Open Access Article
Open Access ArticleNatural defense against multi-drug resistant Pseudomonas aeruginosa: Cassia occidentalis L. in vitro and in silico antibacterial activity†
Raihan Rahman Imona,
Md. Enamul Kabir Talukdera,
Shahina Akhterb,
Md. Saidul Islamc,
Foysal Ahammad de,
K. M. Anis-Ul-Haquef,
Md. Moniruzzamang,
Mirola Afrozeg,
Mala Khang,
Mohammad Abu Hena Mostofa Jamalh,
Tanveer A. Wani
de,
K. M. Anis-Ul-Haquef,
Md. Moniruzzamang,
Mirola Afrozeg,
Mala Khang,
Mohammad Abu Hena Mostofa Jamalh,
Tanveer A. Wani i,
Mohammad Jashim Uddin*j and
Md. Mashiar Rahman
i,
Mohammad Jashim Uddin*j and
Md. Mashiar Rahman *a
*a
aMolecular and Cellular Biology Laboratory, Department of Genetic Engineering and Biotechnology, Jashore University of Science and Technology, Jashore 7408, Bangladesh. E-mail: mm.rahamn@just.edu.bd
bDepartment of Biochemistry and Biotechnology, University of Science and Technology Chittagong, Foy's Lake, Chittagong 4202, Bangladesh
cKorea Institute of Radiological & Medical Sciences, 75, Nowon-ro, Nowon-gu, Seoul, South Korea
dLaboratory of Computational Biology, Biological Solution Centre (BioSol Centre), Jashore 7408, Bangladesh
eDivision of Biological and Biomedical Sciences (BBS), College of Health & Life Sciences (CHLS), Hamad Bin Khalifa University (HBKU), Doha, Qatar
fDepartment of Chemistry, Jashore University of Science and Technology, Jashore 7408, Bangladesh
gBangladesh Reference Institute for Chemical Measurements (BRICM), Bangladesh Council of Scientific and Industrial Research, Dr Qudrat-i-Khuda Road, Dhanmondi, Dhaka 1205, Bangladesh
hDepartment of Biotechnology and Genetic Engineering, Islamic University, Bangladesh
iDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, PO Box 2457, Riyadh 11451, Saudi Arabia
jDepartment of Pharmacy, Jashore University of Science and Technology, Jashore 7408, Bangladesh
First published on 2nd October 2023
Abstract
Cassia occidentalis L. is widely used in indigenous and traditional medicine, but its impact on multi-drug resistant (MDR) bacterial infections mostly remains unknown. Therefore, this study aimed to evaluate the in vitro antibacterial efficiency of methanol and ethyl acetate extracts of C. occidentalis L. leaves (MECOL and EAECOL) against multi-drug resistant Pseudomonas aeruginosa and to identify potential antibacterial agents through computational studies targeting the LasR protein. Initially, 82 compounds were identified using GC-MS analysis, and the functional groups were determined through FT-IR analysis. Both extracts of the plant exhibited dose-dependent antibacterial activity, with MICs of 104.16 ± 36.08 μg mL−1 for MECOL and 83.33 ± 36.08 μg mL−1 for EAECOL, and an MBC of 125 μg mL−1. Among the 82 compounds, 12 potential compounds were identified based on binding scores using molecular docking with the LasR protein and MM-GBSA analysis. Furthermore, screening for ADME properties, including physicochemical features, water solubility, lipophilicity, RO5 compliance, and toxicity, identified the top three compounds: methyl dihydrojasmonate, methyl benzoate, and 4a-methyl-4,4a,5,6,7,8-hexahydro-2(3H)-naphthalenone, which also demonstrated binding affinity with the active site residues of the LpxC protein of the bacteria. Additionally, molecular dynamics (MD) simulations confirmed the binding reliability of these three phytochemicals to LasR's active pocket, comparable to the protein native inhibitory ligands (C12-HSL). The study offers scientific support for the traditional use of C. occidentalis in treating bacterial infections, highlighting the potential of the three compounds as leads for developing LasR inhibitors to combat multi-drug resistant P. aeruginosa.
Introduction
One of the significant biomedical challenges today is developing effective disease-modifying treatments for multidrug-resistant (MDR) bacteria. These MDR pathogens pose a persistent threat to public health and human well-being. Among them, P. aeruginosa infections are particularly concerning, as they have developed resistance against current antibiotics by altering metabolic pathways for survival and persistence.1 The bacteria are classified as a top priority by the World Health Organization for innovative therapeutic approaches, and it is a key concern, according to the U.S. Centers for Disease Control.2 The bacteria cause infections with high mortality rates (up to 61%), posing significant challenges worldwide, especially in intensive care and burn units.3,4 Nosocomial infections are a significant concern, occurring after hospitalization, and P. aeruginosa is known to colonize medical equipment, urinary catheters, and endotracheal tubes.5 The bacteria's infectivity, virulence, and pathogenicity, driven by Quorum Sensing (QS), allow it to function as a unified entity, making it challenging to eradicate even during antimicrobial treatment.6,7The QS systems are encoded by bacterial lasR/lasI and rhlR/rhlI genes.8 The lasI gene encodes LasI, responsible for N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) synthesis, a QS signaling factor. Similarly, rhlI gene translates RhlI, which secretes N-butyryl-L-homoserine lactone (C4-HSL), another QS influencer. These genes also code for the receptors LasR and RhlR, forming complexes that influence biofilm development, virulence, and pathogenicity.7–11 Inhibiting the LasR receptor may prevent P. aeruginosa's virulence and invasiveness, supported by the reduction of virulence in LasR mutants and the suppression of biofilm production by sitagliptin, a medication used for type 2 diabetes (T2D).12
P. aeruginosa's multidrug resistance is driving interest in alternative antimicrobial sources with minimal side effects. Natural plant products, with their diverse structures and multitarget functionality, show promise as potential antibacterial agents.13,14 Medicinal plants contain bioactive phytochemicals with clinical value, and bacteria do not easily develop resistance to them.15,16 For instance, plant-based antibacterial compounds like coumarins demonstrate high activity against Staphylococcus aureus.17 Berberine exhibits potent antibacterial activity against Gram-positive bacteria, including Mycobacterium tuberculosis and methicillin-resistant Staphylococcus aureus.18 Plumbagin acts as a growth inhibitor for Escherichia coli, Enterobacter aerogenes, and Klebsiella pneumoniae.19 C. occidentalis L., an ethnomedicinal herb, has been extensively used in traditional medicine for various ailments.20 It exhibits antibacterial, antifungal, antioxidant, hepatoprotective, and other activities due to its rich phytochemical content.21,22
The antibacterial activity of the plant C. occidentalis L. and its compounds against MDR bacteria remains undiscovered. Therefore, the study aims to explore the antibacterial potential and pharmacological properties of C. occidentalis L. phytochemicals, specifically targeting P. aeruginosa's virulence and pathogenicity through biofilm formation. This comprehensive approach involves in vitro testing, in silico evaluation of pharmacokinetics, toxicity, and drug properties, as well as molecular dynamics simulations to validate lead compounds. The research findings are expected to contribute valuable insights into the potential therapeutic use of C. occidentalis L. leaves against P. aeruginosa and aid in the development of novel treatment strategies for MDR-related infections.
Materials and methods
Chemicals and reagents
Chemicals were sourced from various suppliers: Mayer's and Fehling's solutions obtained from Central Drug House (P) Ltd., pyridine (CAS: 110-86-1) and chloroform (CAS: 67-66-3) from Alfa Aesar, glacial acetic acid from PanReac AppliChem (CAS: 64-19-7), and erythromycin antibiotic discs from Bio-Rad, USA. Additionally, NaCl, H2O2, NaOH from Wako Pure Chemicals Industries, Ltd., and nutrient broth and nutrient agar media from Liofilchem, Italy. Ethyl acetate (CAS: 141-78-6), methanol, HCl (CAS: 7647-01-0), CuSO4 (CAS: 7758-99-8), H2SO4 (CAS: 7664-93-9), dimethyl sulfoxide (CAS: 67-68-5), ferric chloride (CAS: 10025-77-1), lead acetate (CAS: 6080-56-4), and sodium nitroprusside (CAS: 13755-38-9) were purchased from Merck, Darmstadt, Germany.Plant collection
In January 2022, C. occidentalis L. leaves were collected from Churamonkathi, Jashore district, Bangladesh (latitude 23.2238° N, longitude 89.1646° W). The herb's authenticity was verified by Dr Sardar Nasiruddin, a taxonomist at the National Herbarium in Dhaka, Bangladesh. Collected leaves were washed, air-dried at room temperature (∼25 °C), and ground into powder, then stored in an airtight container.23Plant extract preparation
Phytoextracts from C. occidentalis L. leaves were obtained following standard procedures described by Rahman et al. 2020.23 The powdered plant material (100 g) was divided into two 1-liter conical flasks, one with 400 mL of methanol and the other with 400 mL of ethyl acetate. After 72 hours of shaking at 250 rotations per minute and 37 °C, the mixture was filtered using sterile cotton mesh and Whatman filter paper (number 1). The concentrated crude extracts were stored in sterilized tubes at 4 °C, yielding 5.5 g (dry weight, 5.5% w/w) of crude methanol extract and 5.77 g (dry weight, 5.77% w/w) of crude ethyl acetate extract from each 100 g of powdered plant material.Qualitative phytochemical screening
Qualitative analysis of phytochemicals in MECOL and EAECOL solutions was conducted using previously methods describe by Rahman et al. 2023.24 To detect phenolic compounds (flavonoids and tannins), alkaline reagents, FeCl3, and Pb(OAc)2 were employed. Alkaline reagent tests involved dropwise addition of MECOL and EAECOL solutions to NaOH (5%), followed by the addition of 10% HCl solution resulting in color disappearance. FeCl3 (5%) solution was added to MECOL and EAECOL solutions (10 mg mL−1), producing a reddish-black color.25 For tannin detection, 10 mg of MECOL and EAECOL were mixed with chloroform, and 10% conc. Pb(OAc)2 was added. The steroids test showed a dark-black color on the bottom surface. Mayer's test involved dissolving 25 mg of MECOL and EAECOL in 10 mL of aqueous HCl (1%) and adding Mayer's reagent. Legal tests were conducted to identify cardiac glycosides using pyridine, nitroprusside, and sodium hydroxide solution.26 Fehling's assay was used to detect reducing sugars. Foaming and frothing assays were performed to test for the presence of saponin in MECOL and EAECOL solutions.Gas chromatography-mass spectrometry (GC-MS) analysis
The GC-MS analysis followed a previously disclosed methodology described with slight modifications.27 Shimadzu triple-quad GCMS-TQ8040 was used to detect phytochemicals in MECOL and EAECOL. Helium gas was used as the mobile phase, and a Rtx-5MS capillary column (30 m 0.25 mm id, film thickness of 0.25 m) was used as the stationary phase. The column oven temperature was adjusted using dedicated software. The sample injector temperature was maintained at 250 °C for a 40 minute run in splitless mode. The instrument settings included a continuous stream frequency of 1 mL min−1, an interface temperature of 250 °C, an ion source temperature of 230 °C, a scanning range of 50–600 m/z, and an ionization energy of 70 eV with a scan period of 0.3 s. Phytochemicals were identified by contrasting retention time and spectral patterns, and matches were made to the National Institute of Standards and Technology database (NIST) for specific content (%) estimation in MECOL and EAECOL.FT-IR spectroscopic analysis
FT-IR analysis of MECOL and EAECOL followed a previously established method by He et al., (2020).28 Phytoconstituents were loaded as KBr pellets in the FT-IR sample chamber, and the infrared spectra were obtained in the range of 4500 to 400 cm−1 with a resolution of 4 cm−1.Bacterial strains collection
A glycerol stock of a multidrug-resistant P. aeruginosa bacterial strain (Gene Bank Accession Number: OK355439) was collected from the Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia, Bangladesh. This strain was isolated from medical center drainage water and tested for antibiotic susceptibility against various antibiotics, showing multiple antibiotic resistance.29Agar-well diffusion and disc diffusion assay
The antibacterial potential of MECOL and EAECOL was evaluated using agar-well diffusion and disc diffusion techniques against frozen MDR P. aeruginosa stock. Bacterial colonies were grown on LB agar and a confined colony of the bacteria was inoculated into 25 mL of LB broth and cultured for bacterial growth at 37 °C with constant agitation at 250 rpm, until the exponential phase of optical density (OD) reached 0.4 at 600 nm wavelength, as measured using a UV-spectrophotometer. Four wells were made in each LB agar Petri plate containing bacterial culture. Serial dilutions of MECOL and EAECOL (500 μg mL−1) were added to wells. Erythromycin discs served as the standard and the discs of erythromycin in the center of the Petri plates were used as a negative control. Discs with various extract concentrations were placed on the agar plate. Blank discs with DMSO were used as a negative control. Zones of inhibition were measured after incubation at 37 °C for 16 hours, and the experiment was performed in triplicate.MIC and MBC analysis
The MIC and MBC of MECOL and EAECOL were determined using a doubled serial dilution scheme. Stock solutions of MECOL and EAECOL (500 μg mL−1) were diluted with LB broth to concentrations of 250, 125, and 62.5 μg mL−1. Bacterial samples were added to each tube and incubated at 37 °C for 24 hours. The lowest concentration preventing visible bacterial growth was identified as MIC. For MBC, cultures were sub-cultured on LB agar and incubated for 16 hours at 37 °C. The lowest concentration without visible bacterial growth on the agar plate was recorded as MBC. The experiments were conducted thrice for accuracy.Retrieval and preparation of protein structure
The 3D X-ray crystallographic structures of the biofilm-forming LasR (PDB: 3IX3) and lipid A biosynthesis protein LpxC (PDB: 2ves) with their native ligands (C12-HSL and BB-78485, respectively) were obtained from the RCSB-PDB.6,30 The resolution of LasR and LpxC structures was determined to be 1.40 Å and 1.90 Å, respectively. Protein preparation was performed using the Schrodinger protein preparation wizard (version 2020-3), including bond order assignment, creation of zero-order bonds for metals, addition of disulfide bonds, and removal of water molecules.31 The protein structures were further refined, and energy minimized using the OPLS-3e force field.32Compounds preparation
In the GC-MS analysis, a total of 82 different phytochemicals were identified, with seven being identical in both MECOL and EAECOL extracts. The 3D structures of these phytochemicals were retrieved from the PubChem database and processed using the Ligprep wizard in the Maestro Schrodinger suite.31 High-energy ionization states of the ligand molecules were minimized at pH 7.0 using Epik version 5.3. The OPLS3e force field was applied to identify potential chiral centers and generate potential stereoisomers, followed by another minimization step.Molecular docking
Natural phytochemicals identified through GS-MS were analyzed for protein–ligand binding scores using extra precision (XP) molecular docking with LasR. The Glide package in Glide v-8.8 and Maestro v-12.5.139 programs were utilized to assess the best binding scores between the phytochemicals and target proteins.33,34 Docking was performed in the standard precision mode with the OPLS3e force field. Binding positions of reference native inhibitor ligands, C12-HSL, and BB-78485-binding residues in LasR and LpxC active sites, respectively, were determined, and grid boxes were created accordingly. The center coordinates of the grid box were X = 7.25, Y = 2.53, Z = 33.1, X = 9.55, Y = 3.83, Z = 29.1 LasR and LpxC, respectively that determines the area where the ligand docking calculations were taken place. XP molecular docking simulations were conducted on the two proteins with 82 phytochemicals and native ligands, and from these simulations, target proteins and ligand-binding energy were extracted.Molecular mechanics-generalized born surface area (MM-GBSA) analysis
In this study, a MM-GBSA analysis was conducted using the Prime MM-GBSA program package to estimate the binding-free energy of ligands and validate the docking process between the LasR protein and ligands. The analysis considered negative MM-GBSA ΔG bind (NS), ΔG bind Coulomb (Coulomb energy), ΔG bind H-bond (hydrogen bond energy), ΔG bind lipo (lipophilicity energy), and ΔG bind vdW (van der Waals interaction energy) for the 12 highest-interacting ligands and the native ligand of the LasR protein.35,36 These features, representing energy contributions from different terms in the energy expression, provide valuable information on ligand, receptor, and complex structures, as well as energy differences related to strain and binding.Pharmacokinetics (PK) and toxicity (T) analysis
In drug development, investigating absorption, distribution, metabolism, and excretion (ADME), properties including physicochemical properties, lipophilicity, water solubility, drug-likeness, and synthetic accessibility helps identify potential druggable compounds.9 We analyzed the top twelve phyto-ligand molecules with docking scores below five and MM-GBSA profile. To predict the PK properties, we used the SwissADME server (https://www.swissadme.ch/) with the SMILES format data of these molecules.37 Additionally, we assessed toxicity using the ProTox-II web server (https://tox-new.charite.de/).38Molecular dynamics (MD) simulation
To assess protein–ligand interaction stability, a 100 ns MD simulation was conducted for the complex structures using “Desmond v6.3 Program” in Schrodinger 2020-3 on Linux. The simulation focused on LasR-phytocompound complex with a TIP3P water model. An orthorhombic box with 10 Å distance from the center maintained a specific volume, and Na+ and Cl-ions were added to neutralize the system (0.15 M salt concentration). OPLS3e force field was applied.39 The complex system was minimized under isothermal–isobaric (NPT) ensemble (pressure of 101325 pascals and temperature of 300 K). Stability and dynamic characteristics were analyzed using RMSD, RMSF, rGyr, and SASA values.Statistical analysis
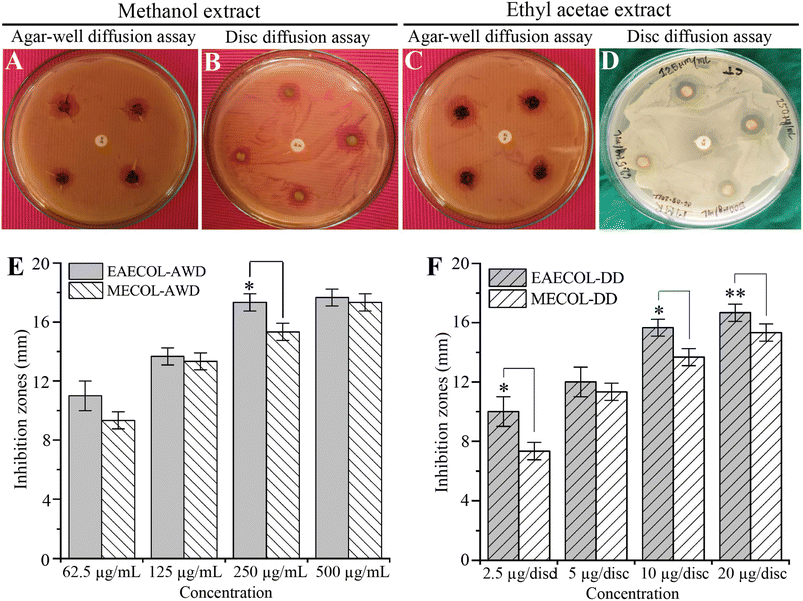
The antibacterial activity test results, performed at varying extract concentrations, were presented as the mean ± standard deviation (STD) of three independent replicates. The statistical analysis comprised one-way ANOVA conducted using Origin Lab 2018 software and Bonferroni's and Tukey's post hoc tests. Significance levels were indicated as *p < 0.05 and **p < 0.01.Results
Phytochemical screening
Phytochemical screening of C. occidentalis L. leaves revealed the presence of various compounds using standard color change methods listed in Table 1 and part of plant as well as primary phytochemical represented in Fig. S1 and S2.† Both MECOL and EAECOL showed light-yellow coloration with 5% NaOH, which faded to colorless with the addition of 10% HCl, indicating the presence of flavonoids.| Test name | Observation | MECOL | EAECOL | Phytochemical nature |
|---|---|---|---|---|
| Alkaline | Colorless solution | + | + | Phenolic (flavonoid) |
| Ferric chloride | Greenish-black coloration | + | + | Phenolic (tannins) |
| Lead acetate | Grey-white precipitation | + | + | Phenolic (tannins) |
| Salkowski's | Brown layer at upper interface | + | + | Terpenoids |
| Salkowski's | Blackish layer in the bottom face | + | + | Steroids |
| Mayer's | Creamy-colored precipitation absent | − | − | Absence of alkaloids |
| Legal's | Blood-red color | + | + | Cardiac glycosides |
| Foaming | Foam formation | + | + | Saponins |
| Fehling | Yellow-lime precipitation | + | + | Reducing sugar |
Tannins were detected by the greenish-black coloration after treatment with 5% FeCl3 and the formation of a grey-white precipitate with lead acetate. Terpenoids were confirmed by Salkowski's test, showing a light-yellow layer in the lower phase and a brown layer at the upper interface. Steroids were indicated by a deep brown layer in the lower phase and a light-yellow layer in the upper phase of Salkowski's test. Alkaloids were absent as Mayer's test did not produce creamy-white precipitation. Legal's test showed a blood-red color, confirming the presence of cardiac glycosides. Reducing sugars were detected with Fehling's test, resulting in a deep green and yellow-lime precipitation. Saponins were present, demonstrated by the 10 minute foaming stability in the foaming and frothing experiments.
FT-IR analysis
FT-IR analysis of MECOL and EAECOL detected various functional groups. Peaks in the spectrum indicated the presence of specific bonds and functional groups listed in Table 2 and shown in Fig. S3.† MECOL exhibited peaks at 3700–3000 cm−1 and 1178.56, 2921.32, 2849.95, 1742.76, 1632.81, 1463.07–1383.96, and 418.57 cm−1, indicating the presence of carboxylic acids, aliphatic hydrocarbons, aldehydes, aldehydes/esters, alkenes, alkanes, and alkyl halides.| Extract | Wavelength (cm −1) | Peak intensity | Functional group |
|---|---|---|---|
| MECOL | 3700–3000 | Broad | Carboxylic acids (O–H stretch) |
| 2921.32 | Weak | Aliphatic hydrocarbon (C–H stretch) | |
| 2849.95 | Weak | Aldehyde (C–H stretch) | |
| 1742.76 | Weak | Aldehyde or ester (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch) O stretch) |
|
| 1632.81 | Strong | Aromatic alkene (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch) C stretch) |
|
| 1463.07 | Weak | Alkane (–CH3– bend) | |
| 1383.96 | Medium | Alkane (–CH3– bend) | |
| 418.57 | Weak | Alkyl halide (C–X) | |
| EAECOL | 3700–3000 | Broad | Carboxylic acids (O–H stretch) |
| 2924.21 | Weak | Aliphatic hydrocarbon (C–H stretch) | |
| 2853.81 | Weak | Aldehyde (C–H stretch) | |
| 1712.86 | Medium | Aldehyde or ketone (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch) O stretch) |
|
| 1639.56 | Strong | Alkene (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch) C stretch) |
|
| 1629.92 | Strong | Primary or secondary amine (N–H bend) | |
| 1510.33 | Weak | Nitro (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch) O stretch) |
|
| 1460.18 | Medium | Alkane (–CH3– bend) | |
| 1401.34 | Medium | Aldehyde/alkane (C–H bend) | |
| 1384.95 | Medium | Alkane (–CH3– bend) | |
| 1178.56 | Weak | Carboxylic acids (C–O stretch) | |
| 1071.5 | Weak | Alcohol/phenol (O–H stretch) | |
| 839.07 | Weak | Aldehyde/alkane (C–H stretch) | |
| 721.41 | Weak | Alkyl halide (C–Cl) | |
| 517.91 | Weak | Alkyl halide (C–X) |
EAECOL showed peaks at 3700–3000 cm−1 and 1178.56, 2924.21, 2853.81, 1712.86, 1639.56, 1629.92, 1510.33, 1460.18, and 1384.95, 1401.34 and 839.07, 1071.50, 839.07, 721.41, 517.91–419.57 cm−1, indicating the presence of carboxylic acids, aliphatic hydrocarbons, aldehydes, aldehyde/ketone, alkene, primary or secondary amine, nitro, alkane, aldehyde/alkane, alcohol/phenol, alkyl chloride, and alkyl halide. A major peak at 1632 and 1639 cm−1 indicated the presence of aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds. The broad peak at 3700–3000 cm−1 may arise from the hydroxyl group (O–H stretch) of carboxylic acids. A trace of carbonyl (C
C bonds. The broad peak at 3700–3000 cm−1 may arise from the hydroxyl group (O–H stretch) of carboxylic acids. A trace of carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bond for aldehydes and esters was found at around 1742 cm−1 in methanol extract and 1712 cm−1 in ethyl acetate extract.
O) bond for aldehydes and esters was found at around 1742 cm−1 in methanol extract and 1712 cm−1 in ethyl acetate extract.
GC-MS analysis of MECOL and EAECOL
MECOL and EAECOL exhibited 46 and 43 peaks respectively in their GC-MS chromatograms shown in Fig. 1(A) and (B). Each peak represented a distinct phytochemical, and we estimated their relative percentage amounts based on average peak areas compared to total areas with retention time (RT) provided in Tables S1 and S2.†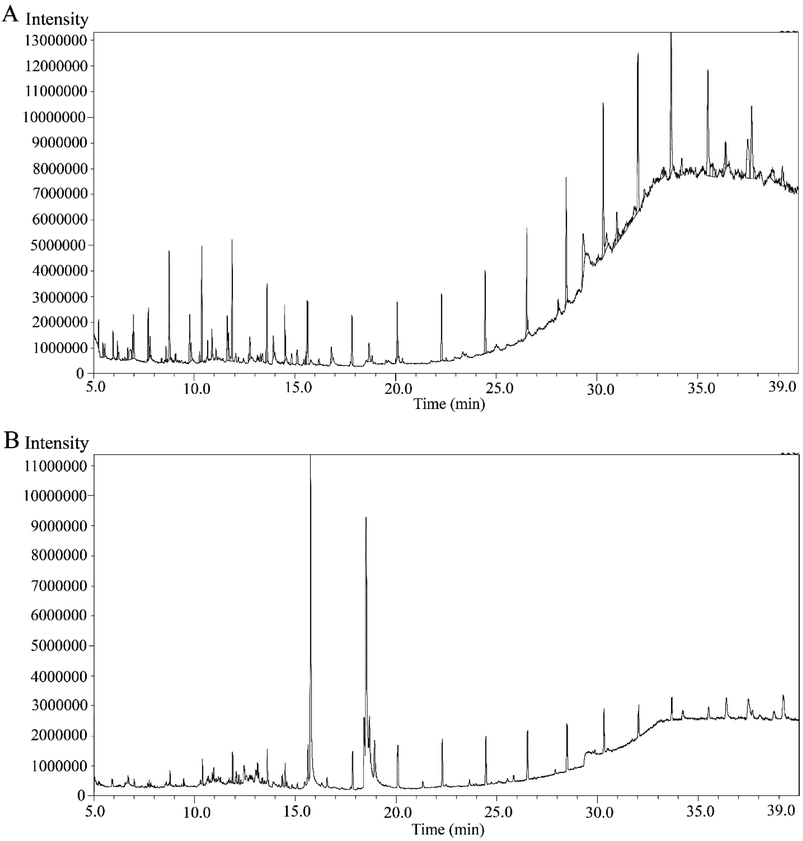 | ||
| Fig. 1 GC-MS spectrometry of C. occidentalis L. leaves extracts, showing different peaks representing various compounds. (A) Chromatogram of MECOL and (B) chromatogram of the EAECOL. | ||
These peaks indicated the presence of specific phytochemicals with unique identities, dependent on their chemical formula, and structure. The phytochemical composition of MECOL consisted of alkane, alcohol, alkene, steroid, terpene, amide, ester, ether, ketone, tocopherols, and phenol, with the major compounds being stigmasterol, 13-docosenamide, neophytadiene, phytol, tetrapentacontane, 1-hexadecanol, and 9-octadecene (Table S1†). Similarly, the phytochemicals identified in EAECOL included esters, carbohydrates, carbonyl compounds, amide, terpenoids, alkane, steroids, alcohol, ketone, phenol, and alkyne, with the primary constituents being esters such as 6-octadecenoic acid and hexadecanoic acid, along with 3-O-methyl-D-glucose and 13-docosenamide (Table S2†). Notably, both MECOL and EAECOL shared seven common phytochemicals (Table S3†) as identified through GC-MS analysis.
Antibacterial activity of C. occidentalis L. leaves extracts
In this study, we investigated the antibacterial potential of MECOL and EAECOL against multi-drug-resistant P. aeruginosa. The agar well diffusion and disc diffusion assays were conducted using various concentrations of MECOL and EAECOL (ranging from 62.55 to 500 μg mL−1).Our results demonstrated that both MECOL and EAECOL inhibited the proliferation of MDR P. aeruginosa, as evidenced by inhibition zones ranging from 9.33 ± 0.57 to 17.33 ± 0.57 mm for MECOL and 13.66 ± 0.57 to 17.66 ± 0.57 mm for EAECOL in the agar well diffusion assay represented in Fig. 2(A), (C) and (E). Similarly, in the disc diffusion assay, inhibition zones ranged from 7.33 ± 1 to 15.33 ± 0.57 mm for MECOL and 12 ± 1 to 16.66 ± 0.57 mm for EAECOL shown in Fig. 2(B), (D), and (F). Comparing the two compounds, EAECOL exhibited superior antimicrobial activity against P. aeruginosa in both the agar well diffusion and disc diffusion assays. Interestingly, erythromycin, a conventional antibiotic, showed no antibacterial activity against MDR P. aeruginosa, which had previously been confirmed to be resistant to erythromycin. The Minimum Inhibitory Concentration (MIC) values for MECOL and EAECOL were determined to be 104.16 ± 36.08 μg mL−1 and 83.33 ± 36.08 μg mL−1, respectively, while the Minimum Bactericidal Concentration (MBC) was found to be 125 μg mL−1 for both compounds presented in Fig. S4.† However, to achieve complete bacterial eradication, a higher concentration of MECOL and EAECOL was required compared to the concentration that inhibited visible bacterial growth in vitro. The experiments were independently repeated three times, and the average values with standard deviations were presented. Statistical analyses indicated that EAECOL showed a significant difference from MECOL in agar well diffusion and disc diffusion assays (*p < 0.05, **p < 0.01). The standard antibiotic erythromycin discs further validated the results.
Molecular docking and MM-GBSA analysis
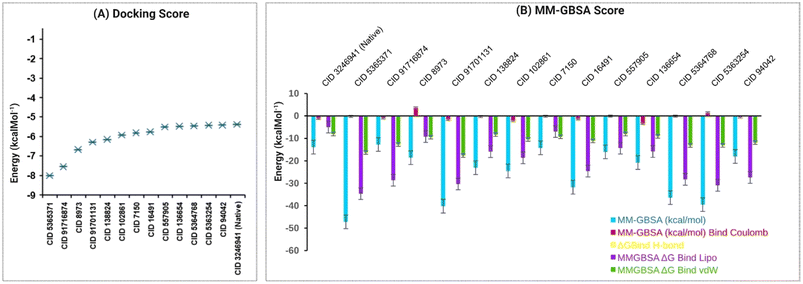
Our study performed molecular docking simulations and post-docking MM-GBSA analysis of 82 phytochemicals derived from C. occidentalis L. leaves against the QS signaling molecule receptor, LasR, of P. aeruginosa. Out of the docked phytochemicals, 13 compounds displayed significant negative binding affinity, ranging from −5.417 to −8.01 kcal mol−1, in comparison to the native LasR ligand, N-3-oxo-dodecanoyl-L-homoserine lactone (CID: 324694) having a binding score −5.375 kcal mol−1 shown in Fig. 3A listed in Table S4.†These 13 compounds were further subjected to MM-GBSA analysis and in silico investigations. Table S5† and Fig. 3B reveal that 12 phytochemical ligands exhibited favorable binding free energy scores compared to the native ligand of LasR (−13.9 kcal mol−1), except CID 91716874, which showed a score of −12.71 kcal mol−1. Among these, CID 91716874 and CID 5365371 demonstrated the lowest and highest negative MM-GBSA ΔG binding (NS) scores of −12.71 and −47.19 kcal mol−1, respectively. The post-docking analysis of LasR–ligand complexes indicated various interaction energies, including ΔG bind Coulomb (Coulomb energy), ΔG bind H-bond (hydrogen bond energy), ΔG bind lipo (lipophilicity energy), and ΔG bind vdW (van der Waals interaction energy), contributing to the overall binding stability of the complexes. Overall, our findings suggest that the 12 identified compounds have a strong binding affinity compared to the native ligand of the LasR protein, indicating their potential as promising candidates for further exploration in inhibiting the QS signaling of P. aeruginosa.
Pharmacokinetics (ADME) and toxicity (T)
The ADME and toxicity assessment is vital for drug development, ensuring safety and efficacy for regulatory approval. Therefore, the ADME and toxicity of the 12 compounds have been identified. Human intestinal absorption (HIA) is essential for drug bioavailability, with seven compounds showing high HIA and 5 exhibiting low HIA. BBB permeability was observed in 7 compounds, indicating CNS targeting, while five compounds lacked this potential. All 12 compounds were non-inhibitors of CYP450 enzymes, ensuring reduced metabolism interference. Nine compounds showed moderate clearance for drug excretion, while 3 had high clearance tendencies. In terms of toxicity, the 12 compounds showed promising results in silico evaluations, being non-hepatotoxic, non-carcinogenic, non-mutagenic, non-immunogenic, and non-cytotoxic. Seven compounds were non-toxic according to EPA toxicity categories, while five were toxic and listed in Table S6.† The physicochemical properties of the 12 compounds were assessed for their ADME influence. All compounds displayed favorable molecular weight, hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), heavy atoms, and topological polar surface area (TPSA) for absorption. Additionally, they had appropriate rotatable bond (RB) counts, indicating good absorption listed in Table S7.† Molecules with cLogP values of ≤5 are considered to have strong absorption, and nine compounds met this criterion. Three compounds were predicted to cross the membrane bilayer based on acceptable LogS values. Five compounds satisfied Lipinski's rule of five for drug-likeness, and eleven compounds showed ease of synthesis. Among these compounds, methyl dihydrojasmonate, methyl benzoate, and 4a-methyl-4,4a,5,6,7,8-hexahydro-2(3H)-naphthalenone were identified as the best-hit compounds based on their favorable pharmacokinetics, lack of toxicity, and suitable drug–ability profiles.Protein–ligands interactions
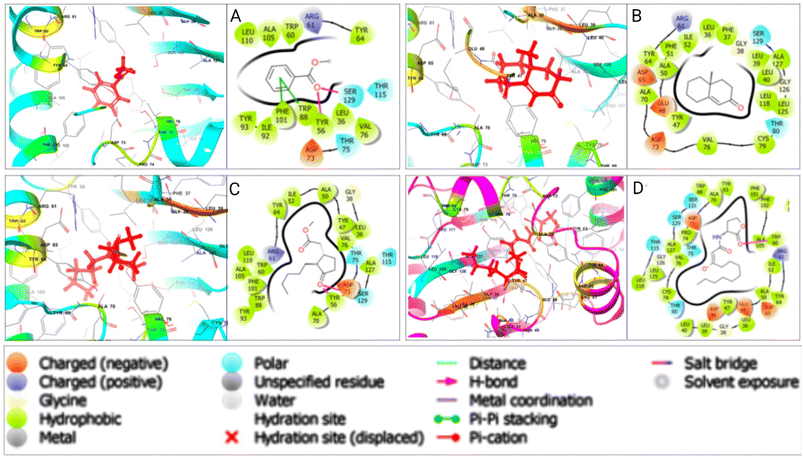
Various non-bonded interactions, such as hydrogen bonds, hydrophobic bonds, and electrostatic bonds, were observed between LasR and the best-hit phytochemicals. Methyl dihydrojasmonate displayed a hydrogen bond with TYR 56 and ASP 73, alongside other interactions, resulting in a binding affinity of −5.923 kcal mol−1. Methyl benzoate showed a hydrogen bond with TYR 56 and SER 129, and other interactions, with a binding affinity of −5.811 kcal mol−1. 4a-Methyl-4,4a,5,6,7,8-hexahydro-2(3H)-naphthalenone had a binding affinity of −5.472 kcal mol−1 with LasR, without a hydrogen bond but containing various other interactions. In contrast, the LasR-native ligand complex formed with a binding energy of −5.375 kcal mol−1, featuring a single TRP 60 hydrogen bond and other interactions. Fig. 4 illustrates the 3D and 2D interactions between the designated three phytocompounds (CID: 7150, CID: 136654, CID: 102861) and LasR, along with the LasR native ligand (CID: 3246941), within the protein's active pocket. The molecular docking interactions and interacting residues of LasR quorum sensing protein with the best phytochemical compounds and the native ligand are listed for each docked complex in Table S7.†Multi-targeting capabilities analysis of the phytochemicals
To explore the inhibitory potential and multi-targeting capabilities of the lead phytochemicals from C. occidentalis, we conducted molecular docking simulations with the LpxC enzyme. LpxC plays a crucial role in the synthesis of toxic bacterial outer lipid A membrane in Gram-negative P. aeruginosa and is a key target for antibiotic development. The phytocompounds CID 102861, CID 136654, and CID 7150 exhibited interactions with the LpxC target protein, with binding energies of −4.772, −4.488, and −3.505 kcal mol−1, respectively. In comparison, the native inhibitor of LpxC, CID 9823454, formed a bond with a binding energy of −4.604 kcal mol−1. This suggests that the potential lead phytocompounds have additional inhibitory interactions with the LpxC enzyme involved in lipid A biosynthesis, as shown in Table S9.†MD simulation
To assess the stability of the three potential hit phytocompounds (CID7150, CID102861, and CID136654) within the protein's binding site, MD simulations of the protein–ligand complex structure were conducted. The analysis considered parameters such as Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of Gyration (Rg), and Solvent Accessible Surface Area (SASA) to evaluate the constancy of the interactions between the LasR protein and the selected phytocompounds.RMSD analysis
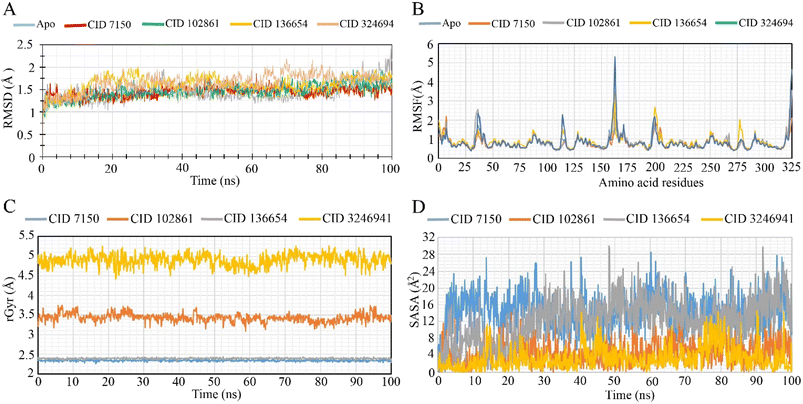
During the 100 ns simulation, the RMSD of Cα atoms was measured to evaluate the stability of the protein–ligand complex structure. Fig. 5A illustrates the average fluctuation of the selected phytocompounds (CID 7150, CID 102861, and CID 136654), which showed values of 1.45 Å, 1.47 Å, and 1.62 Å, respectively. The LasR apoprotein and native ligand complex exhibited average fluctuations of 1.42 Å and 1.65 Å, respectively. These minimal fluctuations, falling well within the acceptable range of 1–3 Å, indicate the conformational stability of the protein–ligand complex structure. Throughout the simulation trajectory, the compounds remained stable, with only minor fluctuations, affirming their structural stability. The highest RMSD fluctuation for CID 7150, CID 102861, and CID 136654 was 1.86 Å, 1.94 Å, and 2.01 Å, respectively, while the lowest RMSD value range was 0.98 Å, 0.87 Å, and 0.939 Å. Similarly, for the apoprotein and the native ligand (CID 3246941), the range was 0.93 Å to 2.40 Å and 0.84 Å to 2.18 Å, indicating good stability, resembling the apoprotein and native compounds as promising lead antibacterial agents.RMSF analysis
To assess the flexibility of the LasR protein in response to specific ligand interactions, RMSF values of compounds CID 7150, CID 102861, and CID 136654 were analyzed (Fig. 5B). The LasR apo protein showed peak fluctuations at HIS 169, GLU 168, ASP 43, PHE 167, and LYS 42 residues, while the native ligand exhibited fluctuations at HIS 169, HIS 169, GLU 168, PHE 7, and GLU 168 residues. The three selected compounds shared a common peak area at the HIS 169-residue location, indicating significant alterations during the simulation. CID 3246941 (native ligand) exhibited a higher RMSF value of 5.308 at residue 162, along with the apoprotein. For CID 7150, CID 102861, and CID 136654, RMSF values were 3.467, 2.368, 3.551, and 4.623, respectively, with CID 7150 showing the highest rigidity.Radius of gyration (Rg)
The radius of gyration (Rg) was analyzed to assess the distribution of atoms around the axis of the protein–compound complexes. CID 102861, CID-7150, CID-136654, and the native ligand (CID 3246941) stability in complex with the LasR protein were examined based on their Rg values over 100 ns simulations (Fig. 5C). The CID 3246941 (native ligand) and CID 102861 showed Rg ranges of 5.265 Å to 4.431 Å (fluctuation of 0.834 Å) and 3.755 Å to 3.097 Å (fluctuation of 0.658 Å), respectively. On the other hand, CID 7150 and CID 136654 exhibited Rg ranges of 2.395 Å to 2.263 Å (fluctuation of 0.132 Å) and 2.441 Å to 2.333 Å (fluctuation of 0.108 Å). Notably, all three phytocompounds demonstrated greater stability in 100 ns simulations, with lower fluctuation ranges compared to native ligand complexes, suggesting minimal conformational changes in the LasR active site due to the binding of the selected phytocompounds.Solvent accessible surface area (SASA)
SASA analysis was performed for the specific phytocompounds to determine the solvent-like characteristics (hydrophobic or hydrophilic) of the LasR protein and protein–compound complexes (Fig. 5D). The native ligand of LasR exhibited a fluctuation range from 0.015 Å2 to 14.608 Å2, with an average fluctuation of 3.287 Å2. In contrast, phytocompounds CID 7150, CID 102861, and CID 136654 showed fluctuation ranges of 0.121 Å2 to 28.514 Å2, 0.004 Å2 to 18.845 Å2, and 0.073 Å2 to 30.044 Å2, respectively, with average fluctuations of 14.14 Å2, 4.74 Å2, and 12.36 Å2. These values indicate the exposure of amino acid residues to the designated phytocompounds in complex systems, suggesting distinct hydrophobic or hydrophilic interactions.Discussion
P. aeruginosa poses a significant threat to public health, especially in hospitals and intensive care units, due to its multidrug-resistant nature.39 Current treatment strategies rely on a combination of antibiotics, but the emergence of resistance and associated side effects has led to a pressing need for innovative antibacterial medications.40,41 In this context, there is growing interest in exploring phytocompounds as potential antimicrobial agents, as they offer diverse chemical structures and biological activities with minimal side effects.Our study focused on the antibacterial potential of C. occidentalis leaf extracts and their phytocompounds against MDR P. aeruginosa. Prior research has explored the antimicrobial activity of these leaf extracts against various pathogens, but there was a lack of information on their activity against MDR P. aeruginosa. Therefore, we aimed to investigate the antibacterial properties of these phytocompounds, particularly targeting LasR, a crucial factor involved in drug resistance.
Initially, GC-MS analysis identified 82 chemical structures, which were docked and analyzed for MM-GBSA to assess their potential as antibacterial agents targeting LasR. Molecular docking simulation identified 13 lead phytocompounds with higher binding affinity, whereas MM-GBSA identified 12 best compounds binding to LasR and evaluated their pharmacokinetics, toxicity, and drug-ability profiles. CID 7150, CID 102861, and CID 136654 showed the most promising characteristics and were selected for further studies. We also evaluated the interaction of these phytocompounds with LpxC, an enzyme involved in lipid A biosynthesis, which is crucial for bacterial outer membrane synthesis.42 The lead phytocompounds demonstrated multitargeting ability and drug-like properties, making them attractive candidates for antimicrobial therapeutics.
Molecular dynamics simulations were performed to assess the stability and structural changes of the protein–ligand complexes. The RMSD, RMSF, Rg, and SASA analyses revealed that the selected phytocompounds formed stable complexes with LasR, indicating their potential as antibacterial agents against MDR P. aeruginosa.
Among the lead phytocompounds, methyl dihydrojasmonate (CID 102861) has been reported for its diverse activities, including anticancer43 and antimicrobial properties.44 Methyl benzoate (CID 7150) has shown promise as an environmentally friendly insecticide45 and an antibacterial agent46 against Gram-negative bacteria. Considering the diverse parameters used for evaluation, CID 7150, CID 102861, and CID 136654 were identified as the most promising lead phytocompounds for developing antibacterial drugs against MDR P. aeruginosa and related infectious diseases. These compounds warrant further investigation in human in vivo studies to validate their potential as antimicrobial therapeutics.
Conclusion
In this study, C. occidentalis L. extract exhibited antibacterial activity against MDR P. aeruginosa, as demonstrated by inhibitory zones in disc diffusion and agar well diffusion tests. Computer-aided drug design identified three lead compounds (methyl dihydrojasmonate, methyl benzoate, and 4a-methyl-4,4a,5,6,7,8-hexahydro-2(3H)-naphthalene) that competitively inhibited LasR, the key signaling molecule receptor responsible for P. aeruginosa's virulence and multi-drug resistance. These findings offer potential for developing new bioactive compounds to combat antibiotic-resistant infections. Further in vivo evaluations are needed to validate our results.Author contributions
Raihan Rahman Imon: writing – original draft, investigation, methodology, formal analysis. EKT: investigation, methodology, formal canalysis. Shahina Akhter: conceptualization, supervision, writing – review & editing. MSI: resources, writing – review & editing. Foysal Ahammad: resources; software; writing – review & editing; visualization. K. M. Anis-Ul-Haque: writing – review & editing; formal analysis. Md. Moniruzzaman: formal analysis; methodology. Mirola Afroze: formal analysis; methodology. Mala Khan: resources, data curation, validation. Mohammad Abu Hena Mostofa Jamal: resources. Tanveer A. Wani: funding acquisition, writing – review & editing. Mohammad Jashim Uddin: conceptualization, funding acquisition, writing – review & editing. Md. Mashiar Rahman: conceptualization, resources, Supervision, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors affirm that they have no known competing financial interests or personal relationships that could have influenced the work presented in this paper.Acknowledgements
This research was supported by the Research Cell of Jashore University of Science and Technology, Bangladesh (grant ID: JUST/Research Cell/22-FoBST-06/2022-2023). The authors extend their appreciation to researchers supporting project number (RSP2023R357) at King Saud University, Riyadh, Saudi Arabia, for funding this research.References
- J. P. Horcajada, M. Montero and A. Oliver, et al., Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections, Clin. Microbiol. Rev., 2019, 32, e000311–e000319 CrossRef PubMed.
- E. Tacconelli, E. Carrara and A. Savoldi, et al., Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed.
- L. Raka, S. Kalenc and Z. Bosnjak, et al., Molecular epidemiology of Acinetobacter baumannii in central intensive care unit in Kosova teaching hospital, Braz. J. Infect. Dis., 2009, 13, 408–413 CrossRef.
- H. Fazzeli, R. Akbari and S. Moghim, et al., Pseudomonas aeruginosa infections in patients, hospital means, and personnel’s specimens, J. Res. Med. Sci., 2012, 17, 332–337 CAS.
- J. A. Driscoll, S. L. Brody and M. H. Kollef, The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections, Drugs, 2007, 67, 351–368 CrossRef CAS PubMed.
- I. Mochalkin, J. D. Knafels and S. Lightle, Crystal structure of LpxC from Pseudomonas aeruginosa complexed with the potent BB-78485 inhibitor, Protein Sci., 2008, 17, 450–457 CrossRef CAS PubMed.
- M. Kostylev, D. Y. Kim and N. E. Smalley, et al., Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy, Proc. Natl. Acad. Sci. U.S.A., 2019, 116, 7027–7032 CrossRef CAS PubMed.
- G. Rampioni, I. Bertani and E. Zennaro, et al, The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter, J. Bacteriol., 2006, 188, 815–819 CrossRef CAS PubMed.
- W. Zhou, Y. Wang and A. Lu, et al., Systems pharmacology in small molecular drug discovery, Int. J. Mol. Sci., 2016, 17, 246 CrossRef.
- S. Mukherjee, D. Moustafa and C. D. Smith, et al., The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer, PLoS Pathog., 2017, 13, e1006504 CrossRef PubMed.
- G. Krishnamoorthy, I. V. Leus and J. W. Weeks, et al., Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria, mBio, 2017, 8, e01172 CrossRef CAS PubMed.
- H. A. Abbas, M. A. Shaldam and D. Eldamasi, Curtailing quorum sensing in Pseudomonas aeruginosa by sitagliptin, Curr. Microbiol., 2020, 77, 1051–1060 CrossRef CAS.
- E. M. Abdallah, Plants: an alternative source for antimicrobials, J. Appl. Pharm. Sci., 2011, 1, 16–20 Search PubMed.
- U. Anand, N. Jacobo-Herrera and A. Altemimi, et al., A comprehensive review on medicinal plants as antimicrobial therapeutics: potential avenues of biocompatible drug discovery, Metabolites, 2019, 9, 258 CrossRef CAS PubMed.
- E. Patridge, P. Gareiss and M. S. Kinch, et al., An analysis of FDA-approved drugs: natural products and their derivatives, Drug Discovery Today, 2016, 21, 204–207 CrossRef CAS PubMed.
- K. Keita, C. Darkoh and F. Okafor, Secondary plant metabolites as potent drug candidates against antimicrobial-resistant pathogens, SN Appl. Sci., 2022, 4, 209 CrossRef CAS PubMed.
- K. Lewis and F. M Ausubel, Prospects for plant-derived antibacterials, Nat. Biotechnol., 2006, 24, 1504–1507 CrossRef CAS.
- M. Tillhon, L. M. G. Ortiz and P. Lombardi, et al., Berberine: new perspectives for old remedies, Biochem. Pharmacol., 2012, 84, 1260–1267 CrossRef CAS PubMed.
- D. Załuski, L. Ciesla and Z. Janeczko, et al., The structure–activity relationships of plant secondary metabolites with antimicrobial, free radical scavenging and inhibitory activity toward selected enzymes, Stud. Nat. Prod. Chem., 2015, 45, 217–249 Search PubMed.
- J. P. Yadav, V. Arya and S. Yadav, et al., Cassia occidentalis L., A review on its ethnobotany, phytochemical and pharmacological profile, Fitoterapia, 2010, 81, 223–230 CrossRef CAS.
- M. Bhagat and A. K. Saxena, et al., Evaluation of Cassia occidentalis for in vitro cytotoxicity against human cancer cell lines and antibacterial activity, Indian J. Pharmacol., 2010, 42, 234–237 CrossRef PubMed.
- Rajni, S. S. Gautam and Navneet, Antibacterial and phytochemical analysis of Cassia occidentalis L. seeds against respiratory tract pathogens, Indian J. Nat. Prod. Resour., 2014, 5, 52–55 Search PubMed.
- M. M. Rahman, M. M. Kabir and M. A. A. Noman, et al., Mikania cordata leaves extract promotes activity against pathogenic bacteria and anticancer activity in EAC cell-bearing swiss albino mice, J. Appl. Pharm. Sci., 2020, 10, 112–122 CrossRef CAS.
- M. M. Rahman, M. A. A. Noman and S. Khatun, et al., Evaluation of Senna tora (L.) Roxb. leaves as source of bioactive molecules with antioxidant, anti-inflammatory and antibacterial potential, Heliyon, 2023, 9, e12855 CrossRef CAS PubMed.
- M. A. Aziz, et al., Qualitative phytochemical screening and evaluation of anti-inflammatory, analgesic and antipyretic activities of Microcos paniculata barks and fruits, J. Integr. Med., 2015, 13, 173–184 CrossRef PubMed.
- S. Akhter, M. W. Hossain and S. Sultana, et al., Ruellia prostrata Poir. activity evaluated by phytoconstituents, antioxidant, anti-inflammatory, antibacterial activity, and in silico molecular functions, J. Saudi Chem. Soc., 2022, 26, 101401 CrossRef CAS.
- R. Alam, R. R. Imon and M. E. K. Talukder, et al., GC-MS analysis of phytoconstituents from Ruellia prostrata and Senna tora and identification of potential anti-viral activity against SARS-CoV-2, RSC Adv., 2021, 11, 40120–40135 RSC.
- Y. He, Y. Jia and F. Lu, et al., New products generated from the transformations of ferulic acid dilactone, Biomolecules, 2020, 10, 175 CrossRef CAS PubMed.
- M. A. H. M. Jamal, M. A. A. Mashud and M. Moinuzzaman, et al., Green synthesis of silver nanoparticles using Cinnamomum tamala (Tejpata) leaf and their potential application to control multidrug resistant Pseudomonas aeruginosa isolated from hospital drainage water, Heliyon, 2022, 8, e09920 CrossRef PubMed.
- P. W. Rose, A. Prlic and A. Altunkaya, et al., The RCSB protein data bank: integrative view of protein, gene and 3D structural information, Nucleic Acids Res., 2017, 45, D271–D281 CrossRef CAS PubMed.
- G. M. Sastry, M. Adzhigirey and T. Day, et al., Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J. Comput.-Aided Mol. Des., 2013, 27, 221–234 CrossRef PubMed.
- E. Harder, W. Damm and J. Maple, et al., OPLS3: a force field providing broad coverage of drug-like small molecules and proteins, J. Chem. Theory Comput., 2016, 12, 281–296 CrossRef CAS PubMed.
- R. A. Friesner, R. B. Murphy and M. P. Repasky, et al., Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- D. Shivakumar, E. Harder and W. Damm, et al., Improving the prediction of absolute solvation free energies using the next generation OPLS force field, J. Chem. Theory Comput., 2012, 8, 2553–2558 CrossRef CAS PubMed.
- P. D. Lyne, M. L. Lamb and J. C. Saeh, et al., Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring, J. Med. Chem., 2006, 49, 4805–4808 CrossRef CAS PubMed.
- F. Ahammad, R. Alam and R. Mahmud, et al., Pharmacoinformatics and molecular dynamics simulation-based phytochemical screening of neem plant (Azadiractha indica) against human cancer by targeting MCM7 protein, Briefings Bioinf., 2021, 22, bbab098 CrossRef PubMed.
- B. G. Gimenez, M. S. Santos and M. Ferrarini, et al., Evaluation of blockbuster drugs under the rule-of-five, Pharmazie, 2010, 65, 148–152 CAS.
- P. Banerjee, A. O. Eckert and A. K. Schrey, et al., ProTox-II: a webserver for the prediction of toxicity of chemicals, Nucleic Acids Res., 2018, 46, W257–W263 CrossRef CAS PubMed.
- P. Pachori, R. Gothalwal and P. Gandhi, Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review, Genes Dis., 2019, 6, 109–119 CrossRef PubMed.
- D. C. Nwobodo, M. C. Ugwu and C. O. Anie, et al., Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace, J. Clin. Lab. Anal., 2022, 36, e24655 CrossRef PubMed.
- J. Murugaiyan, P. A. Kumar and G. S. Rao, et al., Progress in alternative strategies to combat antimicrobial resistance: focus on antibiotics, Antibiotics, 2022, 11, 200 CrossRef CAS PubMed.
- E. M. Fivenson and T. G. Bernhardta, An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli, mBio, 2020, 11, e00939 CrossRef PubMed.
- R. Yehia, R. M. Hathout and D. A. Attia, et al., Anti-tumor efficacy of an integrated methyl dihydrojasmonate transdermal microemulsion system targeting breast cancer cells: in vitro and in vivo studies, Colloids Surf., B, 2017, 155, 512–521 CrossRef CAS PubMed.
- F. Yildirim, Y. Memis and A. Ozturk, et al., Antimicrobial activity of the essential oil and the extracts of Kitaibelia balansae species, J. Essent.Oil Bear. Plants, 2017, 20, 809–819 CrossRef CAS.
- M. M. Mostafiz, E. Hassan and K.-Y. Lee, et al., Methyl benzoate as a promising, environmentally safe insecticide: current status and future perspectives, Agriculture, 2022, 12, 378 CrossRef CAS.
- S. Mohanapriya, M. Muthukumaran and S. Vairam, Synthesis, characterization, thermal behavior and antimicrobial activity of 3-methyl benzoate complexes of transition metal with hydrazine, Bull. Chem. Soc. Ethiop., 2016, 30, 241–252 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra03923d |
| This journal is © The Royal Society of Chemistry 2023 |