reports, topology-controlled platinum35 and gold nanoparticles36,37 were synthesized through electrochemical methods by manipulating the redox reaction time.38 Accordingly, we performed different cycles of voltammetry response in the presence of bacteria. After 10 CV cycles of the redox reaction at the surface of the CuO electrode, we observed the formation of spherical Cu2O nanoparticles. The morphology and size of Cu2O nanoparticles changed upon further increase of CV cycles, with cubic structures being observed after 20 cycles and truncated octahedron after 30 cycles (Fig. 1B).
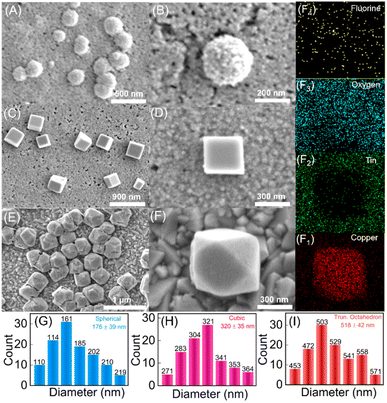
Similar structures of Cu2O nanoparticles have been synthesized previously using wet chemical methods.39,40 However, the bacteria-facilitated topological transformation of Cu2O NPs in has not been reported previously. While such topologies of gold nanoparticles have been reported in the presence of bacteria,41 to the best of our knowledge bacteria-facilitated growth of different topologies of metal oxide nanoparticles has not been reported. The different topologies of Cu2O NPs obtained after 10, 20 and 30 CV cycles were characterized by FESEM (Fig. 2A–F).
The synthesis of the truncated octahedral Cu2O nanostructures was further confirmed by energy dispersive X-ray (EDAX) elemental mappings of Fig. 2F, where copper, tin, oxygen and fluorine ions were evenly distributed on the FTO substrate after performing the redox reaction (Fig. 2(F1–F4)). The spherical, cubic and truncated octahedron Cu2O NPs display progressively greater sizes and broader size distributions with mean diameters of 176 nm, 320 nm and 518 nm, respectively as shown in Fig. 2G–I. Previously reported spherical Cu2O nanoparticles were observed with mean diameter spanning 100–500 nm.42,43 Cubic Cu2O nanoparticles were found to possess sizes in the range of 400–600 nm39,40 while truncated octahedron have been reported with singular methods of synthesis in the size range of 500–700 nm.44 As one would intuitively expect, the formation of truncated octahedra is deeply connected to precursor concentrations, temperature, reaction time, and presence of additives. Nevertheless, the precise conditions and parameters underlying the growth of different topologies of Cu2O NPs themselves vary across different reports.45 In our experiments, the topologies of Cu2O NPs obtained after 10, 20 and 30 cycles were found to be most uniform upon comparing results from variable number of CV cycles (ESI Fig. S4†).
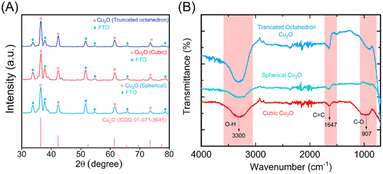
The Cu2O NPs produced after 10, 20 and 30 CV cycles were further analyzed by XRD. As shown in Fig. 3A, the characteristic diffraction peak 2θ of the fresh CuO thin film electrode appears at 35.56°, 38.8°, and 68.03° in the diffraction pattern. These correspond to reference card numbers 00-041-0254 (see ESI†). The XRD spectra of synthesized Cu2O NPs with different morphologies on the FTO working electrode showed the characteristic FTO and Cu2O peaks after the bacteria-mediated redox reaction. XRD spectra of Cu2O NPs produced in presence of bacteria showed significant diffraction peak 2θ at 36.4°, 42.36°, and 73.63° for all (spherical, cubic and truncated octahedron) topologies. These match with reference card numbers 01-071-3645. In addition, FTIR spectra were also recorded for the different topologies of Cu2O NPs. As shown in Fig. 3B, the O–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C–O bond stretching vibrations at ca. 3300, 1647 and 907 cm−1, respectively, become more prominent. These changes confirm the composition and bonding of the Cu2O NPs across all the observed topologies.46,47
C and C–O bond stretching vibrations at ca. 3300, 1647 and 907 cm−1, respectively, become more prominent. These changes confirm the composition and bonding of the Cu2O NPs across all the observed topologies.46,47
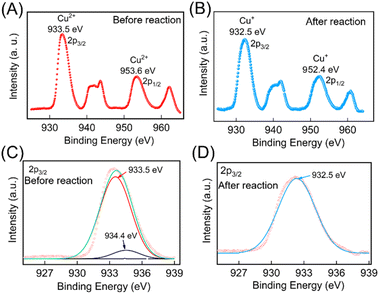
The change in oxidation state of the Cu was further validated through X-ray photoelectron spectroscopy (XPS) studies. The survey scan of the electrodes before and after the redox reaction were performed which contains peaks characteristic of Cu 2p, and O 1s (ESI Fig. S5†). To further confirm the oxidation states of Cu pre- and post-redox reaction, narrow scan XPS spectra were recorded within the BE ranges (925 to 970 eV) and are depicted in Fig. 4A and B. The data were deconvoluted using Fityk software, which identified the presence of two oxidation states of Cu. These peak positions were cross-referenced with the NIST XPS database to identify the oxidation states of the Cu ion. Before the redox reaction, the peaks at BE values 933.5 eV and 934.4 eV in the narrow scan spectra for Cu 2P (Fig. 4C) closely matched the BE of 2P3/2 states of Cu2+. After the redox reaction, a peak at BE values 932.5 eV in the narrow scan spectra for Cu 2P (as depicted in Fig. 4D) closely aligned with the BE of 2P3/2 states of Cu+, thereby confirming the formation of Cu2O NPs. The complete details of the XPS analysis are provided in the ESI.†
Cytochrome c oxidase (CcO)-like activity of the Cu2O nanoparticles
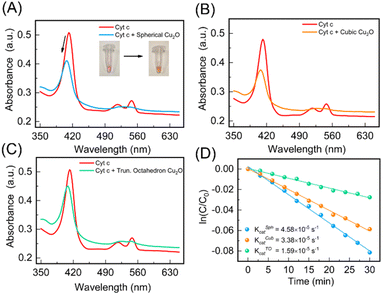
Cu2O NPs demonstrate reversible redox behavior involving Cu+ and Cu2+ species, like enzyme-catalyzed redox reactions. We expected the different topologies of Cu2O NPs to not only mimic cytochrome c oxidase (CcO) enzyme activity, but also display topology-dependent behavior as nanozymes owing to the distinctive structural attributes of each geometry.48 By precisely controlling the shape and size of Cu2O NPs during synthesis, their catalytic performance can be optimized, potentially distinguishing their enzyme-like activity. Therefore, we studied the CcO enzyme-like behavior of the three topologies of the Cu2O NPs against the cytochrome c (Cyt c) substrate. Natural CcO enzyme accepts electrons from the ferrous Cyt c and reduces oxygen O2 to H2O. The UV spectrum of oxidized Cyt c shows absorption peaks at 408 and 530 nm, while reduced Cyt c has absorption peaks at 414, 512 and 550 nm.48 We used the peak at 550 nm to monitor and quantify the amount of reduced Cyt c. We first prepared ferrous Cyt c as per details provided in ESI.† The scrub Cu2O NPs were taken out from the FTO substrate in powder form. The physical appearance of the Cu2O NPs obtained after 10, 20 and 30 CV cycles are shown in ESI (Fig. S4†). Upon addition of a 1 mg mL−1 solution of Cu2O nanoparticles, the absorbance peak of 10 μM of ferrous Cyt c at 414 nm shifted to 408 nm. However, the absorbance peak at 550 nm gradually decreased confirming the catalytic activity of Cu2O NPs (see Fig. 5A). In addition, the color of the Cyt c solution changed from pink to a yellowish red (Fig. 5A, inset), suggesting the conversion of Cyt c from ferrous to ferric form. This conversion is further confirmed by circular dichroism (CD) spectroscopy (see ESI†) and consistent with the previous reports.48The cubic and truncated octahedron topologies of Cu2O NPs displayed an analogous reduction of ferrous Cyt c to ferric Cyt c (Fig. 5B and C). The catalytic activity of the different topologies of Cu2O NPs were quantified by studying the time-resolved reduction of ferrous Cyt c. The absorbance at 550 nm for each of the spherical, cubic and truncated octahedral Cu2O NPs was measured over time (Fig. 5D). The catalytic rate constants of the spherical (KSphcat), cubic (KCubcat) and truncated octahedron (KTOcat) are found to be 4.58 × 10−5 s−1, 3.38 × 10−5 s−1 and 1.59 × 10−5 s−1. These rate constants are consistent with previously reported catalytic rates.48 Our measurements indicate that the oxidation of ferrous Cyt c with cubic and truncated octahedron NPs is slower compared to spherical NPs. This observation aligns with the intuitive expectation that smaller particle sizes are likely to exert higher catalytic activity.49 Smaller nanoparticles have a higher surface-to-volume ratio, providing more active sites for catalytic reactions. Also, increased density of surface atoms and exposure of reactive sites enhance catalytic efficiency. Additionally, smaller nanoparticles offer shorter diffusion distances enabling faster mass transport and more efficient interaction between reactants and the catalyst, leading to increased reaction rates.
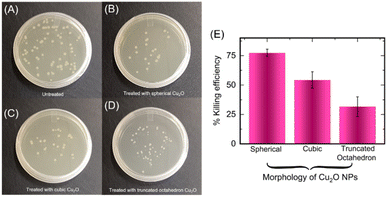
Cuprous oxide has the ability to disrupt various cell functions and can be toxic to cells, making it a promising agent for inhibiting microbial growth.50–52 These nanoparticles can also damage microbial components through various mechanisms such as generation of reactive oxygen species (ROS),53 disruption of cell walls/membranes,54 replacement of native cofactors in metalloproteins55,56 and damage of intracellular components.57 The fabrication of different topologies of copper-containing nanoparticles has been considered for tuning antibacterial effect and enabling broad scope of use in environmental remediation.58 Notably, octahedral and cubic-shaped Cu2O nanoparticles have been used to target specific pollutants and allow for greater control over their activity.59 We were interested in studying antibacterial activity of the different topologies of Cu2O NPs that were produced via bacteria-mediated electrochemical approach. Accordingly, we incubated the spherical, cubic and truncated octahedron of Cu2O NPs (1 mg mL−1) with E. coli bacteria. Interestingly, E. coli are inhibited in presence of all the topologies of Cu2O NPs albeit with different efficiencies (Fig. 6A–D).
The percentage killing efficiency due to spherical, cubic and truncated octahedron nanoparticles were found to be 77%, 54%, and 31%, respectively (Fig. 6D).60 The differing inhibitory effects of the three topologies of Cu2O NPs against E. coli is consistent with previous reports.59,61 The spherical Cu2O NPs display substantially higher antibacterial effect potentially due to smaller size and higher surface area-to-volume ratios, which likely permit better interaction with bacterial cells compared to the other topologies. Two mechanisms of antibacterial activity are likely at play for the Cu2O NPs. Also, as mentioned above, Cu2O NPs are capable of generating reactive oxygen species (ROS) that can cause damage to cellular constituents.62,63 The source of energy for ROS production in our setup is primarily through Fenton type and Haber–Weiss reactions given as below.51
| Cu+ + O2 → Cu2+ + O2˙− |
| 2O2˙− + 2H+ → H2O2 + O2 |
| Cu+ + H2O2 → Cu2+ + OH˙ + OH− |
The unique morphology and surface characteristics of the Cu2O NPs can facilitate redox reactions at the particle–cell interface leading to ROS production, without the need for external energy sources like light, electronic energy, or heat.64–66 Cu2O NPs can also produce disruptions in microbial cell wall and cell membrane thereby exerting bactericidal effects.50
Conclusions
In summary, we have used a bacteria-facilitated redox technique to fabricate different morphologies of the cytochrome c oxidase (CcO)-mimic Cu2O nanozyme and demonstrated their antibacterial effect. The Cu2O NPs were prepared by a hitherto unreported bacteria-mediated redox reaction in an electrochemical setup and involved preferential reduction (Cu(II) to Cu(I)) of the CuO thin film electrode during cyclic voltammetry. The morphologies and size of the Cu2O NPs, depend upon the redox reaction time at the surface of the electrode during CV measurements. After 10, 20, and 30 cycles of CV study, we observed spherical, cubic and truncated octahedron morphologies of the Cu2O nanoparticles, respectively. The catalytic activity of the different morphology of the nanoparticles was calculated against Cyt c substrate and spherical Cu2O NPs showed superior catalytic activity compared to the other two topologies. While all the topologies of Cu2O NPs displayed antibacterial activity against E. coli (DH5-Alpha strain), spherical Cu2O NPs showed highest bacterial killing efficiency compared to the other Cu2O NP topologies. For future prospects, our study suggests the exploration of a generalized approach for the synthesis of different topologies of nanozymes in response to bacteria with specific redox reaction, followed by evaluating the bacterial killing ability of these nanozymes. This methodology can be applied and tested with various types of nanoparticles thereby opening avenues for broader applications and innovative antimicrobial strategies.Author contributions
A. K. S. and B. D. conceived the idea and planned the experiments. A. K. S. designed and performed most of the experiments. V. M. helped with the UV spectroscopy measurements. A. K. S. and B. D. analysed the data and wrote the final draft of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to IIT Gandhinagar for their financial support for this work. Dr Bhaskar Datta thanks to Gujarat Council of Science & Technology for GUJCOST support (project no. GUJCOST/STI/2023-24/254). We acknowledge the help received from Dr Chinmay Ghoroi and CRTDH lab for helping us with instruments (UV-vis spectroscopy and sonicator). We also acknowledge the help received from Dr Biswajit Mondal for helping us with Auto Lab instrument. We also thank the Central Instrument Facility (CIF) at Indian Institute of Technology Gandhinagar, India, for SEM, XRD and EDAX characterization of experimental samples.References
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- H. Wang, K. Wan and X. Shi, Adv. Mater., 2019, 31, e1805368 CrossRef PubMed.
- Y. Zhou, B. Liu, R. Yang and J. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 CrossRef CAS PubMed.
- D. Li, D. Dai, G. Xiong, S. Lan and C. Zhang, Small, 2023, 19, e2205870 CrossRef PubMed.
- X. Wang, Y. Hu and H. Wei, Inorg. Chem. Front., 2016, 3, 41–60 RSC.
- M. Cai, Y. Zhang, Z. Cao, W. Lin and N. Lu, ACS Appl. Mater. Interfaces, 2023, 15, 18620–18629 CrossRef CAS PubMed.
- Y. Wu, D. C. Darland and J. X. Zhao, Sensors, 2021, 21, 5201 CrossRef CAS PubMed.
- W. Lu, Y. Guo, J. Zhang, Y. Yue, L. Fan, F. Li, C. Dong, S. Shuang and A. High, ACS Appl. Mater. Interfaces, 2022, 14, 57206–57214 CrossRef CAS PubMed.
- A. K. Shukla, S. Bhandari and K. K. Dey, Mater. Today Commun., 2021, 28, 102504 CrossRef CAS.
- R. Singh, A. Umapathi, G. Patel, C. Patra, U. Malik, S. K. Bhargava and H. K. Daima, Sci. Total Environ., 2022, 854, 158771 CrossRef PubMed.
- Y. Meng, W. Li, X. Pan and G. M. Gadd, Environ. Sci.: Nano, 2020, 7, 1305–1318 RSC.
- A. K. Shukla, D. Bhatia and K. K. Dey, ACS Appl. Nano Mater., 2023, 6, 8017–8027 CrossRef CAS.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- C. Ge, G. Fang, X. Shen, Y. Chong, W. G. Wamer, X. Gao, Z. Chai, C. Chen and J. J. Yin, ACS Nano, 2016, 10, 10436–10445 CrossRef CAS PubMed.
- M. Eugenio, N. Müller, S. Frasés, R. Almeida-Paes, L. M. T. R. Lima, L. Lemgruber, M. Farina, W. de Souza and C. Sant'Anna, RSC Adv., 2016, 6, 9893–9904 RSC.
- S. Ghosh, R. Ahmad, Md. Zeyaullah and S. K. Khare, Front. Chem., 2021, 9, 626834 CrossRef CAS PubMed.
- N. Shreyash, S. Bajpai, M. A. Khan, Y. Vijay, S. K. Tiwary and M. Sonker, ACS Appl. Nano Mater., 2021, 4, 11428–11457 CrossRef CAS.
- C. A. S. Ballesteros, L. A. Mercante, A. D. Alvarenga, M. H. M. Facure, R. Schneider and D. S. Correa, Mater. Chem. Front., 2021, 5, 7419–7451 RSC.
- S. Nakamura, M. Sato, Y. Sato, N. Ando, T. Takayama, M. Fujita and M. Ishihara, Int. J. Mol. Sci., 2019, 20, 362 CrossRef PubMed.
- M. A. Komkova, K. R. Vetoshev, E. A. Andreev and A. A. Karyakin, Dalton Trans., 2021, 50, 11385–11389 RSC.
- H. Wang, K. Wan and X. Shi, Adv. Mater., 2019, 31, e1805368 CrossRef PubMed.
- N. Goel, R. Ahmad, R. Singh, S. Sood and S. K. Khare, Bioresour. Technol. Rep., 2021, 15, 100753 CrossRef CAS.
- M. T. Yaraki, S. Zahed Nasab, I. Zare, M. Dahri, M. Moein Sadeghi, M. Koohi and Y. N. Tan, Ind. Eng. Chem. Res., 2022, 61, 7547–7593 CrossRef CAS.
- A. Ramesh, M. T. Sundari, and P. E. Thirugnanam, Bio-Nanoparticles, John Wiley & Sons, Ltd, 2015, pp. 53–81 Search PubMed.
- S. Anil Kumar, M. K. Abyaneh, S. W. Gosavi, S. K. Kulkarni, R. Pasricha, A. Ahmad and M. I. Khan, Biotechnol. Lett., 2007, 29, 439–445 CrossRef CAS PubMed.
- C. K. Ng, T. K. C. Cai Tan, H. Song and B. Cao, RSC Adv., 2013, 3, 22498–22503 RSC.
- S. He, Z. Guo, Y. Zhang, S. Zhang, J. Wang and N. Gu, Mater. Lett., 2007, 61, 3984–3987 CrossRef CAS.
- A. K. Shukla, S. Mitra, S. Dhakar, A. Maiti, S. Sharma and K. K. Dey, ACS Appl. Bio Mater., 2023, 6, 117–125 CrossRef CAS PubMed.
- X. Wang, H. F. Wu, Q. Kuang, R. B. Huang, Z. X. Xie and L. S. Zheng, Langmuir, 2010, 26, 2774–2778 CrossRef CAS PubMed.
- L. Rodríguez-Sánchez, M. C. Blanco and M. A. López-Quintela, J. Phys. Chem. B, 2000, 104, 9683–9688 CrossRef.
- Y. Gao and J. Hao, J. Phys. Chem. B, 2009, 113, 9461–9471 CrossRef CAS PubMed.
- L. Jiang, I. Santiago and J. Foord, Langmuir, 2020, 36, 6089–6094 CrossRef CAS PubMed.
- G. Zampardi, J. Thöming, H. Naatz, H. M. A. Amin, S. Pokhrel, L. Mädler and R. G. Compton, Small, 2018, 14, e1801765 CrossRef PubMed.
- L. Wei, Y.-J. Fan, H.-H. Wang, N. Tian, Z.-Y. Zhou and S.-G. Sun, Electrochim. Acta, 2012, 76, 468–474 CrossRef CAS.
- D. Y. Kim, S. H. Im, O. O. Park and Y. T. Lim, CrystEngComm, 2010, 12, 116–121 RSC.
- M. H. Huang and P.-H. Lin, Adv. Funct. Mater., 2012, 22, 14–24 CrossRef CAS.
- L. Wei, N. Tian, Z. Y. Zhou and S. G. Sun, Adv. Electrochem. Sci. Eng., 2017, 59–95 Search PubMed.
- Q. Hua, D. Shang, W. Zhang, K. Chen, S. Chang, Y. Ma, Z. Jiang, J. Yang and W. Huang, Langmuir, 2011, 27, 665–671 CrossRef CAS PubMed.
- A. K. Mishra and D. Pradhan, Cryst. Growth Des., 2016, 16, 3688–3698 CrossRef CAS.
- M. Gericke and A. Pinches, Hydrometallurgy, 2006, 83, 132–140 CrossRef CAS.
- S. Yadav, A. Jain and P. Malhotra, Green Chem., 2019, 21, 937–955 RSC.
- M. A. Bhosale and B. M. Bhanage, Adv. Powder Technol., 2016, 27, 238–244 CrossRef CAS.
- J. Ouyang, H. Yang and A. Tang, Mater. Des., 2016, 92, 261–267 CrossRef CAS.
- S. S. Sawant, A. D. Bhagwat and C. M. Mahajan, J. Nano- Electron. Phys., 2016, 8, 01035 Search PubMed.
- S. Joshi, S. J. Ippolito and M. V. Sunkara, RSC Adv., 2016, 6, 43672–43684 RSC.
- I. Roy, A. Bhattacharyya, G. Sarkar, N. R. Saha, D. Rana, P. P. Ghosh, M. Palit, A. R. Das and D. Chattopadhyay, RSC Adv., 2014, 4, 52044–52052 RSC.
- M. Chen, Z. Wang, J. Shu, X. Jiang, W. Wang, Z. H. Shi and Y. W. Lin, Inorg. Chem., 2017, 56, 9400–9403 CrossRef CAS PubMed.
- P. Suchomel, L. Kvitek, R. Prucek, A. Panacek, A. Halder, S. Vajda and R. Zboril, Sci. Rep., 2018, 8, 4589 CrossRef PubMed.
- K. Sunada, M. Minoshima and K. Hashimoto, J. Hazard. Mater., 2012, 235–236, 265–270 CrossRef CAS PubMed.
- X. Ma, S. Zhou, X. Xu and Q. Du, Front. Surg., 2022, 9, 905892 CrossRef PubMed.
- N. Zhu, S. Zhou, J. Gong, X. Wang, C. Zhang, W. Li, D. Sheng, X. Liu, L. Xia and W. Xu, Ind. Crops Prod., 2022, 187, 115442 CrossRef CAS.
- M. R. Gunther, P. M. Hanna, R. P. Mason and M. S. Cohen, Arch. Biochem. Biophys., 1995, 316, 515–522 CrossRef CAS PubMed.
- D. Mitra, E. T. Kang and K. G. Neoh, ACS Appl. Mater. Interfaces, 2020, 12, 21159–21182 CrossRef CAS PubMed.
- G. Tan, J. Yang, T. Li, J. Zhao, S. Sun, X. Li, C. Lin, J. Li, H. Zhou, J. Lyu and H. Ding, Appl. Environ. Microbiol., 2017, 83, e00867 CrossRef CAS PubMed.
- L. Macomber and J. A. Imlay, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8344–8349 CrossRef CAS PubMed.
- A. K. Chatterjee, R. Chakraborty and T. Basu, Nanotechnology, 2014, 25, 135101 CrossRef PubMed.
- X. Zhu, J. Wang, L. Cai, Y. Wu, M. Ji, H. Jiang and J. Chen, J. Hazard. Mater., 2022, 430, 128436 CrossRef CAS PubMed.
- H. Pang, F. Gao and Q. Lu, Chem. Commun., 2009, 9, 1076–1078 RSC.
- A. M. Ghadiri, N. Rabiee, M. Bagherzadeh, M. Kiani, Y. Fatahi, A. Di Bartolomeo, R. Dinarvand and T. J. Webster, Nanotechnology, 2020, 31, 425101 CrossRef CAS PubMed.
- L. Xiong, H. Yu, C. Nie, Y. Xiao, Q. Zeng, G. Wang, B. Wang, H. Lv, Q. Li and S. Chen, RSC Adv., 2017, 7, 51822–51830 RSC.
- R. L. Davies and S. F. Etris, Catal. Today, 1997, 36, 107–114 CrossRef CAS.
- J. A. Imlay, Annu. Rev. Microbiol., 2003, 57, 395–418 CrossRef CAS PubMed.
- T. Xia, M. Kovochich, J. Brant, M. Hotze, J. Sempf, T. Oberley, C. Sioutas, J. I. Yeh, M. R. Wiesner and A. E. Nel, Nano Lett., 2006, 6, 1794–1807 CrossRef CAS PubMed.
- K. T. Kim, S. J. Klaine, J. Cho, S.-H. Kim and S. D. Kim, Sci. Total Environ., 2010, 408, 2268–2272 CrossRef CAS PubMed.
- E.-J. Park, J. Choi, Y.-K. Park and K. Park, Toxicology, 2008, 245, 90–100 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra05411j |
| This journal is © The Royal Society of Chemistry 2023 |