Open Access Article
Open Access ArticleAffinity- and activity-based probes synthesized from structurally diverse hops-derived xanthohumol flavonoids reveal highly varied protein profiling in Escherichia coli†
Lucas C. Webbera,
Lindsey N. Andersona,
Ines L. Paraisob,
Thomas O. Metz a,
Ryan Bradleycd,
Jan F. Stevensb and
Aaron T. Wright
a,
Ryan Bradleycd,
Jan F. Stevensb and
Aaron T. Wright *aef
*aef
aBiological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA. E-mail: a_wright@baylor.edu
bDepartment of Chemistry, Linus Pauling Institute, Oregon State University, Corvallis, Oregon 97331, USA
cHelfgott Research Institute, National University of Natural Medicine, Portland, Oregon 97201, USA
dHerbert Wertheim School of Public Health and Human Longevity Science, University of California, San Diego, La Jolla, CA 92093, USA
eDepartment of Biology, Baylor University, Waco, Texas 76708, USA
fDepartment of Chemistry & Biochemistry, Baylor University, Waco, Texas 76708, USA
First published on 11th October 2023
Abstract
Xanthohumol, the principle prenylflavonoid found in hops (Humulus lupulus) and a reported anti-inflammatory agent, has great potential for pharmaceutical interventions related to inflammatory disorders in the gut. A suite of probes was prepared from xanthohumol and its structural isomer isoxanthohumol to enable profiling of both protein affinity binding and catalytic enzyme reactivity. The regiochemistry of the reactive group on the probes was altered to reveal how probe structure dictates protein labeling, and which probes best emulate the natural flavonoids. Affinity- and activity-based probes were applied to Escherichia coli, and protein labeling was measured by chemoproteomics. Structurally dependent activity-based probe protein labeling demonstrates how subtle alterations in flavonoid structure and probe reactive groups can result in considerably different protein interactions. This work lays the groundwork to expand upon unexplored cellular activities related to xanthohumol interactions, metabolism, and anti-inflammatory mechanisms.
Introduction
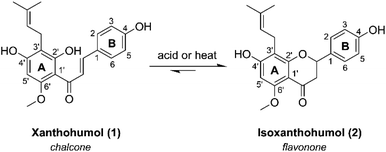
Hops (Humulus lupulus) have been an essential ingredient in the beer making process for over a thousand years.1,2 Beer flavor qualities are due to a variety of biologically relevant secondary metabolites present in hops, including alpha- and beta-humulic acids, terpenes, and flavonoids compounds; all of which have been investigated for potential health-promoting activities.3–5 Of the many flavonoids found in hops, xanthohumol (XN) (Fig. 1), a prenylated (3-methylbut-2-en-1-yl) chalcone, is the most abundant, constituting 0.1–1% of the overall dry weight.6 XN has reported anti-obesity,7–9 anti-cancer,10,11 and anti-inflammatory effects.12,13 Once ingested, XN undergoes numerous transformations, producing various prenylflavonoids through chemical or enzymatic processes, resulting in isoxanthohumol (IXN) (Fig. 1), and side products 8-prenylnaringenin, and dihydroxanthohumol.14XN has shown promise in mediating the treatment of inflammation, a common characteristic of many chronic gut-related diseases, and a key feature of a group of gastrointestinal diseases falling under the umbrella of inflammatory bowel disease (IBD), including Crohn's disease and ulcerative colitis.15 In vitro studies12 suggest XN is able to upregulate several detoxification metabolic proteins to protect against oxidative stress and increased inflammation (e.g. glutathione-S-transferase, heme oxygenase-1, NAD(P)H-quinone oxidoreductase 1), and downregulate pro-inflammatory factors of NF-κB (e.g. inducible nitric oxide synthase, cytokines, cyclooxygenase-2). These related findings coupled with the dietary ubiquity of XN, and a recent clinical trial revealing higher doses of XN is safe and well-tolerated in adult humans,16 suggest that it has significant potential as an oral therapeutic for alleviation of IBD symptoms. Considering its role as a prevalent natural product, as well as interest in potential XN-based therapeutics for treating inflammation in the human gut17–19 in relation to inflammation-induced pathologies, it is crucial to further investigate the interactions related to XN and IXN catabolism resulting in gut-permeable metabolites involved in host-gut microbiome resiliency.
Host microbiome gut bacteria are capable of multiple biochemical transformations altering the activity or toxicity of xenobiotics.20,21 As a result, the gut microbiome community taxonomy and function may fluctuate heavily under extended xenobiotic exposure. Chemical drug transformations can be altered by the microbiome, and in turn the microbiome can be altered by the drug.22,23 Gut microbes such as Eubacterium ramulus and Eubacterium limosum are capable of metabolizing XN into compounds of variable biological activity. Such activity includes decreased biological activity of the reduced chalcone (DXN), increased biological activity of a potent phytoestrogen (8-PN), and unknown biological activity of the demethylated XN (DMX).14 As such, it is critical to gain a better understanding of the microbial mechanisms of the proteins, pathways, and catalytic enzymes involved in the direct and/or indirect metabolic conversions of XN in the human host gut microbiome prior to engineering pharmaceutical interventions for XN-based therapeutics in diseases such as IBD.23,24
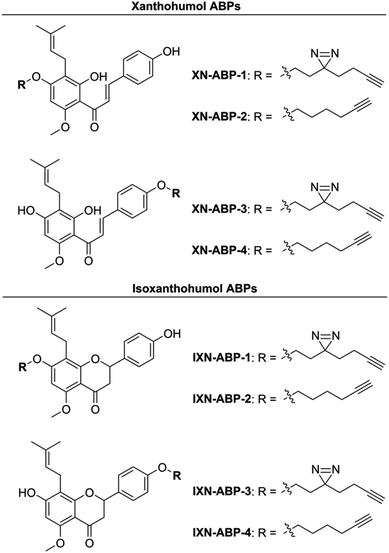
Herein, to determine the scope and range of XN covalent and non-covalent protein interactions, we developed a suite of affinity- and activity-based probes (ABPs) based on the core chemical structures of XN and IXN (Fig. 2). These probes were designed to capture the diverse structural potential of XN conversions, and to improve and expand upon a prior XN-based probe alkylated only on the B ring,25 which was used only for labeling mammalian proteins through Michael-type addition. This suite of ABPs aims to profile the range of non-covalent and covalent xanthohumol–protein interactions (Scheme 1).
Ultimately, to fully understand the role XN has with the human microbiome in IBD, both covalent and non-covalent protein target specificity must be determined. To reach beyond the protein interactions occurring at the Michael-acceptor of XN, we aimed to develop photoaffinity-enabled ABPs through insertion of the minimalist, UV-activatable diazirine moiety, allowing for labeling of non-covalent protein–substrate interactions.26 ABPs based on both the XN and IXN chemical scaffolds allowed us to examine the impact of alkylation site and the binding modes of protein recognition for labeling. We expected the protein labeling affinity for each of the ABPs to vary depending on where the core structure is alkylated through the alteration of hydrogen bond interactions within the binding site of a given protein. From our results, we gained insight into which elements are important for substrate recognition through comparisons of probe protein labeling.
Results and discussion
Probe syntheses
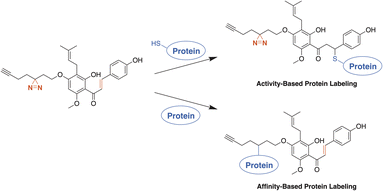
Activity- and affinity-based probes are comprised of three components: (1) a recognition group for targeting specific proteins of interest, (2) a reactive or photoreactive group to covalently label the protein of interest, and (3) a reporter group, which is commonly an alkyne or azide handle enabling biorthogonal click chemistry for downstream fluorescence and enrichment applications. XN contains an intrinsic, although selective, mechanism of covalent protein labeling by way of Michael-type addition at the α,β-unsaturated ketone. To develop ABPs that would profile the range of non-covalent protein targets, we appended a diazirine-alkyne linker to XN and IXN. By alkylating the native core of XN and IXN with either an alkyne linker, or a combination diazirine-alkyne linker, we synthesized a suite of eight ABPs with different protein labeling mechanisms (Fig. 2).We used purified XN (Hopsteiner, Yakima, WA, GAZ:22223928 (https://purl.obolibrary.org/obo/GAZ_22223928)) extracts from spent hops in lieu of total synthesis. Of the three hydroxyls on XN, only the 4 and 4′ are reactive towards alkylation, with little to no substitution observed on the 2′ hydroxyl (Fig. 1). Alkyne linker 3 was readily synthesized in one step from 5-hexynol using a modified Finkelstein reaction. Diazirine-alkyne linker 4 was sourced commercially (Ambeed, Arlington Heights, IL). Using the linkers 3 and 4, XN probes substituted on the 4′-hydroxyl could be synthesized via Williamson ether synthesis to generate ABPs XN-ABP-1 and XN-ABP-2 (Scheme 2). Complete chromatographic separation of the other products alkylated on the 4-OH phenol was not possible. Instead, selective protection of the A ring phenol of XN with methoxymethyl chloride (MOMCl) in N,N-dimethylformamide (DMF) afforded 5, of which the B ring phenol was then selectively alkylated with 3 or 4 using K2CO3 and DMF to afford 6 and 7. Careful acidic deprotection with HCl in MeOH and water gave ABPs XN-ABP-3 and XN-ABP-4.
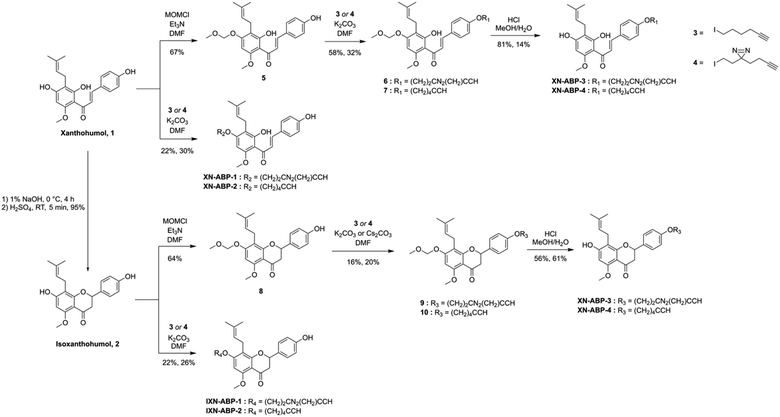 | ||
| Scheme 2 Synthesis of xanthohumol (XN) and isoxanthohumol (IXN) affinity- and activity-based probes. | ||
The general synthetic approach for the corresponding IXN ABPs was like those derived from XN (Scheme 2). Conversion of XN to IXN was achieved using a base-catalyzed Michael-type intramolecular cyclization of the inner ring in aqueous NaOH, followed by precipitation of IXN with concentrated sulfuric acid. IXN was then alkylated on the A ring phenol with 3 or 4, again by way of Williamson ether synthesis conditions, to generate ABPs IXN-ABP-1 and IXN-ABP-2. Similarly, complete chromatographic separation of the other alkylation products was not possible. Instead, IXN was selectively protected using MOMCl in DMF to afford 8. Subsequent alkylation of the protected IXN using similar Williamson ether synthesis conditions afforded compounds 9 and 10. Treatment of 9 and 10 with HCl in MeOH and water generated ABPs IXN-ABP-3 and IXN-ABP-4, respectively.
ABP labeling of chalcone isomerase
To validate ABP labeling of a protein with a known covalent interaction to XN, we labeled the chalcone isomerase N domain-containing protein (U2Q8X2; ∼32.5 kDa, CHI). CHI is a flavonoid degrading enzyme found in the anaerobic gut bacteria Eubacterium ramulus ATCC 29099 (Taxon: 1256908), catalyzing the reversible transformation of a chalcone (i.e. XN) to a flavanone (i.e. IXN).27 As such, both XN and IXN are expected to be recognized as substrates for this enzyme. However, due to the anaerobic nature of E. ramulus and the relative low expression in the native microbe, purified chalcone isomerase N domain-containing protein (HMPREF0373 00112 (https://www.uniprot.org/uniprotkb/U2Q8X2)) was cloned into E. coli BL21-AI.27 ABPs were applied to a bacterial lysate of E. coli BL21-AI over-expressing CHI with incubation at 37 °C, UV-irradiation at 365 nm, click chemistry with the fluorescent reporter picolyl rhodamine-azide, and visualization by fluorescence imaging of proteins separated by SDS-PAGE (Fig. 3). Robust and selective labeling of CHI by XN-ABP-1 and IXN-ABP-1 was confirmed at 5 μM. Notably, labeling by XN-ABP-1 and IXN-ABP-1 is significantly attenuated without UV irradiation, highlighting that covalent labeling of CHI proceeds through the photoactivatable diazirine moiety and confirming the need and applicability of ABPs with a diazirine linker. As expected, minimal labeling is observed with the alkyne-only XN-ABP-2 via Michael addition.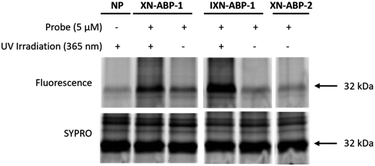 | ||
| Fig. 3 Validation of XN ABP labeling chalcone isomerase overexpressed in E. coli BL21. Samples were labeled, irradiated, then visualized with picolyl rhodamine azide using click-chemistry. Both photoaffinity probes (XN-ABP-1 and IXN-ABP-1) showed specific and robust labeling of CHI and required irradiation to achieve complete labeling. A no probe (NP) sample was included. A general protein stain (SYPRO) was used. Full gel available in ESI Fig. S1.† | ||
To further demonstrate strong probe labeling of CHI, overexpressed CHI in E. coli BL21-AI was diluted into the same, but uninduced, strain. After labeling each sample with XN-ABP-1 or IXN-ABP-1, the samples were irradiated, then visualized with picolyl rhodamine azide using click-chemistry. Separation of proteins by SDS-PAGE followed by fluorescence visualization revealed labeled proteins (Fig. 4). Both XN-ABP-1 and IXN-ABP-1 showed strong labeling (Fig. 4) even as CHI was increasingly diluted into the background lysate of E. coli. Decreased fluorescence was observed concomitant with increased enzyme dilution. Indiscriminate labeling is observed in the heat shock (“HS”, Fig. 4) samples, showing probe selectivity is lost when proteins are denatured demonstrating that enzyme function and/or structure is required for selectivity. Considering the promiscuity of typical diazirine probes, and the polyphenolic structure of XN/IXN, these probes demonstrate strong selectivity for chalcone isomerase when labeling at 5 μM.
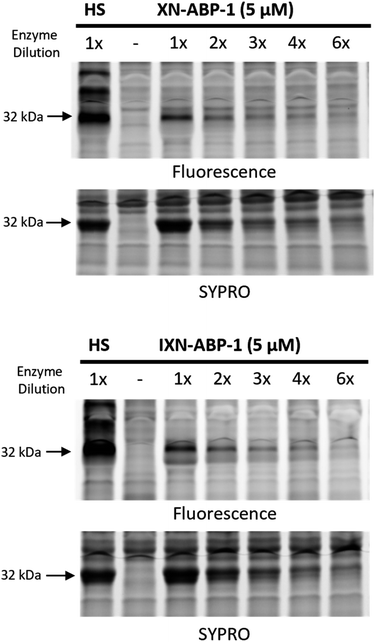 | ||
| Fig. 4 Concentration-dependent photoactivated labelling of CHI overexpressed in E. coli BL21-AI by XN-ABP-1 and IXN-ABP-1. Bacterial lysates containing increasing dilutions of CHI overexpressed in E. coli into uninduced E. coli lysates were co-incubated with XN-ABP-1 and IXN-ABP-1. A probe positive heat shock (HS) sample, and a no probe (NP) sample were included. A general protein stain (SYPRO) was used. ESI Fig. S2a and b† show full gels. | ||
Chemoproteomics profiling of XN–protein interactions
Chemoproteomics was employed to empirically investigate protein labeling by the suite of ABPs. Whole cell lysates from E. coli BL21-AI were individually incubated with each of the eight ABPs, irradiated with UV light for the photoaffinity ABPs, clicked using picolyl biotin-azide, and ABP labeled proteins enriched using streptavidin agarose beads, followed by trypsin digest, and analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The total ion abundances of peptide sequences and identified proteins associated with each ABP enriched sample were compiled and tabulated to generate a dataset (ESI Tables†) detailing the degree to which each ABP and sample type enriched for a specific identified protein, and sorted by iBAQ (Intensity Based Absolute Quantification) score.The photoaffinity probes XN-ABP-1 and IXN-ABP-1 effectively labeled CHI when compared to no probe control samples. Interestingly, none of the other six probes passed statistical filters. Thus, only modification of the A-ring of XN and IXN as ABPs is tolerated by CHI. The probes employing only an alkyne linker (XN-ABP-2, XN-ABP-4, IXN-ABP-2, and IXN-ABP-4) do not show any significant enrichment of CHI. This is predictable, as the only conceivable covalent labeling mechanism of the alkyne-only probes would be a through a Michael-type addition by a free nucleophilic residue near the active site of CHI, of which none are known. Further molecular docking analysis supports that XN and IXN will only bind well to CHI if the B-ring phenol is unmodified.27,28
Despite not all probes effectively labeling CHI, we believed it important to still analyze all ABP protein targets. It is not expected that all proteins that interact with xanthohumols bind through the same mechanism and/or configuration. Looking beyond CHI, a wide variety of protein targets were significantly enriched by the suite of ABPs. Relative quantification of protein targets labeled by the probe suite was calculated by iBAQ using MaxQuant quantification analysis software (v.1.6.17.0).29 732 proteins were identified as having a p value of <0.05 with a 1.5 fold-change versus no probe control samples, and having ≥2 replicate observations across all datasets within each ABP group. Notably, only a single protein was identified as labeled by all eight ABPs, NAD(P)H nitroreductase. More proteins were identified among the photoaffinity ABP subset.
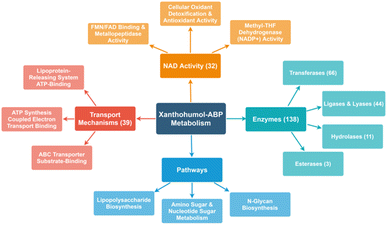
Of the proteins identified by the suite of ABPs, there is a broad range of biological and molecular functions targeted. A significant number of proteins with NAD activity and oxidoreductase activity (32 and 23, respectively) were labeled. Considering the well-established antioxidant activity of xanthohumol, it is unsurprising that numerous oxidoreductases were identified. For instance, prenylated phenols can weakly scavenge reactive oxygen species and may also function as antioxidants through anti-oxidative enzymes.12
Additionally, xanthohumols are known to exhibit a wide range of biological activities, partially due to their relatively small structure, allowing versatility as a potential substrate. Additionally, the Michael acceptor moiety provides an intrinsic protein cross-linking mechanism. As such, chemoproteomics revealed a wide range of targets in E. coli BL21-AI, including at least 39 proteins associated with transport mechanisms, 66 transferases, 44 lyases and ligases, and 11 hydrolases. In addition, some proteins were identified belonging to lipopolysaccharide synthesis, nucleotide sugar metabolism, and N-glycan biosynthesis (Fig. 5).
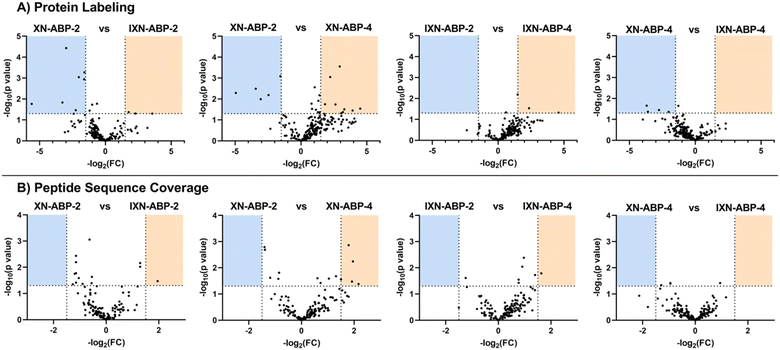
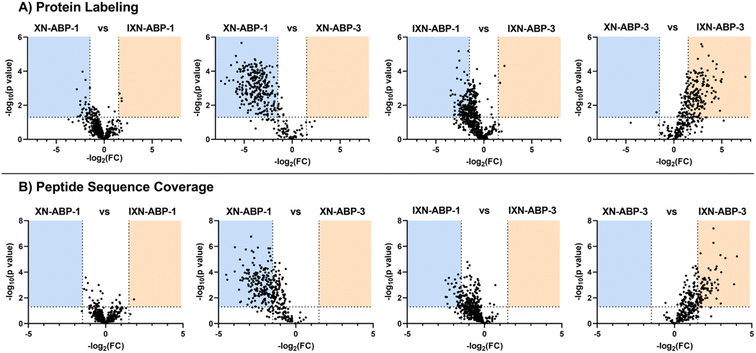
To further examine how structural diversity of the probe suite, comparisons were made between probe pairs with one variable isolated (i.e. core scaffold, alkylation site, or labeling mechanism). By calculating the differential iBAQ scores for each protein shared between each pair of ABPs, a relative measure of labeling performance could be determined. Across the board, XN-ABP-1 performs the best in terms of number of proteins identified, unique proteins, CHI labeling efficiency, and iBAQ score and peptide sequence coverage above similar probes. Conversely, XN-ABP-3 had the fewest number of proteins identified compared to the full set of photoaffinity probes, as well as the fewest uniquely identified proteins, suggesting that XN-ABP-3 could be conformationally distinct from the other 3 photoaffinity probes with regards to biological activity. Thus, this may lead to greater selectivity in identified proteins, and decreased labeling efficiency. When the same analysis is conducted using the peptide coverages, the results are similar (Fig. 6). Comparing probes with the same A-ring alkylation sites but differing scaffolds (XN-ABP-1 and IXN-ABP-1) we see similar performance with regards to peptide coverage as with protein affinity. For the IXN probes, similarly, alkylation on the A ring is strongly preferred, with most shared proteins having greater iBAQ scores and peptide coverage with IXN-ABP-1 than IXN-ABP-3. When comparing probes with the same core structure (chalcone or flavanone), it is evident that probes alkylated on the A ring tend to have greater biological activity, identifying a greater range of proteins, and have greater peptide coverage. All these factors point to a necessity for an unfunctionalized B ring hydroxyl for effective recognition by protein targets. This hypothesis is further substantiated by the fact that flavones and chalcones are most often observed as the 7-O-glycoside, with conjugation of a sugar to the 7-OH of the A ring, leaving the B ring unfunctionalized.30
Furthermore, it is believed that chalcones are typically the end product of plant biosynthesis pathways, and are more chemically reactive, and therefore, likely more biologically active.12 As such, it is unsurprising that the XN based probes would display stronger enrichment and a wider range of protein targets, both shared and unique.
A similar analysis was applied to the set of proteins identified by the alkyne-only ABPs (Fig. 7). However, for each probe-pair comparison, the number of shared proteins was lower, likely because only a few proteins have a nucleophile within the binding site capable of performing the Michael addition. Minimal evidence in support of ubiquitous Michael-type labeling of reactive thiols was observed. iBAQ scores and peptide coverage did not strongly favor either scaffold, suggesting that perhaps most of the labeling in this set comes from only the strongest hydrophobic interactions. As such, those interactions likely will not be affected by the structural nuances between these probe pairs. In this bacterial model, it's apparent that only a portion of the biological significance might come from the alkene moiety of XN, yet the potential for other biology activities is high and should be investigated further.
Conclusions
In summary, we have developed a structurally diverse suite of XN and IXN based ABPs, through attachment of alkyne or diazirine alkyne linkers to the native substrates. By utilizing multiple core scaffolds, alkylation sites, and labeling mechanisms, we were able to examine how each of these variables affects protein binding within a bacterial model. Initial tests with the known flavonoid degrading enzyme CHI showed that the photoaffinity probes are close mimics for the native substrates. ABPP in E. coli BL21 identified a wide range of novel protein targets, and by comparing differential iBAQ scores and peptide coverages between specific probe pairs, we determined XN-ABP-1 and IXN-ABP-1 to be the highest performing ABPs. Despite the importance placed on the intrinsic reactivity of XN towards nucleophilic thiols, we observed minimal evidence in support of widespread protein labeling through a Michael-type addition in a bacterial system. Ongoing work in our group includes application of this refined ABP set to human fecal samples for identification of the molecular interactions of XN and its metabolites with the gut microbiome, and to elucidate how specific bacteria could influence the use of XN as an oral therapeutic for IBD.Data availability
The data described in this study are openly available at MassIVE at https://doi.org/doi:10.25345/C55295 (direct access: ftp://massive.ucsd.edu/MSV000088789/).Author contributions
Conceptualization (LCW, ATW, TOM); data curation (LNA); formal analysis (LCW, LNA, ATW); funding acquisition (TOM, RB, JFS, ATW); investigation (LCW, ILP, LNA, ATW); methodology (LCW, ATW); resources (VLP); supervision (ATW); validation (LCW); visualization (LCW, LNA); writing – draft (LCW, LNA, ATW); writing – editing (LCW, ILP, LNA, TOM, RB, JFS, ATW); data dissemination (LNA).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Center for Complementary and Integrative Health (NCCIH, R01 AT010271). Data acquisition was performed in the Environmental Molecular Sciences Laboratory (EMSL) under the EMSL project award 51663. EMSL is a DOE Office of Science User Facility sponsored by the Biological and Environmental Research program under Contract No. DE-AC05-76RL01830.References
- C. Schönberger and T. Kostelecky, 125th Anniversary Review: the Role of Hops in Brewing, J. Inst. Brew., 2011, 117, 259–267 CrossRef.
- M. Verzele, 100 Years of Hop Chemistry and its Relevance, J. Inst. Brew., 1986, 92, 32–48 CrossRef CAS.
- C. Sharma, J. M. Al Kaabi, S. M. Nurulain, S. N. Goyal, M. A. Kamal and S. Ojha, Polypharmacological Properties and Therapeutic Potential of beta-Caryophyllene: A Dietary Phytocannabinoid of Pharmaceutical Promise, Curr. Pharm. Des., 2016, 22, 3237–3264 CrossRef CAS PubMed.
- A. Y. Benkherouf, K. Eerola, S. L. Soini and M. Uusi-Oukari, Humulone Modulation of GABAA Receptors and Its Role in Hops Sleep-Promoting Activity, Front. Neurosci., 2020, 14, 594708 CrossRef PubMed.
- J. Olšovská, V. Boštíková, M. Dušek, V. Jandovská, K. Bogdanová, P. Čermák, P. Boštík, A. Mikyska and M. Kolář, Humulus lupulus L. (Hops) - a Valuable Source of Compounds with Bioactive Effects for Future Therapies, Mil. Med. Sci. Lett., 2016, 85, 19–30 CrossRef.
- J. F. Stevens and J. E. Page, Xanthohumol and related prenylflavonoids from hops and beer: to your good health, Phytochem, 2004, 65, 1317–1330 CrossRef CAS PubMed.
- I. E. Logan, N. Shulzhenko, T. J. Sharpton, G. Bobe, K. Liu, S. Nuss, M. L. Jones, C. L. Miranda, S. Vasquez-Perez, J. M. Pennington, S. W. Leonard, J. Choi, W. Wu, M. Gurung, J. P. Kim, M. B. Lowry, A. Morgun, C. S. Maier, J. F. Stevens and A. F. Gombart, Xanthohumol Requires the Intestinal Microbiota to Improve Glucose Metabolism in Diet-Induced Obese Mice, Mol. Nutr. Food Res., 2021, 65, e2100389 CrossRef PubMed.
- I. L. Paraiso, T. Q. Tran, A. A. Magana, P. Kundu, J. Choi, C. S. Maier, G. Bobe, J. Raber, C. Kioussi and J. F. Stevens, Xanthohumol ameliorates Diet-Induced Liver Dysfunction via Farnesoid X Receptor-Dependent and Independent Signaling, Front. Pharmacol, 2021, 12, 643857 CrossRef CAS PubMed.
- S. Fukizawa, M. Yamashita, K. I. Wakabayashi, S. Fujisaka, K. Tobe, Y. Nonaka and N. Murayama, Anti-obesity effect of a hop-derived prenylflavonoid isoxanthohumol in a high-fat diet-induced obese mouse model, Biosci. Microbiota, Food Health, 2020, 39, 175–182 CrossRef CAS PubMed.
- C. H. Jiang, T. L. Sun, D. X. Xiang, S. S. Wei and W. Q. Li, Anticancer Activity and Mechanism of Xanthohumol: A Prenylated Flavonoid From Hops (Humulus lupulus L.), Front. Pharmacol., 2018, 9, 530 CrossRef PubMed.
- T. Constantinescu and C. N. Lungu, Anticancer Activity of Natural and Synthetic Chalcones, Int. J. Mol. Sci., 2021, 22, 11306 CrossRef CAS PubMed.
- J. L. Bolton, T. L. Dunlap, A. Hajirahimkhan, O. Mbachu, S. N. Chen, L. Chadwick, D. Nikolic, R. B. van Breemen, G. F. Pauli and B. M. Dietz, The Multiple Biological Targets of Hops and Bioactive Compounds, Chem. Res. Toxicol., 2019, 32, 222–233 Search PubMed.
- G. I. Vazquez-Cervantes, D. R. Ortega, T. Blanco Ayala, V. Perez de la Cruz, D. F. G. Esquivel, A. Salazar and B. Pineda, Redox and Anti-Inflammatory Properties from Hop Components in Beer-Related to Neuroprotection, Nutrients, 2021, 13, 2000 Search PubMed.
- I. L. Paraiso, L. S. Plagmann, L. Yang, R. Zielke, A. F. Gombart, C. S. Maier, A. E. Sikora, P. R. Blakemore and J. F. Stevens, Reductive Metabolism of Xanthohumol and 8-Prenylnaringenin by the Intestinal Bacterium Eubacterium ramulus, Mol. Nutr. Food Res., 2019, 63, e1800923 CrossRef PubMed.
- J. M. Dahlhamer, E. P. Zammitti, B. W. Ward, A. G. Wheaton and J. B. Croft, Prevalence of Inflammatory Bowel Disease Among Adults Aged >/=18 Years - United States, 2015, Morb. Mortal. Wkly. Rep., 2016, 65, 1166–1169 CrossRef PubMed.
- B. O. Langley, J. J. Ryan, D. Hanes, J. Phipps, E. Stack, T. O. Metz, J. F. Stevens and R. Bradley, Xanthohumol Microbiome and Signature in Healthy Adults (the XMaS Trial): Safety and Tolerability Results of a Phase I Triple-Masked, Placebo-Controlled Clinical Trial, Mol. Nutr. Food Res., 2021, 65, e2001170 CrossRef PubMed.
- L. Guthrie and L. Kelly, Bringing microbiome-drug interaction research into the clinic, EBioMedicine, 2019, 44, 708–715 CrossRef PubMed.
- R. Sleha, V. Radochova, A. Mikyska, M. Houska, R. Bolehovska, S. Janovska, J. Pejchal, L. Muckova, P. Cermak and P. Bostik, Strong Antimicrobial Effects of Xanthohumol and Beta-Acids from Hops against Clostridioides difficile Infection In Vivo, Antibiotics, 2021, 10, 392 CrossRef CAS PubMed.
- K. Klimek, K. Tyskiewicz, M. Miazga-Karska, A. Debczak, E. Roj and G. Ginalska, Bioactive Compounds Obtained from Polish “Marynka” Hop Variety Using Efficient Two-Step Supercritical Fluid Extraction and Comparison of Their Antibacterial, Cytotoxic, and Anti-Proliferative Activities In Vitro, Molecules, 2021, 26, 2366 CrossRef CAS PubMed.
- S. P. Claus, H. Guillou and S. Ellero-Simatos, The gut microbiota: a major player in the toxicity of environmental pollutants?, npj Biofilms Microbiomes, 2016, 2, 16003 CrossRef PubMed.
- N. Koppel, V. Maini Rekdal and E. P. Balskus, Chemical transformation of xenobiotics by the human gut microbiota, Science, 2017, 356, eaag2770 CrossRef PubMed.
- D. Dutta and S. H. Lim, Bidirectional interaction between intestinal microbiome and cancer: opportunities for therapeutic interventions, Biomark. Res., 2020, 8, 31 CrossRef PubMed.
- N. Savage, The complex relationship between drugs and the microbiome, Nature, 2020, 577, S10–S11 CrossRef CAS PubMed.
- M. Zimmermann, M. Zimmermann-Kogadeeva, R. Wegmann and A. L. Goodman, Mapping human microbiome drug metabolism by gut bacteria and their genes, Nature, 2019, 570, 462–467 CrossRef CAS PubMed.
- L. Brodziak-Jarosz, Y. Fujikawa, D. Pastor-Flores, S. Kasikci, P. Jirasek, S. Pitzl, R. W. Owen, K. D. Klika, C. Gerhauser, S. Amslinger and T. P. Dick, A click chemistry approach identifies target proteins of xanthohumol, Mol. Nutr. Food Res., 2016, 60, 737–748 CrossRef CAS PubMed.
- L. Dubinsky, B. P. Krom and M. M. Meijler, Diazirine based photoaffinity labeling, Bioorg. Med. Chem., 2012, 20, 554–570 CrossRef CAS PubMed.
- M. Gall, M. Thomsen, C. Peters, I. V. Pavlidis, P. Jonczyk, P. P. Grunert, S. Beutel, T. Scheper, E. Gross, M. Backes, T. Geissler, J. P. Ley, J. M. Hilmer, G. Krammer, G. J. Palm, W. Hinrichs and U. T. Bornscheuer, Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase, Angew. Chem., Int. Ed., 2014, 53, 1439–1442 CrossRef CAS PubMed.
- A. Braune, W. Engst, P. W. Elsinghorst, N. Furtmann, J. Bajorath, M. Gutschow and M. Blaut, Chalcone Isomerase from Eubacterium ramulus Catalyzes the Ring Contraction of Flavanonols, J. Bacteriol., 2016, 198, 2965–2974 CrossRef CAS PubMed.
- J. Cox and M. Mann, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol., 2008, 26, 1367–1372 CrossRef CAS PubMed.
- M. Li, X. Han and B. Yu, Facile synthesis of flavonoid 7-O-glycosides, J. Org. Chem., 2003, 68, 6842–6845 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, preparation of experimental samples for SDS-PAGE fluorescent analysis, chemoproteomics mass spectrometry-based preparations, and proteomics data analysis. See DOI: https://doi.org/10.1039/d3ra05296f |
| This journal is © The Royal Society of Chemistry 2023 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)